Significance
The major viral components of HIV-1 virions are full-length RNA (HIV-1 RNA) and its translation products. Packaged HIV-1 RNA carries viral genetic information, and its translation products Gag and Gag-Pol constitute the virus structure and all the viral enzymes. Using live-cell imaging and biochemical approaches, we found that half of the HIV-1 RNA population is actively translated; cytoplasmic translating and nontranslating RNAs are well mixed, indicating Gag is not synthesized in specialized compartments. Both translating and nontranslating RNAs move dynamically and can reach the major virus assembly site. However, Gag packages nontranslating RNA; thus, although HIV-1 RNA serves two functions, each molecule performs only one function at a time. These studies provide insight into HIV-1 gene expression and virus assembly.
Keywords: HIV, RNA, translation, Gag, genome packaging
Abstract
HIV-1 full-length RNA (HIV-1 RNA) plays a central role in viral replication, serving as a template for Gag/Gag-Pol translation and as a genome for the progeny virion. To gain a better understanding of the regulatory mechanisms of HIV-1 replication, we adapted a recently described system to visualize and track translation from individual HIV-1 RNA molecules in living cells. We found that, on average, half of the cytoplasmic HIV-1 RNAs are being actively translated at a given time. Furthermore, translating and nontranslating RNAs are well mixed in the cytoplasm; thus, Gag biogenesis occurs throughout the cytoplasm without being constrained to particular subcellular locations. Gag is an RNA binding protein that selects and packages HIV-1 RNA during virus assembly. A long-standing question in HIV-1 gene expression is whether Gag modulates HIV-1 RNA translation. We observed that despite its RNA-binding ability, Gag expression does not alter the proportion of translating HIV-1 RNA. Using single-molecule tracking, we found that both translating and nontranslating RNAs exhibit dynamic cytoplasmic movement and can reach the plasma membrane, the major HIV-1 assembly site. However, Gag selectively packages nontranslating RNA into the assembly complex. These studies illustrate that although HIV-1 RNA serves two functions, as a translation template and as a viral genome, individual RNA molecules carry out only one function at a time. These studies shed light on previously unknown aspects of HIV-1 gene expression and regulation.
During HIV-1 infection, viral DNA integrates into the cellular chromosome to form a provirus and becomes a permanent part of the host genome. Host cell RNA polymerase II transcribes the provirus to generate RNA transcripts, which are further modified and processed by the cellular machinery (1). Although driven by a single promoter, multiple HIV-1 RNAs are generated using complex splicing patterns, including full-length RNA, partially spliced RNAs, and fully spliced RNAs (2). Partially and fully spliced RNAs encode various HIV-1 proteins important for virus propagation in the host. The full-length RNA serves at least two roles: It is packaged into nascent particles as the viral genome and it is used as the translation template for the synthesis of Gag/Gag-Pol polyproteins (1, 3). For simplicity, HIV-1 full-length RNA is referred to hereafter as HIV-1 RNA.
Gag and Gag-Pol are translated as polyproteins and processed into mature structural proteins and viral enzymes by HIV-1 protease during or soon after virus assembly (4, 5). Although HIV-1 RNA translation and the regulation of Gag biogenesis are critical steps in viral replication, many aspects of these steps are poorly understood. For example, it is not known whether HIV-1 Gag and Gag-Pol are synthesized in distinct subcellular compartments. It was suggested that the initial loading of the cellular complex that enables translation occurs in specialized RNA granules (6). This hypothesis implies that translating RNA and nontranslating RNA are located in different subcellular compartments. HIV-1 RNA translation may have an additional layer of complexity, as the translational product, Gag, is an RNA-binding protein (7, 8) and Gag–RNA interaction leads to specific packaging of the viral RNA genome (3, 9, 10). Although it has long been speculated that Gag affects RNA translation, and it has been suggested that Gag autoregulates its own translation by binding to HIV-1 RNA (11), the mechanisms by which HIV-1 regulates the functions of its RNA are currently unknown. Furthermore, the relationship between translation and packaging is not defined. It has been hypothesized that translation and packaging are distinct processes and each function is served by a separate RNA pool; alternatively, these two functions are interconnected with Gag specifically packaging the RNA from which it was translated (12, 13). To address these questions, we sought to gain a better understanding of HIV-1 RNA translation and its relationship to RNA packaging.
Live-cell imaging studies have shed light on many aspects of HIV-1 RNA biology. Single-molecule tracking methods were used to show that HIV-1 RNA travels in the cytoplasm mainly by diffusion (14). Total internal reflection fluorescence (TIRF) microscopy was employed to define the behavior of HIV-1 RNA near the plasma membrane, the major site of HIV-1 assembly (15–17). It was shown that HIV-1 RNA can reach the plasma membrane in the absence of Gag, although its residence is transient; the presence of Gag extends the residence time of HIV-1 RNA near the plasma membrane (15, 16). Finally, HIV-1 RNA dimerizes at the plasma membrane and the dimer is stabilized by Gag, allowing the formation of an assembly complex (17). These studies provide insights into aspects of HIV-1 RNA packaging that are difficult to resolve using biochemical assays. However, to our knowledge, live-cell imaging of HIV-1 RNA translation is still lacking, partly because it is difficult to image protein with single-molecule sensitivity. Recent developments have allowed RNA translation in other experimental systems to be studied using imaging approaches (18–21). For example, the translation of a reporter gene has been visualized (18) using the SunTag system (22). The SunTag sequence encodes multiple copies of the GCN4 epitope; each can bind a single-chain antibody tagged with a fluorescent protein. Thus, a SunTag polypeptide can be labeled by multiple fluorescent proteins to allow the detection of a single protein molecule (22). In this recently described system (18), reporter gene RNA contained stem-loop sequences to allow labeling by a tagged RNA-binding protein, whereas a nascently translated polypeptide contained SunTag, which was detected by a single-chain variable fragment of GCN4 antibody fused with a superfolder green fluorescent protein (scFV-sfGFP). When visualized by live-cell imaging, translating RNA was detected as RNA signals colocalizing with GFP signals (18).
In this report, we adapted the aforementioned system to study HIV-1 RNA translation (18). We detected Gag by using the SunTag inserted into the C terminus of the matrix (MA) domain and detected HIV-1 RNA by using tagged bacteriophage MS2 coat protein that specifically binds to RNA sequences engineered into the pol gene of the viral genome. Using these strategies, we were able to visualize HIV-1 RNA translation and Gag biogenesis by live-cell fluorescence microscopy, which provide snapshots of HIV-1 RNA behaviors. We measured the proportion of HIV-1 RNA being actively translated, delineated the subcellular locations of the translation process, defined the mobility of translating and nontranslating RNA, examined the role of Gag in regulating translation, and determined the translation status of RNA being assembled into viral particles. These studies provide insights into Gag biogenesis and how HIV-1 RNA carries out the dual functions of serving as translation template and viral genome.
Results
System Used to Visualize HIV-1 RNA Translation.
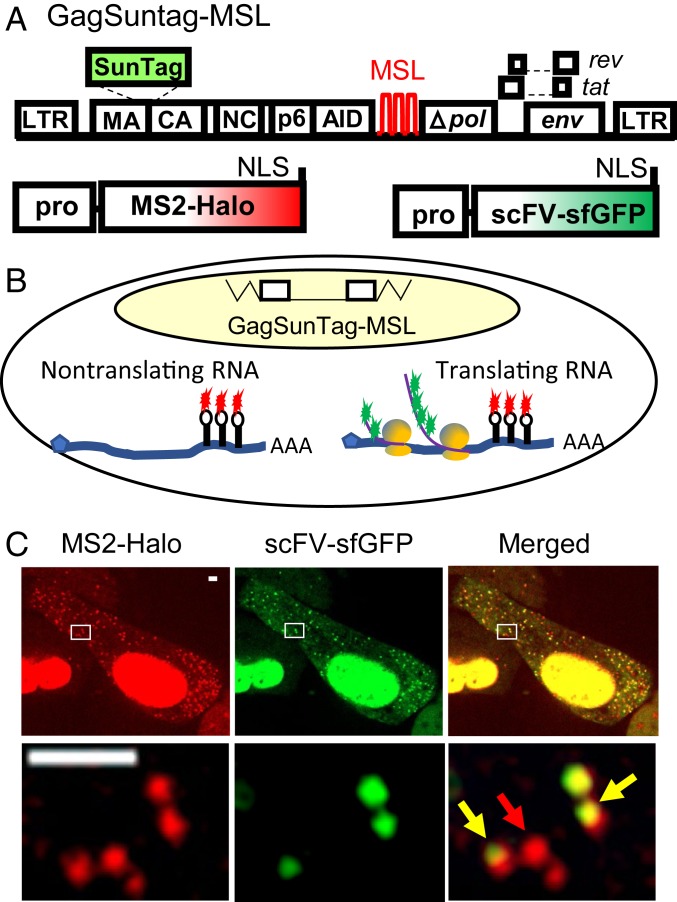
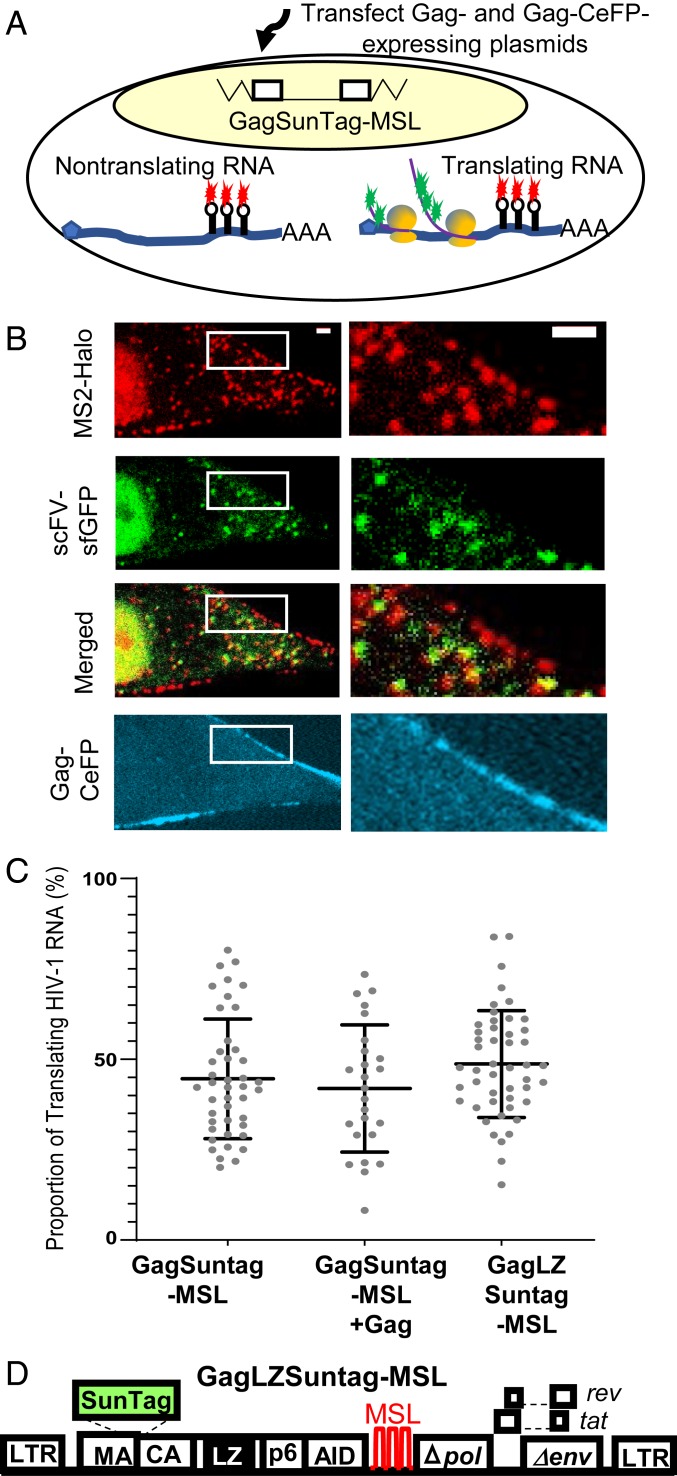
To visualize the biogenesis of Gag, we generated an NL4-3–based HIV-1 construct, GagSunTag-MSL (Fig. 1A), which contains all of the cis-acting elements critical for viral replication, expresses Tat and Rev, and has inactivating deletions in pol, vif, vpr, and vpu. The Gag-coding region of GagSunTag-MSL was modified to contain two in-frame insertions. The SunTag sequence, flanked by viral protease cleavage sites, was inserted into the C terminus of the MA to allow visualization of the polyprotein, and the auxin-inducible degron (AID) domain (23) was inserted into the C terminus of p6 to regulate the stability of the fusion polyprotein. Additionally, 24 copies of stem-loop sequences recognized by the bacteriophage MS2 coat protein (MSL) were inserted into the pol gene to allow the detection of HIV-1 unspliced RNA. Although not shown in Fig. 1A, a mouse heat-stable antigen driven by the internal ribosomal entry site from encephalomyocarditis virus (IRES-HSA) was inserted into the nef gene (24) to allow the enrichment of cells expressing the provirus by cell sorting.
Fig. 1.
System used to visualize the translation of full-length HIV-1 RNA. (A) General structures of constructs used to detect HIV-1 RNA translation. GagSunTag-MSL contains two in-frame insertions in gag, a SunTag sequence and an AID sequence. Stem-loop sequences recognized by MSL were inserted into pol, and a mouse HAS gene is inserted into the nef gene (not illustrated). An NLS is located at the C termini of MS2-Halo and scFV-sfGFP protein. A vector containing an OsTIR1 gene was also expressed but is not illustrated. (B) U2OS cells containing a GagSunTag-MSL provirus were used for imaging studies. Both translating and nontranslating RNAs were labeled by MS2-Halo stained with JF549 dye (red). The nascent polypeptides associated with the translating RNA are detected by scFV-sfGFP (green). Thus, nontranslating RNA exhibits a red signal and translating RNA generats a red-green dual-colored signal. (C) Representative images of a U2OS cell in which both translating and nontranslating RNAs were detected in the cytoplasm. Red arrow, nontranslating RNA; yellow arrow, translating RNA. [Scale bar, 2 μm (both upper panel and enlargement).]
To study translation of HIV-1 RNA, we generated a large pool of cells containing a GagSunTag-MSL provirus. For this purpose, we infected a human osteosarcoma cell line (U2OS) with viruses containing the GagSunTag-MSL genome at low multiplicity of infection (MOI < 0.1) to ensure that most of the infected cells contained only a single provirus. A large pool of cells containing >200,000 independent infection events were enriched by cell sorting based on the expression of the HSA marker. The U2OS cell line (18) was engineered to express three proteins that facilitated our study: An MS2 coat protein fused to a Halo tag (MS2-Halo) (25); an scFV-sfGFP protein that binds to epitopes in SunTag (22, 26); and a plant protein, thermally stable Oryza sativa transport inhibitor response 1 (OsTIR1) (23, 27). In the presence of the auxin indole-3-acetic acid (IAA), OsTIR1 rapidly degrades proteins containing the AID domain; thus, IAA treatment depletes cytoplasmic GagSunTag released from the translational machinery, thereby reducing cytoplasmic GFP background, and facilitating the visualization of partially translated GagSunTag associated with HIV-1 RNA. Both MS2-Halo and scFV-sfGFP contain a nuclear localization signal (NLS) at the C terminus (Fig. 1A). Because MSL is located in the pol gene, only unspliced, full-length RNAs are labeled using MS2-Halo; in our studies, we used Halo-JF549 dyes to label MS2-Halo for RNA detection. The scFV-sfGFP protein binds to SunTag embedded in Gag to allow the detection of translation product. Therefore, in this system, nontranslating RNA is labeled with Halo-JF549, which can be detected in the red channel, whereas translating RNA has both Halo and GFP labels and is red/green dual-colored (Fig. 1B).
Detection of Translating RNA.
To visualize HIV-1 RNA translation, we performed live-cell imaging on cells pretreated with IAA to degrade GagSunTag released from the translational machinery; cells were also stained with Halo-JF549 dyes and washed multiple times to remove unbound dye molecules. The HIV-1 RNA signal in the red channel (MS2-Halo-JF549) and translation signals in the green channel (scFV-sfGFP) were captured simultaneously using two precisely aligned cameras in a spinning-disk microscope imaging system. We observed bright green signals that colocalized and comigrated with red RNA signals in the cytoplasm (Fig. 1C and Movie S1); these dual-colored signals likely represent translating RNA. We also observed molecules with only red signals that likely represent nontranslating RNA. We observed very few green puncta without red signals, likely due to IAA treatment inducing the degradation of the GagSunTag released from the translational machinery.
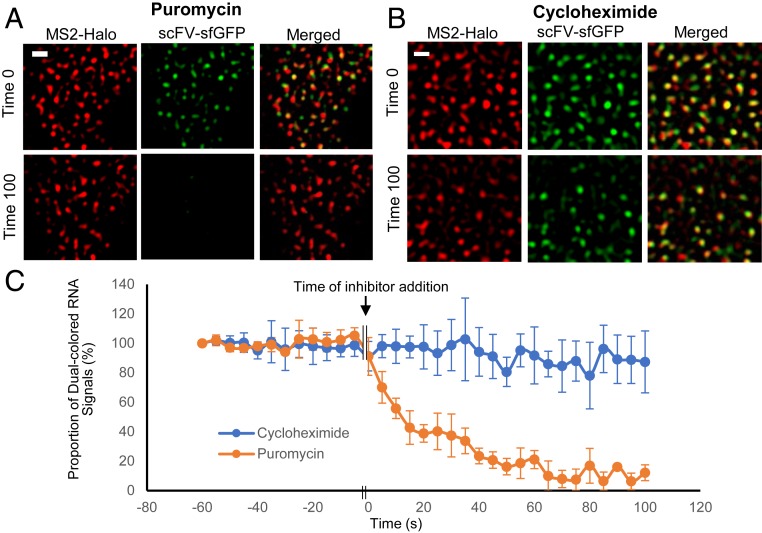
To validate whether the red-green dual-colored signals detected were indeed translating RNA, we examined the effects of translation inhibitors on RNA signals by using puromycin or cycloheximide. These two translation inhibitors have distinct mechanisms of action that lead to different predicted effects on translating RNA upon treatment. Puromycin binds to the ribosome A site and causes premature termination of translation. Thus, it is expected that puromycin treatment will cause nascent Gag polypeptides to dissociate from the RNA, resulting in rapid loss of the number of red/green dual-colored signals. Indeed, we observed rapid disappearance of green signals upon puromycin treatment (Fig. 2A and Movie S2). The proportion of dual-colored RNA signals before and after the addition of puromycin summarized from three cells is shown in Fig. 2C (orange line). In these experiments, images were captured for multiple frames, then paused to allow the addition of inhibitor (defined as time 0) before imaging was resumed. For simplicity, in each cell the proportion of dual-colored signals at the beginning of the observation period (60 s before time 0, or time point −60) was set as 100, and data detected from various time points were standardized to time point −60. As shown in Fig. 2C, the proportion of dual-colored signals remained constant for 60 s before the addition of puromycin. Within seconds of adding puromycin, the proportion of dual-colored signals decreased rapidly and remained very low at the end of the observation time (100 s after adding puromycin). We also examined the effects of adding cycloheximide to the detection of dual-colored signals (Fig. 2 B and C and Movie S3). Cyclohexamide stalls the ribosome on mRNA by binding to the ribosome E site, preventing the translocation step of protein synthesis and thereby interfering with polypeptide elongation. Therefore, it is expected that dual-colored RNA will not decrease drastically upon cycloheximide treatment. The proportion of dual-colored RNA before and after cycloheximide addition from three cells are summarized in Fig. 2C (blue line). In contrast to the results from puromycin treatment, adding cycloheximide did not significantly alter the proportion of dual-colored signals within the observation time. Taken together, these results confirmed that dual-colored signals were indeed translating RNA and that green signals colocalized with red signals were scFV-sfGFP bound to the nascent polypeptides associated with the translational machinery.
Fig. 2.
Validation of translating RNA detection. Images of cells were captured every 5 s for 13 frames, paused to allow the addition of puromycin (A) or cycloheximide (B), and resumed at the same frame rate. Representative images immediately before the pause (time 0) and 100 s after the pause are shown for puromycin (A) or cycloheximide (B) treatments; a Laplacian of Gaussian filter was applied by using ImageJ. (Scale bars, 2 μm.) (C) The proportion of red/green dual-colored signals before and after inhibitor treatment. The proportion of dual-colored signals at −60 s was defined as 100% and used to normalize data from other time points. Results from puromycin or cycloheximide treatment are shown by the orange and blue lines, respectively; the average and SD of three cells are shown.
Determining the Proportion of Translating RNA.
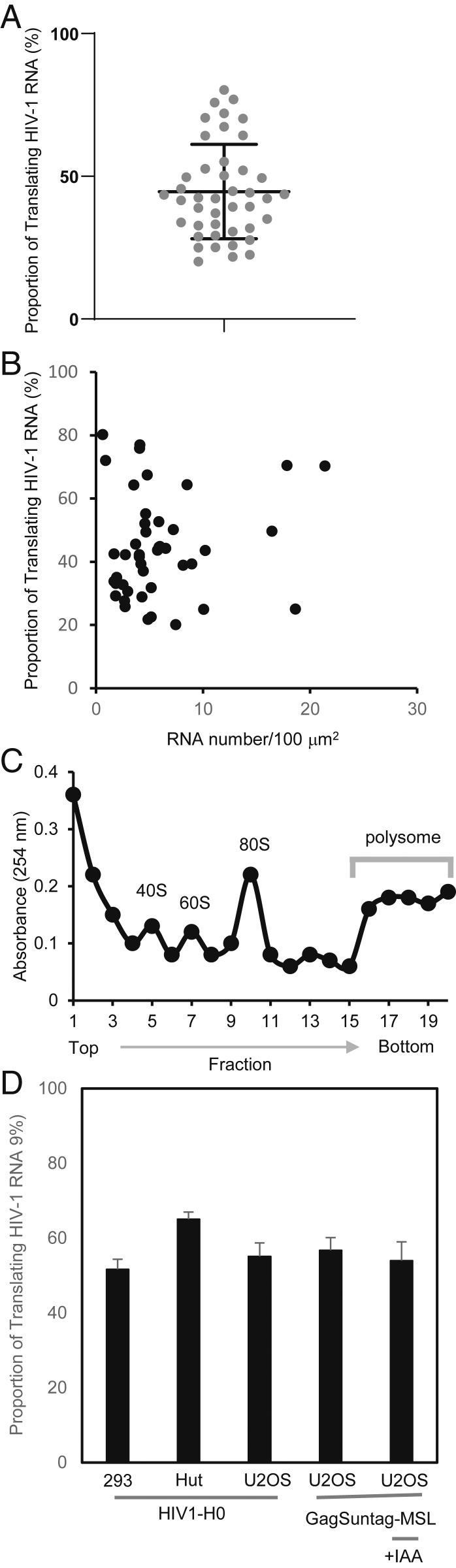
To better understand Gag biogenesis, we quantified the proportion of HIV-1 RNA that was actively translated in individual cells. The proportion of actively translating RNA was calculated as the number of dual-colored signals (translating RNA) divided by the number of total red signals (total HIV-1 RNA). We examined 43 cells and found that, on average, ∼45% of HIV-1 RNA in the cytoplasm was being actively translated during observation time; however, a large variation existed among individual cells (Fig. 3A). To shed light on the possible reason for such a large variation, we examined whether there was a correlation between HIV-1 RNA expression level and translation efficiency. For this purpose, we compared the proportion of translating RNA with the amount of HIV-1 RNA in the cytoplasm. By detecting red puncta in the cytoplasm, we determined the number of cytoplasmic HIV-1 RNA in each cell; we then calculated the number of HIV-1 RNA in 100 μm2 of cytoplasmic area to account for the variation in cell sizes. To visualize the results from individual cells, we plotted the RNA number in the x axis and proportion of translating RNA in the y axis; results from 43 cells are shown in Fig. 3B. However, we found little correlation between the two (R2 = 0.0134), indicating that the HIV-1 RNA expression level had no obvious effect on RNA translation efficiency.
Fig. 3.
Proportion of translating RNA determined using imaging and biochemical methods. (A) The proportion of translating RNA in 43 cells as determined by imaging. Each dot represents data from an individual cell. Mean and SD are shown. (B) The relationship between the amount of cytoplasmic RNA (x axis) and the proportion of translating RNA (y axis). (C) Representative ribosome profile from a fractionation experiment. (D) The proportion of translating RNA was determined using human 293T cells and human U2OS cells containing H0 provirus. Additionally, the proportion of translating RNA was determined in human U2OS cells containing GagSunTag-MSL provirus in the presence or absence of IAA. Averages and SDs from three independent fractionation experiments are shown.
To further examine the proportion of translating RNA, and to validate the results from our imaging studies, we measured the proportion of HIV-1 RNA in polysomes by using a standard biochemical approach (28). For this purpose, we generated provirus-containing cell pools by infecting cells with a previously described near full-length NL4-3–derived virus, ON-H0 (29), at an MOI of ∼1. ON-H0 contains all of the cis-acting elements important for viral replication and expresses functional Gag/Gag-Pol, Tat, and Rev; additionally, ON-H0 expresses an HSA marker gene in nef, which is used to determine the proportion of infected cells. Briefly, provirus-containing cells were lysed and clarified, cytoplasmic lysate was loaded on a sucrose gradient, and 20 fractions were collected after centrifugation. For each fraction, 254-nm absorbance was measured and representative results are shown in Fig. 3C. Additionally, RNA was isolated from each fraction, and quantitative RT-PCR detecting a region in gag was performed to determine the amount of HIV-1 RNA. Using the RNA measurements, we determined the proportion of HIV-1 RNA that were in the polysome fractions.
We examined several cell lines containing ON-H0 provirus (the results are summarized in Fig. 3D). First, we examined provirus-containing human 293T cells and found that ∼48% of the HIV-1 RNA was in polysome fractions. We also examined the proportion of translating ON-H0 RNA in human osteosarcoma U2OS cells and found that the proportion of HIV-1 RNA in polysome fractions was ∼47%. For direct comparison with our imaging studies, we examined cells used for the imaging assay (U2OS cells containing GagSunTag-MSL provirus) under two conditions, either with or without IAA treatment. We found that in both conditions, ∼46% of the RNA was in polysome fractions. Thus, the proportion of translating RNA measured by ribosomal fractionation was similar to that by the imaging assay. Furthermore, IAA treatment did not significantly alter the proportion of translating RNA.
The Mobility of Translating RNA.
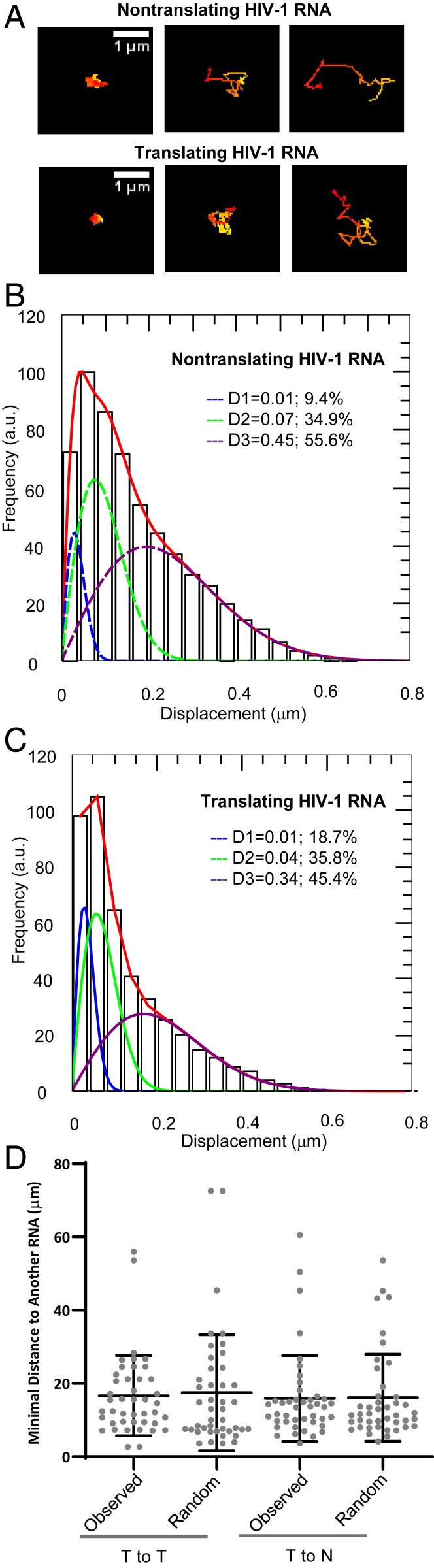
We previously showed that HIV-1 RNA mainly traveled in the cytoplasm by diffusion with heterogenous mobility (14). In the present study, a visual inspection of the entire cell (Movie S1) revealed similar features: Most of the RNA signals exhibited dynamic motion and moved rapidly in a nondirectional, random-walk manner, whereas a small portion of the RNA signals exhibited little movement during the observation time. Furthermore, similar behaviors were observed regardless of whether the RNAs had red/green dual-colored signals (translating RNA) or only red signals (nontranslating RNA); three representative RNA tracks from nontranslating and translating RNAs are shown in Fig. 4A. To determine the effect of translation on HIV-1 RNA mobility, we followed the movement of 550 tracks of translating RNA and 958 tracks of nontranslating RNA from five cells. We used persistent index analysis to quantify the directionality of the RNA movement as described previously (14), and found that only 1 in 550 translating RNA tracks and 1 in 958 nontranslating RNA tracks displayed directional movement. We then performed mean square displacement (MSD) analysis to quantify the overall mobility of these RNAs. Our analyses showed that the diffusion coefficient for translating RNA is 0.11 μm2/s, whereas the diffusion coefficient for nontranslating RNA is 0.16 μm2/s. Thus, these results show dynamic cytoplasmic movement of both translating and nontranslating RNAs; on average, translating RNA diffused slightly slower than nontranslating RNA.
Fig. 4.
Determining the properties of translating and nontranslating RNAs. (A) Representative trajectories of nontranslating and translating RNA tracks; trajectories are depicted with changing colors from start (red) to end (yellow). Distributions of the one-step jump distance of nontranslating (B) and translating RNA (C). To plot the jump distance distribution, data were binned (40-nm bin size) and normalized to the bin that contained the most events, which was set to 100; x axis, one-step jump distance (displacement); y axis, frequency in arbitrary units (a.u.). The distribution was fitted with a three-component model as previously described (14). The solid red line represents the fitted curve and the three dotted lines represent the distribution of each mobility fractions. (D) The spatial relationship between translating and nontranslating RNAs. The distances of individual translating RNA molecules to the nearest translating RNA (T to T) or to the nearest nontranslating RNA (T to N) in a representative cell are shown. To generate distances expected from a cell in which translating and nontranslating RNAs were mixed randomly, we used the spatial information of the RNAs, based on the number of translating RNA molecules in the cell, randomly assigned a subset of RNAs as translating RNAs, and measured the minimal distance of translating RNA to translating or nontranslating RNA; these values are shown as “Random.”
The ensembled MSD analyses demonstrate the average behavior of the nontranslating and translating RNAs. To further analyze whether there were more than one mobility fraction within the viral RNA populations, we calculated the jump distance traveled by each RNA track from one frame to the next within 42 ms. A total of 11,943 one-step jump distances were obtained from 958 nontranslating RNA tracks and a total of 10,041 one-step jump distance were obtained from 550 translating RNA tracks. The distributions of these observed distances are summarized in Fig. 4B (nontranslating RNA) and Fig. 4C (translating RNA); the one-step jump distance (displacement) is shown in the x axis and the frequency is shown in the y axis, with the frequency of distance that was most often traveled defined as 100. Similar to our previous analyses of HIV-1 RNA moblity (14), we found that the distributions of the one-step jump distances for both nontranslating and translating RNAs were too heterogeneous to fit into a single diffusion coefficient, but each could successfully fit into three mobility fractions: Stagnant, intermediate, and fast (Fig. 4 B and C). The stagnant fraction (D1) is defined by the localization precision of our system, 40 nm, which yields a diffusion coefficient of 0.01 μm2/s. These stagnant signals represent jump distances that were either static or moved less than the threshold defined by the localization precision. The stagnant fractions corresponded to 9.4% and 18.7% of the distances generated by the translating and nontranslating RNA (blue lines in Fig. 4 B and C). For the nontranslating RNA, 34.9% and 55.6% of the observed distances corresponded to intermediate and fast fractions (green and purple lines in Fig. 4B, respectively) with diffusion coefficients of 0.07 μm2/s and 0.45 μm2/s, respectively. For the translating RNA, 35.8% and 45.4% of the observed distances were in the intermediate and fast fractions (green and purple lines in Fig. 4C, respectively) with the diffusion coefficient of 0.04 μm2/s and 0.34 μm2/s, respectively. These results are consistent with the ensemble MSD analyses, indicating that the translating HIV-1 RNA on average diffused slower than nontranslating RNAs. Furthermore, both nontranslating and translating HIV-1 RNAs exhibited heterogenous movements, which is similar to those observed using nonviral reporter RNAs (21).
Cytoplasmic Location of Translating RNA.
It was proposed that the translation initiation complex of HIV-1 RNA is assembled in compartmentalized cytoplasmic foci (6), raising the possibility that translating and nontranslating RNAs may occupy distinct subcellular locations. However, visual inspection of the cells suggested that translating and nontranslating RNAs were distributed throughout the cytoplasm in a well-mixed manner, which does not support the hypothesis that Gag is synthesized in a specific subcellular compartment (Figs. 1C and 2A and Movies S1–S3). To determine whether translating and nontranslating RNAs occupy different cytoplasmic locations, we analyzed the relative distribution of these two types of RNAs. We reasoned that, if translating RNA molecules are clustered and located in specific compartments in a cell, then the distance between a translating RNA to the nearest translating RNA should be shorter than that expected from a cell in which translating and nontranslating RNAs are well mixed. For this purpose, we measured the distances between individual translating RNA molecules to the nearest translating or nontranslating RNA in the cells and compared these distances to those expected from these two types of RNAs mixed randomly in the cell (SI Appendix, Fig. S1A). Our results show that the distances between translating RNA molecules to the nearest translating RNA (Fig. 4D) (observed T to T) are not different from those calculated based on random mixing of translating and nontranslating RNAs in a cell using simulation (Fig. 4D) (random T to T; P = 0.78, Welch’s t test). Similarly, measured distances between individual translating RNA molecules to the nearest nontranslating RNA (Fig. 4D) (observed T to N) are not different from those calculated based on random mixing of these two types of RNAs in a cell (Fig. 4D) (random T to N; P = 0.95, Welch’s t test). Additional data from analyses of four other cells are shown in SI Appendix, Fig. S1B. Our results do not support the hypothesis that Gag biogenesis occurs in specific compartments of the cytoplasm separated from nontranslating RNA.
The Effects of Gag Expression on HIV-1 RNA Translation.
Since Gag is an RNA-binding protein and interacts with HIV-1 RNA in the cytoplasm (7, 30, 31), it is possible that Gag can affect HIV-1 RNA translation. Furthermore, it has been suggested that HIV-1 Gag regulates its own translation by competing with the translational machinery for HIV-1 RNA binding (11). To better understand the regulation of Gag biogenesis, we sought to determine the effects of Gag expression on HIV-1 RNA translation. In the imaging experiments described above, we decreased the background signal by using IAA to degrade GagSunTag released from translating RNA. Hence, the level of Gag was reduced in these studies, making it difficult to assess the impact of Gag expression on HIV-1 RNA translation. To study whether HIV-1 Gag–RNA interaction affected RNA translation, we introduced additional Gag proteins that were not sensitive to IAA-induced degradation into GagSunTag-MSL–expressing U2OS cells (Fig. 5A). For this purpose, we transfected into cells two Gag-expressing plasmids at an equal weight ratio: One plasmid expressed untagged Gag and the other plasmid expressed a Gag tagged with cerulean fluorescent protein (CeFP) (Fig. 5A). The expression of equal amounts of Gag and Gag-CeFP allows the production of fluorescent protein-containing HIV-1 particles with normal morphology indistinguishable from immature wild-type particles that can efficiently package the viral RNA genome (32, 33). We then selected cells with sufficient Gag/Gag-CeFP expression to support virus assembly, which is defined by accumulation of Gag-CeFP puncta at the plasma membrane (Fig. 5B; Gag-CeFP panels). In the cytoplasm of these cells, we observed both translating and nontranslating RNA signals (Movie S4), most of which moved in a nondirectional, random-walk manner. We quantified the proportion of translating RNA in 25 different cells as described above and results are summarized in Fig. 5C; for comparison, results from untransfected cells (shown in Fig. 3A) are shown on the left. The proportion of translating RNA in Gag-expressing cells was similar to that in cells without additional Gag/Gag-CeFP expression (Fig. 5C), indicating that expression of Gag/Gag-CeFP in the cytoplasm did not affect RNA translation (P = 0.53, Welch’s t test).
Fig. 5.
The effects of Gag expression on HIV-1 RNA translation. (A) Schematic of the experimental approach. Gag- and Gag-CeFP–expressing plasmids were transfected into GagSunTag-MSL-provirus containing U2OS cells before imaging. (B) Representative images of RNA, translating polypeptide, and Gag-CeFP signals in a cell. Insets shown on the left images are enlarged and shown on the right. (Scale bars, 2 μm.) (C) Quantitation of the proportion of HIV-1 translating RNAs. (D) General structure of the GagLZ-SunTag-MSL construct. The NC domain of Gag was replaced with an LZ motif.
Although unlikely, it is possible that the presence of Gag, even at a very reduced level upon IAA treatment, affects HIV-1 RNA translation. To address this possibility, we examined the effects of abolishing the RNA-binding activity of the GagSunTag polyprotein. It has been shown that when the nucleocapsid (NC) domain of HIV-1 Gag is replaced by a leucine zipper motif (LZ), the GagLZ polyprotein can readily assemble into particles (34); however, the assembled particles do not contain detectable amounts of RNA (35). To further examine the effects of Gag–RNA interactions on the proportion of translating RNA, we replaced the NC domain of GagSunTag with LZ to generate GagLZSunTag-MSL (Fig. 5D). We infected U2OS cells with viruses containing the GagLZSunTag-MSL genome, generated a large pool (>100,000) of independently infected cells, enriched for infected cells by cell sorting, and performed live-cell imaging on these cells. We observed both translating and nontranslating RNA signals (Movie S5); we analyzed 48 cells and found that the proportion of translating RNA was similar to that from RNA derived from the GagSunTag-MSL provirus (Fig. 5C) (P = 0.22, Welch’s t test). Taken together, these studies suggest that the expression of Gag, and the potential interaction of Gag with HIV-1 RNA, did not significantly alter the proportion of translating RNA.
Gag Recruits Nontranslating RNA into Assembly Complex as the Genome for Nascent Particles.
The relationship between HIV-1 translation and packaging has been a long-standing question and it is not known whether the translation status of HIV-1 RNA affects its ability to be packaged. During our studies using cells expressing Gag/Gag-CeFP, we observed that most of the RNA colocalizing with the Gag-CeFP near the plasma membrane contained red signals but not green signals, indicating that most of the RNA molecules colocalizing with Gag were nontranslating RNA (Fig. 5B). These results suggest that nontranslating RNA was recruited by Gag. However, it is also possible that translating RNA was recruited to the plasma membrane; during the assembly process, the translational machinery was stripped off the RNA, resulting in the red signal phenotype.
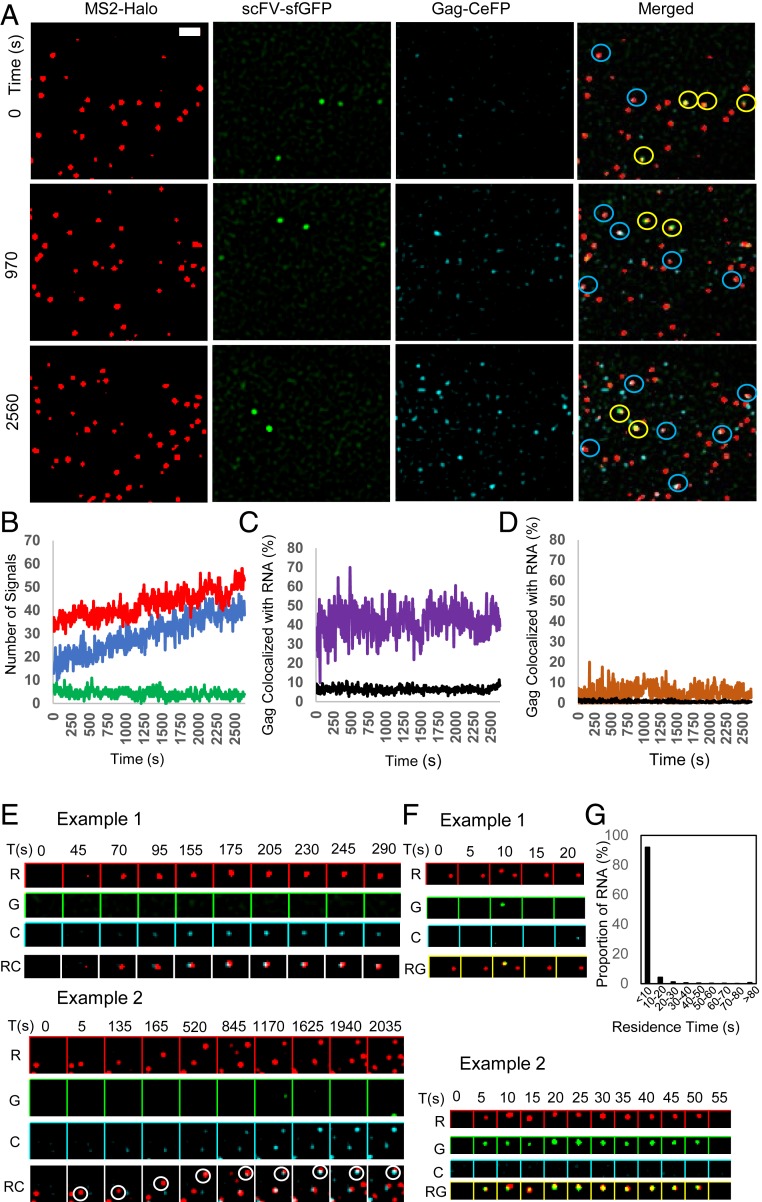
To address this question, we followed the process of HIV-1 particle assembly by using TIRF microscopy, which captures events occurring at or near the plasma membrane. For this purpose, we transfected Gag- and Gag-CeFP–expressing plasmids into cells containing GagSunTag-MSL provirus and examined cells containing some CeFP puncta at the plasma membrane at the beginning of the observation time. We captured the images of red RNA signal, green translating GagSunTag signal, and blue Gag-CeFP signal once every 5 s for 2 to 3 h to follow virus assembly. In these experiments, cells were treated with IAA to degrade the GagSuntag polyprotein released from the translational machinery to prevent GagSunTag from incorporating into the assembly complex and complicating the analyses.
A set of representative images is shown in Fig. 6A and Movie S6. At the beginning of the observation time (time 0), there were several Gag-CeFP puncta and a few more nontranslating RNA signals at the plasma membrane. This result is consistent with the observation from previous studies by us and others that, during HIV-1 assembly, HIV-1 RNA signals were visible before the detection of Gag puncta due to the limited sensitivity of Gag detection (15, 17). As time progressed, we observed a gradual increase of the numbers of nontranslating RNA and Gag-CeFP puncta (Fig. 6B, red and blue lines, respectively). We also observed frequent colocalization of Gag-CeFP puncta with nontranslating RNA signals (Fig. 6A, blue circles and Fig. 6C, purple line) at a rate much higher than expected from random colocalization (Fig. 6C, black line). However, translating RNA signals behaved quite differently from nontranslating RNA signals. We observed a few translating RNA signals at the beginning of and throughout the observation time (Fig. 6A, yellow circles); however, the number of translating RNA signals did not change over time (Fig. 6B, green line). In addition, translating RNA did not colocalize with Gag-CeFP puncta at a frequency higher than expected from random colocalization (Fig. 6D, orange and black lines, respectively). These results suggest that Gag was mainly associated with nontranslating RNA at the plasma membrane.
Fig. 6.
Determining the translation status of HIV-1 RNA assembling with Gag at the plasma membrane by TIRF microscopy. (A) Representative images of RNA (MS2-Halo, red), translating polypeptide (scFV-sfGFP, green), and Gag-CeFP (cyan) signals at the plasma membrane at various times during observation. Blue circles, colocalized red and cyan signals; yellow circles, colocalized red and green signals. A Laplacian of Gaussian filter was applied using ImageJ. (Scale bar, 2 μm.) (B) Proportion of the HIV-1 RNA signals and Gag-CeFP puncta detected near the plasma membrane through observation time. Red, green, and blue lines represent the number of nontranslating RNA molecules with only a red signal, translating RNA with dual-colored signal, and Gag-CeFP puncta, respectively. Proportions of Gag-CeFP puncta colocalized with nontranslating RNA (C) and translating RNA (D) are shown as purple and orange lines, respectively. Black lines represent the proportion of Gag-CeFP expected to colocalize with each type of RNA by random distribution. (E) Representative images of nontranslating RNA (red signal) interacting and colocalizing with Gag (CeFP signal). Images obtained in the red, green, and cyan channels are marked as R, G, and C, respectively. RC, merged images of both the red and cyan channels. Images were captured every 5 s; time (in seconds) of image capture during observation is shown on top of the panels as T(s). (F) Representative images of the arrival and departure of translating RNA near the plasma membrane. RG, merged images of both the red and green channels. Image magnification in E and F is same as in A. (G) Residence time of translating RNA near the plasma membrane. The plasma membrane residence time of 3,263 green signals was separated into multiple bins and displayed. y axis, proportion of RNA (%); x axis, residence time.
To better assess the translation status of the RNA at the beginning of the assembly process, we identified 12 assembly events that resulted in detectable Gag-CeFP:RNA complexes and studied the initial appearance of RNA signals during the assembly process. Two examples are shown in Fig. 6E and Movies S7 and S8. In example 1, we detected the RNA as a red signal at the 45-s time point; a faint Gag-CeFP signal started to appear at the 70-s time pointand became readily detectable after the 155-s time point. In the second example, we detected the RNA as a red signal at the 5-s time point, the RNA signal then migrated toward the upper right-hand corner over time; a faint Gag-CeFP signal first appeared at the 520-s time point and became readily detectable after the 1,170-s time point. Two additional examples are shown in SI Appendix, Fig. S2 and Movies S9 and S10. In all of the 12 examples analyzed, the RNA signals that were initially detected had only red signals and remained as only red signals during the entire observation time.
To better understand the behavior of the translating RNA detected near the plasma membrane, we determined the residence time of 3,263 green signals. We found that most of the dual-colored signals only appeared near the plasma membrane very briefly and disappeared from the field of view within 5 to 10 s (one to two frames) of the imaging time (Fig. 6F, example 1, and Fig. 6G). Occasionally, some translating RNA signals were observed for a longer period of time at the plasma membrane (Fig. 6F, example 2); this translating RNA stayed near the plasma membrane for 50 s before disappearing from the field of view. We then followed 50 individual tracks of translating RNA signals; none of the 50 tracks converted from dual-colored to red-only signal and none was shown to associate with CeFP signals in consecutive frames. Taken together, these results strongly suggest that Gag binds to nontranslating RNA and recruits this RNA into assembly complexes.
Discussion
To generate infectious viruses, HIV-1 RNA must be translated to produce structural proteins and enzymes that form the particle; additionally, HIV-1 RNA needs to be packaged into particles to serve as the viral genome. In this report, we used imaging approaches to visualize HIV-1 RNA and proteins, combined with biochemical experiments, to study HIV-1 translation and its regulation. We showed that translating RNA occupied half of the HIV-1 RNA population and Gag expression did not affect the proportion of translating HIV-1 RNA. Furthermore, translating, and nontranslating RNAs were well mixed in the cytoplasm, and both moved dynamically and could reach the plasma membrane. However, Gag formed assembly complexes with nontranslating RNA. These studies provide insights into Gag biogenesis and how HIV-1 RNA carries out its dual functions as translation template and virion genome.
Our studies showed that, on average, half of the HIV-1 RNAs were being actively translated; however, live-cell imaging results revealed that the proportion of translating RNA varied greatly among individual cells. Furthermore, the proportion of translating RNA was not correlated with the number of cytoplasmic HIV-1 RNA molecules and was not affected by expression of Gag that was capable of binding viral RNA. Such large cell-to-cell variation in translating RNA was also observed in the translation of nonviral reporter RNA (18); thus, we speculate that the cellular state, rather than viral factors, is the main cause for such variations. Our observation that translating and nontranslating RNAs were well mixed in the cytoplasm indicate that Gag and Gag-Pol polyproteins were synthesized throughout the cytoplasm and synthesis did not occur in specific subcellular compartments. How these proteins traffic to the plasma membrane remains an intriguing and unresolved question.
We found that both translating and nontranslating RNAs exhibited heterogenous movements in the cytoplasm; most of the RNAs were highly mobile, although translating RNA moved slightly slower than nontranslating RNA in the cytoplasm. Similar conclusions were reached using nonviral reporter RNAs (18, 21). Although dynamic movement of both translating and nontranslating reporter RNA were observed, translating RNAs exhibited slightly less mobility (18). Heterogeneity in translating RNA movements were also reported with nonviral reporter RNAs, in which RNA tracks with stationary, subdiffusive, and diffusive movements were observed. Furthermore, it was suggested that the movement of translating RNA is more likely to be impeded when the RNA is located at or near subcellular structures (21). We have shown previously that individual HIV-1 RNAs did not remain in a given mobility fraction; instead, the mobility of individual RNA tracks varied as they traveled through the cytoplasm (14). We hypothesize that, in a manner similar to the nonviral reporter RNA study, the local subcellular environment affects HIV-1 RNA mobility and contributes to our observed heterogenous movement of translating and nontranslating RNA.
Our current results are in agreement with our previous studies showing the dynamic movement of cytoplasmic HIV-1 RNA (14). Translating RNA is associated with translational machinery, including ribosomes and nascent polypeptides; thus, it is logical to reason that, compared with nontranslating RNA, translating RNA could be in a larger complex and exhibit a lower mobility. However, the observed differences in mobilities between these two types of RNAs were not drastic; translating RNA diffuses at ∼70% of the rate of the nontranslating RNA. Two factors may contribute to the small difference in the mobility of translating and nontranslating RNA. First, most mRNAs in the cytoplasm are associated with proteins as ribonucleoproteins; thus, the nontranslating RNA is also likely to be associated with proteins. Second, for a diffusive object mobility is proportional to the cube of the mass; thus, an eightfold difference in mass is expected to result in a twofold difference in mobility.
Gag binds to HIV-1 RNA in the cytoplasm (7, 30, 31) and has been postulated to autoregulate its own translation (11). However, we observed that Gag expression did not affect the proportion of translating RNA, suggesting that HIV-1 RNA evades the influence of cytoplasmic Gag. We speculate that the mechanism of this evasion may reside in the cytoplasmic Gag–RNA interactions. Cytoplasmic HIV-1 Gag binds to both cellular RNA and viral RNA; compared with cellular RNA, there is only a small enrichment in cytoplasmic viral RNA bound to Gag (7). Thus, it is likely that the nonspecific binding of Gag to cellular mRNA allows HIV-1 RNA to bypass the influence of cytoplasmic Gag and maintain its proportion of translating RNA.
HIV-1 RNA serves as both the RNA genome and translation template; the relationship between these two roles is complex and ill defined. Actinomycin D treatment of cells infected with murine leukemia virus (MLV) led to the production of MLV particles lacking genomes (36, 37). This finding revealed the presence of two MLV RNA pools, one serving as the template for translation and the other serving as the genome for nascent particles. Actinomycin treatment of HIV-1–infected cells caused a decrease in particle production (38); thus, whether there are two HIV-1 RNA pools is unknown. Recent studies suggested that HIV-1 uses heterogeneous transcription start sites and subpopulations of HIV-1 RNA with different start sites may carry out distinct functions (13, 39). Our imaging assay, similar to our biochemical analyses, provided snapshots of RNA behavior in the cells. Therefore, we determined the proportions of HIV-1 RNA being actively translated during observation time and our studies do not address whether there were one or two pools of HIV-1 RNA. We observed that the proportion of translating RNA was not significantly different when active virus assembly and viral RNA packaging occurred at the plasma membrane. These observations could be interpreted as being consistent with the presence of two RNA populations, each serving a separate function; therefore, virus assembly and genome packaging do not affect the proportion of translating RNA. However, it is also possible that the fraction of RNA bound to Gag at the plasma membrane is only a small population of the HIV-1 RNA and thus could not significantly impact on the proportion of translating RNA. Further studies will be needed to define whether there are distinct pools of HIV-1 RNA serving different functions.
It was postulated that by binding to the RNA template from which it was translated, HIV-1 Gag can select RNAs that encode functional Gag (12). However, HIV-1 RNA lacking a functional gag gene can be packaged efficiently (40); furthermore, results from this study showed that Gag packages nontranslating RNA. Thus, the current study does not support the cis-packaging hypothesis. We observed that both translating and nontranslating RNAs appeared near the plasma membrane. However, only the nontranslating RNA formed stable complexes with Gag puncta, suggesting that Gag recruits nontranslating RNA. What is the molecular mechanism that distinguishes translating and nontranslating RNAs? The 5′UTR of HIV-1 RNA is highly structured and mediates specific genome packaging. It is possible that the structure of the 5′UTR RNA is destroyed during the translation process, reducing the likelihood that Gag forms stable complexes and packages translating RNA. These results suggest that HIV-1 RNA carries out only one function at a time, either as a template for translation or as a packageable RNA.
In a 9-kb RNA, HIV-1 encodes all of the genetic information that allows its propagation in the host; the same RNA species also serves as a translation template to generate most of the viral proteins. In this report, we found that approximately half of the cytoplasmic HIV-1 RNA is being actively translated, and Gag biogenesis occurs throughout the cytoplasm and is not limited to a particular subcellular location. Additionally, our findings have answered two long-standing questions regarding the relationship between Gag and HIV-1 RNA. We have shown that Gag does not significantly affect HIV-1 RNA translation efficiency and Gag recruits nontranslating RNA during virus assembly to be the virion genome. These studies provide insights on the complex roles of the HIV-1 RNA during viral replication and inform possible strategies to interfere with HIV-1 propagation.
Materials and Methods
Construction of HIV-1 Plasmids.
HIV-1 construct GagSunTag-MSL was derived from the NL4-3 molecular clone with inactivating deletions in pol, vif, vpr, vpu, and env. A DNA fragment containing IRES-HSA was inserted into nef. The gag gene contains two in-frame insertions: A SunTag fragment flanked by cleavage site VSQNY/PIVQN, and a sequence containing an AID domain. A fragment containing 24 copies of stem-loop sequences recognized by MSL was inserted into the pol gene. The SunTag sequence was derived from construct pHRdSV40-NLS-dCas9-24xGCN4_v4-NLS-P2A-BFP-dWPRE (Addgene #60910) (22), whereas AID and MSL sequences are based on those described for construct pUbC-FLAG-24xSuntagV4-oxEBFP-AID-baUTR1-24xMS2V5-Wpre (Addgene#84561) (18). Construct GagLZSunTag-MSL was generated by replacing the NC domain of GagSunTag-MSL with an LZ motif from plasmid 1-GagLZ-BSL (14). Molecular cloning was performed using standard techniques; the general structures of all plasmids were verified by restriction enzyme mapping, and DNA sequences generated by PCR were confirmed by DNA sequencing to avoid inadvertent mutations.
Cell Culture, Transfection, and Generation of Provirus-Containing Cell Lines.
Human 293T cells and U2OS osteosarcoma cells were maintained in DMEM supplemented with 10% FBS (HyClone), penicillin (50 U/mL; Gibco), and streptomycin (50 µg/mL; Gibco), and cells were maintained in humidified 37 °C incubators with 5% CO2.
Transient transfection was performed using FuGENE HD transfection reagent (Promega) according to the manufacturer’s recommendation. To generate stable cell lines that contain GagSunTag-MSL or GagLZSunTag-MSL for imaging experiments, we transfected into 293T cells the HIV-1 construct, a helper plasmid (pC-help) that expresses HIV-1 proteins (41), and pHCMV-G, a plasmid that expresses vesicular stomatitis virus G protein (42). Viruses were harvested from transfected cells 24 h later, clarified through a 0.45-μm pore-size filter to remove cellular debris, and used to infect a U2OS cell line stably expressing scFV-sfGFP, MS2-Halo, and OsTIR1 (18). MS2-Halo was referred to as MCP-Halo in a previous publication (18). Infections were performed at low MOI (<0.1) to ensure most infected cells expressed only one provirus. Large pools of cells, estimated to contain >200,000 independent infection events of the GagSuntag-MSL virus or >100,000 independent infection event of the GagLZSuntag-MSL virus, were generated and the infected cells were enriched by repeated cell sorting based on the expression of the HSA marker until more than 90% of the cells in the population expressed HSA.
To generate cell lines containing HIV-1 provirus to be used in biochemical assays, we transfected into 293T cells the near-full-length HIV-1 construct ON-H0 (29) and pHCMV-G. Viruses were harvested, clarified, and used to infect 293T or U2OS cells at various dilutions. In addition to containing all cis-acting elements essential for viral replication, ON-H0 expresses Gag/Gag-Pol, Tat, Rev, and an hsa marker gene in nef. The expression of the HSA marker was used to monitor the level of infection by flow cytometry. Cells infected at MOI ∼ 1 (∼60% cells infected) were selected to be used in polysome fractionation experiments.
Polysome Fractionation Assay, RNA Preparation, and Quantitative Real-Time PCR.
The polysome fractionation assay was performed as previously described (28). Briefly, cells were treated with cycloheximide (100 μg/mL final concentration; Sigma-Aldrich) for 10 min, washed twice with cycloheximide-supplemented ice-cold PBS, and lysed using polysome extraction buffer (20 mM Tris⋅HCl, pH 7.5, 100 mM KCl, 5 mM MgCl2, 1.2% Triton X-100) supplemented with 1× protease inhibitor (Roche), 5 μL/mL RNase Out (Invitrogen), and 100 μg/mL cycloheximide. Cell lysates were collected, incubated on ice for 10 min, and centrifuged at 12,000 × g for 10 min at 4 °C. The cleared cytoplasmic supernatant was transferred to the top of a 10 to 50% sucrose gradient (28), which was then centrifuged in a SW41Ti swinging bucket rotor at 36,000 rpm at 4 °C for 3 h. A total of 20 samples were collected from the bottom of the gradient using a Fraction Recovery System (Beckman Coulter). The 254-nm absorbance of each fraction was measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher) to determine the amount of RNA. TRIzol Reagent (Ambion) was used to extract RNA from each sucrose fraction according to manufacturer’s instruction. RNA was precipitated, dissolved in nuclease-free water, and treated using a TURBO DNA-free Kit (Invitrogen) to avoid DNA contamination. To quantify the amounts of HIV-1 RNA in each fraction, LightCycler 480 RNA Master Hydrolysis Probes (Roche) were used for quantitative RT-PCR. RNA copies were quantified using primers (HIV-gag-F1 and HIV-gag-R1) and probe (P-HUS-103) sets annealed to gag as previously described (43). Polysome fractions were defined as those beyond three fractions past the 80S fraction.
Sample Preparation and Live-Cell Imaging.
The U2OS cells stably expressing GagSunTag-MSL or GagLZSunTag-MSL proviruses also expressed MS2-Halotag, OS-TIR1, and scFV-sfGFP proteins. These cells were plated the day before imaging; IAA (Sigma-Aldrich) was added to the cell culture medium (250 μg/mL, final concentration) 24 h before imaging to degrade the GagSunTag protein released from the translational machinery. To label the MS2-Halo fusion protein, cells were incubated in medium containing 100 nM Halo-JF549 dyes (kindly provided by Luke Lavis, HHMI, Ashburn, Virginia) for 30 min. After the dye-containing medium was removed, cells were washed three times with medium and incubated with supplemented medium for 1 h to remove residual unbound dyes. Before imaging, the medium was replaced with supplemented phenol-red–free DMEM and cells were maintained on a stage incubator at 37 °C with 5% CO2. Because the cell pools used for imaging experiments contain >100,000 independently infected cells, it is most likely that cells used in each set of experiments were derived from different infection events.
Microscopy, Imaging Acquisition, Processing, and Data Analyses.
Spinning-disk confocal imaging was performed using an inverted Nikon Ti microscope with the Yokogawa CSU-X1 confocal scanner unit and a 100× 1.45-NA TIRF oil objective. Simultaneous imaging of GFP and Halo-JF549 was performed by using two precisely aligned cameras (Andor iXon Ultra) on a Cairn image splitter (Optosplit II). Camera alignments were performed using labeled HIV-1 particles as previously described (14, 17). The GFP and Halo-JF549 were excited with 480- and 560-nm lasers, respectively, whereas emission was detected by using 525/50- and 595/50-nm filters, respectively. Rapid HIV-1 RNA movement was acquired by using RAM capture with a 100-ms integration time and ∼2-ms overhead between frames, resulting in an overall 102-ms frame time. Long-term imaging of HIV-1 particle assembly was performed every 5 s with a 100-ms acquisition time for 1 to 2 h, as previously described using TIRF microscopy (17). Subsequent image processing and analyses, including Laplacian of Gaussian filtering and movie encoding, were performed with ImageJ software.
Single-molecule tracking, MSD analysis, persistence index calculation, and one-step jump distance analysis were performed using the MATLAB (MathWorks) program as previously described (14). Directional movement of RNA was defined as a single RNA exhibited persistence index ≥0.7 in ≥18 consecutive frames (14). Different mobility fractions were determined by curve fitting as previously described using the Qtiplot program (14, 44). For the nontranslating and translating RNA jump distance distributions, the one-component fits have χ2 values of 223 and 269, respectively; with 2 df, the probabilities that these fits describe the RNA mobilities are P < 0.0001 and P < 0.0001, respectively. For the nontranslating and translating RNA jump distance distribution, the three-component fits described in Fig. 4 B and C have χ2 values of 0.92 and 1.21, respectively, with 5 df; the probabilities that these fits describe the observed RNA mobility are P = 0.97 and P = 0.94, respectively.
To calculate the proportion of RNA in active translation, we first determined the positions of the diffraction-limited spots of all of the GFP and Halo-JF549 signals in each movie frame using Localize software (45). The GFP and Halo-JF549 signals were defined as colocalized when these two signals were located fewer than four pixels apart (0.13 μm per pixel), and the fraction of colocalized GFP+/Halo-JF549+ signals was calculated in each frame. The expression level of HIV-1 RNA in the cell was calculated by dividing the total number of Halo-JF549 signals by the cytoplasm area of each cell.
To determine whether translating RNA and nontranslating RNAs were well mixed in the cytoplasm, we measured the minimal distance between a translating RNA to another translating RNA or a nontranslating RNA. This was determined by first measuring the distances of each translating RNA to all RNAs in the cytoplasm of the same cells; using these measurements, we identified the nearest translating or nontranslating RNA that occupies the shortest (minimal) distance. The minimal distances of all translating RNA molecules were compiled to generate the observed distribution to translating RNA (T to T) or nontranslating RNA (T to N) described in the text and Fig. 4D. We also calculated the distances between RNAs, assuming the locations of the translating RNA molecules were a random subset of the total RNAs (shown as random in Fig. 4D). For this purpose, we used the spatial information of the RNAs in a cell, based on the number of translating RNA in the cell, randomly assigned a subset of RNAs as translating RNAs using a custom MATLAB program, and measured the minimal distances to the nearest translating or nontranslating RNA. Simulation of random colocalization between Gag-CeFP signals and translating RNA signals or nontranslating RNA signals was performed as previously described with custom MATLAB programs (17).
Data Availability.
All data needed to support the findings of this manuscript are included in the main text and SI Appendix.
Supplementary Material
Acknowledgments
We thank A. Arthur for expert editorial help; E. Freed, R. Burdick, and A. Duchon for discussions and critical reading of the manuscript; Dr. Luke Lavis (HHMI) for the generous gift of Halo-JF549 dye; and Dr. Ron Vale (University of California, San Francisco) for pHRdSV40-NLS-dCas9-24xGCN4_v4-NLS-P2A-BFP-dWPRE (Addgene #60910). This work was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, by NIH Intramural AIDS Targeted Antiviral Program grant funding (to W.-S.H. and to V.K.P.), and by the Innovation Fund, Office of AIDS Research, NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917590117/-/DCSupplemental.
References
- 1.Freed E. O., Martin M. A., “Human immunodeficiency viruses: Replication” in Fields Virology, Knipe D. M., Howley P. M., Eds. (Lippincott, Williams, & Wilkins, Philadelphia, PA, ed. 6, 2013), vol. II, pp. 1502–1560. [Google Scholar]
- 2.Stoltzfus C. M., Chapter 1. Regulation of HIV-1 alternative RNA splicing and its role in virus replication. Adv. Virus Res. 74, 1–40 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Kuzembayeva M., Dilley K., Sardo L., Hu W. S., Life of psi: How full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology 454-455, 362–370 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swanstrom R., Wills J. W., “Synthesis, assembly, and processing of viral proteins” in Retroviruses, Coffin J. M., Hughes S. H., Varmus H. E., Eds. (Cold Spring Harbor Press, Cold Spring Harbor, 1997), pp. 263–334. [PubMed] [Google Scholar]
- 5.Freed E. O., HIV-1 assembly, release and maturation. Nat. Rev. Microbiol. 13, 484–496 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soto-Rifo R., Rubilar P. S., Ohlmann T., The DEAD-box helicase DDX3 substitutes for the cap-binding protein eIF4E to promote compartmentalized translation initiation of the HIV-1 genomic RNA. Nucleic Acids Res. 41, 6286–6299 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kutluay S. B., et al. , Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell 159, 1096–1109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rein A., Datta S. A., Jones C. P., Musier-Forsyth K., Diverse interactions of retroviral Gag proteins with RNAs. Trends Biochem. Sci. 36, 373–380 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bieniasz P, Telesnitsky A, Multiple, switchable protein:RNA interactions regulate human immunodeficiency virus type 1 assembly. Annu. Rev. Virol. 5, 165–183 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Rein A., RNA packaging in HIV. Trends Microbiol. 27, 715–723 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson E. C., Lever A. M., Human immunodeficiency virus type 1 Gag polyprotein modulates its own translation. J. Virol. 80, 10478–10486 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poon D. T., Chertova E. N., Ott D. E., Human immunodeficiency virus type 1 preferentially encapsidates genomic RNAs that encode Pr55(Gag): Functional linkage between translation and RNA packaging. Virology 293, 368–378 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Kharytonchyk S., et al. , Transcriptional start site heterogeneity modulates the structure and function of the HIV-1 genome. Proc. Natl. Acad. Sci. U.S.A. 113, 13378–13383 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J., et al. , Cytoplasmic HIV-1 RNA is mainly transported by diffusion in the presence or absence of Gag protein. Proc. Natl. Acad. Sci. U.S.A. 111, E5205–E5213 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jouvenet N., Simon S. M., Bieniasz P. D., Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc. Natl. Acad. Sci. U.S.A. 106, 19114–19119 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sardo L., et al. , The dynamics of HIV-1 RNA near the plasma membrane during virus assembly. J Virol. 89, 10832–10840 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J., et al. , HIV-1 RNA genome dimerizes on the plasma membrane in the presence of Gag protein. Proc. Natl. Acad. Sci. U.S.A. 113, E201–E208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu B., Eliscovich C., Yoon Y. J., Singer R. H., Translation dynamics of single mRNAs in live cells and neurons. Science 352, 1430–1435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan X., Hoek T. A., Vale R. D., Tanenbaum M. E., Dynamics of translation of single mRNA molecules in vivo. Cell 165, 976–989 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morisaki T., et al. , Real-time quantification of single RNA translation dynamics in living cells. Science 352, 1425−1429 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Wang C., Han B., Zhou R., Zhuang X., Real-time imaging of translation on single mRNA transcripts in live cells. Cell 165, 990–1001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanenbaum M. E., Gilbert L. A., Qi L. S., Weissman J. S., Vale R. D., A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishimura K., Fukagawa T., Takisawa H., Kakimoto T., Kanemaki M., An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6, 917–922 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Dang Q., et al. , Nonrandom HIV-1 infection and double infection via direct and cell-mediated pathways. Proc. Natl. Acad. Sci. U.S.A. 101, 632–637 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimm J. B., et al. , A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250, 3 p following 250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pédelacq J. D., Cabantous S., Tran T., Terwilliger T. C., Waldo G. S., Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Holland A. J., Fachinetti D., Han J. S., Cleveland D. W., Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 109, E3350–E3357 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panda A. C., Martindale J. L., Gorospe M., Polysome fractionation to analyze mRNA distribution profiles. Bio Protoc. 7, e2126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhodes T. D., Nikolaitchik O., Chen J., Powell D., Hu W. S., Genetic recombination of human immunodeficiency virus type 1 in one round of viral replication: Effects of genetic distance, target cells, accessory genes, and lack of high negative interference in crossover events. J. Virol. 79, 1666–1677 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kutluay S. B., Bieniasz P. D., Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 6, e1001200 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hendrix J., et al. , Live-cell observation of cytosolic HIV-1 assembly onset reveals RNA-interacting Gag oligomers. J. Cell Biol. 210, 629–646 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J., et al. , High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl. Acad. Sci. U.S.A. 106, 13535–13540 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larson D. R., Johnson M. C., Webb W. W., Vogt V. M., Visualization of retrovirus budding with correlated light and electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 102, 15453–15458 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., Barklis E., Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J. Virol. 71, 6765–6776 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crist R. M., et al. , Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: Implications for retrovirus assembly. J. Virol. 83, 2216–2225 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levin J. G., Grimley P. M., Ramseur J. M., Berezesky I. K., Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J. Virol. 14, 152–161 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levin J. G., Rosenak M. J., Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin D-treated cells: Evidence for persistence of viral messenger RNA. Proc. Natl. Acad. Sci. U.S.A. 73, 1154–1158 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dorman N., Lever A., Comparison of viral genomic RNA sorting mechanisms in human immunodeficiency virus type 1 (HIV-1), HIV-2, and Moloney murine leukemia virus. J. Virol. 74, 11413–11417 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masuda T., et al. , Fate of HIV-1 cDNA intermediates during reverse transcription is dictated by transcription initiation site of virus genomic RNA. Sci. Rep. 5, 17680 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nikolaitchik O., Rhodes T. D., Ott D., Hu W. S., Effects of mutations in the human immunodeficiency virus type 1 Gag gene on RNA packaging and recombination. J. Virol. 80, 4691–4697 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mochizuki H., Schwartz J. P., Tanaka K., Brady R. O., Reiser J., High-titer human immunodeficiency virus type 1-based vector systems for gene delivery into nondividing cells. J. Virol. 72, 8873–8883 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yee J. K., et al. , A general method for the generation of high-titer, pantropic retroviral vectors: Highly efficient infection of primary hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 91, 9564–9568 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buckman J. S., Bosche W. J., Gorelick R. J., Human immunodeficiency virus type 1 nucleocapsid zn(2+) fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J. Virol. 77, 1469–1480 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grünwald D., et al. , Probing intranuclear environments at the single-molecule level. Biophys. J. 94, 2847–2858 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zenklusen D., Larson D. R., Singer R. H., Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 15, 1263–1271 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to support the findings of this manuscript are included in the main text and SI Appendix.