Significance
CRISPR-Cas9 has been harnessed for genetic editing and other applications that require specific binding to target sites in the genome. The Cas9 enzyme searches for a target by binding to DNA and unwinding the double helix to test for matches to a guide RNA molecule that is carried as a reference. We observe DNA unwinding carried out by individual Cas9 complexes in real time, revealing intermediate steps in this molecular recognition process. We also show how these steps are mechanically sensitive to DNA supercoiling—underwinding or overwinding of the double helix—which can lead to increased binding and cleavage of mismatched off-target sites under biologically relevant conditions.
Keywords: magnetic tweezers, gene editing, torque spectroscopy
Abstract
The CRISPR-Cas9 nuclease has been widely repurposed as a molecular and cell biology tool for its ability to programmably target and cleave DNA. Cas9 recognizes its target site by unwinding the DNA double helix and hybridizing a 20-nucleotide section of its associated guide RNA to one DNA strand, forming an R-loop structure. A dynamic and mechanical description of R-loop formation is needed to understand the biophysics of target searching and develop rational approaches for mitigating off-target activity while accounting for the influence of torsional strain in the genome. Here we investigate the dynamics of Cas9 R-loop formation and collapse using rotor bead tracking (RBT), a single-molecule technique that can simultaneously monitor DNA unwinding with base-pair resolution and binding of fluorescently labeled macromolecules in real time. By measuring changes in torque upon unwinding of the double helix, we find that R-loop formation and collapse proceed via a transient discrete intermediate, consistent with DNA:RNA hybridization within an initial seed region. Using systematic measurements of target and off-target sequences under controlled mechanical perturbations, we characterize position-dependent effects of sequence mismatches and show how DNA supercoiling modulates the energy landscape of R-loop formation and dictates access to states competent for stable binding and cleavage. Consistent with this energy landscape model, in bulk experiments we observe promiscuous cleavage under physiological negative supercoiling. The detailed description of DNA interrogation presented here suggests strategies for improving the specificity and kinetics of Cas9 as a genome engineering tool and may inspire expanded applications that exploit sensitivity to DNA supercoiling.
RNA-guided nucleases of CRISPR-Cas systems protect bacteria and archaea from invading nucleic acids (1–4). The type II CRISPR nuclease Cas9 introduces targeted double-stranded breaks in DNA using a dual guide RNA (gRNA) system, composed of crRNA and tracrRNA (5). When harnessed for genetic engineering (6–9), the crRNA and tracrRNA are often fused into a single guide RNA (sgRNA), providing a convenient two-part programmable set of molecular scissors (10). The nuclease deficient version of the enzyme (dCas9), which can bind stably to targeted sites of the genome without introducing cuts, has also found broad applications in transcriptional regulation, epigenome engineering, and imaging (11, 12). Models for DNA interrogation propose that Cas9 binds at a protospacer adjacent motif (PAM) site and forms an R-loop that extends unidirectionally from the PAM site (13–15). R-loop formation is required for stable specific binding (13) and is used for target discrimination before cleavage. Upon complete R-loop formation, Cas9 undergoes a conformational rearrangement resulting in activation of its HNH and RuvC nuclease domains and subsequent cleavage of both strands of the DNA (16–18).
Precise targeting of double-stranded DNA breaks is critical for successful genome editing, but Cas9 can also cleave DNA at off-target sites (5, 19). The propensity for off-target activity depends on the position of mismatches between the DNA and the guide RNA: the first 8 to 12 PAM-proximal base pairs (bp) of the target sequence form a seed region where mutations are less tolerated compared to mismatches in the PAM-distal region (5). The detailed dynamics of R-loop formation, including the configuration, energetics, and kinetics of partial R-loop intermediates, are still unknown.
Unwinding the DNA duplex, such as occurs during R-loop formation, is facilitated or hindered by negative or positive torsional strain in the DNA, respectively (20). Therefore, it is expected that Cas9 function is affected by DNA supercoiling in the genome. Topoisomerases serve to control supercoiling levels and in bacteria maintain the chromosome at a typical average superhelical density (21, 22). However, local changes in arise dynamically from fundamental cellular processes such as transcription and replication, which can cause buildup of positive torque ahead of the transcription and replication machinery and negative torque behind a transcribing RNA polymerase (23). Studies in eukaryotes have identified gene- and locus-specific variations in supercoiling, with regions of high negative superhelicity strongly correlated with promoter sites (24–26). The response of Cas9 to torsional strain is thus important to understanding its function in cellular contexts.
Previous single-molecule studies (27) have indicated that Cas9 can be affected by mechanical perturbations: conventional magnetic tweezers measurements showed that R-loop stability is torque-dependent (14) as expected, and recent optical tweezers measurements showed that Cas9 cleavage specificity is affected by high stretching forces (28). However, high-resolution and multimodal approaches are needed to elucidate how multistep Cas9 DNA interrogation dynamics are affected by physiologically relevant supercoiling and sequence mismatches, including unambiguous identification and characterization of visited R-loop intermediates and torque-dependent rates of transitions between states.
Rotor bead tracking (RBT) is a single-molecule technique that can monitor conformational dynamics of nucleoprotein complexes in real time under controlled force and torque on the nucleic acid substrate (29, 30). In recent technical advances, high resolution in measuring DNA twist and torque was achieved by using small light-scattering probes in an implementation called AuRBT (31). Additionally, RBT was combined with single-molecule fluorescence and FRET to detect local conformational and compositional changes in an adaptation called FluoRBT (32). Previously, RBT and its extensions have been used to study structural transitions in DNA under torque and to construct a detailed model of the mechanochemical cycle of DNA gyrase (31–36).
Here we use RBT methods, including AuRBT and FluoRBT, to study dCas9 R-loop formation and dissociation dynamics under torque and in the presence of sequence mismatches. We observe that R-loop formation leads to the unwinding of 20 bp of DNA and that the rates of R-loop formation and dissociation are dependent on torque. We detect a discrete intermediate, consistent with DNA:RNA hybridization through the seed region, that is transiently visited on the way to complete R-loop formation or collapse. In the presence of PAM-distal mismatches, we observe that dCas9 can achieve stable binding at the intermediate state and can reversibly visit the fully unwound state under negative torque. PAM-proximal mismatches destabilize the intermediate state and permit R-loop formation only under negative torque. Mechanical stabilization of the complete R-loop in negatively supercoiled substrates can lead to cleavage even in the presence of extensive mismatches.
Results
We used AuRBT to determine how torsional strain in DNA affects dCas9 binding and release from its target site. We assembled a DNA tether containing a PAM site followed by the λ1 protospacer sequence (13) below the rotor bead (Fig. 1A). We modulated the total twist in the DNA by rotating the magnetic bead and measured the applied torque based on the angular deflection of the rotor bead (Materials and Methods) (31, 34). We introduced a low concentration of Streptococcus pyogenes dCas9 to observe binding and unwinding events without cleavage of the DNA tether.
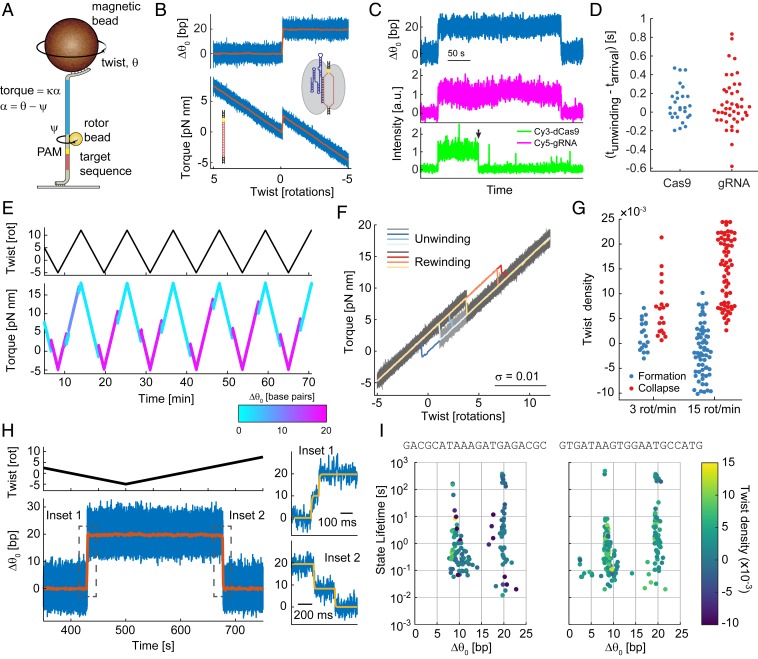
Fig. 1.
dCas9 R-loop detection using RBT. (A) Schematic representation of the experimental setup. A magnetic bead is used to apply tension and modulate the total twist, θ, of a DNA tether. The applied torque is measured via the angle ψ of a small rotor bead (d = 100 nm) that reports on the deflection, α, of the transducer segment (blue) with torsional stiffness κ. A Cas9 target sequence (red) and TGG PAM site (yellow) are positioned below the rotor bead. (B) dCas9 R-loop formation is observed as a jump in the torque-twist curve, corresponding to a change in the equilibrium twist, , consistent with unwinding of 19.8 bp of DNA (Materials and Methods; blue, 5-kHz raw data; orange, 1-s block average). (C and D) FluoRBT detects arrival of the dCas9-gRNA complex concurrent with R-loop formation. (C) Fluorescence arrival of Cy3-labled dCas9 (green) and Cy5-labeled gRNA (magenta) is observed at R-loop formation events. The two dyes are excited in alternating camera frames (Materials and Methods). The Cy3 dye bleaches (black arrowhead) before departure of the dCas9-gRNA complex. (D) R-loop formation and dCas9 and gRNA arrival coincide within the 100-ms resolution of this assay (SI Appendix, Figs. S2 and S3). (E–G) dCas9 R-loop formation and dissociation are dependent on torque. (E) By cycling between positive and negative twist values, we observe multiple R-loop formation and dissociation events, shown here in color corresponding to the number of base pairs unwound. (F) Overlay of unwinding and rewinding portions of the torque-twist curves (gray, 5-kHz raw data; color, 1-s [0.05 rotations] block average) shows transitions between two dominant states and hysteresis between twist of R-loop formation and collapse. (G) The characteristic twist density of R-loop formation and collapse depends on the rate at which twist across the DNA is changed. (H and I) dCas9 R-loop formation and collapse proceed via a transient intermediate. (H) Magnification at the time of R-loop formation and collapse (Insets 1 and 2, respectively) shows that a transient intermediate is visited on the way to complete hybridization or dissociation of the fully matched guide RNA and the target DNA. Blue, 5-kHz raw data; orange, 1-s block average. State transitions were identified using the change-point detection algorithm Steppi (43, 44) (SI Appendix, Fig. S8); fit results are given in yellow. (I) Plot of state position and lifetime for the λ1 sequence (Left), as well as for the distinct λ2 guide RNA with a matching DNA target site (Right). The twist density of state formation is given in color.
We initiated the experiment at positive torque to suppress R-loop formation. When we rotated the magnetic bead to decrease the torque and permit R-loop formation, the linear torsional response of the DNA was interrupted by a torque jump corresponding to a change in equilibrium twist , consistent with DNA unwinding below the rotor bead (34). The magnitude of corresponds to the unwinding of 19.8 ± 0.3 bp (mean ± SD, n = 32 long dwells; Materials and Methods) of B-form DNA (Fig. 1B), as expected based on the 20-nucleotide (nt) region of complementarity between the sgRNA and the target DNA. This signature of R-loop formation was not observed in control experiments using dCas9 without sgRNA, dCas9 in complex with nontargeting sgRNA, or on-target sgRNA alone (SI Appendix, Fig. S1). In FluoRBT experiments using labeled protein or gRNA, we observed arrival and departure of the dCas9-gRNA complex concurrent with R-loop formation and collapse (Fig. 1C). Limited by the lower temporal resolution of the FluoRBT assay, we can conclude that R-loop formation occurs within ∼100 ms of dCas9 binding (Fig. 1D and SI Appendix, Figs. S2 and S3), consistent with PAM surveillance kinetics reported previously (13, 15, 37–39).
We also found that dCas9 R-loops can be dissociated under positive torque. We developed a protocol of cycling between positive and negative torque to observe repeated R-loop formation and dissociation events (Fig. 1E). Overlay of the resulting torque-twist curves shows that the system primarily exists in two states, corresponding to the free DNA and the fully formed R-loop (Fig. 1F). In our assay, R-loop formation occurs readily on DNA that is approximately torsionally relaxed; R-loop collapse is observed primarily under positive torque, at levels achievable physiologically during transcription (40, 41). The characteristic twist densities required for R-loop formation and collapse depend on the rate of imposed DNA twisting, as expected for an out-of-equilibrium system where transition kinetics are slow in comparison with mechanical loading (42). At a faster cycling rate, we observe increased hysteresis, with R-loop formation and collapse occurring at higher negative and positive torques, respectively (Fig. 1G). Our results are in agreement with previous magnetic tweezers studies showing that efficient R-loop formation by Streptococcus thermophilus dCas9 requires DNA underwinding and that R-loop collapse is dependent on torque (14). Our measurements extend these results by showing that S. pyogenes dCas9 R-loop formation is also dependent on torque and that the free energy of tertiary complex formation can overcome low positive torques in DNA to form an R-loop.
We next asked whether R-loop formation proceeds via discrete partially unwound intermediates, taking advantage of the high spatiotemporal resolution of AuRBT (31) (SI Appendix, Fig. S2) and applying automated change-point detection (43, 44) to score dynamic transitions between R-loop states. We observed a well-defined intermediate state equivalent to unwinding 9.3 ± 0.7 bp (n = 21 long dwells) of DNA (Fig. 1 H and I and SI Appendix, Fig. S4). This intermediate is visited transiently prior to complete R-loop formation and on the way to R-loop collapse (Fig. 1H), typically lasting from tens of milliseconds to seconds (Fig. 1I). The same intermediate is also observed with a different DNA target sequence and matching guide RNA (Fig. 1I; 8.5 ± 0.3 bp unwound, n = 35). In nearly all cases (90 out of 92), R-loop formation and collapse proceed via a scored intermediate (SI Appendix, Fig. S5); we also observe dynamic reversible transitions between the intermediate and open states. We sometimes detected long-lived dwells (10 to 100 s) at the intermediate state (Fig. 1I and SI Appendix, Fig. S6), which might correspond to nonproductive binding (37, 38, 45). The intermediate is consistent with DNA unwinding localized to the ∼8- to 12-bp seed region and may correspond to an intermediate previously proposed on the basis of single-molecule FRET measurements (46, 47), which lacked the resolution to distinguish numbers of base pairs unwound. An analogous intermediate has previously been observed in single-molecule and cryo-EM studies of the Cascade complex (14, 48, 49).
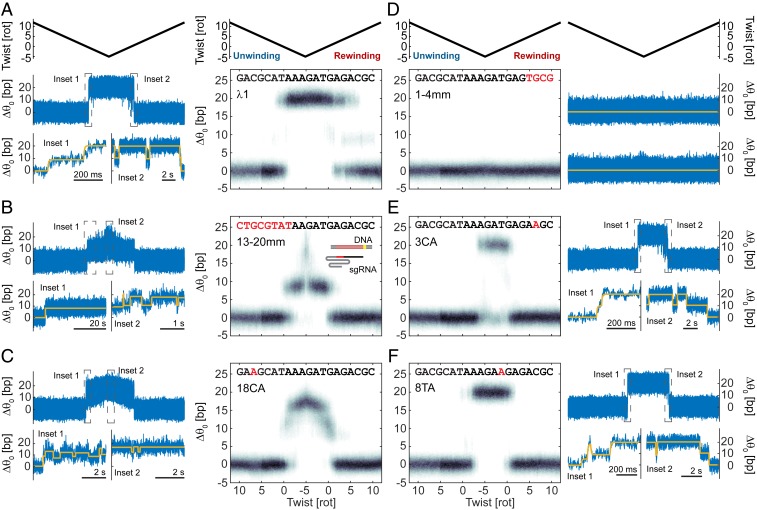
In vitro and in vivo results show that Cas9 can bind and cleave DNA at off-target sites, particularly when the mismatches are PAM-distal (5, 19, 50). We sought to probe how the position of sequence mismatches and assisting or opposing torque in the DNA may reshape the energy landscape of R-loop formation. We repeated our cycling protocol using a series of partially mismatched guide RNA sequences and found that mismatches between the guide RNA and the target DNA affect the stability of R-loop intermediates and the positions of dominant states as a function of twist (Fig. 2 and SI Appendix, Fig. S4). R-loop formation with a fully matched guide RNA only transiently visits the intermediate before reaching the open state, which then remains stable over long timescales on relaxed or negatively supercoiled DNA (Fig. 2A). In contrast, mutating the last 8 PAM-distal nucleotides of the spacer region of the guide RNA (13-20mm) prevents full R-loop formation on relaxed DNA and leads to stable dwells at the intermediate (Fig. 2B). These observations are consistent with previous results showing that Cas9 in complex with guide RNA carrying mismatches at these positions can stably bind but does not cleave DNA (5, 13, 15, 16). At negative twist densities, we observed that the 13-20mm sequence makes quick transitions to more fully unwound states (Fig. 2B). A more conservative single mutation at the 18th base of the spacer region (18CA) leads to frequent transitions between the intermediate and a new ∼17-bp unwound state in near-relaxed DNA (Fig. 2C).
Fig. 2.
Sequence mismatches and torque affect the stability of R-loop intermediates. (A) (Left) Example trace showing R-loop formation with λ1 sgRNA. Blue, raw 5-kHz data; yellow, change-point analysis (43, 44) fit. (Right) Histogram of change in equilibrium twist (, in base pairs) as a function of imposed twist. Data from multiple cycles using measurement protocols as in Fig. 1E are block averaged in 20-ms bins and aligned in twist; plots contain data from multiple experiment repetitions and duplicate sgRNA preparations (Materials and Methods). (B–F) Data as in A for 13-20mm, 18CA, 1-4mm, 3CA, and 8TA sgRNAs. Mismatched bases are highlighted in red. Cartoon in B illustrates the orientation of the DNA and sgRNA segments.
Distinct from the behavior of PAM-distal mismatches, we find that PAM-proximal mismatches reduce the stabilities of both the intermediate and open states, such that the closed state remains favored over a wider range of mechanical conditions. When we mutate the first four nucleotides of the spacer region (1-4mm), we observe no R-loop formation under the conditions of the assay (Fig. 2D). With a single mutation at the third base (3CA), R-loop formation occurs only at negative torque (Fig. 2E). As with the fully matched sequence, R-loop formation goes through the transient intermediate before reaching a long-lived open state, but more negative torque is required before any R-loop is formed. A mutation at base 8 (8TA) has an intermediate effect, permitting R-loop formation at lower negative torque compared to the 3CA sequence (Fig. 2F). This mutation destabilizes the intermediate, and we also see more frequent dwells at otherwise atypical positions corresponding to ∼5 or ∼12 to 13 bp unwound (SI Appendix, Fig. S4).
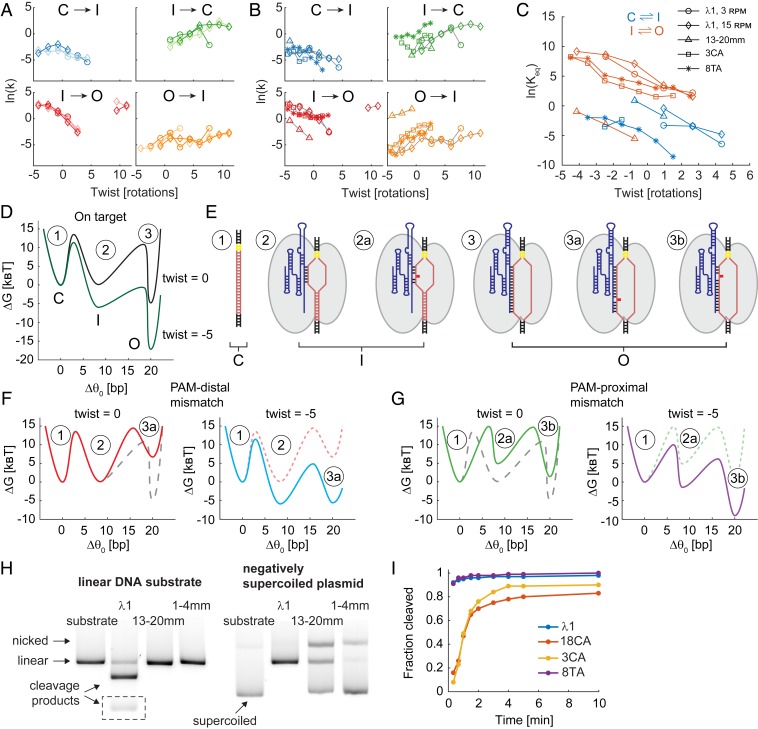
We quantified the transition rates between R-loop conformations as a function of imposed twist and reconstructed an approximate energy landscape for dCas9 R-loop formation (Fig. 3 and SI Appendix, Figs. S7 and S8). R-loop formation proceeds through three major conformations corresponding to closed unbound DNA (C), intermediate (I), and fully open (O) states. Transition rates between these states calculated from the unwinding and rewinding portions of the torque-twist curve, as well as rates derived from data collected using the slow and fast cycling protocols, collapse onto the same master curves (42) (Fig. 3A). We find that sequence mismatches can perturb transition rates and that for all sequences the closed to intermediate (C→I) and intermediate to open (I→O) transitions are accelerated under negative torque, as expected (Fig. 3 B and C). The interplay between torque and sequence mismatches can be visualized using an energy landscape (Fig. 3 D–G). PAM-distal mismatches destabilize the O state (Fig. 3F), but this state can still be reached under negative torque, which tilts the energy landscape (51). PAM-proximal mismatches destabilize both the I and the O states and may preclude dCas9 binding and R-loop formation (Fig. 3G). Negative torque can tilt the energy landscape and permit R-loop formation in the presence of PAM-proximal sequence mismatches. For fully matched guide RNAs, a comparison of the two different target sequences shown in Fig. 1I suggests that sequence can also modestly affect the kinetics and energetics of state transitions (SI Appendix, Fig. S7).
Fig. 3.
Energy landscape of dCas9 R-loop formation. (A) Rates of transition between the closed (C), intermediate (I), and open (O) R-loop states as a function of twist. Data are for the λ1 sgRNA sequences, collected using the 3 and 15 rot/min protocols shown with circle and diamond markers, respectively. Lighter color is used for transitions during the unwinding phase of the torque-twist curve, and darker color is used for transitions during the rewinding phase. (B) Transition rates as a function of twist for mismatched sequences. Data from unwinding and rewinding curves were processed together. Marker symbols are as in C. (C) Apparent equilibrium constant as a function of twist for the C–I transition (blue) and I–O transition (orange). (D) Depiction of the free energy landscape of dCas9 R-loop formation with λ1 sgRNA at varying DNA superhelical density (black, relaxed DNA; green, twist = −5 rotations) based on data in A–C. (E) Cartoon representations of R-loop states. (F) Energy landscapes of R-loop formation in the presence of PAM-distal mismatches (using 13-20mm sgRNA) under imposed twist of 0 (Left) and −5 rotations (Right). (G) Free energy landscapes as in F for sequences with PAM-proximal mismatches (here using 8TA sgRNA). Free energy landscapes from previous plots are shown as dashed lines for reference. (H) Cas9 cleavage of linearized (Left) and negatively supercoiled plasmid DNA (Right) for subset of sgRNAs. The linear DNA template was constructed by treating the plasmid DNA with DraIII endonuclease. Images were tiled to show the short product of λ1 cleavage (dashed box) and to exclude intervening lanes. See Table 1 for data quantification. (I) Cleavage time course for subset of sgRNAs using shorter linear DNA template (Materials and Methods).
The ability of torsional stress in DNA to modulate access to R-loop intermediates suggests that DNA superhelical density will also modulate the efficiency of Cas9 cleavage. We tested this hypothesis by measuring Cas9 cleavage efficiency on torsionally relaxed and negatively supercoiled DNA templates (Fig. 3H and Table 1). We focused our comparison on the most extreme mismatches since some of the mismatch sequences used in this study are still able to efficiently cleave linear DNA, albeit typically at lower rates than the fully matched λ1 sequence (Fig. 3I and Table 1). As predicted, the 13-20mm sgRNA showed partial cleavage of the negatively supercoiled plasmid DNA, despite extensive mismatches with the target; we also observed nicking of this substrate. Cleavage activity was not detected when the same template was first torsionally relaxed by nicking or linearization. Using the 1-4mm sgRNA, we observed 2% cleavage and 18% DNA nicking of negatively supercoiled DNA in the course of 30 min, suggesting that even this sequence can form infrequent R-loops and reach cleavage-competent states. Our results complement previous measurements of RNA-guided nuclease activity on supercoiled DNA: cleavage and nicking of targets that contain distal mismatches was measured for S. thermophilus Cas9 (14) in a study that relied exclusively on supercoiled substrates for bulk cleavage assays, and a very recent study (52) showed that negative supercoiling strongly accelerates the cleavage reaction for Cas12a.
Table 1.
Cas9 cleavage of torsionally relaxed and negatively supercoiled DNA templates
| Linear | Nicked | Negatively supercoiled | ||
| Cleavage | Cleavage | Cleavage | Nicking | |
| λ1 | 0.91 ± 0.01 | 0.92 ± 0.01 | 1.00 | n.d. |
| 13-20mm | n.d. | n.d. | 0.35 ± 0.04 | 0.34 ± 0.01 |
| 18CA | 0.88 ± 0.01 | 0.90 ± 0.02 | 1.00 | n.d. |
| 3CA | 0.97 ± 0.01 | 0.94 ± 0.03 | 1.00 | n.d. |
| 8TA | 0.98 ± 0.01 | 1.00 | 1.00 | n.d. |
| 1-4mm | n.d. | n.d. | 0.02 ± 0.01 | 0.19 ± 0.05 |
Linear and nicked reaction templates were generated by treating the negatively supercoiled plasmid DNA with endonucleases DraIII and Nb.BsrDI, respectively. Fractional cleavage is given as mean ± SD for n = 3 replicates. The detection limit of the assay is ∼1% cleavage or nicking; SD is not given when all three measurements agree within <1%. n.d., not detected.
Discussion
The position of the intermediate we identified and the effect of sequence mismatches on its stability are consistent with DNA:RNA hybridization in the seed region. This state may be stabilized by the preordering of the first 10 bp of the guide RNA spacer region when in complex with Cas9, prior to binding DNA (53). Our results suggest that while Cas9 can remain stably bound to DNA in the intermediate state, access to more fully unwound states is required for cleavage; such states can be reached under negative torque, even in the presence of many sequence mismatches. Docking of the HNH nuclease domain, which is sensitive to mismatches and is required before Cas9 can cleave DNA, is likely coupled to R-loop formation (16, 45). We propose that the HNH domain is undocked in the intermediate state but can become docked upon reaching the fully open R-loop state, consistent with the timescale (∼1.5 s) for reaching the docked state in a single-molecule FRET study of HNH docking dynamics (45). This timescale is also consistent with the ∼1-s delay between binding and R-loop formation in a separate single-molecule FRET study that used dye pairs located in the DNA duplex in order to monitor unwinding (37).
Our data are consistent with a model in which Cas9 R-loop formation goes through two distinct states: the partial R-loop intermediate and the cleavage-competent open state (Fig. 3). PAM-distal mismatches destabilize the open state. Negative torque can tilt the energy landscape and allow access to the fully unwound state in the presence of mismatches. PAM-proximal mismatches destabilize both the intermediate and open states. Consequently, Cas9 binding and stable R-loop formation require higher negative torque. The position of mismatches within the target sequence may give rise to other stable intermediates, as seen with the 18CA and 8TA sgRNAs. However, our data are not consistent with models which propose R-loop formation proceeding via a sequence of increasingly more stable energetic states at every base pair (13, 37). Instead, we observe two-step R-loop formation and dissociation, as some groups have proposed before (14, 15, 46). While the structural details of the discrete R-loop intermediate reported here remain to be elucidated, it could correspond to an intermediate previously described on the basis of FRET measurements; the authors concluded that this intermediate was stabilized by interactions between the nontarget strand and the REC2 domain, suppressing the formation of a cleavage-competent state when mismatches are present (46, 47).
We observed a high level of Cas9 promiscuity at target sites with many PAM-distal mismatches under physiological levels of negative supercoiling. However, such promiscuity is greatly suppressed on torsionally relaxed DNA. Thus, the local superhelical density of the genome plays an important role in the success of genetic engineering efforts and might be controlled to improve the fidelity of genome editing. As has been noted (28), off-target editing appears to be enriched in promoters and transcribed regions (54, 55), where torsional strain may accumulate (24, 25). Large effects on Cas9 function are seen under torques of a magnitude that can readily be generated in biological contexts. For example, the maximum underwinding used in our experiments corresponds to the accumulation of approximately −7.5 pN nm of torque in the bare DNA tether, whereas RNA polymerase can generate approximately −10 pN nm before stalling (41). The ability to program Cas9 to respond to different levels of torque in DNA may further create new opportunities for constructing local sensors of supercoiling based on fluorescent Cas9 fusions (56) or for engineering supercoiling-dependent transcriptional control by adapting existing regulation approaches (57). The physical mechanisms of DNA interrogation described here may apply to other RNA-guided nucleases and could reflect general features of sequence homology searches across biology, which may be further investigated using the methods presented in this study.
Materials and Methods
Protein and Guide RNA Samples.
AuRBT studies of R-loop formation (Figs. 1 B and E–I and 2) were conducted with 2 nM nuclease deficient S. pyogenes dCas9 (D10A, H840A) and 10 nM chimeric sgRNA (SI Appendix, Table S3). FluoRBT experiments correlating arrival of the dCas9-gRNA complex with R-loop formation (Fig. 1 C and D) were conducted using either 0.5 nM dCas9 and 0.6 nM Cy3-labeled gRNA, constructed by annealing Cy3 λ1 crRNA to tracrRNA (SI Appendix, Table S3), or 0.5 nM Cy3-labeled dCas9 (D10A, H840A, C80S, C574S, S867C) and 0.6 nM Cy5-labeled gRNA, constructed by annealing λ1 crRNA 1–34 to Cy5 tracrRNA 11–75 (SI Appendix, Table S3). dCas9-gRNA complexes were preformed by incubating 100 nM dCas9 and excess gRNA, as described, for 10 min at 37 °C in C9T buffer (20 mM Tris⋅HCl, pH 7.5, 100 mM KCl, 5 mM MgCl2, 1 mM TCEP, 0.1 mM EDTA) supplemented with 5% glycerol. Proteins were expressed and purified as previously described (5).
DNA Tether Construction.
DNA tethers for single-molecule experiments were assembled by ligation of restriction enzyme digested PCR products, as previously described (31, 32, 34). See SI Appendix, Note S1 and Table S1, for details on tether building blocks. Tethers containing the SPD280 unit were used for studying the λ1 target sequence and mutations to that sequence (Figs. 1 and 2), tethers containing the SPD280–lam2 unit were used for studying on-target R-loop formation by the λ2 sgRNA (Fig. 1I), tethers containing the SPD640 unit were used in FluoRBT experiments and control experiments using 300 nm rotor beads, and tethers containing the SPD640–no PAM unit were used in control experiments of dCas9 R-loop formation at a mutated PAM site.
Transcription of Guide RNA.
DNA templates for sgRNA transcription were constructed by PCR of synthetic oligonucleotide pieces (SI Appendix, Table S2) using Phusion DNA polymerase (Thermo Fisher Scientific, F530S). PCR products were purified by phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation. tracrRNA was transcribed from template constructed by annealing T25 and tracrRNA_template (SI Appendix, Table S2). Transcription reactions containing 1.5 μM DNA template, 4 U/μL T7 RNA polymerase (Thermo Fisher Scientific, AM2085), and 2 × 10−3 U/μL yeast inorganic pyrophosphatase (New England Biolabs, M2403S) were incubated overnight. The DNA template was digested using 0.1 U/μL TURBO DNase (Thermo Fisher Scientific, AM2238), and RNA was purified by electrophoresis on 10% polyacrylamide gels. After gel extraction, RNA was precipitated in isopropyl alcohol and refolded in buffer containing 20 mM Hepes, pH 7.5, 100 mM KCl, and 5 mM MgCl2 by heating and cooling slowly.
Two preparations of each RNA transcript were made. The two preparations of the λ1 sgRNA differed in the measured DNA cleavage efficiency—preparation A led to 89% DNA cleavage after 30 min incubation (see below), while preparation B led to 98% DNA cleavage. Preparation A was used in two single-molecule imaging sessions (see below) and in data presented in Fig. 3H and Table 1. Preparation B was used in one single-molecule imaging session and in data presented in Fig. 3I. We did not observe substantial differences in Cas9 cleavage efficiency between preparations of other sgRNA sequences used in this study.
Instrumentation.
Experiments were performed on custom-built FluoRBT microscopes (31, 32). Rotor bead scattering was recorded at 5 kHz when using 102 ± 6 nm [mean ± SD, n = 10 measurements (31)] gold rotor beads (Cytodiagnostics, AC-70-04-10) or at 250 Hz when using 300 ± 8 nm [mean ± SD, n = 10 measurements (31)] polystyrene rotor beads (Thermo Fisher Scientific, 29000701011150). We used 300 nm rotor beads in FluoRBT experiments and for control experiments, unless otherwise specified. High-resolution gold rotor beads were used for all other studies. Fluorescence intensity was recorded at 10 Hz when observing a single fluorophore or at 20 Hz using synchronized alternating laser excitation when observing two fluorophores. For experiments with Cy3-labeled gRNA (e.g., SI Appendix, Fig. S3B), we used 19 mW of 552-nm excitation (OBIS 552 LS, Coherent; illumination area ∼9 × 10−5 cm2) for an estimated irradiance at the sample of ∼200 W/cm2. For two-color experiments with Cy3-labeled dCas9 and Cy5-labeled gRNA, we combined 552 nm excitation with 637 nm excitation (OBIS 637 LX, Coherent; illumination area ∼4 × 10−4 cm2) and chose laser powers to give approximately equal irradiances of ∼50 W/cm2 (Fig. 1C and SI Appendix, Fig. S3A; 4.6 mW at 552 nm and 21 mW at 637 nm) or ∼15 W/cm2 (SI Appendix, Fig. S3C; 1.4 mW at 552 nm and 5.4 mW at 632 nm) between the two wavelengths, accounting for the differing illumination areas.
Imaging Conditions.
Single-molecule experiments were performed in C9T buffer supplemented with 0.2% Tween-20 and 0.2 mg/mL BSA; the buffer was further supplemented with 0.4% glucose, 1,250 U/mL glucose oxidase (Sigma-Aldrich, G2133), 100,000 U/mL catalase (Sigma-Aldrich, C100), and 2 mM Trolox (Sigma-Aldrich, 238813) for combined fluorescence experiments.
Experiments were performed under 5 pN of tension. The total twist in the tether was changed by rotating the magnetic tweezers at 3 rpm following the protocol illustrated in Fig. 1E. Cycling at 15 rpm was used to measure the effect of torque ramp rate on the kinetics and thermodynamics of R-loop formation and collapse (Figs. 1G and 3A). Torque was calculated from the angular deflection, α, of the transducer segment with torsional stiffness [mean ± SD, n = 6 measurements (34)], i.e., . The angle of the magnetic tweezers corresponding to zero twist was set by maximizing the extension of the DNA tether under low force (33, 34). AuRBT datasets were each compiled from three imaging sessions using two replicates of each sgRNA preparation.
Data Analysis.
Rotor bead tracking was performed as previously described (31, 32). The extent of DNA unwinding was calculated from the change in equilibrium twist assuming that any deviations from the torsional response of B-DNA arise from DNA unwinding below the rotor bead and using B-DNA helicity of 10.5 bp per turn, i.e.,
| [1] |
where is the torsional stiffness of the full DNA tether (SI Appendix, Note S1), obtained from the slope of the torque-twist curve, and is the expected torque for unperturbed B-DNA. The measured unwinding angle may also include contributions from residual helicity of separated strands within the complex.
R-loop states were identified using the Steppi change-point analysis tool (43, 44). We modeled the data as originating from an Ornstein–Uhlenbeck process. We fixed the stiffness and coupling parameters using a segment of data containing only thermal fluctuations of the rotor bead. Thus, the only free model parameters were the change point time and mean position of the new state. When displaying idealized traces overlaid on data, we used unmodified idealizations directly generated by Steppi, except in cases where Steppi’s plotting convention generates a spike at the boundary between states that could be confused for an additional scored state; in those cases a single data point corresponding to the spike was omitted from the plot.
For data collected using the λ1, λ2, 13-20mm, 3CA, and 8TA sgRNA, identified R-loop states were assigned into one of three clusters: intermediate (I), open (O), and other (SI Appendix, Fig. S4). For the λ1 and λ2 sequences we further designated a long-lived intermediate state cluster which included I states longer than 10 s or shorter visits to I states during binding events in which a long-lived I state was observed. R-loop formation in the long-lived intermediate state proceeded to the O state only transiently under high negative torque and may represent nonproductive dCas9 binding (37, 38, 45). Consecutive states from the I and O clusters were merged (SI Appendix, Fig. S9). The number of base pairs unwound in the merged states was calculated as the average of contributing states, weighted by their lifetimes. We did not carry out state assignment for the 18CA sequence due to the wide distribution of observed states (SI Appendix, Fig. S4).
The reported mean position of the intermediate (including long-lived intermediates) and open states was calculated by averaging states in that cluster longer than 1 s. The contribution of individual states was not weighted by their lifetimes for this calculation.
Twist-dependent rates of transition between R-loop intermediates were calculated using a procedure analogous to a previously described method (58). In brief, we constructed histograms of the number of transitions between states and as a function of twist normalized by the bin width. To calculate the rate constant , the twist density of the number of transitions in each bin was divided by the number of trajectories contributing to that bin and multiplied by the twist ramp rate , i.e.,
| [2] |
Using the absolute value of makes the expression applicable to both the unwinding and rewinding cycles of the torque-twist curve. Transition rates from unwinding and rewinding curves or from protocols with different twist ramp rates were also calculated separately as a demonstration that in all cases the same rate constant is derived (Fig. 3 A and B). Apparent equilibrium constants (Fig. 3C) were calculated as . We modeled the transition between R-loop intermediates under fixed total twist as transition between states with difference in equilibrium twist and difference in free energy in a polymer with a torsional stiffness , as previously described (33). The model parameters were derived from a linear fit of as a function of twist (SI Appendix, Fig. S8):
| [3] |
where .
Free energy differences between states were determined as for constructing the free energy landscapes depicted in Fig. 3 D, F, and G.
Fluorophore intensities in FluoRBT traces were calculated as the sum of pixel intensities over an 8 × 8 pixel region (corresponding to ∼1 μm2) centered at the DNA tether, and may include contributions of nonspecific binding events. Intensity traces were corrected for background signal based on a nearby region lacking fluorophores; background intensity was block-averaged in 20-s windows to remove noise. For display of the traces shown in SI Appendix, Fig. S3 B and C, the observation region was further restricted to 8 × 5 pixels in order to exclude spurious fluorescence due to binding adjacent to the tether.
Cas9 DNA Cleavage.
The negatively supercoiled pII-lam1 plasmid DNA used in bulk cleavage experiments (Fig. 3H and Table 1) was isolated from Escherichia coli. Linear and nicked versions of the template were generated by treatment with DraIII (New England Biolabs, R3510S) and Nb.BsrDI (New England Biolabs, R0648S), respectively. The SPD640 template used in time course experiments (Fig. 3I) was generated by PCR with Phusion DNA polymerase from the pII-lam1 plasmid using primers TGACGCTTCAGTCAGAACTAC and TCTGTCATCATGCACAGCAC.
Cleavage reactions contained 1 nM DNA template, 50 nM Cas9, and 100 nM guide RNA in C9T buffer with 5% glycerol. The protein-RNA complexes were preformed by incubating at 37 °C for 10 min. DNA was then added, and the reaction was incubated at 37 °C for 30 min. In time-course experiments (Fig. 3I), aliquots of the reaction were taken at the designated time points. Reactions were stopped and Cas9 was denatured by adding 25 mM EDTA, 1× TBE-Urea sample buffer (Thermo Fisher Scientific, LC6876), and heating to 65 °C for 5 min. Cleavage products were resolved on 10% TBE-Urea polyacrylamide gels when using the SPD640 template or on 0.8% agarose gels when using pII-lam1-based templates. Gels were stained with SYBR Gold (Thermo Fisher Scientific, S11494) and imaged on a Typhoon FLA 9500 gel scanner (GE Healthcare).
Data Availability Statement.
Data and code are available to readers at the Stanford Digital Repository, https://purl.stanford.edu/ks501bq7804.
Supplementary Material
Acknowledgments
This work was supported by a Tusher Family Stanford Interdisciplinary Graduate Fellowship to I.E.I., National Institutes of Health grants to Z.B. and J.A.D., and a Stanford Discovery Innovation Fund award to Z.B. J.A.D. is an Investigator of the Howard Hughes Medical Institute and receives funding from the Defense Advanced Research Projects Agency, the Paul G. Allen Frontiers Group, and the National Science Foundation.
Footnotes
Competing interest statement: J.A.D. is a cofounder of Caribou Biosciences, Editas Medicine, Intellia Therapeutics, Scribe Therapeutics, and Mammoth Biosciences. J.A.D. is a scientific advisory board member of Caribou Biosciences, Intellia Therapeutics, eFFECTOR Therapeutics, Scribe Therapeutics, Synthego, Mammoth Biosciences, and Inari. J.A.D. is a director at Johnson & Johnson and has research projects sponsored by Biogen and Pfizer. The Regents of the University of California have patents issued and pending for CRISPR technologies on which J.A.D. is an inventor. A.V.W. is an employee of Scribe Therapeutics.
Data deposition: Data and code are available to readers at the Stanford Digital Repository, https://purl.stanford.edu/ks501bq7804.
This article is a PNAS Direct Submission. D.S.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913445117/-/DCSupplemental.
References
- 1.Wiedenheft B., Sternberg S. H., Doudna J. A., RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331–338 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Sorek R., Lawrence C. M., Wiedenheft B., CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 82, 237–266 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Barrangou R., Marraffini L. A., CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol. Cell 54, 234–244 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marraffini L. A., CRISPR-Cas immunity in prokaryotes. Nature 526, 55–61 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Jinek M., et al. , A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doudna J. A., Charpentier E., Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Hsu P. D., Lander E. S., Zhang F., Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mali P., Esvelt K. M., Church G. M., Cas9 as a versatile tool for engineering biology. Nat. Methods 10, 957–963 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrangou R., Horvath P., A decade of discovery: CRISPR functions and applications. Nat. Microbiol. 2, 17092 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Urnov F. D., Genome editing B.C. (Before CRISPR): Lasting lessons from the “Old Testament”. CRISPR J 1, 34–46 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Knott G. J., Doudna J. A., CRISPR-Cas guides the future of genetic engineering. Science 361, 866–869 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H., La Russa M., Qi L. S., CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 85, 227–264 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Sternberg S. H., Redding S., Jinek M., Greene E. C., Doudna J. A., DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szczelkun M. D., et al. , Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc. Natl. Acad. Sci. U.S.A. 111, 9798–9803 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh D., Sternberg S. H., Fei J., Doudna J. A., Ha T., Real-time observation of DNA recognition and rejection by the RNA-guided endonuclease Cas9. Nat. Commun. 7, 12778 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sternberg S. H., LaFrance B., Kaplan M., Doudna J. A., Conformational control of DNA target cleavage by CRISPR-Cas9. Nature 527, 110–113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang F., et al. , Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 351, 867–871 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J. S., et al. , Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550, 407–410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X.-H., Tee L. Y., Wang X.-G., Huang Q.-S., Yang S.-H., Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids 4, e264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bustamante C., Bryant Z., Smith S. B., Ten years of tension: Single-molecule DNA mechanics. Nature 421, 423–427 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Wang J. C., Cellular roles of DNA topoisomerases: A molecular perspective. Nat. Rev. Mol. Cell Biol. 3, 430–440 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Sinden R. R., Carlson J. O., Pettijohn D. E., Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: Analogous measurements in insect and human cells. Cell 21, 773–783 (1980). [DOI] [PubMed] [Google Scholar]
- 23.Ma J., Wang M. D., DNA supercoiling during transcription. Biophys. Rev. 8 (suppl. 1), 75–87 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouzine F., et al. , Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat. Struct. Mol. Biol. 20, 396–403 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naughton C., et al. , Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat. Struct. Mol. Biol. 20, 387–395 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corless S., Gilbert N., Investigating DNA supercoiling in eukaryotic genomes. Brief. Funct. Genomics 16, 379–389 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh D., Ha T., Understanding the molecular mechanisms of the CRISPR toolbox using single molecule approaches. ACS Chem. Biol. 13, 516–526 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Newton M. D., et al. , DNA stretching induces Cas9 off-target activity. Nat. Struct. Mol. Biol. 26, 185–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryant Z., et al. , Structural transitions and elasticity from torque measurements on DNA. Nature 424, 338–341 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Bryant Z., Oberstrass F. C., Basu A., Recent developments in single-molecule DNA mechanics. Curr. Opin. Struct. Biol. 22, 304–312 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lebel P., Basu A., Oberstrass F. C., Tretter E. M., Bryant Z., Gold rotor bead tracking for high-speed measurements of DNA twist, torque and extension. Nat. Methods 11, 456–462 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivanov I. E., et al. , Multimodal measurements of single-molecule dynamics using FluoRBT. Biophys. J. 114, 278–282 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oberstrass F. C., Fernandes L. E., Bryant Z., Torque measurements reveal sequence-specific cooperative transitions in supercoiled DNA. Proc. Natl. Acad. Sci. U.S.A. 109, 6106–6111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oberstrass F. C., Fernandes L. E., Lebel P., Bryant Z., Torque spectroscopy of DNA: Base-pair stability, boundary effects, backbending, and breathing dynamics. Phys. Rev. Lett. 110, 178103 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basu A., Schoeffler A. J., Berger J. M., Bryant Z., ATP binding controls distinct structural transitions of Escherichia coli DNA gyrase in complex with DNA. Nat. Struct. Mol. Biol. 19, 538–546, S1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basu A., et al. , Dynamic coupling between conformations and nucleotide states in DNA gyrase. Nat. Chem. Biol. 14, 565–574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh D., et al. , Mechanisms of improved specificity of engineered Cas9s revealed by single-molecule FRET analysis. Nat. Struct. Mol. Biol. 25, 347–354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gong S., Yu H. H., Johnson K. A., Taylor D. W., DNA unwinding is the primary determinant of CRISPR-Cas9 activity. Cell Rep. 22, 359–371 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones D. L., et al. , Kinetics of dCas9 target search in Escherichia coli. Science 357, 1420–1424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harada Y., et al. , Direct observation of DNA rotation during transcription by Escherichia coli RNA polymerase. Nature 409, 113–115 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Ma J., Bai L., Wang M. D., Transcription under torsion. Science 340, 1580–1583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dudko O. K., Hummer G., Szabo A., Intrinsic rates and activation free energies from single-molecule pulling experiments. Phys. Rev. Lett. 96, 108101 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Wiggins P. A., An information-based approach to change-point analysis with applications to biophysics and cell biology. Biophys. J. 109, 346–354 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LaMont C. H., Wiggins P. A., The development of an information criterion for change-point analysis. Neural Comput. 28, 594–612 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Dagdas Y. S., Chen J. S., Sternberg S. H., Doudna J. A., Yildiz A., A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Sci. Adv. 3, eaao0027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim Y., et al. , Structural roles of guide RNAs in the nuclease activity of Cas9 endonuclease. Nat. Commun. 7, 13350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sung K., Park J., Kim Y., Lee N. K., Kim S. K., Target specificity of Cas9 nuclease via DNA rearrangement regulated by the REC2 domain. J. Am. Chem. Soc. 140, 7778–7781 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Xiao Y., et al. , Structure basis for directional R-loop formation and substrate handover mechanisms in type I CRISPR-Cas system. Cell 170, 48–60.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rutkauskas M., et al. , Directional R-loop formation by the CRISPR-Cas surveillance complex cascade provides efficient off-target site rejection. Cell Rep. 10, 1534–1543 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Boyle E. A., et al. , High-throughput biochemical profiling reveals sequence determinants of dCas9 off-target binding and unbinding. Proc. Natl. Acad. Sci. U.S.A. 114, 5461–5466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tinoco I. Jr, Bustamante C., The effect of force on thermodynamics and kinetics of single molecule reactions. Biophys. Chem. 101-102, 513–533 (2002). [DOI] [PubMed] [Google Scholar]
- 52.van Aelst K., Martínez-Santiago C. J., Cross S. J., Szczelkun M. D., The effect of DNA topology on observed rates of R-loop formation and DNA strand cleavage by CRISPR Cas12a. Genes (Basel) 10, 169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang F., Zhou K., Ma L., Gressel S., Doudna J. A., STRUCTURAL BIOLOGY. A Cas9-guide RNA complex preorganized for target DNA recognition. Science 348, 1477–1481 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Tsai S. Q., et al. , GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Geen H., Henry I. M., Bhakta M. S., Meckler J. F., Segal D. J., A genome-wide analysis of Cas9 binding specificity using ChIP-seq and targeted sequence capture. Nucleic Acids Res. 43, 3389–3404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Knight S. C., Tjian R., Doudna J. A., Genomes in focus: Development and applications of CRISPR-Cas9 imaging technologies. Angew. Chem. Int. Ed. Engl. 57, 4329–4337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu X., Qi L. S., A CRISPR-dCas toolbox for genetic engineering and synthetic biology. J. Mol. Biol. 431, 34–47 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y., Dudko O. K., A transformation for the mechanical fingerprints of complex biomolecular interactions. Proc. Natl. Acad. Sci. U.S.A. 110, 16432–16437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and code are available to readers at the Stanford Digital Repository, https://purl.stanford.edu/ks501bq7804.