Significance
Nutrient requirements play an important role in host–microbe interactions, and can heavily affect the composition of microbial communities by setting hard constraints on microbe–microbe interactions. The auxotrophic capabilities of strains and their underlying genetic and metabolic basis have rarely been studied on a large scale. This paper presents a computational approach using genome-scale models of metabolism and genomic sequences to predict auxotrophies in over 1,300 Gram-negative strains. Our network-based approach identifies auxotrophs, their nutrient requirements, and the corresponding causal genetic and metabolic basis, making it one of the most comprehensive efforts in large-scale strain-specific auxotrophy prediction.
Keywords: systems biology, mathematical modeling, auxotrophy, pangenome, comparative genomics
Abstract
Auxotrophies constrain the interactions of bacteria with their environment, but are often difficult to identify. Here, we develop an algorithm (AuxoFind) using genome-scale metabolic reconstruction to predict auxotrophies and apply it to a series of available genome sequences of over 1,300 Gram-negative strains. We identify 54 auxotrophs, along with the corresponding metabolic and genetic basis, using a pangenome approach, and highlight auxotrophies conferring a fitness advantage in vivo. We show that the metabolic basis of auxotrophy is species-dependent and varies with 1) pathway structure, 2) enzyme promiscuity, and 3) network redundancy. Various levels of complexity constitute the genetic basis, including 1) deleterious single-nucleotide polymorphisms (SNPs), in-frame indels, and deletions; 2) single/multigene deletion; and 3) movement of mobile genetic elements (including prophages) combined with genomic rearrangements. Fourteen out of 19 predictions agree with experimental evidence, with the remaining cases highlighting shortcomings of sequencing, assembly, annotation, and reconstruction that prevent predictions of auxotrophies. We thus develop a framework to identify the metabolic and genetic basis for auxotrophies in Gram-negatives.
Host–pathogen interactions and pathogen–microbiota interactions are dictated by the availability of nutrients as well as the metabolic capability of each participant to transform the nutrients into metabolic energy and biomass components. Many bacterial strains (both commensal and pathogenic) lose the capability to synthesize essential biomass precursors and become dependent on extracellular resources for survival despite having prototrophic ancestors. Some auxotrophies arise as a result of the strain’s adaptation to a specific host or environment through the formation of small-scale deleterious mutations leading to gene loss (1). For example, a methionine requirement is common among Pseudomonas aeruginosa strains isolated from cystic fibrosis patients (2–4), a requirement likely satisfied by the high concentration of amino acids in the patient’s sputum (5). Nutrient obligate pathogens are often host-associated (6), have a reduced genome (7), and retain a fitness advantage over the free-living bacteria in their specific niche (8). Additionally, auxotrophs dictate the carbon and energy flow as well as the stability of endosymbiotic communities (9). Auxotrophies have been exploited 1) as markers for strain detection and identification (10–12), 2) for the elucidation of their lifestyle and microenvironment (13–17), 3) for the design of microbial ecosystems (18, 19), 4) for the design of attenuated live vaccines (20, 21), and 5) for molecular therapy and tumor targeting (22–24).
The identification of an auxotroph’s nutrient requirements experimentally is difficult, occurs on a strain-by-strain basis, and is rarely accompanied with an identified causative genetic or genomic lesion. Comparative genomic analyses suggest that deleterious disruption of biomass precursor biosynthetic pathways exist in most free-living microorganisms, indicating that they rely on cross-feeding (25). However, it has been demonstrated that amino acid auxotrophies are predicted incorrectly as a result of the insufficient number of known gene paralogs (26). Additionally, these methods rely on the identification of pathway completeness, with a 50% cutoff used to determine auxotrophy (25). A mechanistic approach is expected to be more appropriate and can be achieved using genome-scale models of metabolism (GEMs). For example, requirements can arise by means of a single deleterious mutation in a conditionally essential gene (CEG), or as a result of a combination of deletions, in which case they would escape detection via comparative genomics. Given the high interconnectedness of metabolic networks, finding such sets manually is not a trivial task but can be efficiently approached computationally using GEMs.
GEMs are assembled based on genome annotation and curation of published literature (27, 28). They contain the most up-to-date metabolic networks linking reactions with genes according to experimentally validated mechanisms (28–30). Once they are converted into a mathematical format (27, 31), flux balance analysis (32) can be used to identify essential genes and auxotrophies that result from gaps in the network (15, 31, 33, 34). However, a workflow for this purpose has yet to be formalized into a mathematical problem and developed into a fully fledged algorithm that can be reused across the community. Here, we develop a custom algorithm (AuxoFind) using comparative genomics coupled with metabolic modeling to computationally predict auxotrophies and pinpoint the corresponding genetic basis.
Results
AuxoFind Predicts Auxotrophy from Genomic Sequences Using GEMs.
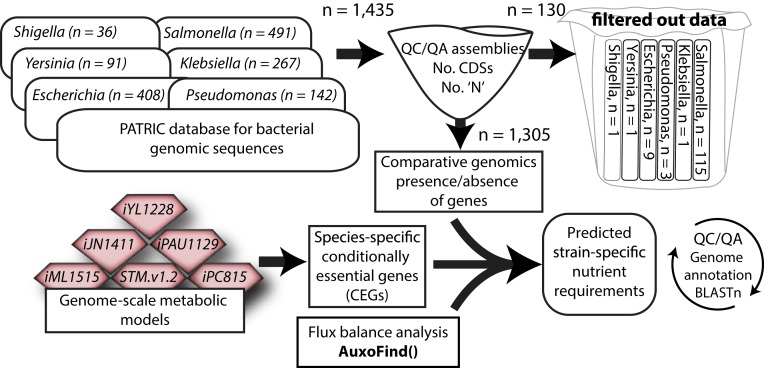
As a first step toward determining strain-specific auxotrophy, we collected available curated GEMs for Gram-negative bacteria from the Biochemically, Genetically and Genomically structured genome-scale metabolic network reconstructions (BiGG) database (15, 31, 35–37). We proceeded to download and quality check genomic sequences from the PATRIC database (38), including 408 Escherichia, 491 Salmonella, 91 Yersinia, 142 Pseudomonas (39), 267 Klebsiella, and 36 Shigella sequences (Fig. 1, Dataset S1, and SI Appendix, SI Materials and Methods). For each of the six GEMs, one for each genus, we predicted CEGs for aerobic growth on minimal medium. CEGs differ from absolutely essential genes in that their absence can be compensated for by the addition of an extracellular nutrient. In other words, if a strain is missing a CEG, it is auxotrophic for one or more nutrients. In contrast, a strain cannot survive without any one of its essential genes, regardless of the nutritional background. We then homology mapped all of the modeled genes to other strains within the same genus to identify the strains which are lacking one or more CEG, and developed a custom algorithm (AuxoFind) which predicts nutrient requirements from a list of present and absent metabolic genes using flux balance analysis (SI Appendix, SI Materials and Methods) (32). AuxoFind exploits the mechanistic link between enzymatic functions and prototrophy encoded in GEMs, taking into account metabolic and genetic redundancy, and using the genomic background of each strain as input. Applying AuxoFind allows the user the flexibility to choose a growth medium and a biomass objective function to take into full account the strain’s metabolic environment. This, in turn, allows for the analysis of auxotrophies as a result of changing nutrient sources, or biomass requirements. In addition, instead of returning a single solution, AuxoFind can be set to output multiple alternative solutions as well as suboptimal solutions. Applying AuxoFind to the strains collected from PATRIC, we predicted a total of 58 strains to be auxotrophic for at least one nutrient, 4 of which (Salmonella enterica serovar Bovismorbificans str. 3114, serovar Enteritidis str. EC20090884, serovar Ouakam, and serovar Paratyphi A strain A73-2) were subsequently selected out [see Experimental Validation of Auxotrophies Highlight Technological Shortcomings at Multiple Levels for filtering criteria through Basic nucleotide Local Alignment Search Tool (BLASTn)]. The final results included 11 Salmonella strains, 18 Yersinia strains, 15 Escherichia strains, 5 Pseudomans putida, and 5 Klebsiella strains. The predicted auxotrophies in these 54 strains are analyzed in detail below.
Fig. 1.
Workflow chart: Genomes were downloaded from PATRIC (38) and quality controlled based on completeness, number of annotated coding DNA sequences, and percentage of unassigned nucleotide sequences. Manually curated GEMs were queried from BiGG (35), and used to identify CEGs in minimal medium. Each GEM was used across strains of the same genus, except for iML1515, which was used for both Escherichia and Shigella strains, iJN1411, which was only used for P. putida, and iPAU1129, which was only used for P. aeruginosa. Next, we identified the list of missing metabolic genes in each strain through comparative genomics using genomic sequences as an input. We used AuxoFind to predict auxotrophies and their genetic basis using, as input, the identified list of missing genes. Finally, when a missing gene was linked to a predicted auxotrophy, we verified its absence algorithmically using BLASTn.
The Majority of Predicted Nutrient Requirements Were Specific.
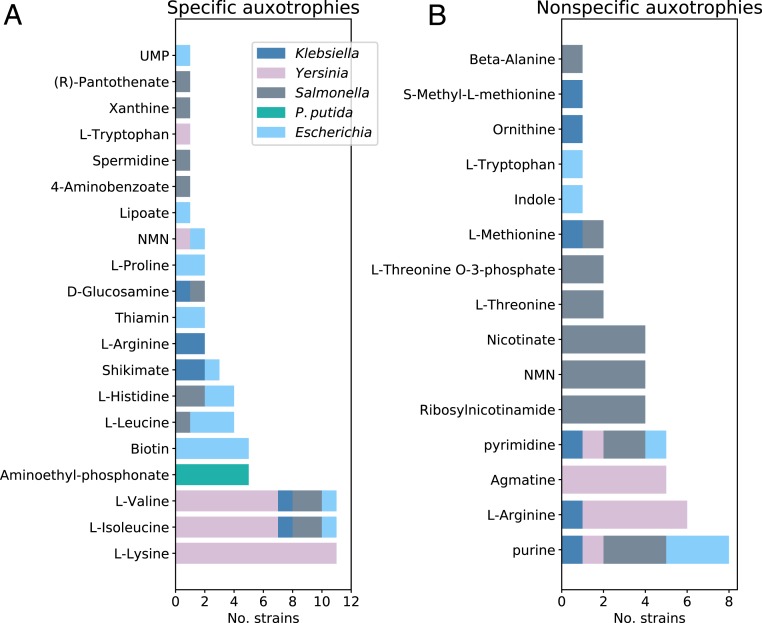
We classified the predicted nutrient auxotrophies into two categories: specific and nonspecific. Specific auxotrophies occur when the strain requires a specific nutrient to be added to minimal medium in order to grow, while a strain with a nonspecific auxotrophy can grow when any of a selection of nutrients is added to minimal medium. The requirement for amino acids was found to be predominantly specific (Fig. 2A), while requirements for nucleotides were nonspecific (Fig. 2B). The specificity of amino acid auxotrophy is due to the structure of the metabolic pathways, and the irreversibility of intermediate steps. In contrast, nucleotide biosynthesis can be achieved via multiple routes (including purine and pyrimidine biosynthesis), as well as nucleotide salvage and interconversion. In the latter subsystem, there are multiple redundant pathways, few of which are irreversible, resulting from the promiscuity of participating enzymes. Interestingly, multiple auxotrophies which were predicted across isolates involved nutrients known to be important in host–pathogen interactions, suggesting that these auxotrophies may give selective advantage during host–pathogen interactions. For example, specific auxotrophies for branched chain amino acids (BCAAs: L-isoleucine, L-leucine, and L-valine) were shared across 11 strains, 5 of which were isolated from human samples. Intracellular levels of BCAAs play a critical role in host–pathogen interactions, affecting both pathogenicity and immune activation (40, 41). Similarly, L-tryptophan (n = 2) constitutes a resource over which the host and the pathogen compete (42), niacin (n = 5) affects the pathogen’s virulence and its detection by the immune system (43), and tetrathionate (n = 1) is a gut inflammation by-product which is known to provide a respiratory electron acceptor in Salmonella (44). In total, 72% (107 out of 149) of predicted auxotrophies were specific, with some strains having multiple specific and/or nonspecific predicted requirements (Dataset S2). Notably, we observed a specific L-lysine requirement due to the absence of argD across 11 Yersinia pestis strains, a specific biotin requirement (due to the absence of either bioAB, fabH, or fabI) across 5 Escherichia coli strains, multiple amino acid auxotrophies in Yersinia ruckeri strains, and an L-leucine requirement in 3 E. coli K-12 strains. To determine which strains were closely related, we constructed phylogenetic trees from the nucleotide polymorphism (single-nucleotide polymorphism [SNP]) count from the concatenation of all core genes using rapid core genome multi-alignment (ParSNP) (45). While the Y. pestis L-lysine auxotrophs were not clustered together in a single subclade, the L-leucine E. coli auxotrophs, Y. ruckeri amino acid auxotrophs, and E. coli K-12 strains were (SI Appendix, Figs. S1 and S2). Otherwise, predicted auxotrophs were generally spread across the phylogenetic tree, with many subclades containing only one auxotrophic strain.
Fig. 2.
In silico predictions of metabolic nutrient requirements across multiple Gram-negative species. (A) Nutrients for which a specific auxotrophy was predicted in at least one strain. (B) Top 15 metabolites for which a nonspecific auxotrophy was predicted (see Dataset S2 for the full results). Nutrient requirements were predicted for a total of 54 strains across Escherichia coli, Salmonella, Klebsiella, and Yersinia, with amino acids auxotrophies appearing with the highest frequency.
Amino Acid and Vitamin Auxotrophies Confer a Fitness Advantage In Vivo.
We next sought to identify the effect of metabolic requirements on the fitness of auxotrophic strains in their native environment. We evaluated published fitness profiles for E. coli strains UTI89 and EC958 and S. enterica subsp. enterica ser. Typhimurium str. SL1344 mutants across various in vivo and in vitro environments (including cattle, pig and chicken intestine, mouse spleen, human serum, and bladder cell infection model) (46–51). Briefly, fitness profiles are derived through transposon-directed insertion-site sequencing (TRADIS), and a fitness measure is calculated by comparing the number of reads across mutants between the inocula and output samples. We posit that a strain with a disrupted CEG (as defined in AuxoFind Predicts Auxotrophy from Genomic Sequences Using GEMs) has an increased fitness, that is, when the gene’s function is dispensable. In this case, it is evident that the mutant’s auxotrophy is at least partially compensated for by a favorable nutritional background. Conversely, loss of function (and therefore nutrient dependence) resulting in reduced fitness indicates an unfavorable environment and/or insufficient access to important metabolites. Nutrient dependence was predicted in both aerobic and anaerobic conditions for each transposon mutant by AuxoFind, by knocking out the disrupted gene in silico and simulating for auxotrophy (Datasets S3 and S4). TRADIS yields a fitness measure (log2 fold change) for each mutant, which we filtered for adjusted P value smaller than 0.05. A total of 960 measured log2 fold changes passed these thresholds. We considered log2 fold changes in fitness smaller than −1 to be detrimental and those larger than 1 to be beneficial. Only 25 CEGs yielded increased fitness upon disruption in at least one condition, while 70 were detrimental (in ≥1 conditions) (Fig. 3). At the gene level, 5 out of 15 in E. coli and 6 out of 12 in S. enterica of the CEGs whose disruption increased fitness were lost in one or more natural isolates (highlighted in bold in Fig. 3; Datasets S3 and S4).
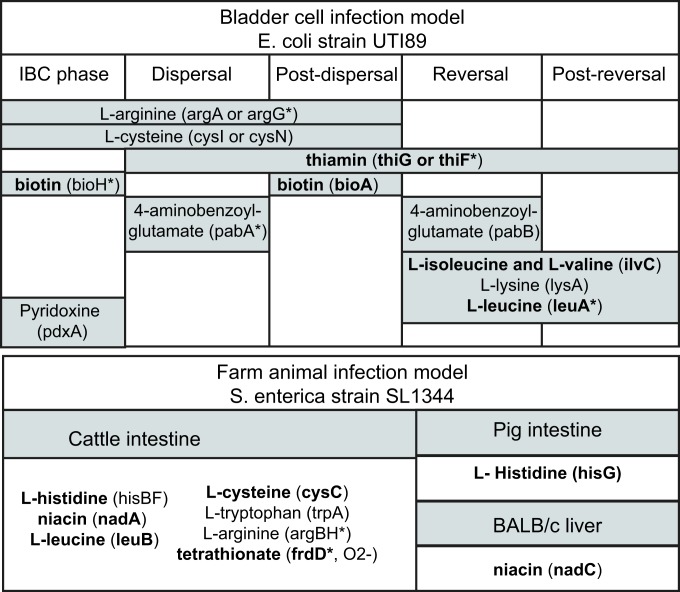
Fig. 3.
Computationally predicted CEGs that were found to result in increased fitness (more than one log2 fold change, P value < 0.05) in mutant screens. For each condition in which the mutant fitness was tested, we list out both the nutrient for which it is predicted to be auxotrophic and the gene which has been disrupted. The fitness profiles were obtained from various sources in which the TRADIS workflow was applied. The bladder cell infection model was designed as a proxy for urinary tract infection. Genes highlighted in bold are lost across natural isolates. An asterisk (*) indicates that CEG knock-out yields a detrimental effect on fitness in other conditions. See Datasets S3 and S4 for the full dataset.
Of note, the disruption of two out of five and five out of six CEGs in S. enterica and E. coli, respectively, yielded increased fitness in one condition but decreased fitness in another, suggesting that the conferred fitness advantage is niche-specific. For example, argH and frdD disruption in S. enterica were beneficial in chicken intestine but detrimental in cattle intestine. Additionally, fitness change upon CEG disruption varied across phases of infection. For example, the disruption of bioH in E. coli was advantageous in the intracellular bacterial communities (IBC) phase but detrimental in later phases (dispersal and postdispersal phase). In contrast, leuA disruption yielded decreased fitness in the dispersal phase but increased fitness in the reversal and postreversal phases. Some beneficial nutrient requirements carried over from one stage of infection to the next. For example, L-arginine and L-cysteine auxotrophic mutants exhibited elevated fitness in three consecutive phases including IBC, dispersal, and postdispersal phases. In addition, a thiamin requirement was beneficial in four out of the five tested phases of infection. Other auxotrophic mutants with increased fitness included biotin, 4-aminobenzoyl glutamate, L-isoleucine and L-valine, L-lysine, and L-leucine (49). In an intestinal infection of cattle with S. enterica, auxotrophic mutants with elevated fitness were auxotrophic for nicotinate (nadA), L-histidine (hisBF), L-cysteine (cysC), L-arginine (argBH), L-leucine (leuB), tetrathionate (frdD only under anaerobic conditions), and L-tryptophan (trpA).
The metabolic basis for auxotrophies diverges across species as a function of their metabolic network topology and systems-level metabolic capabilities.
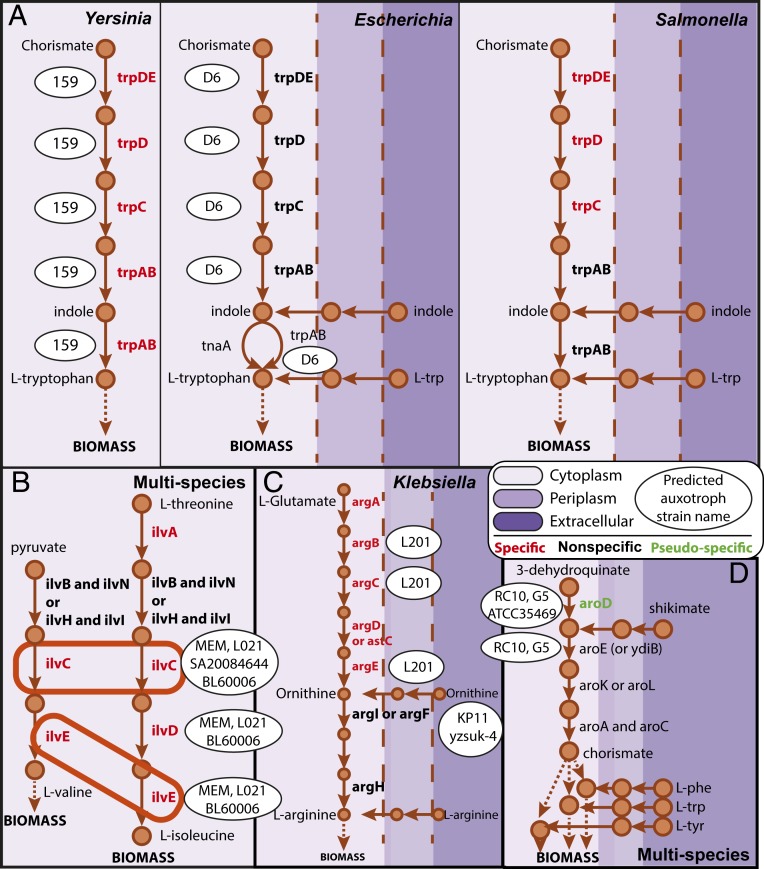
A subset of genes conferred both specific and nonspecific auxotrophies, depending on both the location of the missing enzymatic function in the strain-specific metabolic pathway and the species-specific local structure of the network. For example, L-tryptophan biosynthesis can be achieved via three different routes in Escherichia, two in Salmonella, and only one in Yersinia (Fig. 4A). Strains across all three genera have the capability to synthesize L-tryptophan from chorismate (via trpABCDE), while only Salmonella and Escherichia strains are capable of indole transport and utilization, and only Escherichia strains can synthesize L-tryptophan via both tnaA and trpAB pathways. As a result, loss of trpCDE confers a nonspecific auxotrophy in Escherichia and Salmonella but a specific auxotrophy in Yersinia, while the loss of trpAB confers a specific auxotrophy in Salmonella and Yersinia but is not conditionally essential in Escherichia. In our dataset, both E. coli str. D6 and Yersinia aleksiciae str. 159 were missing the full trp operon. However, strain D6 was predicted to be a nonspecific L-tryptophan auxotroph, while strain 159 was predicted to have an L-tryptophan-specific requirement.
Fig. 4.
(A) Specificity of L-tryptophan requirement as a function of species-specific systems-level pathway structure. (B) Multiple simultaneous specific auxotrophies as a result of single gene deletion. (C) Auxotrophies as a result of deletion of multiple isozymes. In K. pneumoniae str. KP11, argI and argF are both absent. (D) Pseudospecific auxotrophies in which the alternative to one nutrient requirement (e.g., shikimate) is the simultaneous requirement for multiple nutrients (e.g., L-phenylalanine, L-tryptophan, and L-tyrosine).
We observed cases in which the simultaneous supplementation of multiple nutrients was required to support growth. The multiplicity of nutrient requirements was caused either by the absence of multiple CEGs distributed across different pathways or by the participation of a CEG in multiple essential biosynthetic pathways. For example, ketol-acid reductoisomerase (ilvC) is essential for the biosynthesis of both L-valine and L-isoleucine. In the absence of ilvC alone, supplementation of both L-valine and L-isoleucine is required (Fig. 4B). Interestingly, ilvC, ilvD, and ilvE were lost across strains in multiple species including Klebsiella pneumoniae, S. enterica, and E. coli. There were also cases in which only the simultaneous absence of two or more genes (e.g., encoding isozymes) was predicted to confer an auxotrophy. For example, K. pneumoniae strain L201 and S. enterica ser. Newport str. 0307-213 were predicted to require L-arginine supplementation due to the absence of both acetylornithine deacetylase and ornithine carbamoyltransferase isozyme (Fig. 4C). Finally, we observed instances in which the alternative to supplementing with one nutrient was to supplement with multiple nutrients. For example, a shikimate auxotrophy was predicted for Klebsiella G5, Klebsiella michiganensis str. RC10, and Escherichia fergusonii str. ATCC35469 due to the absence of 3-dehydroquinate dehydratase (aroD). If shikimate is excluded from the set of acceptable supplementations, a requirement for multiple nutrients (including L-tyrosine, L-tryptophan, and L-phenylalanine) is predicted, making the shikimate requirement pseudospecific (Fig. 4D).
Small-Scale Mutations Constitute the Genetic Basis for Auxotrophies in P. aeruginosa and Shigella.
Among the species studied, none of the P. aeruginosa or Shigella species in our dataset were predicted to be auxotrophic, despite extensive reports for amino acid auxotrophy across P. aeruginosa strains isolated from cystic fibrosis patients and a predominant niacin auxotrophy in Shigella strains (3, 14). Instead, we found that CEGs were highly conserved. Niche adaptation through small-scale loss-of-function mutations has been observed in strains including P. aeruginosa and Shigella (52–54). This result emphasizes that, in order to study auxotrophy development in host-adapted strains, future efforts should expand our workflow for the prediction of bacterial nutrient requirements (which is currently limited to the identification of genetic lesions at the gene level) to account for smaller-scale deleterious mutations. Here, we do not attempt to predict pseudogenization events, as this would constitute an effort of its own. However, for proof of concept, we demonstrate one such analysis for the well-known case of niacin auxotrophy in Shigella, extending it to all strains in our dataset.
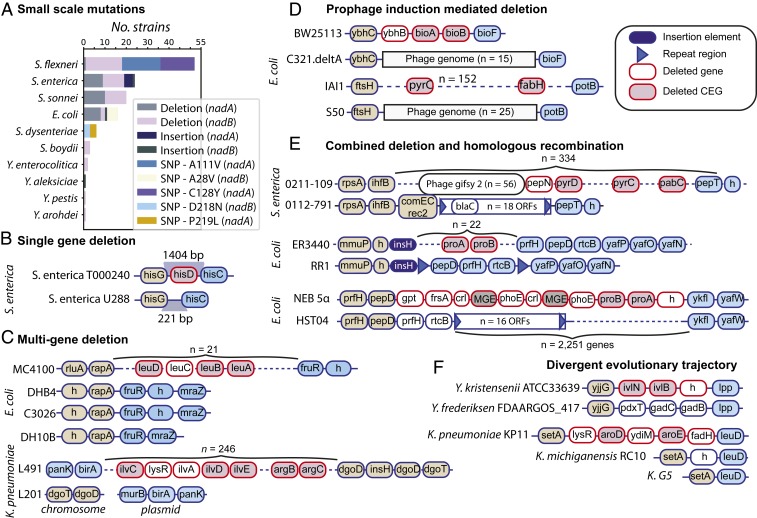
Causal loss of function mutations in nadB (including A28V, D218N, and G74E) and in nadA (including W299X, P219L, C128Y, C113A, C200A, C297A, and A111V) result in a niacin requirement in natural strains of E. coli, Shigella, and S. enterica (14, 43, 55). We searched our dataset for these mutations and found a total of 71 strains carrying at least one of the validated SNPs and/or an indel or deletion of more than 10 amino acids (which are likely to result in protein structural variations and which we assume to be deleterious) (56, 57). The affected species were Shigella flexneri, S. enterica, Shigella sonnei, E. coli, Shigella dysenteriae, Shigella boydii, Yersinia enterocolitica, Y. pestis, and Yersinia rohdei, and the SNPs found included A111V, C128Y, and P219L (nadA) and A28V and D218N (nadB). (Fig. 5A). Interestingly, subsets of deleterious mutations were restricted to different species. For example, C128Y and A111V mutations were restricted to S. flexneri strains, D218N and P219L were restricted to S. dysenteriae strains, and A28V was restricted to E. coli strains (Dataset S5). While the A111V mutation was initially described in S. enterica serovar Dublin, we only identify it here in strains of S. flexneri (55). Additionally, we observe 16 S. enterica serovar Enteritidis strains to carry large deletions in either nadA or nadB (or both in the case of 5 strains). These results demonstrate that convergent evolution may have led to loss of the nicotinate biosynthesis capability across species. That the deleterious mutations were maintained across descendants indicates that a niacin requirement confers a selective advantage, an observation which is also supported in Amino Acid and Vitamin Auxotrophies Confer a Fitness Advantage In Vivo.
Fig. 5.
The genetic basis for nutrient auxotrophy spans various levels of complexity. (A) Niacin auxotrophy due to known loss-of-function mutations in nadA and nadB as well as large in-frame deletions/insertions (>30 amino acids). (B) Single gene deletion of hisD in S. enterica strain T000240. (C) Simple multigene deletion with rejoining of deletion edges. (D) Phage insertion and phage-mediated multigene deletion. (E) Multigene deletion coupled with homologous recombination mediated by prophages and insertion sequences. (F) Divergent and ancient evolutionary trajectory across species.
Genomic Basis of Auxotrophy.
Next, we proceeded to examine the genomic basis of the auxotrophies predicted from our workflow. To assess the genetic changes at the strain level, we compared the genomic region of each predicted auxotroph with a closely related strain (SI Appendix, SI Materials and Methods). We observed multiple cases in which a missing CEG constituted a part of a larger deleted genomic fragment, in which multiple syntenic operons were lost simultaneously. We found that the number of missing syntenic genes surrounding an absent CEG varied from n = 1 to n = 251 open reading frames (ORFs) averaging 49 genes. S. enterica str. U288 was the only strain for which the missing chromosomal region contained only one CEG (hisD) (Fig. 5B). Conversely, S. enterica str. 0112-791 was missing two genomic regions (n = 251 ORFs and n = 28 ORFs) and did not carry a total of four CEGs.
When the conserved genes flanking the missing genomic fragment were adjacent in the auxotroph, we classified the observed loss as a simple deletion. There were 15 cases of simple multigene deletion events (Fig. 5C). For example, the deleted region in E. coli strains DHB4, C3026, and DH10B consisted of 21 genes, and the genes upstream and downstream of the deletion endpoints in E. coli MC4100 (rapA and fruR) are adjacent in the three auxotrophs. Conversely, K. pneumoniae strain L201 is missing a total of 246 contiguous ORFs with respect to strain L491. One deletion edge was located at position 4,943,680, marking the end of the chromosomal GenBank file, while the other was located at position 371, marking the start of the sequence for plasmid p1-L201. We suspect that an assembly error may explain this observation.
In the remaining instances, the genes which were located at the edges of the missing fragment were separated by multiple ORFs in the auxotroph. Interestingly, these ORFs constituted a prophage in four auxotrophs, suggesting that insertion of viral DNA may have mediated the deletion event (Fig. 5D). In particular, E. coli strain C321.A, which is a genomically recoded organism lacking bioAB and ybhB, carried enterobacteria phage [containing 15 genes, predicted by PHAge Search Tool (PHAST) (58)] in the deletion locus. Similarly, E. coli strain S50 (isolated from forest soil) carried a prophage sharing three ORFs with phage Stx2 at the locus for 152 missing contiguous genes.
A slightly more complex sequence of events affected S. enterica ser. Newport strain 0211-109 which was isolated from a cow with gastroenteritis (Dataset S6 and Fig. 5E). At the locus of deletion (consisting of 252 genes), we found an insertion sequence cluster likely conferring beta-lactam resistance (18 ORFs, containing two copies of Class C beta-lactamase, three copies of small multidrug efflux transporters, and three copies of mobile element proteins). The auxotroph also carried a DNA internalization-related competence protein ComEC/Rec2 (involved in binding and uptake of transforming DNA) directly upstream of the insertion sequence cluster. Notably, the deletion region (spanning genes between pepN and potA) was located immediately downstream of prophage gifsy 2 (with 56 ORFs) in a close relative (strain 0112-791), but was relocated elsewhere in strain 0211-109. We hypothesize that genomic rearrangement was caused by the inserted cluster of genes. Similarly, 14 genes are deleted in E. coli strain RR1 (a derivative of K-12), including proA and proB, and 9 genes upstream of the deleted fragment (including 3 transposases) are redistributed across the genome. At the locus of deletion, RR1 carries multiple repeat regions denoting that transposition may have occurred. In E. coli strain HST04, the genes flanking the deletion region (pepD and ykfl) are located 2,251 ORFs apart, with an insertion sequence cluster consisting of 16 ORFs located downstream of pepD. Insertion elements can promote the rearrangement of bacterial genomes (59).
Finally, we observed four instances in which the predicted auxotrophs corresponded to species for which there was only one representative genome in our dataset. For example, Yersinia frederiksenii carries pdxT and gadCB between yjjG and lpp, while Yersinia kristensenii (which shares the largest number of gene families) carries a hypothetical protein ilvNB (involved in L-isoleucine biosynthesis). The absence of CEGs in these cases are likely a result of evolutionary events occurring after speciation (Fig. 5F), and a larger number of pertinent genomic sequences would be necessary to retrace the evolutionary history of these chromosomal regions.
Genome streamlining is often associated with niche adaptation and evolution toward symbiosis, and massive gene losses can occur on a small evolutionary timescale as a result of population bottlenecks (60). We asked whether the predicted auxotrophs had a reduced genomic sequence length with respect to other strains of the same genus. For each genus, we collected the strains’ sequence length and fit the observed distribution to a generalized extreme value distribution using the block maxima approach. We calculated the probability of a genome length to be less than or equal to each value in our dataset, and found that a total of 41 strains fell under a probability of 5%. Of those, six were predicted auxotrophs (including K. michiganensis strain RC10, K. pneumoniae strains KP11 and yzusk-4, K. G5, S. enterica str. 0112-791, and S. enterica str. 9-65), further supporting the hypothesis that these strains have developed auxotrophy as a result of niche adaptation. However, a Fisher’s exact test reveals that there is no significant enrichment of auxotrophs among the population of strains with reduced genomes (P value = 0.6), indicating that genome streamlining is not a predominant phenomenon across the six genera.
Experimental Validation of Auxotrophies Highlight Technological Shortcomings at Multiple Levels.
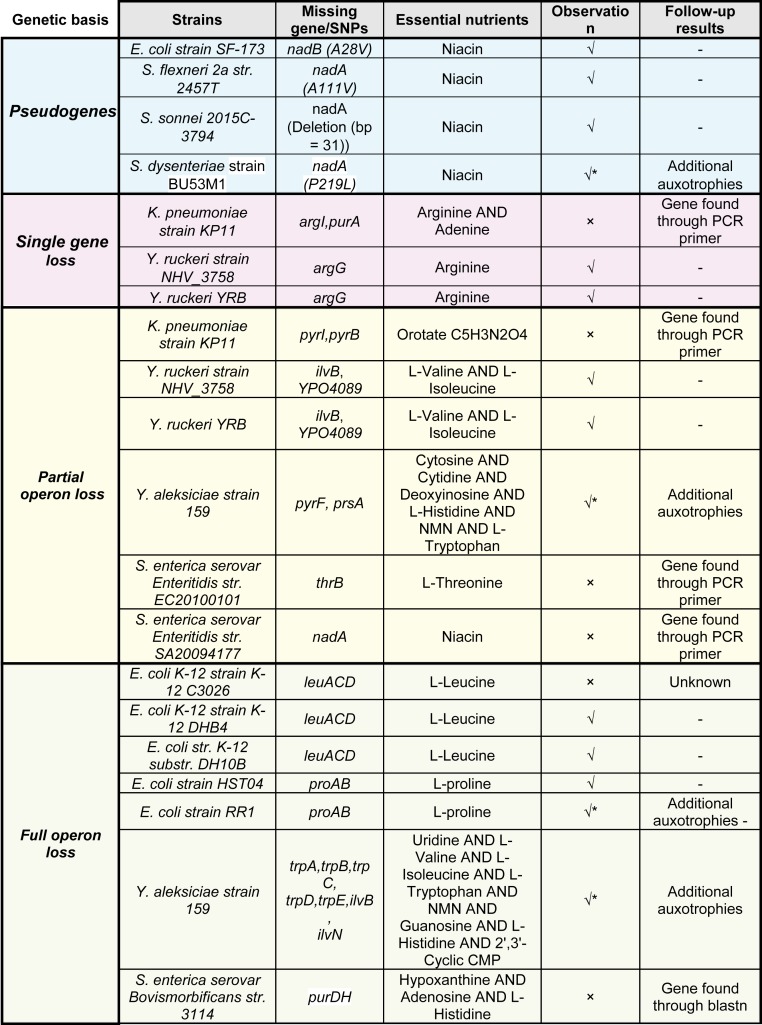
We proceeded to validate our predictions experimentally by evaluating growth requirements of the predicted auxotrophs. We were able to obtain six E. coli (61–64), three Yersinia (65), three Salmonella (66, 67), three Shigella (68, 69), and one Klebsiella (70) strain (Fig. 6). Out of 16 strains that we experimentally tested, 8 (4 E. coli, 2 Y. ruckeri, 1 S. flexneri, and 1 S. sonnei) grew only when glucose + M9 media was supplemented with the predicted essential nutrient(s). The confirmed auxotrophies included 1) an L-proline requirement in E. coli strain HST04; 2) an L-leucine requirement in E. coli strains DHB4 and DH10B; 3) a niacin requirement in E. coli strain SF-173, S. flexneri strain 2457T, and S. sonnei strain 2015C-3794; and 4) an L-valine, L-isoleucine, and L-arginine requirement in Y. ruckeri strains YRB and NHV_3758. In addition, we found literature evidence for a biotin requirement in E. coli strain C321.A and its two genomically recoded derivatives (CP006698.1, CP010455.1, and CP010456.1) (71). Three predicted auxotrophs could grow neither in minimal medium nor in minimal medium supplemented with the corresponding predicted nutrient. For example, E. coli strain RR1 is a predicted L-proline auxotroph, S. dysenteriae strain BU53M1 is a predicted niacin auxotroph, and Y. aleksiciae strain 159 was predicted to have multiple auxotrophies. However, neither exhibited any growth upon supplementation, suggesting that they may have additional nutrient requirements.
Fig. 6.
Results of in-house experimental validations for nutrient requirements across 16 Gram-negative strains and outcome of follow-up experiments and analysis for failure cases. Growth curves, PCR primers, and details regarding the list of strains can be found in SI Appendix. An asterisk (*) denotes that these strains couldn’t grow upon predicted nutrient supplementation, suggesting that they have additional nutrient requirements. Additionally, we found literature evidence for a biotin requirement in E. coli strain C321.A and its two genomically recoded derivatives. Note that one strain may have multiple genetic basis of auxotrophy, for example, Yersinia strains. (71).
Notably, we tested growth of two Y. ruckeri strains on a reduced chemically defined medium. While there is no precedent for such an effort for this species, a chemically defined medium was nonetheless derived for multiple clinical Y. enterocolitica (which included L-methionine, L-glutamate, glycine, and L-histidine) (72), and another for Y. pestis strains (with 12 amino acids, 3 vitamins, and citrate) (73, 74). Our strain-specific models predicted an auxotrophy for six amino acids: L-phenylalanine, L-methionine, L-cysteine, L-arginine, L-valine, and L-proline, with the first three carrying over from the reference reconstruction for Y. pestis strain CO92. However, the supplementation of M9 with all six nutrients alone could not support growth unless R-pantothenate was also added. This result came as a surprise, since both strains seem to carry an intact R-pantothenate biosynthetic pathway. Consequently, we found severe growth limitations to arise in both strains in the absence of L-methionine, R-pantothenate, and L-isoleucine, with intermediate growth obtained in the absence of L-valine, L-cysteine, or L-arginine. These validated modeling predictions confirm that the approach suggested by D’Souza et al. (8) may miss a few cases due to conservative thresholding (SI Appendix, SI Text).
Follow-up analyses and experiments highlighted links between the remaining four erroneous predictions and technological shortcomings at multiple levels, including 1) wrong sequence annotation (which was corrected by running BLASTn directly on the assembly, S. enterica ser. Bovismorbificans strain 3114), 2) localized low sequencing quality (identified by gene-specific primers, K. pneumoniae strain KP11, S. enterica strains EC20100101 and SA20094177), 3) erroneous assemblies (verified through manual analysis of the deletion regions in S. enterica ser. Bovismorbificans strain 3114), 4) truncated assembly with genes missing at the origin of sequencing (e.g., verified through manual analysis of the deletion regions, S. enterica ser. Bovismorbificans strain 3114), and 5) potential reconstruction knowledge gaps (experimental trial and error and intermediate growth observed for Y. ruckeri strains). In particular, while S. enterica strain 3114 had a high-quality assembly, genes that should have been located near the origin of sequencing were absent, and could only be found via BLASTn. As a result of these observations, we subsequently added one quality control check consisting of a search for the missing CEG in the assembly file via BLASTn. The results are shown in Dataset S2.
Discussion
In this study, we devise an algorithm (AuxoFind) which bypasses user-defined thresholds and pathway definitions using reconstructed genome-scale networks of metabolism and 1,305 quality controlled/quality assured publicly available complete genomic sequences to 1) computationally predict auxotrophies, 2) identify the corresponding metabolic basis, and 3) explore the underlying genetic basis. We further verify 16 of our predictions experimentally and identify the basis for inconsistencies between predictions and observations.
We predict auxotrophies for several amino acids, nucleotides, and vitamins, distinguishing specific from nonspecific nutrient dependencies. Surprisingly, only 38% of predicted auxotrophies were nonspecific. Nonspecific auxotrophs should have a more relaxed flexibility in their ability to grow across nutritional environments with respect to specific auxotrophs while still relying on external nutrient sources. However, such a view does not take into account the strain’s phylogeny which indicates that the strain’s ancestor was prototrophic, and that auxotrophy likely developed as a result of selection pressure directed toward the utilization of a key nutrient in its immediate niche. Indeed, we predict specific auxotrophies for multiple nutrients previously found to be involved in host–pathogen interactions (including BCAAs, L-tryptophan, niacin, and tetrathionate), or which seem to provide a fitness advantage in various niches in vivo (including L-histidine, L-cysteine/tetrathionate, L-tryptophan, niacin, L-glutamine, L-arginine, and L-leucine). Strikingly, we observe that auxotrophies that are beneficial in one environment are detrimental in another. In addition, while the fitness benefits of some auxotrophies carries over multiple stages of bladder infection (such as L-arginine, L-cysteine, and thiamin), that of others (L-leucine and biotin) varies across stages. We hypothesize that these variations reflect differences in nutritional availability between niches and suggest that the context-specific nutritional background likely plays a role in auxotrophy development.
We found that the metabolic basis (including specificity/nonspecificity and multiplicity) of auxotrophies depends on 1) the entire structure of the metabolic pathway, 2) the promiscuity of a protein’s enzymatic activity, and 3) functional or pathway redundancy, and therefore varied in a strain-specific fashion. CEGs carrying out the same function in two different strains can confer a specific auxotrophy in one species but a nonspecific auxotrophy in the other. Additionally, two CEGs participating in the same biosynthetic pathways confer different simulated specificity upon deletion, depending on the position of alternative pathways with respect to that of the CEG. We therefore suggest that selective pressures for auxotrophy development leading to loss of function may affect paralogs differently across strains and vary across CEGs participating in the same pathway as a function of a strain’s full reactome.
We observed a continuity in the complexity of the genetic basis for auxotrophy, ranging from single nucleotide polymorphism causing a loss of function mutation to large multigene and multioperon deletions coupled with extensive homologous recombination events. Interestingly, the only case of a single gene deletion event affected hisD, a gene which was observed to have the largest number of alleles in a pangenome analysis of E. coli strains (36). There were multiple instances in which the loss of CEGs was likely mediated and/or accompanied by prophage insertion and/or insertion sequence movement across the genome, with one strain losing four CEGs due to the insertion of a cluster of genes conferring beta-lactam resistance. In particular, 6 of the 54 predicted auxotrophs had significantly smaller genomes, suggesting that they are niche adapted; this is indeed the case for both S. enterica serovar Newport strain 0112-791 and serovar Paratyphi A strain 9-65. Overall, auxotrophies arising from large-scale deletions (one or more ORFs) are rare (3.8%) in our dataset. They could perhaps be reversed under the right conditions when their genetic basis constitutes small variations such as SNPs (75). However, major events such as full gene deletion and full operon removal are likely to be more permanent and highly constrain the strain’s colonization space and bacterial social network.
Finally, we experimentally verified our predictions for nutrient requirements in 16 strains and observed that 11 strains were auxotrophs, but that minimal media could support growth of 5 mutants. The latter strains served to highlight technological shortcomings at multiple levels. The challenges behind calling genes/functions absent from a genomic sequence became apparent, and the identification of deletions/missing genes is hampered, even in complete sequences, by 1) uneven sequencing quality across the genome, 2) incorrect genome assembly, and 3) erroneous genome annotation. We observed that pangenome alignment (at the ORF level) of closely related prototrophs can be used to overcome these technological shortcomings and distinguish between true and false positives. Knowledge gaps in amino acid biosynthesis of Y. ruckeri, and the presence of unknown in-frame loss of function mutations affecting three strains, constituted additional sources of inconsistency between in silico predictions and experimental observations. These contradictions generate testable hypotheses for follow-up studies (76).
Altogether, our results constitute the most comprehensive systems biology effort aimed at predicting and understanding nutrient auxotrophies using mechanistic models of metabolism. The approach developed can be applied to quickly and systematically predict nutrient requirements from genomic sequences.
Materials and Methods
The specific procedure of data collection and quality control, prediction of CEGs, and homologous gene identification is described in SI Appendix,SI Materials and Methods. The detailed workflow for prediction of nutrient auxotrophy is described in SI Appendix, SI Materials and Methods. Determination of gene neighborhood and synteny is described in SI Appendix, SI Materials and Methods. Sixteen strains were tested as a part of this study. The experimental validation methods and conditions are described in SI Appendix, SI Materials and Methods.
Data Availability.
All data generated in this study are included in this published article (and in SI Appendix and Datasets S1–S6). AuxoFind is available on GitHub, with an example notebook. All genomic sequences analyzed in this study are publicly available on PATRIC (38), and accession numbers are available in Dataset S1.
Supplementary Material
Acknowledgments
We thank Dr. Karsten Zengler and Marc Abrams for reviewing the manuscript and providing constructive suggestions. This work was supported by NIH Grants AI124316 and GM057089, and Novo Nordisk Foundation Grant NNF10CC1016517. We are grateful to Drs. Rebecca Lindsey, Nancy Strockbine, Shi Chen, Sang Jun Lee, Dana Boyd, Mehmet Berkmen, Henning Sørum, David Rozak, Shannon Lyn Johnson, Craig Winstanely, Roger Johnson, and Weihua Huang for generously providing bacterial strains for this study.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: AuxoFind is available on GitHub, with an example notebook. https://github.com/yseif/AuxoFind. All genomic sequences analyzed in this study are publicly available on The Pathosystems Resource Integration Center (PATRIC).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910499117/-/DCSupplemental.
References
- 1.Low B. K., Auxotroph in Encyclopedia of Genetics, Brenner S., Miller J. H., Eds. (Academic, New York, NY, 2001). [Google Scholar]
- 2.Agarwal G., Kapil A., Kabra S. K., Das B. K., Dwivedi S. N., Characterization of Pseudomonas aeruginosa isolated from chronically infected children with cystic fibrosis in India. BMC Microbiol. 5, 43 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barth A. L., Pitt T. L., Auxotrophic variants of Pseudomonas aeruginosa are selected from prototrophic wild-type strains in respiratory infections in patients with cystic fibrosis. J. Clin. Microbiol. 33, 37–40 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barth A. L., Woodford N., Pitt T. L., Complementation of methionine auxotrophs of Pseudomonas aeruginosa from cystic fibrosis. Curr. Microbiol. 36, 190–195 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Barth A. L., Pitt T. L., The high amino-acid content of sputum from cystic fibrosis patients promotes growth of auxotrophic Pseudomonas aeruginosa. J. Med. Microbiol. 45, 110–119 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Yu X. J., Walker D. H., Liu Y., Zhang L., Amino acid biosynthesis deficiency in bacteria associated with human and animal hosts. Infect. Genet. Evol. 9, 514–517 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giovannoni S. J., Cameron Thrash J., Temperton B., Implications of streamlining theory for microbial ecology. ISME J. 8, 1553–1565 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D’Souza G., et al. , Less is more: Selective advantages can explain the prevalent loss of biosynthetic genes in bacteria. Evolution 68, 2559–2570 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Embree M., Liu J. K., Al-Bassam M. M., Zengler K., Networks of energetic and metabolic interactions define dynamics in microbial communities. Proc. Natl. Acad. Sci. U.S.A. 112, 15450–15455 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lv J., Wang S., Wang Y., Huang Y., Chen X., Isolation and molecular identification of auxotrophic mutants to develop a genetic manipulation system for the haloarchaeon Natrinema sp. J7-2. Archaea 2015, 483194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pronk J. T., Auxotrophic yeast strains in fundamental and applied research. Appl. Environ. Microbiol. 68, 2095–2100 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ulfstedt M., Hu G. Z., Johansson M., Ronne H., Testing of auxotrophic selection markers for use in the moss physcomitrella provides new insights into the mechanisms of targeted recombination. Front. Plant Sci. 8, 1850 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boer V. M., Amini S., Botstein D., Influence of genotype and nutrition on survival and metabolism of starving yeast. Proc. Natl. Acad. Sci. U.S.A. 105, 6930–6935 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouvet O., Bourdelier E., Glodt J., Clermont O., Denamur E., Diversity of the auxotrophic requirements in natural isolates of Escherichia coli. Microbiology 163, 891–899 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Seif Y., et al. , Genome-scale metabolic reconstructions of multiple salmonella strains reveal serovar-specific metabolic traits. Nat. Commun. 9, 3771 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seif Y., Monk J. M., Machado H., Kavvas E., Palsson B. O., Systems biology and pangenome of Salmonella O-antigens. MBio 10, e01247-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang X., et al. , Escherichia coli B2 strains prevalent in inflammatory bowel disease patients have distinct metabolic capabilities that enable colonization of intestinal mucosa. BMC Syst. Biol. 12, 66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lloyd C. J., et al. , Model-driven design and evolution of non-trivial synthetic syntrophic pairs. bioRXiv:10.1101/327270 (21 May 2018).
- 19.Mee M. T., Wang H. H., Engineering ecosystems and synthetic ecologies. Mol. Biosyst. 8, 2470–2483 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cabral M. P., et al. , Design of live attenuated bacterial vaccines based on D-glutamate auxotrophy. Nat. Commun. 8, 15480 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoiseth S. K., Stocker B. A., Aromatic-dependent salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291, 238–239 (1981). [DOI] [PubMed] [Google Scholar]
- 22.Hoffman R. M., Tumor-targeting amino acid auxotrophic Salmonella typhimurium. Amino Acids 37, 509–521 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Juliao P. C., Marrs C. F., Xie J., Gilsdorf J. R., Histidine auxotrophy in commensal and disease-causing nontypeable Haemophilus influenzae. J. Bacteriol. 189, 4994–5001 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaccaro D. E., Symbiosis therapy: The potential of using human protozoa for molecular therapy. Mol. Ther. 2, 535–538 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Mee M. T., Collins J. J., Church G. M., Wang H. H., Syntrophic exchange in synthetic microbial communities. Proc. Natl. Acad. Sci. U.S.A. 111, E2149–E2156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price M. N., et al. , Filling gaps in bacterial amino acid biosynthesis pathways with high-throughput genetics. PLoS Genet. 14, e1007147 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thiele I., Palsson B. Ø., A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 5, 93–121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seif Y., et al. , A computational knowledge-base elucidates the response of Staphylococcus aureus to different media types. PLoS Comput. Biol. 15, e1006644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kavvas E. S., et al. , Updated and standardized genome-scale reconstruction of mycobacterium tuberculosis H37Rv, iEK1011, simulates flux states indicative of physiological conditions. BMC Syst. Biol. 12, 25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norsigian C. J., Kavvas E., Seif Y., Palsson B. O., Monk J. M., iCN718, an updated and improved genome-scale metabolic network reconstruction of Acinetobacter baumannii AYE. Front. Genet. 9, 121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Brien E. J., Monk J. M., Palsson B. O., Using genome-scale models to predict biological capabilities. Cell 161, 971–987 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orth J. D., Thiele I., Palsson B. Ø., What is flux balance analysis? Nat. Biotechnol. 28, 245–248 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bosi E., et al. , Comparative genome-scale modelling of Staphylococcus aureus strains identifies strain-specific metabolic capabilities linked to pathogenicity. Proc. Natl. Acad. Sci. U.S.A. 113, E3801–E3809 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monk J. M., et al. , Genome-scale metabolic reconstructions of multiple Escherichia coli strains highlight strain-specific adaptations to nutritional environments. Proc. Natl. Acad. Sci. U.S.A. 110, 20338–20343 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.King Z. A., et al. , BiGG models: A platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 44, D515–D522 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monk J. M., et al. , iML1515, a knowledgebase that computes Escherichia coli traits. Nat. Biotechnol. 35, 904–908 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schellenberger J., Park J. O., Conrad T. M., Palsson B. Ø., BiGG: A biochemical genetic and genomic knowledgebase of large scale metabolic reconstructions. BMC Bioinf. 11, 213 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wattam A. R., et al. , PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 42, D581–D591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartell J. A., et al. , Reconstruction of the metabolic network of Pseudomonas aeruginosa to interrogate virulence factor synthesis. Nat. Commun. 8, 14631 (2017). [DOI] [PMC free article] [PubMed]
- 40.Brenner M., Lobel L., Borovok I., Sigal N., Herskovits A. A., Controlled branched-chain amino acids auxotrophy in listeria monocytogenes allows isoleucine to serve as a host signal and virulence effector. PLoS Genet. 14, e1007283 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tattoli I., et al. , Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe 11, 563–575 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Ren W., et al. , Amino acids as mediators of metabolic cross talk between host and pathogen. Front. Immunol. 9, 319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Martino M. L., et al. , Molecular evolution of the nicotinic acid requirement within the Shigella/EIEC pathotype. Int. J. Med. Microbiol. 303, 651–661 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Winter S. E., et al. , Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Treangen T. J., Ondov B. D., Koren S., Phillippy A. M., The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15, 524 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaudhuri R. R., et al. , Comprehensive identification of Salmonella enterica serovar typhimurium genes required for infection of BALB/c mice. PLoS Pathog. 5, e1000529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaudhuri R. R., et al. , Comprehensive assignment of roles for Salmonella typhimurium genes in intestinal colonization of food-producing animals. PLoS Genet. 9, e1003456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grant A. J., et al. , Genes required for the fitness of Salmonella enterica serovar typhimurium during infection of immunodeficient gp91-/- phox mice. Infect. Immun. 84, 989–997 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mediati D. G., “Identification of Escherichia coli genes required for bacterial survival and morphological plasticity in urinary tract infections,” PhD thesis, University of Technology Sydney, Sydney, Australia (2018).
- 50.Phan M. D., et al. , The serum resistome of a globally disseminated multidrug resistant uropathogenic Escherichia coli clone. PLoS Genet. 9, e1003834 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vohra P., et al. , Retrospective application of transposon-directed insertion-site sequencing to investigate niche-specific virulence of Salmonella Typhimurium in cattle. BMC Genom. 20, 20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Foley S. L., Johnson T. J., Ricke S. C., Nayak R., Danzeisen J., Salmonella pathogenicity and host adaptation in chicken-associated serovars. Microbiol. Mol. Biol. Rev. 77, 582–607 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hilliam Y., et al. , Pseudomonas aeruginosa adaptation and diversification in the non-cystic fibrosis bronchiectasis lung. Eur. Respir. J. 49, 1602108 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.La Rosa R., Johansen H. K., Molin S., Convergent metabolic specialization through distinct evolutionary paths in Pseudomonas aeruginosa. mBio, 10.1128/mBio.00269-18. (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bergthorsson U., Roth J. R., Natural isolates of Salmonella enterica serovar Dublin carry a single nadA missense mutation. J. Bacteriol. 187, 400–403 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin M., et al. , Effects of short indels on protein structure and function in human genomes. Sci. Rep. 7, 9313 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nuccio S. P., Bäumler A. J., Comparative analysis of Salmonella genomes identifies a metabolic network for escalating growth in the inflamed gut. MBio 5, e00929–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou Y., Liang Y., Lynch K. H., Dennis J. J., Wishart D. S., PHAST: A fast phage search tool. Nucleic Acids Res. 39, W347–W352 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nyman K., Nakamura K., Ohtsubo H., Ohtsubo E., Distribution of the insertion sequence IS1 in Gram-negative bacteria. Nature 289, 609–612 (1981). [DOI] [PubMed] [Google Scholar]
- 60.Nilsson A. I., et al. , Bacterial genome size reduction by experimental evolution. Proc. Natl. Acad. Sci. U.S.A. 102, 12112–12116 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anton B. P., Fomenkov A., Raleigh E. A., Berkmen M., Complete genome sequence of the engineered Escherichia coli SHuffle strains and their wild-type parents. Genome Announc. 4, e00230-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boyd D., Manoil C., Beckwith J., Determinants of membrane protein topology. Proc. Natl. Acad. Sci. U.S.A. 84, 8525–8529 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen C., et al. , Convergence of DNA methylation and phosphorothioation epigenetics in bacterial genomes. Proc. Natl. Acad. Sci. U.S.A. 114, 4501–4506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jeong H., Sim Y. M., Kim H. J., Lee S. J., Unveiling the hybrid genome structure of Escherichia coli RR1 (HB101 RecA+). Front. Microbiol. 8, 585 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wrobel A., Ottoni C., Leo J. C., Gulla S., Linke D., The repeat structure of two paralogous genes, Yersinia ruckeri invasin (yrInv) and a “Y. ruckeri invasin-like molecule”, (yrIlm) sheds light on the evolution of adhesive capacities of a fish pathogen. J. Struct. Biol. 201, 171–183 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Bronowski C., et al. , Genomic characterisation of invasive non-typhoidal Salmonella enterica subspecies enterica serovar bovismorbificans isolates from Malawi. PLoS Negl. Trop. Dis. 7, e2557 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Labbé G., et al. , Complete genome sequences of 17 Canadian isolates of Salmonella enterica subsp. enterica serovar Heidelberg from human, animal, and food sources. Genome Announc. 4, e00990-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lindsey R. L., et al. , High-quality draft genome sequences for four drug-resistant or outbreak-associated Shigella sonnei strains generated with PacBio sequencing and whole-genome maps. Genome Announc. 5, e00906-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim J., et al. , High-quality whole-genome sequences for 59 historical Shigella strains generated with PacBio sequencing. Genome Announc. 6, e00282-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang W., et al. , Emergence and evolution of multidrug-resistant Klebsiella pneumoniae with both blaKPC and blaCTX-M integrated in the chromosome. Antimicrob. Agents Chemother. 61, e00076-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wannier T. M., et al. , Adaptive evolution of genomically recoded Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 115, 3090–3095 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Amirmozafari N., Robertson D. C., Nutritional requirements for synthesis of heat-stable enterotoxin by Yersinia enterocolitica. Appl. Environ. Microbiol. 59, 3314–3320 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brownlow W. J., Wessman G. E., Nutrition of Pasteurella pestis in chemically defined media at temperatures of 36 to 38 C. J. Bacteriol. 79, 299–304 (1960). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fowler J. M., Brubaker R. R., Physiological basis of the low calcium response in Yersinia pestis. Infect. Immun. 62, 5234–5241 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wright B. E., Minnick M. F., Reversion rates in a Leub auxotroph of Escherichia coli K-12 correlate with ppGpp levels during exponential growth. Microbiology 143, 847–854 (1997). [DOI] [PubMed] [Google Scholar]
- 76.Monk J., Nogales J., Palsson B. O., Optimizing genome-scale network reconstructions. Nat. Biotechnol. 32, 447–452 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study are included in this published article (and in SI Appendix and Datasets S1–S6). AuxoFind is available on GitHub, with an example notebook. All genomic sequences analyzed in this study are publicly available on PATRIC (38), and accession numbers are available in Dataset S1.