Significance
Developmental signaling pathways play important roles in cancer initiation and therapeutic resistance. Poorly differentiated hepatocellular carcinomas (HCCs) usually show phenotypic resemblance to their ancestral precursor cells and are associated with poor clinical outcome. In a recent study, a dynamic oncofetal-like gene expression signature during liver development was identified from an in vitro hepatocyte differentiation model. Two subtypes of liver cancer with different oncofetal properties and upstream oncogenic drivers were found in clinical patients with poor prognosis. Clinically available small-molecule inhibitors HLM6474 and SIS3 specifically targeting the oncofetal drivers extensively down-regulated subtype-specific developmental signaling and significantly inhibited tumorigenicity. Further treatment of HCC patients with specified small-molecular inhibitors according to subtype-specific biomarkers might accurately block the tumor-initiating steps and eliminate tumor progression.
Keywords: hepatocyte differentiation, oncofetal properties, liver development, oncogenic driver, cancer subtype
Abstract
Clinical observation of the association between cancer aggressiveness and embryonic development stage implies the importance of developmental signals in cancer initiation and therapeutic resistance. However, the dynamic gene expression during organogenesis and the master oncofetal drivers are still unclear, which impeded the efficient elimination of poor prognostic tumors, including human hepatocellular carcinoma (HCC). In this study, human embryonic stem cells were induced to differentiate into adult hepatocytes along hepatic lineages to mimic liver development in vitro. Combining transcriptomic data from liver cancer patients with the hepatocyte differentiation model, the active genes derived from different hepatic developmental stages and the tumor tissues were selected. Bioinformatic analysis followed by experimental assays was used to validate the tumor subtype-specific oncofetal signatures and potential therapeutic values. Hierarchical clustering analysis revealed the existence of two subtypes of liver cancer with different oncofetal properties. The gene signatures and their clinical significance were further validated in an independent clinical cohort and The Cancer Genome Atlas database. Upstream activator analysis and functional screening further identified E2F1 and SMAD3 as master transcriptional regulators. Small-molecule inhibitors specifically targeting the oncofetal drivers extensively down-regulated subtype-specific developmental signaling and inhibited tumorigenicity. Liver cancer cells and primary HCC tumors with different oncofetal properties also showed selective vulnerability to their specific inhibitors. Further precise targeting of the tumor initiating steps and driving events according to subtype-specific biomarkers might eliminate tumor progression and provide novel therapeutic strategy.
The resemblance between cancer cells and embryonic stem cells (ESs) has aroused great interest in the investigation of cancer cellular origins (1). Although it is still under debate whether cancer originates from ESs or undergoes dedifferentiation from terminally differentiated cells, the critical roles of developmental signaling pathways in cancer initiation and progression have no doubt been widely accepted (2, 3). In clinical pathology, the poorly differentiated tumors usually show stem cell-like traits and have poor clinical outcomes (4, 5). Further molecular investigation revealed that signaling pathways regulating the properties of ESs or cell lineage differentiation are highly active and might be oncogenic drivers in the initiation and progression of poorly differentiated tumors (6). For instance, the embryonic stem cell pluripotency transcriptional factors Oct4, Sox2, Nanog, and c-myc were found to be activated in certain tumor types and to regulate tumor differentiation (7–9). Developmental signaling pathways which regulate normal organogenesis and cellular differentiation were also found to be frequently altered during malignant transformation of tumors with stem cell-like properties (10–14).
Liver cancer ranks fifth among the most prevalent cancers in the world and is the third leading cause of cancer death. Lack of suitable biomarkers for early detection and limited treatment strategies are the major causes of high mortality (15). Although it is becoming clear that liver cancer might originate from chronic inflammation induced by viral infection or fatty liver disease, the cellular origins and the molecular driving events are still unclear. Oncofetal proteins such as SALL4, which are absent in the normal differentiated hepatocytes, reappear in aggressive hepatocellular carcinoma (HCC) and drive tumorigenesis (16). Comprehensive cross-talk between the Yes-associated protein, the Notch signaling pathway, and the transforming growth factor type β (TGF-β) signaling pathway was found during liver development and cancer progression (17–19). Aberrant activation of those developmental networks can also induce retrodifferentiation or transdifferentiation between different cellular lineages including liver progenitors, hepatocytes, and cholangiocytes, which constitute the cellular heterogeneity of liver cancer (20–22). Thus, a systemic characterization of the liver developmental landscape might help in understanding liver carcinogenesis and identifying driver events.
By comparing the gene expression signatures between different liver developmental stages and HCC clinical samples, we identified a previously undescribed gene expression pattern of liver cancer with different oncofetal properties. Network analysis further revealed subgroup-specific biomarkers and potential oncogenic drivers ideal for therapeutic targeting.
Results
Establishment of an In Vitro Hepatocyte Differentiation Model.
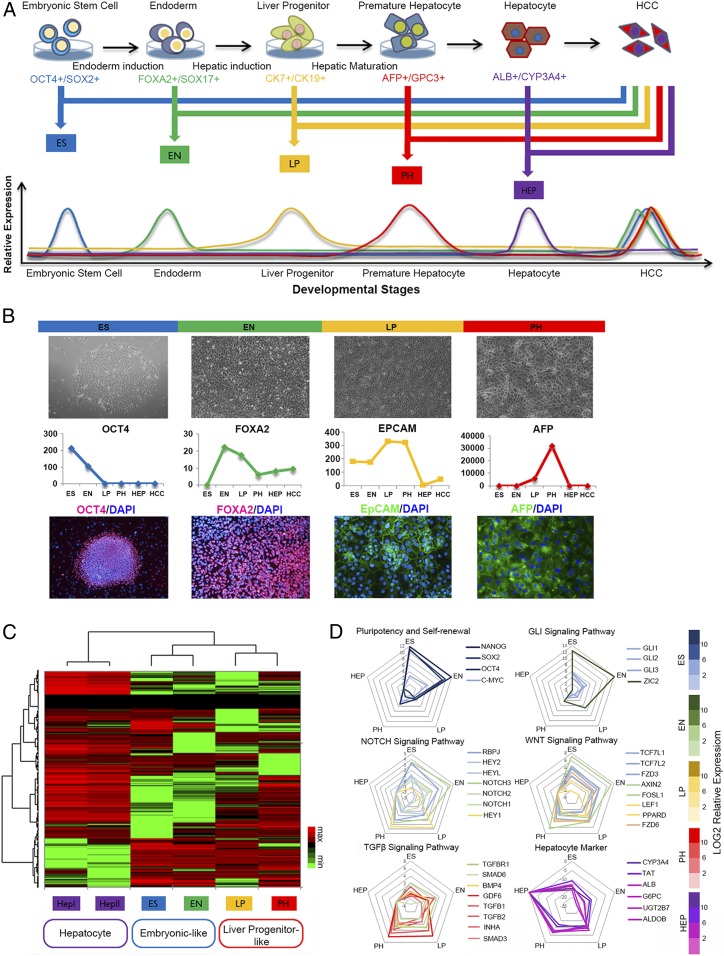
Human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs) have been intensively investigated in the last decades for their potentially valuable applications in studying human organogenesis. With the advance of stem cell technology it is now possible to direct hESC or iPSC differentiation into defined cellular lineages, for example human hepatocytes (23). With input of the transcriptomic information from the liver tumor tissues and their paired nontumor counterparts, the model was ideal for studying the link between developmental signaling and liver carcinogenesis. In this study, hESCs were differentiated into human hepatocytes. The whole differentiation process was defined by four stages, including embryonic stem cell (ES), endoderm (EN), liver progenitor cell (LP), and premature hepatocytes (PH). Cells from the four developmental stages as well as two paired HCC clinical tissues were collected for further transcriptomic profiling (Fig. 1A).
Fig. 1.
Establishment of an in vitro hepatocyte differentiation model and identification of oncofetal gene expression patterns. (A) hESCs were induced to differentiate along hepatic lineages into adult hepatocytes. Cells from different developmental stages, including ES, EN, LP, and PH, as well as paired clinical HCC tissues, were selected for transcriptomic sequencing. Genes which both reached their peak expressions at different liver developmental stages and significantly up-regulated in the tumor tissues were selected for further analysis. (B) Morphologies of the cells at different hepatocyte developmental stages (Upper). Representative biomarkers at different hepatocyte developmental stages were selected to test the validity of the hepatic differentiation model (Middle). Immunoflorescent staining of representative biomarkers at different hepatocyte developmental stages (Lower). (C) Hierarchical clustering analysis was used to examine the link between the four developmental subgroups, as well as the adult hepatocytes. (D) Representative molecules from several cancer-related canonical developmental signaling pathways were selected, and their distributions among different hepatocyte differentiation stage were tested in the hepatic differentiation model.
Selection of Potential Oncofetal Gene Sets Reflecting Different Liver Developmental Stages.
In order to identify oncofetal genes and their molecular characteristics in HCC, we used our hepatocyte differentiation model to select genes that reached their peak expression level in the intermediate liver developmental stages. Selected genes were then overlapped with genes significantly up-regulated in the tumor tissue. The final list was distributed into four subgroups according to developmental stages. Cell morphologies at different developmental stages are shown in Fig. 1B. Representative biomarkers at different hepatocyte developmental stages were selected to test the validity of our model and selection criteria. As expected, the molecules which mark different hepatocyte developmental stages (24, 25) all showed their unique expression pattern in our model (Fig. 1B). Immunoflorescent staining of the selected biomarkers further confirmed the successful establishment of the in vitro hepatocyte differentiation model.
Hierarchical Clustering Analysis Reveals an Oncofetal Gene Expression Pattern in Liver Cancer.
To look into the relationship between different oncofetal gene sets, hierarchical clustering analysis was used to examine the link among the four developmental subgroups, as well as the adult hepatocytes. As shown in Fig. 1C, the selected oncofetal genes were clustered into two major subtypes. We named the genes from ES and EN groups as embryonic-like subtype (ES+ subtype) and genes from the LP and PH groups as liver progenitor-like subtype (LP+ subtype). Genes from two adult hepatocytes were clustered into a hepatocyte-like subtype. These findings indicated that there might be two subtypes of liver cancer with different oncofetal properties (Fig. 1C). Representative molecules from several cancer-related canonical developmental signaling pathways were selected, and their distributions among different hepatocyte differentiation stage are shown in Fig. 1D. Interestingly, we found genes in the pluripotency and stem cell self-renewal signaling pathway and the Gli signaling pathway are mainly restricted to the ES and EN stages, while genes in the Notch signaling and the Wnt signaling pathway span from the ES and EN stages to the LP and PH stages. However, for the TGF-β signaling pathway, most of the molecules fall in the LP and PH stages. This indicated that different hepatocyte differentiation stages might be characterized by their specific developmental signaling pathways.
Two Subtypes of Liver Cancer with Oncofetal Properties Associate with Different Molecular Traits and Upstream Activators.
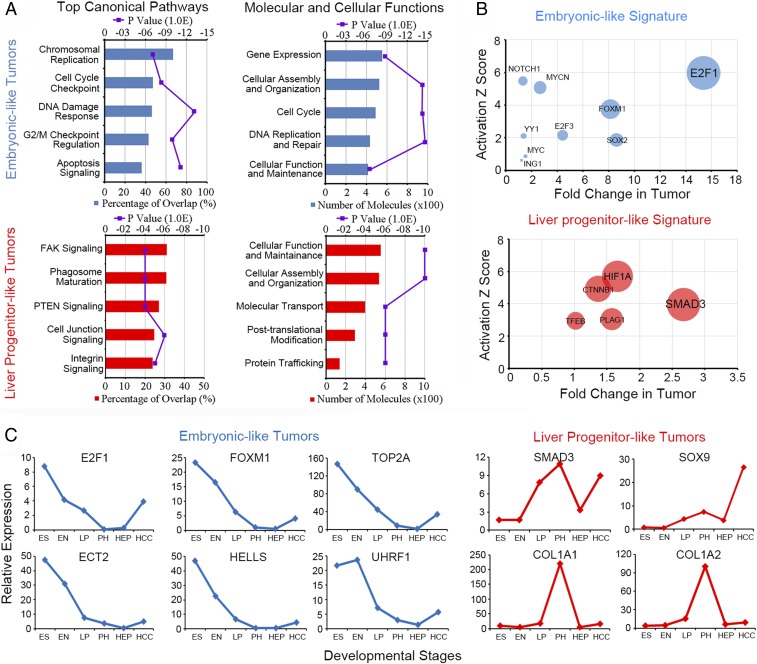
Gene ontology analysis was performed to characterize the molecular features of the two subtypes of liver cancer. We found totally different molecular signatures between the ES+ tumors and the LP+ tumors. The genes from the ES+ subtype were enriched in signaling pathways related to chromosomal replication, cell cycle checkpoint, and DNA damage response. However, for the LP+ tumors, the genes were enriched in cell adhesion signaling pathways. (Fig. 2A). To further identify the oncogenic drivers during liver tumorigenesis, Ingenuity Pathway Analysis (IPA) upstream activator analysis was used to identify the common transcriptional activators of the oncofetal genes isolated from the two subtypes of liver cancer. For our selection criteria, the transcriptional activators themselves should possess expression patterns similar to their downstream genes according to the hepatocyte differentiation model. Transcriptional factors both gain high activation z-scores and fold change in tumors were regarded as potential oncogenic drivers in the two subtypes of liver cancer with oncofetal properties (SI Appendix, Table S1). E2F1 and SMAD3 got the highest score and were selected as the potential upstream activators of ES+ tumors and LP+ tumors, respectively (Fig. 2B).
Fig. 2.
Two subtypes of liver cancer with oncofetal properties associate with different molecular traits and upstream activators. (A) Gene ontology analysis was used to characterize the molecular features of the two subtypes of liver cancer. The top-ranked canonical pathways and the molecular and cellular functions for ES+ tumors (Upper) and liver-progenitor-like tumors (Lower) are listed. (B) IPA upstream activator analysis was used to identify the common transcriptional activators of the oncofetal genes isolated from the two subtypes of liver cancer. Transcriptional factors both gain high activation z-score and fold change in tumor were regarded as potential oncogenic drivers. (C) A six-gene core module was selected to represent the ES+ tumors, and a four-gene core module was selected to represent the LP+ tumors. The gene expression pattern is shown according to the different liver developmental stages.
Characterization of the Core Molecular Module Reflecting the Two Subtypes of Liver Cancer.
To further understand the molecular features of the two subtypes of liver cancer, the predicted downstream targets of E2F1 and SMAD3 which gained high activation scores in HCC were selected (SI Appendix, Table S1). The integrated cancer genomics platform cBioportal was used to analyze the genomic characteristics and clinical significance of those selected genes. A tight connection of those genes which further forms a comprehensive interacting network was predicted to exist in the two subtypes of liver cancer (SI Appendix, Fig. S1A). Since the small-molecular inhibitors specific for E2F1 and SMAD3 are already commercially available, we treated several HCC cell lines with the inhibitors and screened for the responsive targets from the above signature genes. At the same time, the expression of the candidate genes was also examined in paired HCC clinical samples. Only the top-ranked predicted targets which both significantly up-regulated in HCC clinical samples and responded to small-molecular inhibitors in HCC cell lines were further selected as the core module genes of the two subtypes of liver cancer (SI Appendix, Fig. S1 B–D). According to the selection criteria, a six-gene core module was selected to represent the ES+ tumors and a four-gene core module was selected to represent the LP+ tumors. The expression of E2F1 and SMAD3 were further detected by quantitative real-time PCR (qPCR) in series of HCC cell lines as well as immortalized liver cell lines (SI Appendix, Fig. S2A). Relative expressions of the oncofetal core module genes in the cell lines are shown in SI Appendix, Fig. S2B. The relative expression of LP markers and hepatocyte differentiation markers was also examined (SI Appendix, Fig. S2C). All of the selected core module genes showed both their subtype-specific peak expressions among four hepatocyte differentiation stages and overexpression in the HCC tumor tissues compared to their normal counterparts (Fig. 2C). E2F1-specific inhibitor HLM6474 and SMAD3-specific inhibitor SIS3 significantly inhibited the expression of the selected core module genes in HCC cell lines (SI Appendix, Fig. S2D).
Validation of the Subtype-Specific Gene Signature In Vivo and Clinical Significance in HCC Patients.
To further validate whether the newly identified gene signature really exists in true liver development in vivo, we examined the expressions of the signature genes in fetal mouse liver at different developmental stages in vivo. According to the process of mouse liver development, the morphogenesis of the liver bud, which is derived from the ESs and subsequent EN formation, was finished before embryonic day 12 (e12). Starting at e13, the bipotential hepatoblasts begin to activate and differentiate into hepatocytes or biliary epithelial cells. At e17, the immature hepatocytes acquire their characteristic epithelial morphology and begin to form mature hepatocytes until the perinatal stage (26). The selected biomarkers of different developmental stages and oncofetal signature genes showed similar expression pattern in mice fetal liver development in vivo and our in vitro hepatocyte differentiation model (SI Appendix, Fig. S3). These results further indicated that the specified developmental stages and oncofetal gene signatures in our in vitro model could well represent their actual physiological conditions in vivo.
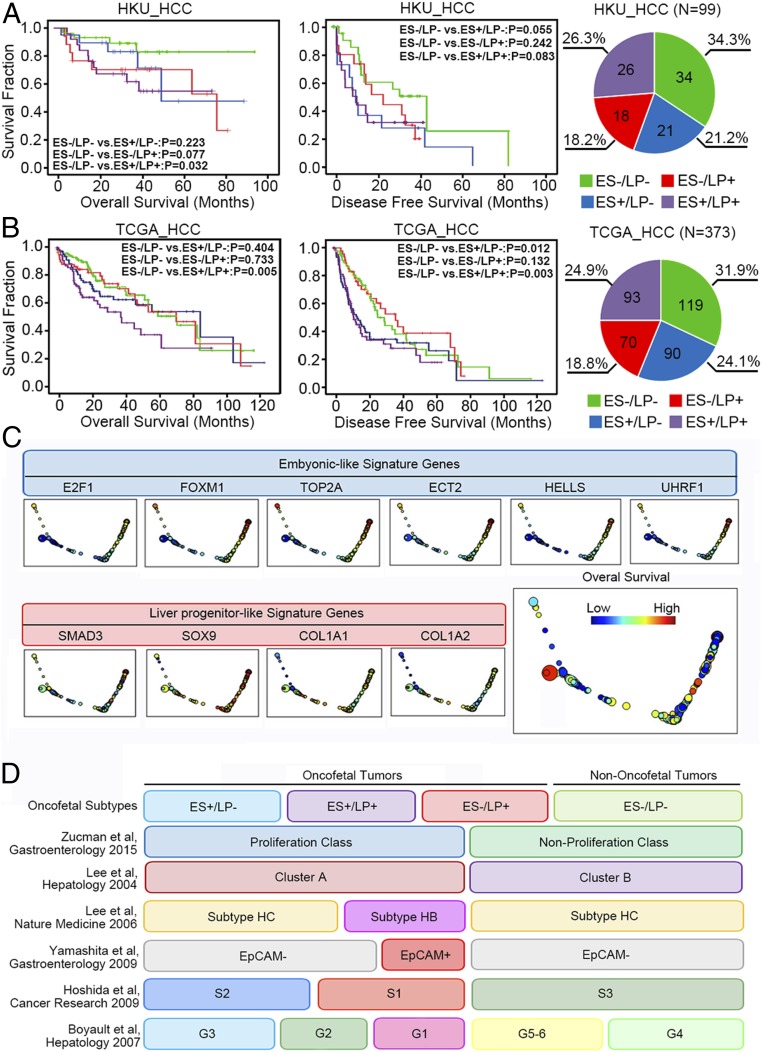
The relative expression of the selected core module genes was examined in an independent cohort of HCC patients. As expected, hierarchical clustering analysis divided the core module genes into two major subtypes. (SI Appendix, Fig. S4). Significant up-regulation of each selected core module gene was also found in the tested cohort as well as in The Cancer Genome Atlas (TCGA) database (SI Appendix, Fig. S5 A and B). In addition, the expression of the upstream activators and their downstream targets closely associated with each other in the tested clinical HCC tissues and the TCGA database (SI Appendix, Fig. S5 C and D). These indicated the clinical existence of the subtype-specific gene signatures derived from our in vitro hepatocyte differentiation model. To distinguish the two subtypes of liver cancer in the clinical sample, we further established a score system. Each patient was assigned an ES+ feature score and an LP+ feature score according to the extent of respective subtype-specific core module gene activation as compared to their normalized distribution in the normal tissues. Kaplan–Meier survival analysis was performed among different subtypes of liver cancer patients. Patients characterized with mixed signatures consistently showed significant poor prognosis in both HKU_HCC cohort and TCGA_HCC cohort (Fig. 3 A and B). The associations of the oncofetal properties with different etiologies and clinical pathological features of HCC are shown in SI Appendix, Table S2. Topological data analysis (TDA) further distinguished the two subtypes of signature genes in HCC patients. The expression of ES-like genes showed high expression in the nodes on the far right side, while LP-like genes showed a different expression pattern with high expression in the nodes on the lower left side (Fig. 3C).
Fig. 3.
Validation of the subtype-specific gene signatures and their clinical significance in HCC patients. (A) Kaplan–Meier plots and log-rank tests were used for survival analysis of HCC patients with different oncofetal signatures in the HKU_HCC cohort (n = 99). The number of patients belong to different subgroups and their distribution are shown on the right (ES−/LP−: n = 34; ES+/LP−: n = 21; ES−/LP+: n = 18; ES+/LP+: n = 26). (B) Kaplan–Meier plots and log-rank tests were used for survival analysis of HCC patients with different oncofetal signatures in the TCGA_HCC cohort (n = 373). The number of patients belong to different subgroups and their distribution are shown on the right (ES−/LP-: n = 119; ES+/LP−: n = 90; ES−/LP+: n = 70; ES+/LP+: n = 93). (C) TDA of the TCGA tumor samples. The nodes (circles) are sets of samples with similar expression levels of indicated genes, and the sizes correspond to number of samples in that set. Edges (lines) connect the nodes that have at least one sample in common. The color corresponds to the expression of specific genes of the sample set. The figure that is colored by the overall survival is shown at the bottom right. (D) Integration of the oncofetal liver cancer subtypes with previously reported molecular classifications.
Integration of the Oncofetal Liver Cancer Subtypes with Previously Reported Molecular Classifications.
Previous reports have classified HCC patients into different molecular subtypes based on stem cell-like expression patterns (27, 28). It is interesting to know the relationship between the oncofetal subtypes and the previous molecular classifications. The molecular features of the HCC patients from the TCGA cohort were analyzed according to different classification criteria described previously in the literature. As summarized in Fig. 3D, most of the patients with oncofetal properties overlapped with the “proliferation class” in Zucman et al.’s study (29). The nononcofetal tumors and a small fraction of ES−/LP+ patients overlapped with the “nonroliferation class.” A similar pattern was observed in a study by Lee et al. (30), which divided the patients into the proliferative “cluster A” and the nonproliferative “cluster B.” In a study by Lee et al. (31) they identified a subgroup of HCC patients with hepatoblast-like (HB subtype) features, such as activation of oval cells and elevated expression of KRT7, KRT19, and VIM. The HB subtype patients matched with part of the LP+ tumors (ES−/LP+, ES+/LP+) in our study. This is consistent with our findings that the LP+ tumors are characterized with high expression of KRT7 and KRT19. The EpCAM+ subgroup of patients in Yamashita et al.’s study (32) showed an expression pattern similar to the HB subgroup in our study. In Hoshida et al.’s study (33) they classified the HCC patients into S1, S2, and S3 subgroups according to the extent of tumor differentiation. Most of the patients with oncofetal properties in our study were matched with the poorly differentiated S1 and S2 subgroups. Accordingly, the nononcofetal tumors were matched with the well-differentiated S3 subgroup. In Boyault et al.’s study (34) they divided the patients into G1 through G6 subgroups according to different molecular features. The G1 and G2 tumors characterized with AKT activation and fetal liver properties were mostly overlapped with the LP+ tumors (ES−/LP+, ES+/LP+) in our study. The G3 tumors with activation of cell cycle genes were matched with the ES+/LP− subgroups. The G5 and G6 tumors with WNT signaling activation were mostly overlapped with the ES−/LP+ subgroup, and the heterogeneous G4 tumors were mostly overlapped with the nononcofetal tumors in our study. Detailed distribution of the patients in previous classification criteria and their overlapping with current subclassification are shown in SI Appendix, Table S3. Pearson’s χ2 test was used for the association study.
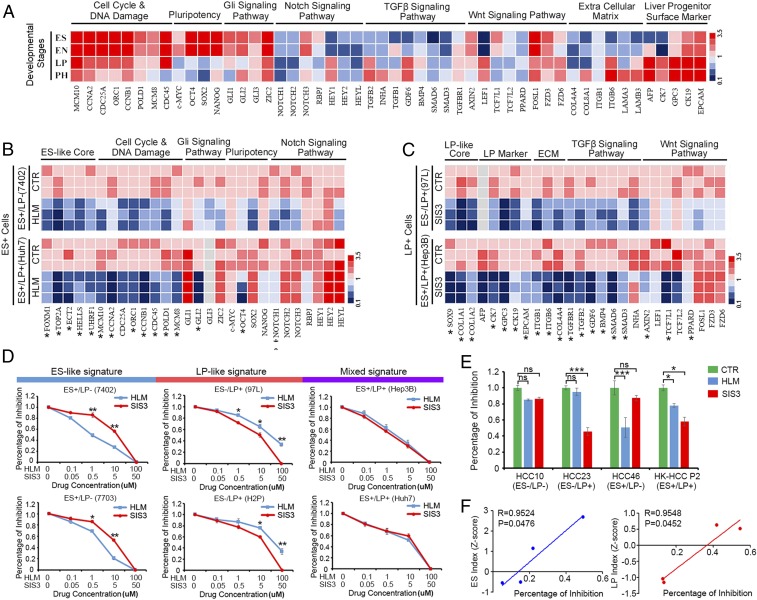
Inhibition of E2F1 and SMAD3 Extensively Down-Regulated Subtype-Specific Signature Genes and Developmental Signaling Pathways.
The expressions of subtype-specific signature genes and the featured molecules belonging to several developmental signaling pathways were detected at different liver developmental stages. As shown in Fig. 4A, the activation of cell cycle and DNA damage, pluripotency, and Gli signaling pathway were associated with ES+ features, and the activation of LP surface marker, extracellular matrix, and the Wnt and TGF-β signaling pathway were associated with LP+ features. To test whether inhibition of E2F1 or SMAD3 could affect the subtype-specific developmental signaling pathways, small-molecule inhibitors HLM6474 and SIS3 were used to treat HCC cell lines with different oncofetal signatures. We found inhibition of E2F1 significantly down-regulated the ES signature molecules, while inhibition of SMAD3 extensively down-regulated the LP signature genes (Fig. 4 B and C). These results indicated that E2F1 and SMAD3 might lie upstream of those developmental signaling pathways and be potential oncofetal drivers of liver tumors with defined gene signature.
Fig. 4.
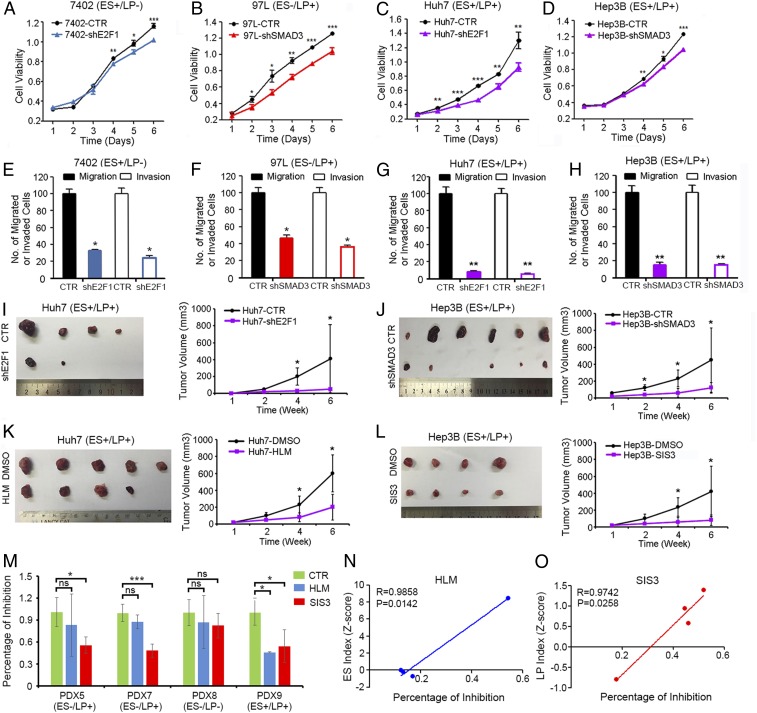
Targeting E2F1 and SMAD3 could extensively down-regulate subtype-specific signature genes and exert selective vulnerability on different subtypes of liver cancer. (A) The expressions of subtype-specific signature genes and the featured molecules belonging to several developmental signaling pathways were detected at different liver developmental stages. (B) E2F1- and (C) SMAD3-specific small-molecule inhibitors were used to treat HCC cell lines. The relative expression of their subtype-specific signature genes and developmental signaling pathways were detected by qPCR. The heat map represents three independent experiments. *P < 0.05. (D) HCC cells with ES-like signature (ES+/LP−), LP-like signature (ES−/LP+), or mixed signature (ES+/LP+) were treated with E2F1 and SMAD3 inhibitors at indicated concentrations. The percentage of cell growth inhibition was detected. *P < 0.05, **P < 0.01 by independent t test. (E) Tumor organoids derived from primary HCC patients with different oncofetal properties were treated with E2F1 or SMAD3 inhibitors, and their sensitivities to drug treatment were examined. *P < 0.05, ***P < 0.001, independent t test. ns, not significant. (F) The correlations of ES index (z-score) and LP index (z-score) of the primary HCC organoids with percentage of drug inhibitory rates.
Two Subtypes of Liver Cancer Showed Selective Vulnerability to Their Specific Inhibitors.
HCC cell lines with different oncofetal properties (ES+/LP−: 7402, 7703; ES−/LP+: 97L, H2P; ES+/LP+: Hep3B, Huh7) were treated with E2F1 and SMAD3 inhibitors at indicated concentrations. Interestingly, for the ES+/LP− group, the cells were more sensitive to HLM6474 as compared to SIS3. Conversely, for the ES−/LP+ group, the cells showed higher sensitivity to SIS3. In the ES+/LP+ group, the cells were sensitive to either inhibitor but showed no selective vulnerability (Fig. 4D). To further confirm our findings, primary tumor tissues derived from HCC patients were cultured to form organoids and treated with either HLM6474 or SIS3. Drug sensitivity assays were used to check the responses of primary HCC tissues with different oncofetal properties to therapeutic drugs. In accordance with the results from cell lines, tumor organoids derived from primary HCC patients with an ES-like signature were more sensitive to E2F1 inhibitor HLM6474, while HCC tumors with an LP-like signature were more sensitive to SMAD3 inhibitor SIS3 (Fig. 4E). The ES index which represents the extent of ES-like properties significantly correlated with the percentage of inhibition from HLM drug treatment. Accordingly, the LP index which represents the extent of LP-like properties significantly correlated with the percentage of inhibition from SIS3 drug treatment in the tested HCC organoids (Fig. 4F). These results indicated that different subtypes of liver cancer might rely on different oncogenic signaling pathways and have their subtype-specific vulnerable targets. Targeting the oncofetal drivers precisely according to the specified biomarkers might enhance drug selectivity further in the clinic.
Targeting Oncofetal Drivers Might Be a Novel Therapeutic Strategy in HCC Treatment.
To further evaluate whether targeting the oncofetal drivers might be effective in HCC treatment, HCC cells with ES-like oncofetal properties (ES+/LP−: 7402) were stably transfected with short hairpin RNAs (shRNAs) specifically targeting E2F1. Compared with control groups, targeting E2F1 dramatically inhibited cell viability (Fig. 5A), colony formation abilities in soft agar (SI Appendix, Fig. S6A), and cell migration and invasion (Fig. 5E). Accordingly, HCC cells with LP-like oncofetal properties (ES−/LP+: 97L) were stably transfected with shRNAs specifically targeting SMAD3. Tumor inhibitory functions were also observed (Fig. 5 B and F and SI Appendix, Fig. S6B). Interestingly, for the cells with mixed signature (ES+/LP+: Huh7 and Hep3B), targeting either E2F1 or SMAD3 significantly inhibited the oncogenic phenotypes (Fig. 5 C, D, G, and H and SI Appendix, Fig. S6 C and D). This indicated that both oncofetal signaling pathways might be indispensable and substantially contributed to the malignant phenotypes of HCC patients with mixed signature. Furthermore, in vivo targeting the oncofetal drivers with either shRNAs or small molecular inhibitors dramatically inhibited the tumor growth in the xenograft mouse model (Fig. 5 I–L). The expression of E2F1 and the activated form of SMAD3 at the protein level was detected by immunoblot/immunohistochemistry in relevant cell lines and xenograft tissues before and after drug treatment (SI Appendix, Fig. S6 E and F). To further test the inhibitors on primary tumors, patient-derived HCC tissues were inoculated into the left dorsal side of immune-deficient mice and treated with vehicle control, HLM6474, and SIS3. As shown in Fig. 5M, HCC tumors with an ES-like signature were more sensitive to E2F1 inhibitor HLM6474, while HCC tumors with an LP-like signature were more sensitive to SMAD3 inhibitor SIS3. The ES index which represents the extent of ES-like properties significantly correlated with the percentage of inhibition from HLM drug treatment (Fig. 5N). Accordingly, the LP index which represents the extent of LP-like properties significantly correlated with the percentage of inhibition from SIS3 drug treatment in the tested HCC primary tumors (Fig. 5O). This evidence supports the future clinical use of E2F1 inhibitors and SMAD3 inhibitors in treatment of HCC patients with specified oncofetal properties.
Fig. 5.
Targeting oncofetal drivers might be a novel effective strategy in HCC treatment. (A) HCC cells with ES-like signature were stably transfected with shRNAs specifically targeting E2F1 (shE2F1) or control shRNAs (CTR). The cell growth rates were determined using an MTT assay. (B) HCC cells with LP-like signature were stably transfected with shRNAs specifically targeting SMAD3 (shSMAD3) or control shRNAs (CTR). The cell growth rates were determined using an MTT assay. (C and D) HCC cells with mixed signature were stably transfected with shE2F1 or shSMAD3, and the cell growth rates were determined using an MTT assay. (E–H) Cells were seeded in the cell migration or invasion chamber at a density of 5,000 cells per well. The migrated or invaded cells were counted 48 h later. (I) Huh7-CTR or Huh7-shE2F1 cells were subcutaneously injected into the left dorsal and right dorsal flank of nude mice. The tumor volumes were monitored for 6 wk. (J) Xenograft tumor assays were also performed under similar conditions in Hep3B-CTR and Hep3B-shSMAD3 cells. (K) Huh7 xenograft tumors were implanted in to the left dorsal flank of nude mice, and E2F1 inhibitor HLM6474 (20 mg/kg) were intraperitoneally injected into the tumor-bearing mice. (L) Hep3B xenograft tumors were implanted in to the left dorsal flank of nude mice, and SMAD3 inhibitor SIS3 (2 mg/kg) were intraperitoneally injected into the tumor-bearing mice. Tumor volumes were monitored for 6 wk. (M) Patient-derived HCC tissues with different oncofetal properties were inoculated into the left dorsal flank of immune-deficient mice and treated with vehicle control, HLM6474, or SIS3 and their sensitivities to drug treatment were examined. *P < 0.05, **P < 0.01, ***P < 0.001, independent t test. ns, not significant. (N) The correlations of ES index (z-score) and (O) LP index (z-score) of the primary HCC tissues with percentage of drug inhibitory rates were examined.
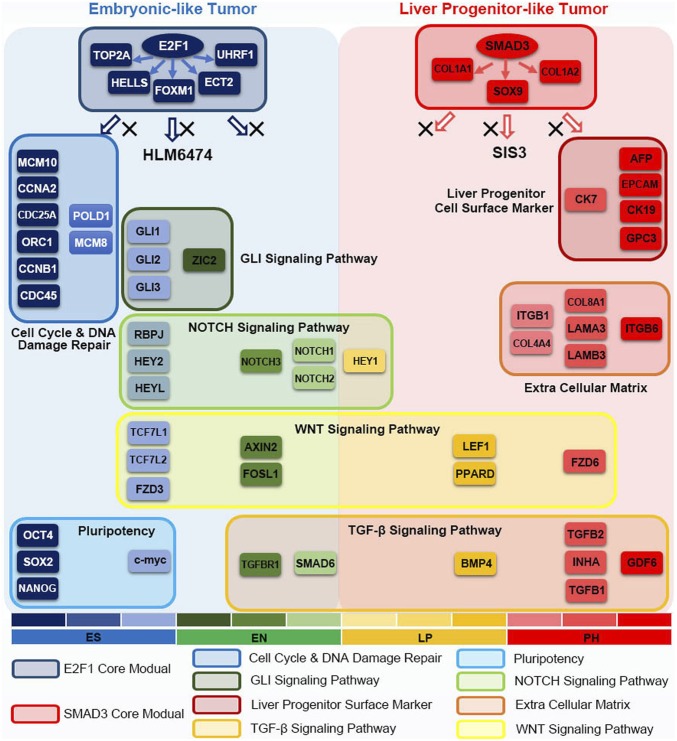
In summary, we identified gene expression signatures and liver cancer subtypes through linking hepatocyte differentiation with liver tumorigenesis. Activation of specified developmental signaling pathways along different stages of hepatocyte differentiation revealed two major subtypes of liver cancer with different oncofetal properties and molecular traits. Targeting the master transcriptional activators significantly suppressed their subtype-specific developmental signaling pathways and effectively eliminated tumorigenicity, which provided a therapeutic strategy in HCC treatment (Fig. 6).
Fig. 6.
Model of the developmental hierarchy with different oncofetal properties and potential oncogenic drivers in HCC.
Discussion
Clinical observation of poorly differentiated tumors preserving lineage characteristics of their developmental precursor cells indicated the strong link between tumor aggressiveness and embryonic development (35). Signaling pathways maintaining embryonic stem cell pluripotency or governing pattern formation during organogenesis may also induce malignant transformation of normal cells. Our recent study also suggested the important linkage between embryonic genes and cancer progression (36). Further investigation of embryonic development and organ differentiation might help in understanding the molecular mechanisms of cancer initiation and cellular origins (37). Hyperactivation of embryonic genes in the tumor cells might be the driving events during malignant transformation and provide ideal therapeutic targets for cancer treatment (13).
In this study, we established an in vitro hepatocyte differentiation model to investigate the association between liver development and carcinogenesis. Through analyzing the combined data from the hepatocyte differentiation model and HCC clinical samples, we identified a gene expression signature of liver cancer indicating different oncofetal properties. Recent data from genomic profiling enabled the proposals of different molecular clusters of HCCs according to their proliferation index, cellular origins, and immune responses (38, 39). Interestingly, all of the newly established classification models mentioned the evidence of a stem cell or progenitor cell-like properties of poor prognostic liver tumors. However, detailed subclassification of the stem cell-like tumors is lacking, and the upstream master regulators controlling stem cell properties are still not clear. In this study, we classified liver tumors according to the timeline of liver development. Moreover, we identified the core module biomarkers to distinguish the stem-like tumor subtypes and upstream drivers for therapeutic intervention. Inhibition of the master transcriptional regulators of oncofetal signaling might attack the tumor-initiating steps and efficiently eliminate tumor growth. The classification also distinguished the vulnerabilities of different subtypes of liver cancer with stem cell properties, which might guide precision medicine further in the clinic. Genomic copy number gain of E2Fs including E2F1 and E2F3 has been found to account for E2F activation and drive HCC tumorigenesis in a dose-dependent manner (40). In contrast, the activity of SMAD3 is mainly regulated by the classical TGF-β signaling pathway, which is constantly activated in the inflammatory liver tumor microenviroment (41). In addition, epigenetic regulation has also been proposed to activate SMAD3 during malignant transformation, indicating novel strategies for cancer therapeutics (42). We noticed that gain of a single oncofetal property might not necessarily cause worse prognosis for the patients. However, patients with mixed signature (ES+/LP+) showed consistent significant poor clinical outcome in both HCC cohorts. We believe that a hierarchy of cancer stem cells reflecting different developmental stages might exist and constitute the heterogeneity of the tumor. The poor prognostic patients underwent accumulation of different oncofetal properties during malignant progression, and this process substantially contributed to tumor heterogeneity, which further led to malignant progression such as tumor dissemination, metastasis, and resistance to chemotherapeutic drugs. We divided HCC cell lines as well as primary HCC tumors into different subgroups according to their oncofetal properties. Interestingly, HCC cell lines which are highly tumorigenic in a xenograft mouse model, such as Huh7 and Hep3B, showed mixed oncofetal signature compared to other less tumorigenic cell lines with only ES-like or LP-like signatures. Both HCC cells and primary tumors with different oncofetal properties showed selective vulnerability to their specific inhibitors. This indicated that further treatment of HCC patients according to subtype-specific biomarkers might precisely target the vulnerability of tumor cells and enhance treatment efficiency.
Above all, we investigated the potential oncofetal genes derived from an in vitro hepatocyte differentiation model. A previous undescribed gene expression pattern was identified, and core molecular biomarkers were selected to represent specified liver cancer subtypes. Discovery of the upstream master transcriptional regulators, followed with functional and mechanical testing, provided a therapeutic strategy and molecular targets. Further precise targeting of the tumor-initiating steps and driving events according to subtype-specific biomarkers might provide novel therapeutic strategy in HCC treatment.
Materials and Methods
Cell Culture.
The derivation of hESCs and their use for research was approved by the ethical committee of the CITIC-Xiangya Reproductive & Genetic Hospital. The chHES-90 cells were established as previously described (43). Briefly, hESCs were derived and cultured on a feeder layer of mitotically inactivated human embryonic fibroblasts at a density of ∼2,500 cells per cm2. The hESCs cells were cultured in the medium consisting of knockout DMEM/F12 medium supplemented with 15% knockout serum replacement (SR), 2 mM nonessential amino acids, 2 mM l-glutamine, 0.1 mM β-mercaptoethanol, and 4 ng/mL of basic fibroblast growth factor (Invitrogen). The medium was changed daily and hESCs were routinely passaged every 6 or 7 d.
Generation of Hepatocyte-Like Cells.
The differentiation protocol for obtaining hepatocyte-like cells was conducted as described in previous studies (44). In brief, hESCs were passaged onto a feeder free system until a confluence of 50 to 70% was attained. Then cells cultured in RPMI-1640 (Life Technologies) supplemented with 100 ng/mL activin A (R&D Systems) and 25 ng/mL Wnt3 a (R&D Systems) for 3 d. To induce hepatic EN, cells were grown in KO/DMEM (Life Technologies) supplemented with 25 nm/mL keratinocyte growth factor (R&D Systems) and 2% fetal bovine serum (FBS) (Gibco) for 2 d and then further cultured in the KO/DMEM containing 20% SR, 1 mM glutamine, 1% nonessential amino acids, 0.1 mM 2-mercaptoethanol, and 1% dimethyl sulfoxide (DMSO) for 4 to 7 d. The final maturation step to obtain hepatocyte-like cells involved culturing the cells in mature medium containing 10% FBS, 10 ng/mL hepatocyte growth factor (R&D Systems), 20 ng/mL oncostatin M (R&D Systems), and 0.5 μM dexamethasone (R&D Systems) for seven more days.
Mice, Tumor Specimens, and Cell Lines.
Mice were housed in pathogen-free laboratory animal unit at The University of Hong Kong. The endpoint of the in vivo study was set at the end of the sixth week after tumor inoculation. All of the animal experiments were approved by the review board of The University of Hong Kong. Studies using human tissues were reviewed and approved by the Committees for Ethical Review of Research involving Human Subjects of Sun Yat-Sen University and The University of Hong Kong. All patients gave written informed consent for the use of their clinical specimens for medical research. The HKU-HCC cohort, which contains 99 patients, was used as the initial discovery cohort. The TCGA-HCC cohort, which contains 373 patients, was used as the validation cohort. Detailed clinical pathological variables of the HKU-HCC cohort according to the reporting guidelines REMARK (Reporting Recommendations for Tumor Marker Prognostic Studies) and the EASL (European Association for the Study of the Liver) guidelines are listed in SI Appendix, Table S2. Kaplan–Meier curves and log-rank analyses were used in all survival analysis. Pearson’s χ2 test was used for the analysis of clinical pathological features. HCC cell lines Huh7, Hep3B, and PLC-8024 were bought from ATCC, and the rest of the cell lines were bought from the cell bank of the Chinese Academy of Sciences. All of the cells used were passaged for fewer than 6 mo after initial resuscitation.
RNA Extraction and qPCR.
Total RNA was extracted using TRIzol Reagent (Life Technologies), and reverse transcription was performed using an Advantage RT-for-PCR Kit (Clontech Laboratories) according the manufacturer’s instructions. For qPCR analysis, aliquots of double-stranded complementary DNA were amplified using a SYBR Green PCR Kit (Life Technologies) and an ABI PRISM 7900 sequence detector. Sequences of primers used in this study are listed in SI Appendix, Table S4. For cell lines, the relative gene expression is given as 2−ΔCT [ΔCT = CT(gene) – CT(18S)] and normalized to the relative expression that was detected in the corresponding control cells. For clinical samples, we calculated the relative expressions of target genes in clinical HCCs and their matched nontumor specimens by the formula 2−ΔCT [ΔCT = CT(target genes) – CT(18S)] and normalized to the average relative expression in all of the nontumor tissues, which was defined as 1.0.
RNA Sequencing and Data Analysis.
RNA sequencing was done according to previous descriptions (45). IPA software (QIAGEN) was used for gene ontology analysis, upstream activator analysis, and gene network analysis. XLSTAT software (Addinsoft) was used for heat-map analysis. The integrated cancer genomics platform cBioportal was used to analyze the genomic characteristics and clinical significance of selected genes (46). The GEPIA (Gene Expression Profiling Interactive Analysis) web server was used to validate the expressions as well as gene expression correlations (47). Z-score calculation was used to assign an embryonic -like feature score and a LP+ feature score. The z-score values reflect the expression difference between tumor and normal and are calculated as z = (x = lg2[tumor RSEM]), where x is log2(tumor tissue expression value) and and are the mean and SD of the fitted normal distribution in normal tissue population. These z-scores were standardized again by individual signature genes (z-scores of z-scores) and were used to calculate the average value for each of the two subgroups. If the average value for ES-like signature genes for a sample is same or above the cutoff, it was considered as ES-positive (ES+), and vice versa. The same method was used to define LP+ feature (LP+, LP−). The cutoff value 0.1 was used for both features and cohorts. For TDA of the TCGA tumor samples, single-cell TDA was used (48). Multidimensional scaling projection was used to generate a mapper representation for TDA analysis, using an 18 × 18 bin with an average 45% overlap. The nodes in each figure are sets of samples with similar expression level of all 10 genes, and the sizes correspond to the number of samples in that set. Edges connect the nodes that have at least one sample in common. The color corresponds to the expression of a specific gene, except the overall survival, where the color corresponds to the patient’s survival. Statistical analysis was performed using SPSS version 16. (SPSS, Inc.). A Pearson’s χ2 test was used for the categorical variables, and an independent Student’s t test was used for continuous data. Kaplan–Meier plots and log-rank tests were used for overall survival and disease-free survival analysis, respectively. A P value less than 0.05 was considered statistically significant.
Immunohistochemical Staining and Antibodies.
Paraffin-embedded tissue sections were deparaffinized and rehydrated. Slides were immersed in 10 mM citrate buffer and boiled for 15min in a microwave oven and then incubated with primary antibody at 4 °C overnight in a moist chamber and then sequentially incubated with biotinylated general secondary antibody for 1 h at room temperature, streptavidin–peroxidase conjugate for 15 min at room temperature. Finally, a 3, 5-diaminobenzidine Substrate Kit (Dako) was used for color development followed by Mayer’s hematoxylin counterstaining. The antibodies used in this study included anti-OCT4 (SC-5279; Santa Cruz Biotechnology), anti-FOXA2 (07-633; Millipore), anti-EpCAM (SAB4200690; Sigma), anti-AFP (SAB4200746; Sigma), anti-E2F1 (ab79445; Abcam), anti-Smad3 (phosphor S423+S425) (ab52903; Abcam), Alexa Fluor 488 donkey anti-mouse (A21202; Life Technologies), and Alexa Fluor 594 donkey anti-rabbit (A21207; Life Technologies).
HCC Patient-Derived Organoid Cultures and Cell Viability Assay.
HCC tissues used for organoid establishment were obtained from HCC patients undergoing hepatectomy or liver transplantation at Queen Mary Hospital, Hong Kong, with informed consent obtained from all patients and protocol approved by the Institutional Review Board of The University of Hong Kong/Hospital Authority Hong Kong West Cluster. Samples were collected from patients who had not received any previous local or systemic treatment prior to operation. Cells were isolated and cultured as organoids according to published protocol (49). Cell viability of organoid cultures treated with specified concentrations of inhibitors was evaluated using CellTiter-Glo Luminescent Cell Viability Assay (Promega) according to the manufacturer’s protocol.
In Vitro and In Vivo Functional Assays for Tumorigenicity.
For the cell proliferation assay, HCC cells stabling expressing shE2F1, shSMAD3, and nontargeting control shRNA were seeded in 96-well plates at a density of 1,000 cells per well. The cell growth rate was detected using a cell proliferation MTT kit (Sigma-Aldrich). For the foci formation assay, cells were seeded in six-well plates at a density of 1,000 per well. For the soft agar assay, cells were seeded in 0.4% bactoagar on a bottom layer of solidified 0.6% bactoagar in six-well plates at a density of 5,000 per well. For all in vitro assays using small-molecular inhibitors, the drug vehicle DMSO was used as a control. To avoid vehicle toxicity, we dissolved different concentrations of drugs in the same volume of DMSO. No significant toxicity of DMSO was found in the experiments. For the xenograft tumor growth assay, E2F1-specific inhibitor HLM6474 (20 mg/kg) or SMAD3-specific inhibitor SIS3 (2 mg/kg) dissolved in 10% 2-hydroxypropyl-β-cyclodextrin was intraperitoneally injected into the tumor-bearing mice, respectively. Tumor volume was measured weekly and calculated by the formula V = 0.5 × L × W2. For patient-derived primary tumor transplantation, fresh human HCC tissues were cut into pieces equally (∼80 mm3) with scalpels and subcutaneously transplanted into the left dorsal side of severely immune-deficient NSG mice. After the xenograft tumors were stably formed, the mice were intraperitoneally injected with therapeutic drugs with the same dosage used in the cell xenograft model and the tumor volume was monitored for 2 wk. HCC tissues were obtained from patients undergoing hepatectomy at Sun Yat-sen University Cancer Center, with informed consent obtained from all patients and protocol approved by the Institutional Review Board of the Sun Yat-sen University. Samples were collected from patients who had not received any previous local or systemic treatment prior to operation.
Data Availability.
All data are included in the paper and SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Meritxell Huch (The Gurdon Institute, University of Cambridge) for sharing HCC organoids. This work was supported by the National Natural Science Foundation of China (NSFC) (Grants 81272416 and 81702400), the National Basic Research Program of China (Grant 2012CB967001), the Hong Kong Research Grant Council (RGC) General Research Fund (Grant 767313), Collaborative Research Funds (Grants C7027-14G, HKU3/CRF/11R, C7038-14G, and C6002-17G), the Theme-Based Research Scheme Fund (Grant T12-403/11), The University of Hong Kong Small Project Fund (Grant 104003640), the NSFC-RGC Joint Research Scheme (Grant N_HKUST606/17), Shenzhen Peacock Team Project Grants KQTD2015033117210153 and KQTD2018041118502879, Guangzhou Key medical discipline construction project fund, and the Guangdong Province Universities and Colleges Pear River Scholar Funded Scheme (2018). X.-Y.G. is Sophie Y. M. Chan Professor in Cancer Research.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
3Department of Biology, Guangdong Provincial Key Laboratory of Cell Microenvironment and Disease Research, Shenzhen Key Laboratory of Cell Microenvironment, Southern University of Science and Technology, Shenzhen 518055, China.
4State Key Laboratory of Oncology in Southern China, Sun Yat-sen University Cancer Center, 510000 Guangzhou, China; and Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen University Cancer Center, 510000 Guangzhou, China.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912146117/-/DCSupplemental.
References
- 1.Reya T., Morrison S. J., Clarke M. F., Weissman I. L., Stem cells, cancer, and cancer stem cells. Nature 414, 105–111 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Lobo N. A., Shimono Y., Qian D., Clarke M. F., The biology of cancer stem cells. Annu. Rev. Cell Dev. Biol. 23, 675–699 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Marquardt J. U., Andersen J. B., Thorgeirsson S. S., Functional and genetic deconstruction of the cellular origin in liver cancer. Nat. Rev. Cancer 15, 653–667 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Ben-Porath I., et al. , An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 40, 499–507 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu R., et al. , The prognostic role of a gene signature from tumorigenic breast-cancer cells. N. Engl. J. Med. 356, 217–226 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Shachaf C. M., et al. , MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature 431, 1112–1117 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Bass A. J., et al. , SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat. Genet. 41, 1238–1242 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hochedlinger K., Yamada Y., Beard C., Jaenisch R., Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 121, 465–477 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Kim J., et al. , A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 143, 313–324 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takebe N., et al. , Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: Clinical update. Nat. Rev. Clin. Oncol. 12, 445–464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villanueva A., et al. , Notch signaling is activated in human hepatocellular carcinoma and induces tumor formation in mice. Gastroenterology 143, 1660–1669.e7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao C., et al. , Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature 458, 776–779 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anastas J. N., Moon R. T., WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer 13, 11–26 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Wakefield L. M., Hill C. S., Beyond TGFβ: Roles of other TGFβ superfamily members in cancer. Nat. Rev. Cancer 13, 328–341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Serag H. B., Hepatocellular carcinoma. N. Engl. J. Med. 365, 1118–1127 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Yong K. J., et al. , Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N. Engl. J. Med. 368, 2266–2276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tschaharganeh D. F., et al. , Yes-associated protein up-regulates Jagged-1 and activates the Notch pathway in human hepatocellular carcinoma. Gastroenterology 144, 1530–1542.e12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee D. H., et al. , LATS-YAP/TAZ controls lineage specification by regulating TGFβ signaling and Hnf4α expression during liver development. Nat. Commun. 7, 11961 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fitamant J., et al. , YAP inhibition restores hepatocyte differentiation in advanced HCC, leading to tumor regression. Cell Rep. 10, 1692–1707 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tarlow B. D., et al. , Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell 15, 605–618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yimlamai D., et al. , Hippo pathway activity influences liver cell fate. Cell 157, 1324–1338 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yanger K., et al. , Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 27, 719–724 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaffer C. L., et al. , Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 154, 61–74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D’Amour K. A., et al. , Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 23, 1534–1541 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Kubo A., et al. , Development of definitive endoderm from embryonic stem cells in culture. Development 131, 1651–1662 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Ober E. A., Lemaigre F. P., Development of the liver: Insights into organ and tissue morphogenesis. J. Hepatol. 68, 1049–1062 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Llovet J. M., Villanueva A., Lachenmayer A., Finn R. S., Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat. Rev. Clin. Oncol. 12, 408–424 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Llovet J. M., Montal R., Sia D., Finn R. S., Molecular therapies and precision medicine for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 15, 599–616 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zucman-Rossi J., Villanueva A., Nault J. C., Llovet J. M., Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 149, 1226–1239.e4 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Lee J. S., et al. , Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology 40, 667–676 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Lee J. S., et al. , A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat. Med. 12, 410–416 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Yamashita T., et al. , EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology 136, 1012–1024 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoshida Y., et al. , Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 69, 7385–7392 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyault S., et al. , Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology 45, 42–52 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Caramel J., et al. , A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell 24, 466–480 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Liu M., et al. , CHD1L promotes lineage reversion of hepatocellular carcinoma through opening chromatin for key developmental transcription factors. Hepatology 63, 1544–1559 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Ben-David U., Benvenisty N., The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat. Rev. Cancer 11, 268–277 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Sia D., Villanueva A., Friedman S. L., Llovet J. M., Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology 152, 745–761 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sia D., et al. , Identification of an immune-specific class of hepatocellular carcinoma, based on molecular features. Gastroenterology 153, 812–826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kent L. N., et al. , Dosage-dependent copy number gains in E2f1 and E2f3 drive hepatocellular carcinoma. J. Clin. Invest. 127, 830–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Majumdar A., et al. , Hepatic stem cells and transforming growth factor β in hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 9, 530–538 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang Y. N., et al. , Epigenetic regulation of Smad2 and Smad3 by profilin-2 promotes lung cancer growth and metastasis. Nat. Commun. 6, 8230 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Lin G., et al. , HLA-matching potential of an established human embryonic stem cell bank in China. Cell Stem Cell 5, 461–465 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Xie P., et al. , Physiological oxygen prevents frequent silencing of the DLK1-DIO3 cluster during human embryonic stem cells culture. Stem Cells 32, 391–401 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Chen L., et al. , Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 19, 209–216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cerami E., et al. , The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang Z., et al. , GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rizvi A. H., et al. , Single-cell topological RNA-seq analysis reveals insights into cellular differentiation and development. Nat. Biotechnol. 35, 551–560 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huch M., et al. , Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160, 299–312 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the paper and SI Appendix.