Significance
NF1 is a monogenic disorder caused by mutations in the NF1 gene. While it affects multiple organs and tissues, how brain symptoms arise has been complicated to uncover. Since brain glial and inhibitory neuron cells have been implicated in NF1, we conditionally deleted the mouse Nf1 gene in progenitors that give rise to both of these cell types. We found that glial cells were impacted when both copies of Nf1 were deleted, while properties of a specific group of inhibitory neurons were altered when either one or both copies of Nf1 were deleted. These data demonstrate phenotypes that may contribute to NF1 and uncover roles for the Nf1 gene in brain development.
Keywords: cortical interneuron, oligodendrocyte, MGE
Abstract
Neurofibromatosis 1 (NF1) is caused by mutations in the NF1 gene, which encodes the protein, neurofibromin, an inhibitor of Ras activity. Cortical GABAergic interneurons (CINs) are implicated in NF1 pathology, but the cellular and molecular changes to CINs are unknown. We deleted mouse Nf1 from the medial ganglionic eminence, which gives rise to both oligodendrocytes and CINs that express somatostatin and parvalbumin. Nf1 loss led to a persistence of immature oligodendrocytes that prevented later-generated oligodendrocytes from occupying the cortex. Moreover, molecular and cellular properties of parvalbumin (PV)-positive CINs were altered by the loss of Nf1, without changes in somatostatin (SST)-positive CINs. We discovered that loss of Nf1 results in a dose-dependent decrease in Lhx6 expression, the transcription factor necessary to establish SST+ and PV+ CINs, which was rescued by the MEK inhibitor SL327, revealing a mechanism whereby a neurofibromin/Ras/MEK pathway regulates a critical CIN developmental milestone.
Neurofibromatosis 1 (NF1) is caused by mutations in the gene NF1, which encodes the neurofibromin protein (1). Neurofibromin contains a centrally located GTPase-activating protein (GAP) domain, which inhibits RAS activity (2) as well as domains that mediate protein interactions or regulate other signaling events (3, 4). NF1 is a RASopathy (5), which can impact multiple organs and tissues, including the brain, thereby leading to cognitive changes (i.e., increased incidence of autism spectrum disorder [ASD] and learning disabilities) in a subset of those diagnosed (6–8). While the cellular and molecular mechanisms underlying these cognitive changes are poorly understood, they are likely due to haploinsufficiency of the NF1 gene, suggesting that phenotypes uncovered in models where one allele is lost would be more relevant to understanding the underlying biology.
In animal models, loss of Nf1 leads to altered myelination, increased oligodendrocyte precursors in the spinal cord, and preferential gliogenesis at the expense of neurogenesis in the brain (9–12). While most brain cell types likely contribute to the cognitive changes in NF1, pan-GABAergic deletion of one Nf1 allele in mice impairs cognitive performance and alters excitatory/inhibitory balance (13, 14). These studies suggest glia and/or GABAergic neurons as potential modulators of Nf1’s function in the brain.
The median ganglionic eminence (MGE) gives rise to multiple cell types, including the majority of GABAergic cortical interneurons (CINs) (15) and the first wave of oligodendrocytes to populate the brain (16). MGE-derived oligodendrocytes die off after birth, and later waves of oligodendrocytes that are derived from Gsh2 and Emx1 lineages are the dominant populations in the adult cortex (16). MGE-derived CINs primarily express the molecular markers somatostatin (SST) or parvalbumin (PV), which are determined by the transcription factor Lhx6 after the MGE is regionally patterned by Nkx2.1 (17–19). Lhx6 is a cardinal cell fate determination factor expressed in the MGE, and when deleted, SST+ and PV+ CINs do not acquire their mature cell fates, and a subset assume properties of caudal ganglionic eminence-derived CINs (17, 19–21). While recent studies have examined the role of transcription factors that influence MGE and caudal ganglionic eminence CIN development (22–24), few reports have assessed how cellular signaling proteins regulate these processes (25–27). After their programming in the MGE, CINs tangentially migrate into and laminate the neocortex. As they mature, they become diversified into further subclasses with distinct molecular, anatomical, and physiological properties (28, 29). These diverse properties contribute unique roles to brain function, but whether distinct CIN groups are altered in NF1 is unknown.
The MGE provides an ideal opportunity to probe both oligodendrocytes and GABAergic CINs after Nf1 deletion. Here, we show that loss of Nf1 in mouse MGE-lineages results in a persistence of immature oligodendrocytes in the adult neocortex, preventing later-generated oligodendrocyte establishment. Moreover, PV expression is lost in half of the CINs, and molecular and cellular properties unique to this CIN group are depleted in a dose-dependent manner, without any changes in the SST group. Finally, Lhx6 expression was lost after one or both copies of Nf1 was deleted, and this loss can be rescued by inhibiting MEK, thus implicating a RAS/MAPK mechanism. This provides insights into glial as well as CIN molecular and cellular alterations that could occur in NF1, and links Nf1 loss to alterations in a cardinal CIN developmental program.
Results
Conditional Loss of Nf1 from the MGE Results in a Persistence of Immature MGE Lineage Oligodendrocytes.
To assess Nf1’s role in MGE lineages, we crossed Nf1Floxed mice (30) with Nkx2.1-Cre, which begins to express approximately embryonic day 9.5 (E9.5) in MGE progenitors (31) and Ai14, which expresses tdTomato after cre-recombination (32). Conditional Nf1 heterozygous (cHet) and Nf1 knockout (cKO) mice were born at Mendelian ratios and survived into adulthood. Fluorescent in situ hybridization detected Nf1 in wild-type (WT) MGE-lineage CINs (i.e., SST+) in the somatosensory cortex at postnatal day 30 (P30), but not in cKOs (SI Appendix, Fig. S1A).
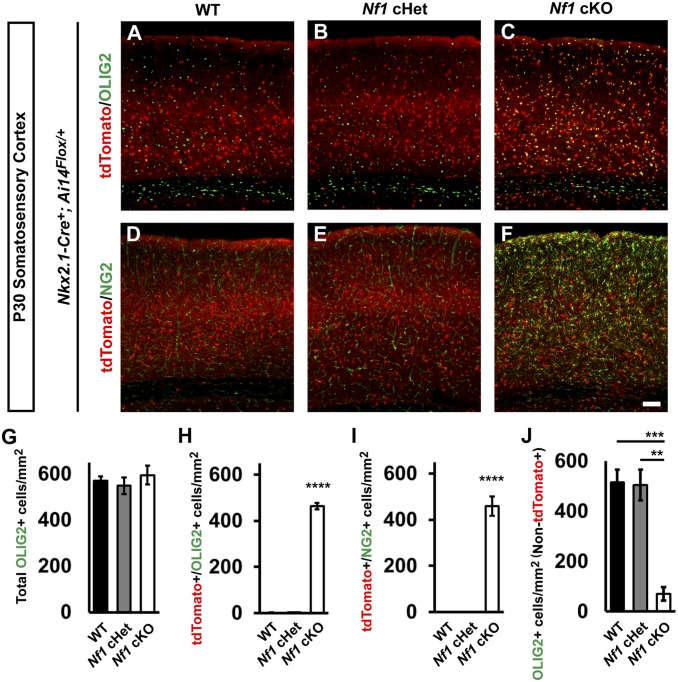
Next, Nkx2.1-Cre-lineage cells from WT, Nf1 cHet, and Nf1 cKO brains were assessed at P30 in the neocortex. While WT and Nf1 cHet brains looked equivalent, Nf1 cKO brains had an increase in Nkx2.1-Cre-lineage cells (tdTomato+) with small cell bodies (Fig. 1 A–F). Previous studies have indicated that glial numbers can increase after Nf1 deletion (9–11). Since the early wave of oligodendrocytes is derived from the MGE (16), we explored whether oligodendrocytes may represent this population. We probed for OLIG2 (i.e., a general marker of oligodendrocytes). While the total number of OLIG2+ cells did not change (Fig. 1G), there was a ∼200-fold increase in the number of tdTomato+ cells colabeled for OLIG2 in the cKOs (Fig. 1 A–C and H; P < 0.0001).
Fig. 1.
Persistence of immature Nkx2.1-Cre lineage and depletion of later-generated oligodendrocytes in Nf1 cKOs. Immunofluorescent images from the somatosensory cortex of P30 Nkx2.1-Cre+; Ai14Flox/+ mice that were WT (A and D), Nf1Flox/+ (cHet; B and E), or Nf1Flox/Flox (cKO; C and F). Images show Nkx2.1-Cre lineages (tdTomato+) colabeled for the oligodendrocyte markers OLIG2 (all oligodendrocytes) or NG2 (oligodendrocyte progenitor cells). (G) Quantification of the cell density of total OLIG2+ cells in the somatosensory cortex at P30. (H) Quantification of double-labeled tdTomato+/OLIG2+ cells in the somatosensory cortex at P30. (I) Quantification of the cell density of double-labeled tdTomato+/NG2+ cells in the somatosensory cortex at P30. (J) Quantification of OLIG2+ cells that were not tdTomato+ in the somatosensory cortex at P30. Data are presented as the mean ± SEM. All groups, n = 4. **P < 0.01; ***P < 0.001; ****P < 0.0001. (Scale bar in F, 100 µm.)
We next probed for NG2, a marker of oligodendrocyte precursor cells that are still immature. Interestingly, many Nf1 cKO MGE-lineage oligodendrocytes were NG2+, suggesting they were still oligodendrocyte precursor cells; there were no NG2+ cells in the WTs or Nf1 cHets (Fig. 1 D–F and I; P < 0.0001). Finally, we asked whether these immature oligodendrocytes influenced later-generated waves of oligodendrocytes in the somatosensory cortex. Thus, we counted tdTomato−/OLIG2+ cells and found ∼84% reduction in these cells in Nf1 cKOs (Fig. 1J; WT [P = 0.0009] and Nf1 cHet [P = 0.001]). Thus, the persistence of early-generated MGE lineage oligodendrocytes prevented later-generated waves from occupying the cortex.
Molecular and Cellular Properties of PV+ CINs Are Specifically Reduced in Nf1 cKOs.
We first asked whether Nf1 was expressed in different CIN groups via assessing single-cell levels of Nf1 transcripts from ∼P30 CINs in the cortex (33). Nf1 transcripts were found in all CIN cell types examined (SI Appendix, Fig. S1B).
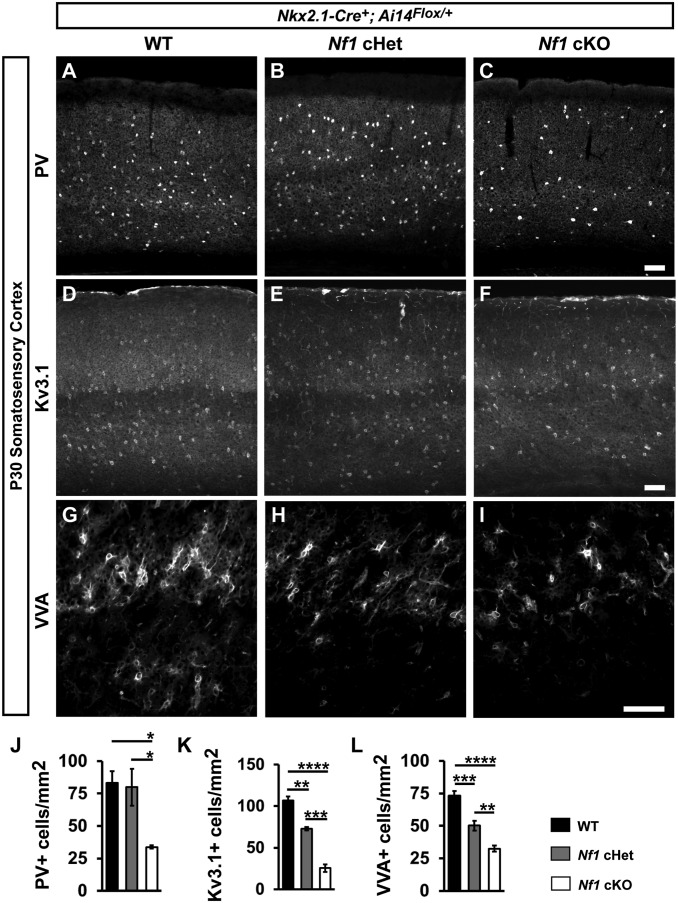
We probed for the CIN marker, PV, in the somatosensory cortices of WTs, Nf1 cHets, and Nf1 cKOs at P30 (Fig. 2 A–C); an example of PV/tdTomato colabeled CINs is shown (SI Appendix, Fig. S2 A–C). While there was no difference between WT and Nf1 cHet PV+ CINs, Nf1 cKOs had a ∼60% decrease in PV+ CINs (Fig. 2J; vs. WT [P = 0.03] and Nf1 cHet [P = 0.04]). In addition, there was an increased percentage of PV+ CINs in cortical layers ii/iii/iv and less in layer v in both Nf1 cHets and Nf1 cKOs (SI Appendix, Fig. S2D; layers ii/iii/iv, WT vs. Nf1 cHet [P = 0.0007] and WT vs. Nf1 cKO [P = 0.002]; layer v, WT vs. Nf1 cHet [P = 0.005] and WT vs. Nf1 cKO [P = 0.01]). In contrast, we did not detect any changes in SST cell density or lamination between genotypes (SI Appendix, Fig. S3).
Fig. 2.
Molecular and cellular impacts on PV+ CINs in Nf1 cHets and cKOs. Immunofluorescent images from the somatosensory cortex of P30 Nk x2.1-Cre+; Ai14Flox/+ mice that were WT, Nf1Flox/+ (cHet), or Nf1Flox/Flox (cKO), stained for PV (A–C), Kv3.1 (D–F), or VVA (G–I). Quantification of the cell density of PV+ cells (J), Kv3.1+ cells (K), or VVA+ cells (L) in the somatosensory cortex at P30. Data are presented as the mean ± SEM. All groups, n = 4. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (Scale bars in C, F, and I, 100 µm.)
PV+ CINs exhibit fast-spiking properties, which are primarily mediated by the fast-inactivating potassium channel, Kv3.1, which is highly expressed within PV+ CINs (26). We thus examined whether Kv3.1 expression was altered in Nf1 cHets and cKOs (Fig. 2 D–F) and found that this channel was reduced in both genotypes (Fig. 2K; WT vs. Nf1 cHet [P = 0.002], WT vs. Nf1 cKO [P < 0.0001], and Nf1 cHet vs. Nf1 cKO [P = 0.0003]). In addition, PV+ CINs develop perineuronal nets composed of various extracellular matrix proteins at more mature stages of development (34). We found that in both Nf1 cHets and cKOs, the density of perineuronal nets was decreased in a dose-dependent manner (Fig. 2 G–I and L; WT vs. Nf1 cHet [P = 0.0005], WT vs. Nf1 cKO [P < 0.0001], and Nf1 cHet vs. Nf1 cKO [P = 0.005]). Thus, while PV expression was only decreased in cKOs, cHets had less severe but significant molecular and cellular changes induced by the loss of Nf1.
We also examined SST+ and PV+ cells in the hippocampus and striatum. Notably, a similar increase in OLIG2+ cells was observed (SI Appendix, Fig. S4 [P < 0.0001] and SI Appendix, Fig. S5 [P = 0.0004]), and PV+ cells decreased in each region (SI Appendix, Fig. S4 [P = 0.01] and SI Appendix, Fig. S5 [P = 0.04]). However, no changes in SST+ cells in each region or choline acetyltransferase-positive cells in the striatum were seen (SI Appendix, Figs. S4 and S5). Thus, PV+ CINs are particularly sensitive to Nf1 loss.
No Change in PV+ CINs When Nf1 Is Deleted from Postmitotic PV-Cre-Lineage CINs.
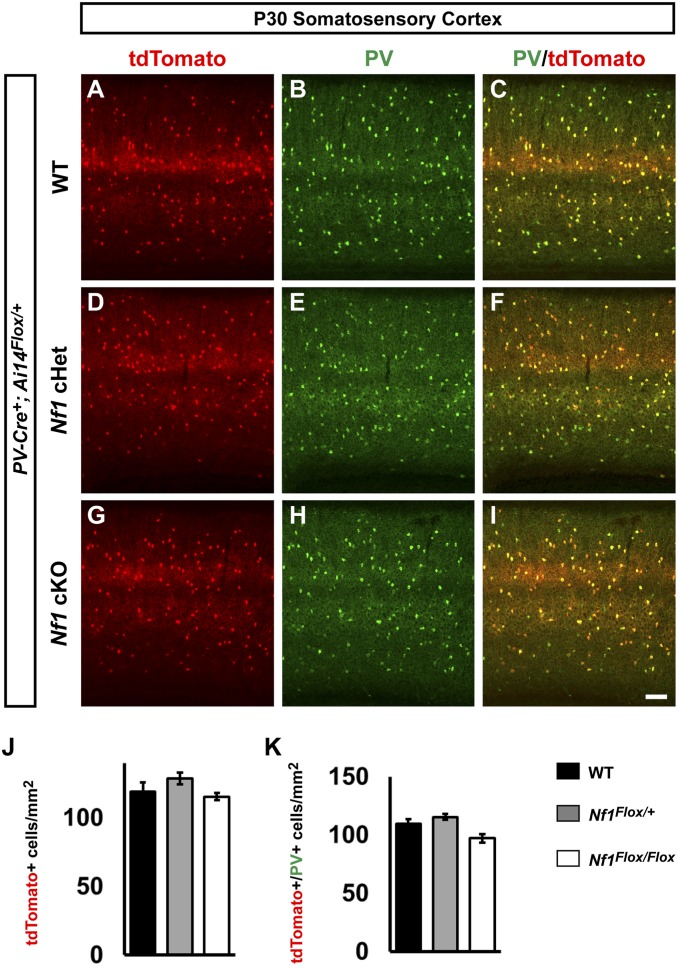
To determine whether Nf1 was necessary in postmitotic PV+ CINs, we crossed our Nf1Floxed; Ai14 mice with a PV-Cre line (35). We found no differences in the cell density of PV-Cre-lineage CINs (tdTomato+) in the somatosensory cortex at P30 (Fig. 3 A, D, G, and J). We also colabeled these cells with PV and found no difference in the number of CINs that expressed PV (Fig. 3 A–I and K). Thus, loss of Nf1 in postmitotic PV+ CINs does not recapitulate the phenotypes observed by deletion in developing CINs or progenitors, suggesting a role during development.
Fig. 3.
Lack of overt phenotypes after conditional loss of Nf1 in postmitotic PV-Cre lineages. P30 WT, Nf1 cHet, and Nf1 cKO coronal issue was visualized for tdTomato; that is, PV-Cre lineages (A, D, and G), PV (B, E, and H), or merged (C, F, and I). (J) Quantification of the cell density of PV-Cre lineages at P30 in the somatosensory cortex. (K) Quantification of the cell density of tdTomato+/PV+ colabeled cells. Data are expressed as the mean ± SEM. WT, n = 3; cHet, n = 3; cKO, n = 4. (Scale bar in I, 100 µm.)
Nf1 Loss in the MGE Does Not Alter Cell Proliferation.
To probe how the loss of PV expression could occur, we assessed whether loss of Nf1 altered cell proliferation in the MGE. MGE-derived CINs are generated throughout midgestation, with peak production occurring ∼E13.5 (36, 37). Moreover, since some PV+ CINs are generated later than SST CINs, we also assessed proliferation at E15.5. We first pulsed pregnant dams with EdU that had either E13.5 or E15.5 embryos for 30 min to label proliferating cells in (S) synthesis phase of the cell cycle. We also costained these brains for the (M) mitosis phase marker, phospho-histone 3 (PH3). Labeled cells were counted in both the ventricular zone and subventricular zones of the MGE, which are hypothesized to differentially generate SST+ and PV+ CINs (38). We did not detect any differences (SI Appendix, Fig. S6 A–V). Thus, loss of Nf1 does not alter cell proliferation during peak CIN proliferation.
The Lhx6 Transcription Factor Is Reduced in Both Nf1 cHets and Nf1 cKOs.
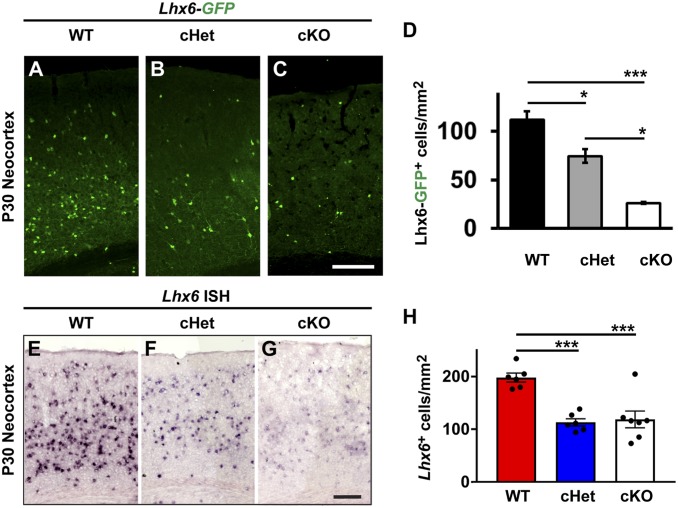
Since Lhx6 is a cardinal transcription factor that is the primary determinant of MGE-specific CIN properties (21), we wanted to assess whether Lhx6 expression was altered in Nf1 mutants. To this end, we first crossed a Lhx6-GFP-expressing mouse, which drives GFP to a greater extent in PV+ MGE-lineage CINs than in SST+ (24), with our Nkx2.1-Cre; Nf1Floxed mice to examine MGE-derived CINs in the somatosensory cortex at P30.
We first assessed the cell density of Lhx6-GFP+ CINs in the somatosensory cortex at P30. Interestingly, the number of GFP+ cells was decreased in both Nf1 cHets, ∼37% reduced vs. WT, and the Nf1 cKOs, ∼77% reduced vs. WT (Fig. 4 A–D; WT vs. Nf1 cHet [P = 0.02], WT vs. Nf1 cKO [P = 0.0008], Nf1 cHet vs. Nf1 cKO [P = 0.01]). Considering there were no changes in SST+ CINs, or any other marker in Nf1 cHets, we wanted to assess Lhx6 expression in another manner. Thus, we performed in situ hybridization at P30 and counted the number of cells expressing Lhx6 transcript in the neocortex. Notably, the number of Lhx6+ cells was also reduced by half in both the Nf1 cHets and Nf1 cKOs (Fig. 4 E–H; WT vs. Nf1 cHet [P = 0.0004] and WT vs. Nf1 cKO [P = 0.006]). While the decrease in total Lhx6 expression was not as severe as the loss of GFP expression, there was a reduction in the intensity of Lhx6 expression in both the Nf1 cHets and Nf1 cKOs in CINs that was more noticeable in the Nf1 cKOs. Together, these data reveal that loss of Nf1 results in decreased Lhx6 levels in a dose-dependent manner.
Fig. 4.
Loss of Lhx6 expression at P30. GFP expression from a Lhx6-GFP reporter was probed in the neocortices of WT, Nf1 cHet, and Nf1 cKO mice (A–C). (D) Quantification of Lhx6-GFP+ cell density. In situ hybridization to detect Nf1 RNA in P30 neocortices in the same genotypes at P30 (E–G). (H) Quantification of Nf1+ cells in the neocortex at P30. Data are presented as the mean ± SEM. For Lhx6-GFP counts, n = 3 (WT), n = 6 (cHet) and n = 3 (cKO); in situ counts, n = 3, all groups. *P < 0.05; ***P < 0.001. (Scale bars in C, 100 µm and G, 200 µm.)
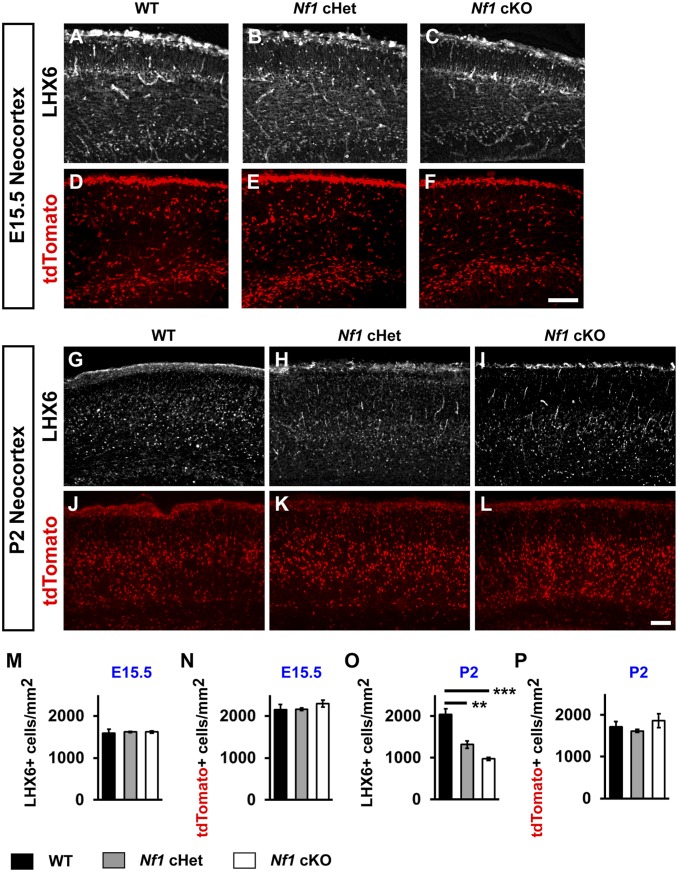
We next examined whether expression from the Lhx6-GFP reporter was altered during development by first examining Lhx6-GFP+ embryos at E15.5, a point when CINs are tangentially migrating through the developing neocortex, but before large numbers of OLIG2+ cells enter the neocortex (39). As in the P30 brains, the expression of Lhx6-driven GFP in E15.5 neocortices was decreased in a Nf1 dose-dependent manner (SI Appendix, Fig. S7 A–C and G; WT vs. Nf1 cHet [P = 0.0003], WT vs. Nf1 cKO [P < 0.0001], and Nf1 cHet vs. Nf1 cKO [P = 0.009]); the same patterns were observed at P2 (SI Appendix, Fig. S7 D–F and H; WT vs. Nf1 cHet and Nf1 cKO, P < 0.0001). While this GFP reporter provided an inroad into a potential molecular change in Nf1-depleted CINs, it was unknown whether the LHX6 protein was still present and/or whether a loss of MGE-derived CINs had occurred.
LHX6 Protein Persists Embryonically in Nf1 cHets and cKOs.
To first understand whether the LHX6 protein was lost in Nf1 loss-of-function mutants, we probed E15.5 and P2 brains with a LHX6 antibody to examine the protein in the neocortex. Unlike the GFP reporter, there was no change in LHX6 protein at E15.5 (Fig. 5 A–C and M). However, by P2, there was a dramatic decrease in LHX6 protein in both the Nf1 cHets and cKOs (Fig. 5 G–I and O; WT vs. Nf1 cHet [P = 0.004] and WT vs. Nf1 cKO [P = 0.0006]). Thus, while the GFP reporter showed faster dynamics after Nf1 depletion, the LHX6 protein remained in developing CINs embryonically. Importantly, at both E15.5 and P2, there was no change in tdTomato+ cell density (Fig. 5 D–F, J–L, N, and P). These data demonstrate that LHX6 expression is decreased over a long developmental window, and that a decrease in LHX6 expression occurs, rather than a loss of CIN cells.
Fig. 5.
Persistence of LHX6 protein embryonically but loss postnatal. LHX6 protein was probed in E15.5 WT Nf1 cHet and Nf1 cKO neocortices (A–C). TdTomato+ cells (Nkx2.1-Cre-lineage) were probed at E15.5 from the same genotypes (D–F). LHX6 protein and tdTomato expression was probed at P2 from the same brains (G–L). Quantification of the cell density of E15.5 LHX6+ CINs (M), tdTomato+ CINs (N) or P2 LHX6+ CINs (O) and tdTomato+ CINs (P). Data are expressed as the mean ± SEM n = 3, all groups. **P < 0.01; ***P < 0.001. (Scale bars in F and L, 100 µm.)
We also probed for OLIG2 and the early oligodendrocyte marker PDGFR-alpha at E15.5. Consistent with earlier experiments (SI Appendix, Fig. S6), OLIG2+ cells in the MGE ventricular zone (VZ) of Nf1 cKOs were not altered (SI Appendix, Fig. S8 A, B, and I; SVZ1, P = 0.01; SVZ2, P = 0.03); these are MGE progenitors. While there was an increased trend in OLIG2+ cells in the neocortex, these did not reach significance (SI Appendix, Fig. S8 C, D, and J). Interestingly, PDGFRa+ cells were also increased in the MGE in Nf1 cKOs and trended up in the neocortex (SI Appendix, Fig. S8 E–H, K, and L; SVZ1, P = 0.002; SVZ2, P = 0.005). Thus, immature oligodendrocytes are elevated in Nf1 cKOs as early as E15.5.
LHX6 Is Lost in a MEK-Dependent Manner.
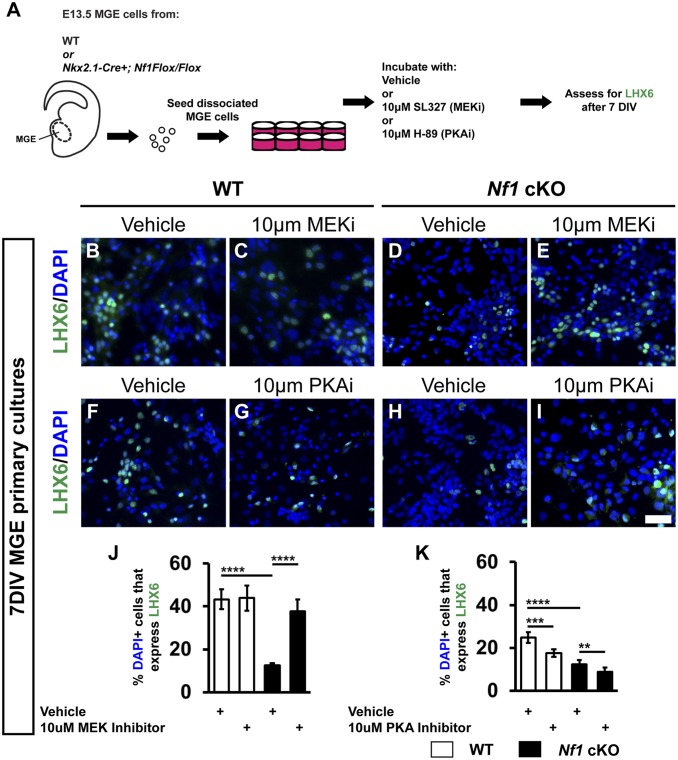
To uncover the mechanism leading to the loss of LHX6, we asked whether core signaling pathways downstream of Nf1 may be responsible (i.e., MEK or PKA) (3). To this end, we generated primary MGE cultures and found that both the loss of LHX6 and increase in oligodendrocyte phenotypes could be recapitulated in vitro (Fig. 6). Next, we applied vehicle, the MEK inhibitor SL327 (40), or the PKA inhibitor H-89 (41) to E13.5 MGE primary cultures and then assessed the proportion of LHX6+ cells after 7 d of culture (Schema, Fig. 6A).
Fig. 6.
LHX6 expression is rescued by the MEK inhibitor SL327. E13.5 MGE cells were cultured and either vehicle (DMSO) or the MEK inhibitor (SL327, MEKi) or PKA inhibitor (H-89, PKAi) were added every other day. Cells were cultured for 7 d and assessed for LHX6 (A–I). Quantification of the %DAPI+ cells that express LHX6 treated with vehicle or MEKi (J) or vehicle or PKAi (K). Data are expressed as the mean ± SEM n = 4, all groups. **P < 0.01; ***P < 0.001; ****P < 0.0001. (Scale bar in I, 100 µm.)
The MEK inhibitor (MEKi) did not alter LHX6 expression in WT CINs; however, it restored expression of LHX6 in Nf1 cKO CINs (Fig. 6 B–E and J; WT vehicle vs. Nf1 cKO vehicle and Nf1 cKO vehicle vs. Nf1 cKO MEKi; P < 0.0001). However, PKA inhibition not only failed to rescue the LHX6 phenotype but also exacerbated the decreased expression in both WTs and Nf1 cKOs (Fig. 6 F–I and K; WT vehicle vs. WT PKAi [P = 0.0002], WT vehicle vs. Nf1 cKO vehicle [P < 0.0001], and Nf1 cKO vehicle vs. Nf1 cKO PKAi [P = 0.001]). Unlike the specific effect of MEK inhibition on LHX6 expression, both the MEKi and PKAi decreased OLIG2 expression in both WTs and Nf1 cKOs (SI Appendix, Fig. S9 A–J; MEKi: between WTs [P = 0.0001], WT vehicle vs. drug and Nf1 cKO vehicle vs. Nf1 cKO drug [P < 0.0001]; PKAi: between WTs [P = 0.0002], WT vehicle vs. drug and Nf1 cKO vehicle vs. Nf1 cKO drug [P < 0.0001]). Thus, LHX6 expression is dependent on MEK activity downstream of Nf1, providing an initial inroad and mechanism to this observation in NF1 biology.
Discussion
We explored how cells derived from the MGE are altered by mouse Nf1 loss. Notably, both GABAergic CINs and oligodendrocytes are implicated in NF1 (9–14), yet the molecular and cellular impacts had not been explored. We found that immature oligodendrocytes appear embryonically, but LHX6 protein is not depleted until early postnatal stages, which may be why SST+ CINs are spared in this model but late-developing PV+ CINs are impacted. Importantly, while PV expression is only altered in Nf1 cKOs, there are intermediate phenotypes affecting Kv3.1 expression and the formation of perineuronal nets in cHets as well, which are important for their fast-spiking properties and mature phenotypes. Importantly, since these properties, as well as the depletion of LHX6, occurs in cHets but only cKOs exhibit the oligodendrocyte phenotype, we propose these phenotypes are independent of each other. Given these findings, it is important to remember that cognitive changes in those diagnosed with NF1 are caused by haploinsufficiency of NF1 and likely involve the dysfunction of other brain cell types. Moreover, while phenotypes uncovered in our Nf1 cHets are more likely to reflect what could occur in NF1 CINs, the loss of both Nf1 alleles in the cKOs is valuable to understanding the biological function of neurofibromin.
We first discovered a glial phenotype that was different from the competition between gliogenesis and neurogenesis previously reported in the SVZ of the dorsal LGE, where progenitors preferentially generated oligodendrocytes at the expense of neurons (11). Here, MGE-derived oligodendrocytes that lacked Nf1 expressed the immature marker, NG2; persisted into adult stages; and competed out later-generated oligodendrocytes. This occurred without changes in MGE proliferation or changes in embryonic CIN numbers. This is interesting, as ventral-derived oligodendrocytes of the spinal cord outnumber their later-generated dorsal counterparts and loss of Nf1 in the spinal cord results in elevated immature markers (9, 10), similar to what we observe in MGE-derived oligodendrocytes that also have ventral origins. Moreover, early-generated oligodendrocytes may noncell-autonomously influence the development of their later-generated counterparts, suggesting a form of glial communication that may help us understand how oligodendrocytes establish proper numbers in the brain.
Since GABAergic neurons have been implicated in ASD and NF1 (13, 42, 43), we wanted to explore these cells in more detail. Our study demonstrates that loss of Nf1 in MGE-derived CINs, a deletion more spatially restrictive than a previous report (13), results in a specific loss of PV expression. This is interesting, as the expression of PV is uniquely reduced in the cortices of humans with ASD (44). Thus, manipulating PV+ CIN function and numbers may be a potential therapeutic inroad in the future. However, it is also important to consider that many other cells in the brain will be impacted in those diagnosed with NF1, and the role of GABAergic CINs will need to be considered in this context.
While our deletion strategy led to a loss of Lhx6 and depleted molecular and cellular properties of PV+ CINs, future studies are needed to determine the net effect on brain physiology and function. The loss of PV+ CINs is likely to lead to a net increase in excitability. This raises an interesting point, which is that while our more restrictive manipulation may reveal an important role for PV+ CIN development, there may be other GABAergic neurons responsible for the increased GABAergic tone previously reported (14), and potentially those derived from the caudal ganglionic eminence.
Lhx6 expression in MGE-derived CINs is necessary for many of their molecular properties (17, 19, 20, 45, 46). Moreover, Lhx6 is repressed in several types of cancers via hypermethylation of its regulatory elements (47–50). While the signaling events repressing Lhx6 are not known, it is possible that a RAS/MEK pathway mediates these effects and could potentially lead to Lhx6 repression in CINs. Our primary culture experiments highlight the role of MEK in regulating LHX6 expression, which we hypothesize is the mechanism for this repression. Interestingly, another group found that a constitutively active MEK1 mouse model also exhibits a selective loss of PV+ CINs in the neocortex (51). This suggests that PV+ CIN development may be regulated through the core RAS/MAPK pathway and may not be due to other Nf1-regulated signaling events. However, oligodendrocyte numbers responded moderately to both MEK and PKA inhibition, suggesting that this may be primarily due to another mechanism, or this phenotype is regulated by multiple functions of Nf1.
The selective loss of PV+ CINs and the preservation of the SST+ CIN numbers in our mutants raises important questions regarding the role of Lhx6 in the regulation of CIN phenotypes. While GFP expression from a Lhx6-reporter line was decreased by E15.5, LHX6 protein was present at this age and did not decrease until early postnatal stages. We propose that SST development may be mostly complete by early postnatal ages, but PV+ CINs may need additional time for LHX6 to instruct their development. This is in line with a recent report proposing that SST lineages are preferentially determined very early in the VZ of the MGE, while PV lineages are more likely to be committed at later stages (38).
The persistence of immature oligodendrocytes and loss of PV+ CINs in Nf1 mutants opens avenues for probing potential mechanisms underlying Nf1 dysfunction. Our studies provide insights into some molecular and cellular alterations that may occur in those diagnosed with NF1. Moreover, these findings may have broader implication for other RASopathies and associated neurodevelopmental disorders, such as ASD. Finally, how changes in GABAergic CINs work with alterations in other brain cell types will be important to uncover in the future.
Experimental Procedures
Animals.
All mouse strains have been published: Ai14 Cre-reporter (32), Nkx2.1-Cre (31), Lhx6-GFP (Gensat), Nf1flox (30), PV-Cre (35). Nf1flox mice were initially on a mixed C57BL6/J, CD-1 background and then backcrossed to CD-1 for at least four generations before analysis. For timed pregnancies, noon on the day of the vaginal plug was counted as E0.5. All animal care and procedures were performed according to the Michigan State University and University of California San Francisco Laboratory Animal Research Center guidelines.
Immunofluorescent Tissue Staining.
Brains were harvested either from embryos and then fixed overnight in 4% paraformaldehyde (PFA) or from postnatal mice that were perfused with saline followed by 4% PFA; the latter group was postfixed in 4% PFA for 30 min. The brains were then sunk in 30% sucrose, embedded in OCT, and sectioned using a cryostat (Tissue-Tek Cryo-3). Immunofluorescent labeling was performed on 25-µm (P30) or 20-µm (embryonic and P2) cryosections with the primary antibodies listed in supplemental information. The appropriate 488, 594, or 647 Alexa-conjugated secondary antibodies were from Thermo Fisher Scientific. Sections were coverslipped with Vectashield containing DAPI (Vector Laboratories).
In Situ Hybridization.
For standard in situ hybridization, a Lhx6 Digoxigenin-labeled riboprobe was used as previously described (52). In situ RNA hybridization was performed as previously described (53). Imaging was performed with a 60× objective (Nikon Apo 1.4 oil) under Nikon Ti microscope with DS-Qi2 color camera.
Primary MGE Cultures and Drug Treatments.
MGE tissue was harvested from E13.5 WT or Nf1 cKO brains and then mechanically dissociated via trituration with a P1000 pipette tip as previously described (54). Roughly 100,000 cells were seeded into individual wells of a 24-well plate that was coated with poly-l-Lysine and laminin containing DMEM with 10% FBS. The following day, the media was replaced with Neurobasal media containing glutamax, glucose, B27, and penicillin/streptomycin. In addition, wells were incubated with vehicle (DMSO), 10 µM SL327 in DMSO or 10µM H-89 dissolved in DMSO. Media was replaced every 2 days with either vehicle or drugs added fresh each time. Cells were fixed in 4% PFA on day 7 and probed for either LHX6 or OLIG2. NucBlue (Thermo Fisher) was added to label DAPI+ nuclei. For analysis, random images were taken from each well, and either the number of LHX6+ or OLIG2+ cells were counted using Image-J software and normalized to the number of DAPI+ cells in each image.
Data Availability Statement.
All data are available upon request from the corresponding author.
Supplementary Material
Acknowledgments
E.L.-L.P. was funded by University of California, San Francisco, neuroscience graduate program and National Institute of Mental Health (NIMH) Grant R01 MH081880. J.L.R. was funded by Nina Ireland, NIMH R01 MH081880 and NIMH Grant R37/R01 MH049428. D.V. was funded by a pilot grant from Clinical and Translational Science Institute (#1111111). K.A., A.M.S., J.T.N., K.X.L., and D.V. were supported by the Spectrum Health-Michigan State University Alliance Corporation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915458117/-/DCSupplemental.
References
- 1.Mattocks C., et al. , Automated comparative sequence analysis identifies mutations in 89% of NF1 patients and confirms a mutation cluster in exons 11-17 distinct from the GAP related domain. J. Med. Genet. 41, e48 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klose A., et al. , Selective disactivation of neurofibromin GAP activity in neurofibromatosis type 1. Hum. Mol. Genet. 7, 1261–1268 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Gutmann D. H., Parada L. F., Silva A. J., Ratner N., Neurofibromatosis type 1: Modeling CNS dysfunction. J. Neurosci. 32, 14087–14093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ratner N., Miller S. J., A RASopathy gene commonly mutated in cancer: The neurofibromatosis type 1 tumour suppressor. Nat. Rev. Cancer 15, 290–301 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rauen K. A., et al. , Proceedings from the 2009 genetic syndromes of the Ras/MAPK pathway: From bedside to bench and back. Am. J. Med. Genet. A. 152A, 4–24 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garg S., et al. , Neurofibromatosis type 1 and autism spectrum disorder. Pediatrics 132, e1642–e1648 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Morris S. M., et al. , Disease Burden and symptom structure of autism in neurofibromatosis type 1: A study of the international NF1-ASD consortium team (INFACT). JAMA Psychiatry 73, 1276–1284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogel A. C., Gutmann D. H., Morris S. M., Neurodevelopmental disorders in children with neurofibromatosis type 1. Dev. Med. Child Neurol. 59, 1112–1116 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Bennett M. R., Rizvi T. A., Karyala S., McKinnon R. D., Ratner N., Aberrant growth and differentiation of oligodendrocyte progenitors in neurofibromatosis type 1 mutants. J. Neurosci. 23, 7207–7217 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J.-S., et al. , Oligodendrocyte progenitor cell numbers and migration are regulated by the zebrafish orthologs of the NF1 tumor suppressor gene. Hum. Mol. Genet. 19, 4643–4653 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., et al. , ERK inhibition rescues defects in fate specification of Nf1-deficient neural progenitors and brain abnormalities. Cell 150, 816–830 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayes D. A., et al. , Nf1 loss and Ras hyperactivation in oligodendrocytes induce NOS-driven defects in myelin and vasculature. Cell Rep. 4, 1197–1212 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa R. M., et al. , Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature 415, 526–530 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Cui Y., et al. , Neurofibromin regulation of ERK signaling modulates GABA release and learning. Cell 135, 549–560 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wonders C. P., Anderson S. A., The origin and specification of cortical interneurons. Nat. Rev. Neurosci. 7, 687–696 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Kessaris N., et al. , Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat. Neurosci. 9, 173–179 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liodis P., et al. , Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J. Neurosci. 27, 3078–3089 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sussel L., Marin O., Kimura S., Rubenstein J. L., Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: Evidence for a transformation of the pallidum into the striatum. Development 126, 3359–3370 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y., et al. , Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. J. Comp. Neurol. 510, 79–99 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vogt D., et al. , Lhx6 directly regulates Arx and CXCR7 to determine cortical interneuron fate and laminar position. Neuron 82, 350–364 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silberberg S. N., et al. , Subpallial enhancer transgenic lines: A data and tool resource to study transcriptional regulation of GABAergic cell fate. Neuron 92, 59–74 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu J. S., et al. , Coup-TF1 and Coup-TF2 control subtype and laminar identity of MGE-derived neocortical interneurons. Development 144, 2837–2851 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayer C., et al. , Developmental diversification of cortical inhibitory interneurons. Nature 555, 457–462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pai E. L.-L., et al. , Mafb and c-maf have prenatal compensatory and postnatal antagonistic roles in cortical interneuron fate and function. Cell Rep. 26, 1157–1173.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kepecs A., Fishell G., Interneuron cell types are fit to function. Nature 505, 318–326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malik R., et al. , Tsc1 represses parvalbumin expression and fast-spiking properties in somatostatin lineage cortical interneurons. Nat. Commun. 10, 4994 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenzie M. G., et al. , Non-canonical Wnt signaling through Ryk regulates the generation of somatostatin- and parvalbumin-expressing cortical interneurons. Neuron 103, 853–864.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kessaris N., Magno L., Rubin A. N., Oliveira M. G., Genetic programs controlling cortical interneuron fate. Curr. Opin. Neurobiol. 26, 79–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu J. S., Vogt D., Sandberg M., Rubenstein J. L., Cortical interneuron development: A tale of time and space. Development 144, 3867–3878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Y., et al. , Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 15, 859–876 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Q., Tam M., Anderson S. A., Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J. Comp. Neurol. 506, 16–29 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Madisen L., et al. , A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paul A., et al. , Transcriptional architecture of synaptic communication delineates GABAergic neuron identity. Cell 171, 522–539.e20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myers A. K., Ray J., Kulesza R. J. Jr, Neonatal conductive hearing loss disrupts the development of the Cat-315 epitope on perineuronal nets in the rat superior olivary complex. Brain Res. 1465, 34–47 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Hippenmeyer S., et al. , A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 3, e159 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyoshi G., Butt S. J. B., Takebayashi H., Fishell G., Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J. Neurosci. 27, 7786–7798 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inan M., Welagen J., Anderson S. A., Spatial and temporal bias in the mitotic origins of somatostatin- and parvalbumin-expressing interneuron subgroups and the chandelier subtype in the medial ganglionic eminence. Cereb. Cortex 22, 820–827 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petros T. J., Bultje R. S., Ross M. E., Fishell G., Anderson S. A., Apical versus basal neurogenesis directs cortical interneuron subclass fate. Cell Rep. 13, 1090–1095 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tekki-Kessaris N., et al. , Hedgehog-dependent oligodendrocyte lineage specification in the telencephalon. Development 128, 2545–2554 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Satoh Y., et al. , ERK2 contributes to the control of social behaviors in mice. J. Neurosci. 31, 11953–11967 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chijiwa T., et al. , Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J. Biol. Chem. 265, 5267–5272 (1990). [PubMed] [Google Scholar]
- 42.Hussman J. P., Suppressed GABAergic inhibition as a common factor in suspected etiologies of autism. J. Autism Dev. Disord. 31, 247–248 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Robertson C. E., Ratai E.-M., Kanwisher N., Reduced GABAergic action in the autistic brain. Curr. Biol. 26, 80–85 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Hashemi E., Ariza J., Rogers H., Noctor S. C., Martínez-Cerdeño V., The number of parvalbumin-expressing interneurons is decreased in the prefrontal cortex in autism. Cereb. Cortex 27, 1931–1943 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elbert A., et al. , CTCF governs the identity and migration of MGE-derived cortical interneurons. J. Neurosci. 39, 177–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neves G., et al. , The LIM homeodomain protein Lhx6 regulates maturation of interneurons and network excitability in the mammalian cortex. Cereb. Cortex 23, 1811–1823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Estécio M. R. H., et al. , LHX6 is a sensitive methylation marker in head and neck carcinomas. Oncogene 25, 5018–5026 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Jung S., et al. , The role of hLHX6-HMR as a methylation biomarker for early diagnosis of cervical cancer. Oncol. Rep. 23, 1675–1682 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Jung S., et al. , Epigenetic regulation of the potential tumor suppressor gene, hLHX6.1, in human cervical cancer. Int. J. Oncol. 38, 859–869 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Liu W.-b, et al. , LHX6 acts as a novel potential tumour suppressor with epigenetic inactivation in lung cancer. Cell Death Dis. 4, e882 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holter M. C., et al. ,Hyperactive MEK1 signaling in cortical GABAergic neurons causes embryonic parvalbumin-neuron death and defects in behavioral inhibition. bioRxiv:10.1101/748087 (27 August 2019). [DOI] [PMC free article] [PubMed]
- 52.Long J. E., Cobos I., Potter G. B., Rubenstein J. L. R., Dlx1&2 and Mash1 transcription factors control MGE and CGE patterning and differentiation through parallel and overlapping pathways. Cereb. Cortex 19 (suppl. 1), i96–i106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cobos I., Borello U., Rubenstein J. L. R., Dlx transcription factors promote migration through repression of axon and dendrite growth. Neuron 54, 873–888 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vogt D., et al. , Viral-mediated labeling and transplantation of medial ganglionic eminence (MGE) cells for in vivo studies. J. Vis. Exp. 98, e52740 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available upon request from the corresponding author.