Significance
Exosomes have emerged as extracellular organelles controlling cell-cell communication in health and disease. Poor knowledge of the molecular mechanisms supporting exosome formation limits rational exploitation of exosomes in diagnostics and therapeutics. Tetraspanins and syndecans, two families of membrane scaffold proteins, compose characteristic exosomal cargo. Syndecans are versatile coreceptors and tetraspanins are well known to control the traffic of various signaling molecules, but the interconnection between these two families remains largely unexplored. Here we identify a functional crosstalk between specific members of the tetraspanin and syndecan families in exosome formation. Specifically, we demonstrate the importance of tetraspanin-6-controlled syndecan-4-syntenin trafficking in the balance between exosomal secretion and lysosomal degradation. This work reveals that syndecan-tetraspanin membrane scaffolds cooperate to dictate exosome formation.
Keywords: tetraspanin, syndecan, syntenin, exosomes
Abstract
Exosomes, extracellular vesicles (EVs) of endosomal origin, emerge as master regulators of cell-to-cell signaling in physiology and disease. Exosomes are highly enriched in tetraspanins (TSPNs) and syndecans (SDCs), the latter occurring mainly in proteolytically cleaved form, as membrane-spanning C-terminal fragments of the proteins. While both protein families are membrane scaffolds appreciated for their role in exosome formation, composition, and activity, we currently ignore whether these work together to control exosome biology. Here we show that TSPN6, a poorly characterized tetraspanin, acts as a negative regulator of exosome release, supporting the lysosomal degradation of SDC4 and syntenin. We demonstrate that TSPN6 tightly associates with SDC4, the SDC4-TSPN6 association dictating the association of TSPN6 with syntenin and the TSPN6-dependent lysosomal degradation of SDC4-syntenin. TSPN6 also inhibits the shedding of the SDC4 ectodomain, mimicking the effects of matrix metalloproteinase inhibitors. Taken together, our data identify TSPN6 as a regulator of the trafficking and processing of SDC4 and highlight an important physical and functional interconnection between these membrane scaffolds for the production of exosomes. These findings clarify our understanding of the molecular determinants governing EV formation and have potentially broad impact for EV-related biomedicine.
Exosomes, a class of extracellular vesicles (EVs), are now considered as important organelles for intercellular communication, allowing cells to exchange proteins, lipids, and genetic material. They are present in biological fluids and are involved in a plethora of physiological and pathological processes (1–3). Knowledge of the molecular processes that govern the formation, identity, and fate of exosomes is essential for any rational clinical applications involving their use or analysis. Yet our understanding of such processes is still in its infancy.
Nascent exosomes are formed in endosomes after inward budding of the endosomal membrane to generate the so-called intraluminal vesicles (ILVs) present in multivesicular bodies (MVBs). To a certain extent, this process of membrane budding and abscission is reversible (back-fusion), but ILVs and their cargo are thought to have two main alternative outcomes, either 1) degradation upon fusion of MVBs with lysosomes or 2) liberation in the cell exterior (where they are called exosomes) upon fusion of MVBs with the plasma membrane. It is clear that multiple molecular mechanisms operate to support the formation of exosomes, and it is equally clear that several subpopulations, with different functional properties, exist within this type of extracellular vesicle (4, 5). In general, it is thought that the mechanisms of vesicle loading (the sorting and sequestration of particular cargo into budding membrane domains) and vesicle biogenesis (the bending and abscission of membrane) are intimately linked to each other, and are largely initiated by signaling (5, 6). The termination of the signaling and down-regulation of EGFR (epidermal growth factor receptor) by sequestration of the endocytosed receptor in ILVs that ultimately are degraded in lysosomes or, alternatively, transmitted to other cells as “exosomes” (even initiating EGFR signaling in these recipients) illustrates this notion well (7, 8).
The budding of intraluminal vesicles is largely dependent on the endosomal sorting complex required for transport (ESCRT) (9–11). ESCRT consists of four multimeric complexes that are assembled in an orderly manner at the endosome, recognizing and sequestering ubiquitinated membrane proteins for ultimate budding and incorporation into ILVs (12, 13). ESCRTs operate in both “degradative” and “secretory” MVBs (14). Yet, ILV formation inside MVBs also implicates specific lipids and scaffold proteins such as ceramide, syndecans (SDCs), and syntenin (15, 16). As we have shown previously, syndecans and their cytosolic adaptor protein syntenin, linking syndecans to ALIX, support the formation of ILVs destined for exosomal secretion, in a manner that depends at least in part on ESCRT (16). Strikingly, we found that syndecans are—and possibly need to be—proteolytically processed during ILV biogenesis. Consequently, the main form of syndecan present on exosomes corresponds to a syndecan C-terminal fragment (SDC-CTF) comprising the transmembrane and intracellular domains of the protein. In some cell lines (e.g., MCF-7 cells) the syndecan-syntenin pathway accounts for a major part (up to 50%) of the total pool of exosomes. More recently, we demonstrated that syndecan-syntenin control the promigratory activity of exosomes (17).
Tetraspanins (TSPNs) compose a family of membrane proteins (with four membrane spans), comprising 33 members in mammals, that have been shown to regulate the trafficking of selective associated proteins, as well as their function, possibly through membrane compartmentalization (18). Several tetraspanins such as CD9, CD81, and CD63 are major constituents of EVs, including exosomes, and therefore serve as canonical markers for EVs. Surprisingly little is known about the function of tetraspanins in EV formation (19). Noteworthy, like SDCs, the tetraspanin CD63 directly interacts with syntenin (20). We previously established that (in MCF-7 cells) syntenin and syndecans control the exosomal release of CD63; CD63, in turn, does not impact on exosomal syntenin and syndecan (16). The exosomal accumulations of CD9 and CD81, in contrast, are not affected by syndecan-syntenin (17, 21). Tetraspanin-6 (TSPN6) also directly interacts with syntenin (22). Yet its function in exosome formation was incompletely characterized. Here, we show that low cellular levels of TSPN6 are permissive for exosomal syntenin and SDC4 secretion while high levels of TSPN6 direct the same cargos to lysosomal degradation. Moreover, TSPN6 also controls SDC4 proteolytic cleavage that leads to ectodomain shedding. Thus, pending on the levels of TSPN6 in cells, the sorting of specific endosomal cargo segregates between distinctive routes, with alternative outcomes.
Results
Tetraspanin-6 Specifically Reduces the Exosomal Releases of Syntenin and Syntenin Cargo.
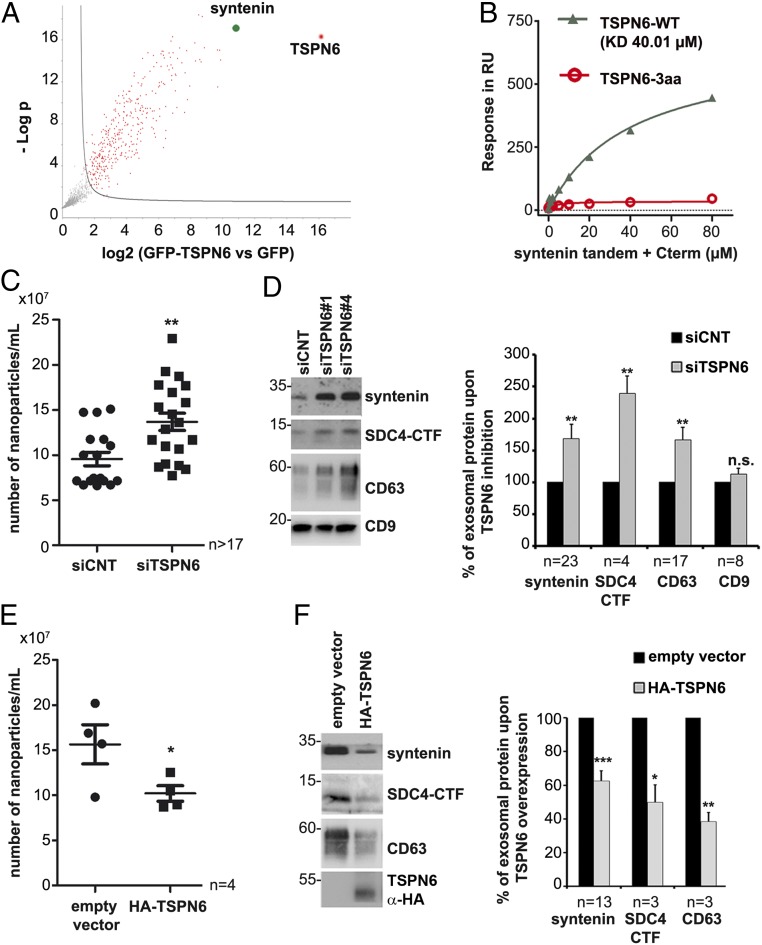
To determine the TSPN6 interactome in MCF-7 cells, we transiently overexpressed GFP-TSPN6 (versus solely GFP [green fluorescent protein]), and analyzed immunoprecipitates (using nanobodies against GFP) by mass spectrometry. Syntenin was identified as the cellular protein bound most preferentially to TSPN6 (Fig. 1A). Further surface plasmon resonance (SPR) experiments showed that TSPN6 can directly interact with the PDZ domains of recombinant syntenin and that this interaction depends on its canonical PDZ-binding motif, since abolished by the deletion of the last three C-terminal residues (Fig. 1B). Since we previously demonstrated the important role of syntenin in exosomal release, we then tested the potential impact of this tetraspanin on exosome composition and production. Nanoparticle tracking analyses (NTA) revealed that TSPN6-loss (mediated by the administration of validated short interfering RNA [siRNA], SI Appendix, Fig. S1 A and B) increases the total number of secreted particles (Fig. 1C), but has no effect on their size (SI Appendix, Fig. S1C). Inversely, TSPN6-gain (mediated by the transient overexpression of hemagglutinin [HA]-tagged TSPN6) decreases the total number of secreted particles (Fig. 1E). More specifically, using two different siRNAs, we observed that TSPN6-loss stimulates the exosomal secretion of syntenin. TSPN6-loss also stimulates the exosomal releases of syndecan-4 C-terminal fragment (SDC4-CTF) and CD63, both composing bona fide syntenin cargo; i.e., cargo binding directly to the syntenin PDZ domains, while leaving intact the exosomal levels of “nonbinders” like CD9 and CD81 (Fig. 1D and SI Appendix, Fig. S1D). Interestingly, TSPN6-loss also stimulates the vesicular/exosomal release of TSG101 (SI Appendix, Fig. S1E), a component of ESCRT that is required for epidermal growth factor (EGF)-initiated/stimulated MVB formation and EGFR degradation (23). Thus, loss of TSPN6 affects at least one population of “canonical” exosomes, but might also result in the inadvertent “exosomal” release of ILV that are normally or in major part destined for lysosomal degradation. Conversely, TSPN6-gain resulted in a loss of exosomal SDC4-CTF and CD63 (Fig. 1F). Of note, the inhibition of CD63, a tetraspanin that is classically used as exosome marker and that, like TSPN6, also binds directly to the PDZ domains of syntenin, had no significant effect on exosome number and size (SI Appendix, Fig. S2 A and B). Consistently, exosomal cargo such as syntenin, SDC4-CTF, and CD81 remained also unaffected (SI Appendix, Fig. S2C). Since CD63 mRNA (messenger RNA) is prominent in MCF-7 cells and by far exceeds the levels of TSPN6 (24), this suggests that TSPN6 effects on exosomal releases are specific (rather than a matter of TSPN abundance). Taken together, these data show that TSPN6 down-regulates the production of exosomes; in particular, exosomes that contain SDC4, CD63, and syntenin.
Fig. 1.
Tetraspanin-6 interacts with syntenin and restricts the formation of syntenin/syndecan-4/CD63 exosomes. (A) Volcano plot showing the significance two-sample t test (−Log P value) vs. iBAQ intensity (Log2 [GFP-TSPN6 vs. GFP as control]) on the y- and x-axes, respectively. Data result from four different experiments processed three times. TSPN6 (the bait) is represented in red and syntenin, one of the most specifically TSPN6-associated proteins, is represented in green. Proteins showing nonsignificant differences are represented in gray. (B) Direct syntenin-TSPN6 interaction as detected by surface plasmon resonance. Syntenin construct (comprising only the tandem-PDZ domains + C-terminal domain) was injected over an immobilized peptide corresponding to the last 22 C-terminal amino acids of wild-type TSPN6 (gray triangles) or a similar peptide deleted from its last three amino acids (red circles), abolishing the PDZ-binding motif. (C–F) Exosomes secreted by TSPN6-depleted cells (siTSPN6, C and D) or TSPN6-overexpressing cells (HA-TSPN6, E and F), versus corresponding controls, were isolated by ultracentrifugation. Exosomes were analyzed by Nanosight (C and E) or by Western blot, testing for several markers, as indicated (D and F). TSPN6 was depleted using two different RNAi (siTSPN6#1 and siTSPN6#4). In the Nanosight analyses, each point represents the number of exosomes from three independent measurements. The two TSPN6 siRNAs were tested separately, in parallel cultures, and the results of both were pooled. Histograms represent mean signal intensities ± SEM in exosomes relative to signals obtained for control exosomes. SDC4-CTF; syndecan-4 C-terminal fragment. *P < 0.05, **P < 0.01, ***P < 0.001, n.s. nonsignificant (Student’s t test). The value “n” indicates the number of independent experiments.
Tetraspanin-6 Addresses Syntenin to Lysosomal Degradation.
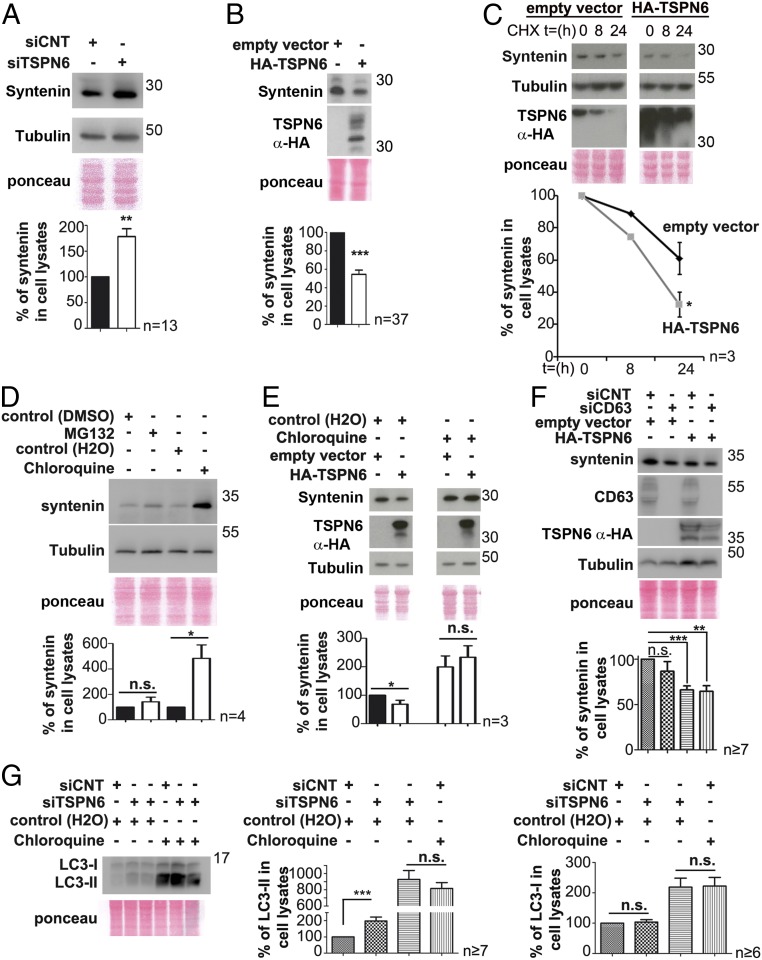
In MCF-7 cells, syntenin is a limiting factor for exosome production (16). We therefore investigated the impact of TSPN6 on the cellular levels of syntenin. TSPN6-loss (Fig. 2A) and TSPN6-gain (Fig. 2B), respectively, increase and decrease the levels of syntenin in cells. Moreover, when cells are treated with cycloheximide, the cellular levels of syntenin are decreasing more rapidly in TSPN6-overexpressing cells than in control cells (Fig. 2C). We thus concluded that TSPN6 addresses syntenin to degradation. Assessing different possible syntenin degradation routes, we observed that the cellular levels of syntenin were significantly increased upon chloroquine treatment, whereas they remained nearly unchanged upon MG132 treatment (Fig. 2D). Thus, at least in MCF-7 cells, syntenin is not turned over by the proteasome, but primarily follows lysosomal degradation routes. Consistent with that notion, we observed that syntenin and TSPN6 are found together in the lumen of RAB5-Q79L-endosomes, indicating they can join during the process of endosomal budding (SI Appendix, Fig. S2D). Noteworthy, in cells treated with chloroquine, TSPN6-gain had no significant effect on the (elevated) syntenin cellular levels (Fig. 2E). Moreover, CD63, a TSPN that also binds to syntenin, whose exosomal secretion is also negatively affected by TSPN6, and which has been shown to promote the lysosomal degradation of particular protein partners (25), is not required for TSPN6-mediated syntenin degradation (Fig. 2F). Turnover by autophagy, also sensitive to chloroquine, may provide an alternative path upstream of lysosomal degradation. Of note, syntenin was previously shown to maintain protective autophagy (26). TSPN6-depleted cells show a significant increase in LC3-II levels, but upon chloroquine treatment these levels are similarly elevated in TSPN6-depleted and in control cells, suggesting that TSPN6 has no impact on the biogenesis of autophagosomes but enhances or accelerates their fusion with lysosomes (Fig. 2G). Altogether, these data indicate that TSPN6 addresses syntenin to lysosomal degradation. Thus, TSPN6 seems to direct some specific endosomal assemblies and compartments toward lysosomal degradation routes.
Fig. 2.
Tetraspanin-6 addresses syntenin to lysosomal degradation. (A and B) TSPN6-depleted MCF-7 cells (siTSPN6, the four TSPN6 siRNAs were tested separately, in parallel cultures, and the results were pooled) and MCF-7 cells overexpressing TSPN6 (HA-TSPN6) with their respective control cells (siCNT and empty vector) were evaluated for syntenin accumulation in cells. Total cell lysates were analyzed by Western blot (Upper). Histograms (Lower) represent the mean signal intensity for syntenin ± SEM, relative to the signal in control cells. (C) Cells overexpressing TSPN6 (HA-TSPN6) and control cells (empty vector) were incubated with cycloheximide (CHX; 20 µg/mL) for the indicated lengths of time (hours). Syntenin cellular levels were analyzed by Western blot (Upper). In the corresponding plots (Lower), each point represents syntenin mean signal intensity at the corresponding time point ± SEM relative to time 0. (D) MCF-7 cells were treated overnight with MG132 10 µM (inhibitor of proteasomal degradation) or with chloroquine 100 µM (inhibitor of lysosomal degradation) or with respective controls (DMSO [dimethyl sulfoxide] 10 µM or H2O). The total cellular levels of syntenin were analyzed by Western blot (Upper). Histograms (Lower) represent syntenin mean signal intensities in cells after drug treatments relative to the respective controls. (E) MCF-7 cells overexpressing TSPN6 (HA-TSPN6) and control cells (empty vector) were incubated with chloroquine (100 µM) or control (H2O) overnight. Syntenin and TSPN6 total cellular levels were analyzed by Western blot. Corresponding histograms (Lower) represent the mean signal intensity ± SEM obtained for syntenin in the total cell lysate of cells transfected with empty vector or overexpressing TSPN6 and treated with chloroquine relative to signals in cells transfected with empty vector and nontreated with chloroquine. Note that, by itself, and like TSPN6 suppression, chloroquine markedly enhances (nearly doubling) the levels of syntenin in cells and prevents TSPN6 overexpression to have negative effects on cellular syntenin levels. (F) Cells first silenced for CD63 expression (siCD63) and control cells (treated with siCNT) were maintained for 72 h and were then transfected for 24 h with HA-TSPN6 or the empty vector (control). Histograms (Lower) represent mean signal intensity of syntenin ± SEM in total cell lysates relative to signals in control cells. (G) Cells silenced for TSPN6 expression (siTSPN6, the four TSPN6 siRNAs were tested separately, in parallel cultures, and the results were pooled) and control cells (siCNT) were treated overnight with chloroquine 100 µM (inhibitor of lysosomal degradation) or with control agent (H2O). The total cellular levels of LC3 were analyzed by Western blot. Histograms represent LC3-I and LC3-II mean signal intensities in cells after drug treatments, relative to the respective controls. *P < 0.05, **P < 0.01, ***P < 0.001, n.s. nonsignificant (Student’s t test).
Tetraspanin-6-Dependent Degradation of Syntenin Requires Syndecan-4.
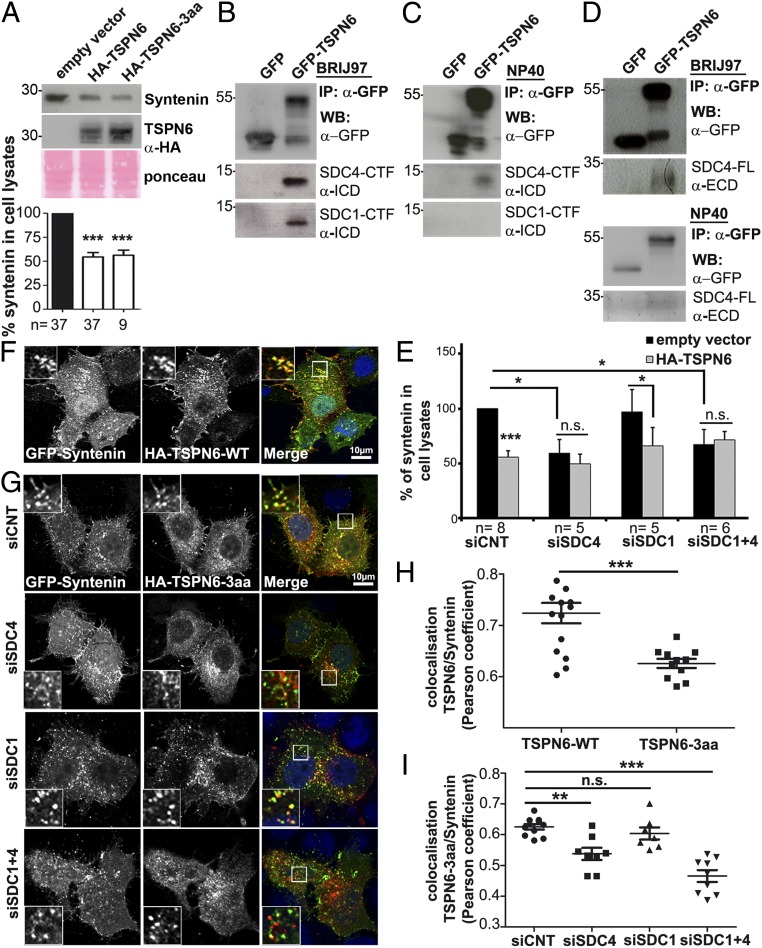
We next investigated the role of the PDZ-binding motif (PDZ-BM) of TSPN6. Surprisingly, both the gain of wild-type TSPN6 and TSPN6-3aa (TSPN6 with a mutant PDZ-BM) significantly decreased syntenin cellular levels (Fig. 3A). Thus, while the TSPN6 PDZ-BM is essential for a direct interaction between TSPN6 and syntenin, this motif is not required for TSPN6 to promote syntenin degradation. In an effort to clarify this apparent paradox, we investigated whether TSPN6 effects might depend on syndecans (SDCs), major alternative PDZ-binding partners of syntenin (27). SDC1 and SDC4 are the major (if not sole) SDC family members expressed in MCF-7 cells (SI Appendix, Fig. S3A). We investigated whether SDC4, SDC1, or both are associated with TSPN6. We overexpressed GFP or GFP-tagged TSPN6 in MCF-7 cells and performed anti-GFP immunoprecipitation, followed by Western blot analysis of the precipitate (Fig. 3 B–D). We used two alternative detergents; namely, BRIJ97, known to preserve TSPN-TSPN interactions and Nonidet P-40 (NP-40) disrupting TSPN-TSPN webs, thereby allowing to identify closely associated partners (28). In mild detergent, TSPN6 associated with endogenous SDC4- and SDC1-CTFs (Fig. 3B). Interestingly, the interaction of TSPN6 with SDC4-CTFs resists NP-40 (Fig. 3C), indicative of a tight interaction. TSPN6 also tightly associates with endogenous full-length SDC4 (Fig. 3D). Then, we tested for the effect of TSPN6-gain in MCF-7 cells with down-regulated expressions of SDC1, SDC4, or both by siRNAs (Fig. 3E and SI Appendix, Fig. S3B). Surprisingly, we observed that in the absence of SDC4, syntenin cellular levels were decreased by nearly half (Fig. 3E). Overexpression of TSPN6 in SDC4-depleted cells did not further decrease the cellular levels of syntenin in significant ways. This effect was SDC4 specific. Indeed, a similar drop in the cellular levels of syntenin, and a similar resistance of the residual syntenin to TSPN6 overexpression were not observed for the knock-down of SDC1. Neither did the additional knock-down of SDC1 further enhance the effects of a SDC4 knock-down (Fig. 3E). These results suggest that in the absence of SDC4 syntenin might be degraded and imply that TSPN6 acts upon SDC4-associated syntenin. Furthermore, confocal imaging revealed that HA-tagged wild-type TSPN6 colocalizes with syntenin (GFP-Syntenin; construct containing the PDZ-tandem and C terminus of the protein; yellow staining, Fig. 3 F and H). A direct PDZ-mediated interaction of TSPN6 with syntenin is not mandatory for such in cellulo colocalization, as it is also observed with the TSPN6 with a mutant PDZ-BM (HA-TSPN6-3aa) (Fig. 3 G and I). Yet, the TSPN6 PDZ-BM mutant (HA-TSPN6-3aa) was not colocalizing with syntenin in cells inhibited for SDC4 expression (siRNA targeting SDC4 [siSDC4]), in striking contrast to what is observed in control cells (control siRNA [siCNT]) and in cells inhibited for SDC1 expression only (siRNA targeting SDC1 [siSDC1]) (Fig. 3 G and I). Altogether, these results suggest that TSPN6 associates with SDC4 and (SDC4 with) syntenin, forming a tripartite complex, and that, in the presence of SDC4, the TSPN6 PDZ-BM is neither required for TSPN6 colocalization with syntenin nor required for TSPN6 addressing syntenin to degradation. This further underscores the importance of the SDC4-TSPN6 association in the functional effects of TSPN6.
Fig. 3.
Tetraspanin-6-dependent degradation of syntenin does not depend on the syntenin-binding site of tetraspanin-6 but requires syndecan-4. (A) MCF-7 cells transiently transfected with empty vector, or an expression vector encoding either wild-type TSPN6 (HA-TSPN6) or a mutant TSPN6 lacking its last three amino acids and defective for direct syntenin binding (HA-TSPN6-3aa) were tested for syntenin expression levels, analyzing total cell lysates by Western blot (Upper). Histograms (Lower) represent mean signal intensities of syntenin ± SEM in the total cell lysate of TSPN6 overexpressing cells relative to signals in control cells (transfected with the empty vector). (B) Coimmunoprecipitation of endogenous full-length syndecan-4 (SDC4-FL) with overexpressed GFP-TSPN6, but not GFP, from MCF-7 cell lysates. Cells were lysed with BRIJ97 detergent. The precipitates (IP) were subjected to heparitinase/chondroitinase (H/C) digestion before analysis by Western blot with the indicated antibodies. (C and D) Coimmunoprecipitation of endogenous syndecan-4 (SDC4-CTF) and syndecan-1 (SDC1-CTF) with overexpressed GFP-TSPN6, but not GFP (used as a control), from MCF-7 cell lysates. Cells were lysed with two different detergents: BRIJ97 that conserves tetraspanin-tetraspanin interactions (C) or NP-40 that disrupts tetraspanin-tetraspanin interactions (D). The GFP precipitates (IP) were subjected to Western blot with the indicated antibodies (α-). ECD, extracellular domain; ICD, intracellular domain; SDC4-FL, full length syndecan-4; SDC4-CTF, syndecan-4 C-terminal fragment; SDC1-CTF, syndecan-1 C-terminal fragment. (E) MCF-7 cells treated with siRNA downregulating SDC4 (siSDC4), SDC1 (siSDC1), or both SDC1 and SDC4 (siSDC1+4), and control cells (treated with siCNT) were transfected with TSPN6 (HA-TSPN6) or empty vector, and syntenin total cellular levels were analyzed by Western blot. Histograms represent mean signal intensity of syntenin ± SEM in total cell lysates relative to signals in control cells, transfected with empty vector. (F) Representative confocal micrographs showing MCF-7 cells transiently overexpressing wild-type TSPN6 (HA-TSPN6-WT; red in merge) together with GFP-Syntenin (green in merge). See Insets for high magnification; zoom × 3. (G) Representative confocal micrographs showing MCF-7 cells down-regulated for SDC4 expression (siSDC4), SDC1 expression (siSDC1), or both SDC4 and SDC1 expression (siSDC1+4), and control cells (siCNT) transiently overexpressing HA-TSPN6-3aa (red in merge) together with GFP-Syntenin (green in merge). See Insets for high magnification; zoom × 3. Note that TSPN6-3aa colocalizes with the syntenin construct on intracellular vesicular structures, except in cells depleted for SDC4 expression. (H and I) Syntenin colocalization with TSPN6-WT or TSPN6-3aa (H) or syntenin colocalization with TSPN6-3aa in cells depleted for SDC4, SDC1, both SDC4+1 or control cells (I) was assessed by calculating the Pearson correlation coefficient on at least 10 cells per condition using the JACoP plugin on ImageJ. Histograms represent the mean Pearson coefficient ±SEM. The “n” value indicates the number of independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, n.s. nonsignificant (Student’s t test).
Tetraspanin-6 Directs Syndecan-4 C-Terminal Fragment to Lysosomal Degradation.
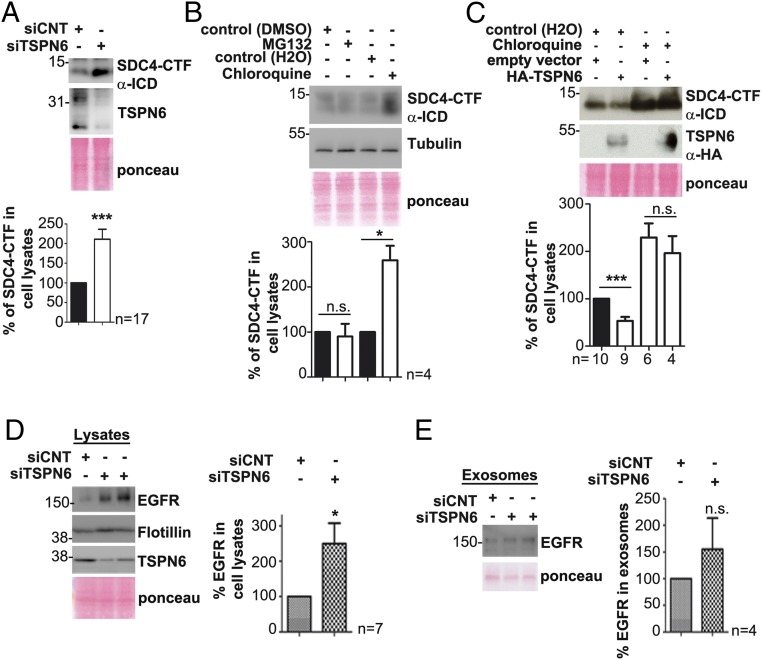
To better understand the function of TSPN6:SDC4 complexes, we investigated the impact of TSPN6 on SDC4 turnover (modeled in SI Appendix, Fig. S3C). Upon TSPN6 depletion, the levels of SDC4-CTF in cells increased by a factor of 2 (Fig. 4A). Chloroquine treatment (and not the proteasomal inhibitor MG132) enhances the levels of SDC4-CTF in cells, consistent with a mostly lysosomal mode of degradation for SDC4-CTF (Fig. 4B). Importantly, after chloroquine treatment, the levels of SDC4-CTF were similar in TSPN6-transfected cells and in control cells (Fig. 4C). We then investigated whether TSPN6 might also address potential SDC cargo toward degradation. Upon TSPN6-loss, EGFR levels were significantly increased in cells, but less significantly (or not) in exosomes (Fig. 4 D and E), suggesting that TSPN6 also supports the degradation of specific signaling receptors that are—at least in part—SDC4 associated (29). Together with the negative effects of TSPN6 on the exosomal levels of SDC4-CTFs (Fig. 1), these data clearly indicate that TSPN6 supports the lysosomal degradation of SDC4 (at the same time subtracting SDC4 and SDC cargo from “exosomal secretory” routes).
Fig. 4.
Tetraspanin-6 addresses syndecan-4 C-terminal fragment to lysosomal degradation. (A) Cells silenced for TSPN6 expression (siTSPN6, the four TSPN6 siRNAs were tested separately, in parallel cultures, and the results were pooled) and control cells (siCNT) were analyzed for SDC4-CTF and TSPN6 expression in total cell lysates, by Western blot (Upper). Histograms (Lower) represent mean signal intensity of SDC4-CTF ± SEM in total cell lysates relative to signals in control cells. (B) MCF-7 cells were treated overnight with MG132 10 µM (inhibitor of proteasomal degradation) or with chloroquine 100 µM (inhibitor of lysosomal degradation) or with respective controls (DMSO 10 µM or H2O). The total cellular levels of SDC4-CTF were analyzed by Western blot (Upper). Histograms (Lower) represent SDC4-CTF mean signal intensities in cells after drug treatments relative to the respective controls. (C) MCF-7 cells overexpressing TSPN6 (HA-TSPN6) and control cells (empty vector) were incubated with chloroquine (100 µM) or control agent (H2O) overnight. SDC4-CTF and TSPN6 total cellular levels were analyzed by Western blot. Corresponding histograms (Lower) represent the mean signal intensity ± SEM obtained for SDC4-CTF in the total cell lysate of cells transfected with empty vector or overexpressing TSPN6 and treated with chloroquine relative to signals in cells transfected with empty vector and nontreated with chloroquine. Note that, by itself, and like TSPN6 suppression, chloroquine markedly enhances (nearly doubling) the levels of SDC4-CTF in cells and prevents TSPN6 overexpression to have negative effects on cellular SDC4-CTF levels. (D and E) TSPN6-depleted MCF-7 cells (siTSPN6) and control cells (siCNT) were evaluated for EGFR accumulation in cells (D) and in exosomes (E). Total cell lysates (D) and exosomes (E) were analyzed by Western blot (Left). Histograms (Right) represent the mean signal intensity for EGFR ±SEM, relative to the signal in control cells. Values were calculated from n independent experiments as indicated. *P < 0.05, ***P < 0.001, n.s. nonsignificant (Student’s t test). SDC4-CTF; syndecan-4 C-terminal fragment.
Tetraspanin-6 Prevents Syndecan-4 Ectodomain Cleavage and Shedding.
We next investigated the impact of TSPN6 on the abundance of the full-length form of SDC4 (SDC4-FL) in cells. Similarly to what we observed for SDC4-CTF, TSPN6 depletion increases SDC4-FL by a factor of 1.5 (Fig. 5A). Yet, TSPN6 overexpression also increases the cellular levels of full-length SDC4 (Fig. 5B). This paradoxical result suggests heterogeneity among the full-length forms of SDC4, and opposing effects of TSPN6 on the turnover of distinctive pools of SDC4-FL. Similar observations were made for CD63 (SI Appendix, Fig. S4 A and B).
Fig. 5.
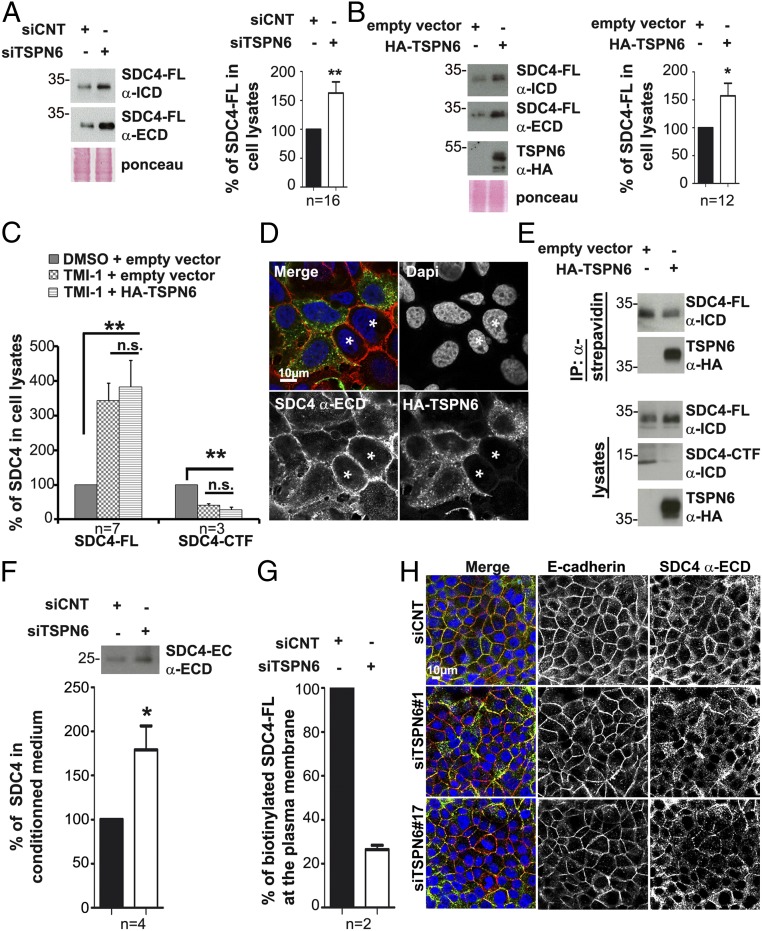
Tetraspanin-6 prevents syndecan-4 ectodomain cleavage and shedding. (A) Cells silenced for TSPN6 expression (siTSPN6, the four TSPN6 siRNAs were tested separately, in parallel cultures, and the results were pooled) and control cells (siCNT) were evaluated for SDC4-FL expression using two different antibodies recognizing SDC4 extracellular domain, (α-ECD) or intracellular domain (α-ICD). SDC4 in total cell lysates was analyzed by Western blot after heparitinase/chondroitinase (H/C) digestion. Histograms represent mean signal intensity of SDC4-FL ± SEM in total cell lysates relative to signals in control cells. (B) MCF-7 cells overexpressing TSPN6 (HA-TSPN6) and control cells (empty vector) were similarly analyzed for SDC4-FL expression. Histograms represent relative mean signal intensities of SDC4-FL ± SEM in total cell lysates, relative to controls. (C) Cells overexpressing TSPN6 (HA-TSPN6) for 24 h and control cells (empty vector) were treated with TMI-1 (inhibitor of matrix metalloproteinase [MMPs] and ADAM17; 10 µM) or DMSO as control. Histograms represent mean signal intensities of SDC4-FL and SDC4-CTF ± SEM in total cell lysates relative to signals in control cells. (D) Representative confocal micrographs showing the steady-state distribution of endogenous SDC4 in HA-TSPN6-overexpressing cells, compared to control cells (asterisk). MCF-7 cells were transiently transfected with expression vector for HA-TSPN6 for 24 h. In merge, HA-TSPN6 is in green, SDC4 (extracellular domain [ECD] epitope) is in red, and nuclei are in blue (4′,6-diamidino-2-phenylindole [DAPI]). Note that the cells overexpressing TSPN6 feature an intracellular distribution of SDC4-ECD compared to surrounding cells (asterisks) where SDC4-ECD is localized predominantly at the plasma membrane. Cells are confluent to visualize SDC4 membrane-accumulation. (E) MCF-7 cells transiently transfected with expression vector for TSPN6 (HA-TSPN6) or the empty vector as control were cell surface-biotinylated. Biotin-labeled proteins present in total cell lysates, captured with streptavidin beads, were analyzed for SDC4 content, by Western blot. Note that cells overexpressing TSPN6 present less SDC4 at the plasma membrane. (F) Culture media from cells inhibited for TSPN6 expression (siTSPN6) and from control cells (siCNT) were analyzed for the presence of shed cleaved SDC4 (25 kDa form) by immunoprecipitation experiments using the antibody recognizing the ECD. TSPN6 was down-regulated using two different RNAi (siTSPN6, siTSPN6#1 or siTSPN6#4). Histograms represent SDC4 (25 kDa) mean signal intensity ±SEM after immunoprecipitation from cell media originating from TSPN6 depleted cells relative to signals originating from controls. (G) MCF-7 cells depleted for TSPN6 (siTSPN6#1) and control cells (siCNT) were biotinylated at the cell surface. Biotin-labeled proteins were analyzed for SDC4 content, by Western blot. Histograms represent SDC4-FL mean signal intensities ±SEM normalized to control, calculated for two independent experiments. Note that in TSPN6-depleted cells (and as in TSPN6-overexpressing cells, see E, Right) less SDC4 is present at the plasma membrane. (H) Representative confocal micrographs showing the steady-state distribution of endogenous SDC4 (using an antibody directed against its extracellular domain; α-ECD) and endogenous E-cadherin in TSPN6-depleted cells (siTSPN6#1 and siTSPN6#17) compared to control cells (siCNT). In merge, nuclei are stained with DAPI (blue), SDC4 is in green, and E-cadherin is in red. Values were calculated from n independent experiments as indicated. *P < 0.05, **P < 0.01, n.s. nonsignificant (Student’s t test).
Shedding (directly, or following recycling) represents an alternative for endocytosis and lysosomal degradation in clearing SDCs from cell surfaces (SI Appendix, Fig. S3C). Noteworthy, only signals for epitopes present in the SDC4 intracellular domain, exposed to cytosol (but not epitopes present in the extracellular domain, exposed in the luminal spaces) were observed to undergo endosomal budding in the large vesicles that are observed after transfection of Rab5Q79L, suggesting that SDC4 is proteolytically cleaved prior to its sorting into ILVs (SI Appendix, Fig. S3D). We therefore also tested for a possible role of TSPN6 as regulator of the cleavage of SDC4. For this purpose we used TMI-1 drug to inhibit matrix metalloproteinases (MMPs) and ADAM17. In cells treated with TMI-1, by itself markedly enhancing the levels of SDC4-FL in cells, the overexpression of TSPN6 does not significantly further enhance the SDC4-FL levels (Fig. 5C). Moreover, in cells overexpressing TSPN6, endogenous SDC4 ectodomain is less concentrated at the plasma membrane and predominates in the cell interior (Fig. 5D). Cell surface-biotinylation experiments, using an antibody directed against the intracellular domain of SDC4, indicated that TSPN6-overexpressing cells present less full-length SDC4 at their plasma membrane, complementing and validating the confocal imaging observations (Fig. 5E). We also measured the levels of SDC4 ectodomain released in the culture medium (Fig. 5F and SI Appendix, Fig. S4E). Upon the loss of TSPN6, the levels of secreted SDC4 ectodomain rose by almost a factor of 2 (Fig. 5F), an increase of the same order of magnitude as the concomitant increase of SDC4-FL (Fig. 5A) and SDC4-CTF (Fig. 4A) in cells. Consistent with a negative effect of TSPN6 on cell surface SDC4 cleavage, the loss of TSPN6 (enhancing shedding) diminishes the levels of plasma membrane SDC4-FL in biotinylation experiments (Fig. 5G). Even at high confluency, when E-cadherin concentrates at cell-cell junctions, loss of TSPN6 disrupts the SDC4 “honey comb” pattern (Fig. 5H). Noteworthy, TSPN6-loss did not appear to influence the endoplasmic reticulum–golgi-plasma membrane transport of SDC4 as detected by RUSH (retention using selective hooks) experiments (SI Appendix, Fig. S4F). Taken together, these results highlight a role for TSPN6 in regulating SDC4 localization and cleavage. Altogether, these results indicate that TSPN6 addresses SDC4-syntenin to lysosomal degradation, restricting SDC4 ectodomain shedding and inhibiting the role of this syndecan in syntenin-supported exosome formation (Fig. 6).
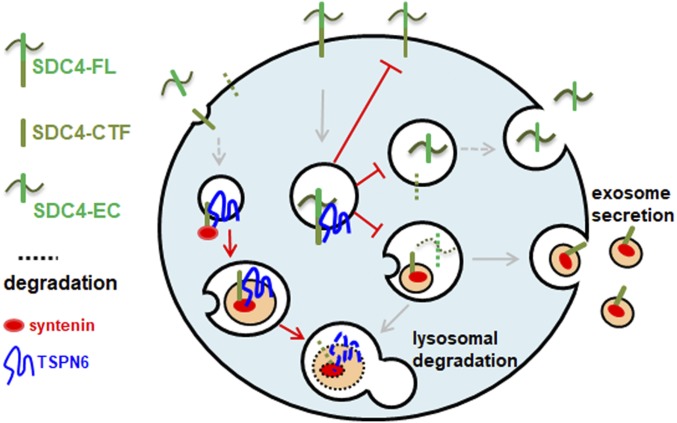
Fig. 6.
Model recapitulating the findings of the present work. TSPN6 forms tight complexes with SDC4-FL and SDC4-CTF, and blocks exosomal release, likely by blocking initial cleavage of endosomal SDC4-FL by MMPs and ADAM17, addressing SDC4 and syntenin to lysosomal degradation. SDC4-FL, syndecan-4 full length; SDC4-CTF, syndecan-4 C-terminal fragment; SDC4-EC, SDC4 extracellular part.
Discussion
Here, by gain- and loss-of-expression studies in MCF-7 cells, we show that TSPN6 negatively regulates the production of exosomes. Indeed, TSPN6-loss increases the number of released nanoparticles pelleting at 100,000 × g, while TSPN-gain decreases their number. These opposing effects on particle number are accompanied, respectively, by an increased or a decreased exosomal release of syntenin, SDC4-CTF, and CD63, three bona fide exosomal markers (16, 30, 31). The exosomal releases of CD9 and CD81, in contrast, are not affected. This identifies TSPN6 as a negative regulator of a specific subclass of exosomes (Fig. 1). TSPN6 directly binds syntenin. Yet, our further observations suggest direct TSPN6-syntenin binding is not mandatory for the effect of TSPN6 on syntenin (Fig. 3 A and G). In cells, TSPN6-syntenin interactions instead might depend on lateral TSPN6 interactions with SDC4 and, in turn, on SDC4 interacting with syntenin. Both TSPN6 and SDC4 featuring syntenin-compatible PDZ-binding motifs, and with TSPN6 binding preferentially to PDZ1 (22) and SDCs binding preferentially to PDZ2 (32) in the syntenin PDZ-tandem, TSPN6-SDC4 complexes would seem well suited for recruiting syntenin to endosomal membranes.
TSPN6 is the second tetraspanin shown to interact with syntenin in a PDZ-dependent manner. In 2006, a study by Latysheva et al. (20) first identified CD63. Yet, while both CD63 and TSPN6 interact directly with syntenin, their functional relationships in MCF-7 cells are quite different. Indeed, CD63 has no significant impact on the exosomal secretion of syntenin, or on syntenin intracellular levels (SI Appendix, Fig. S2 and ref. 16), while TSPN6 brings syntenin to lysosomal degradation (Fig. 2). TSPN6 expression (as inferred from mRNA levels) being far less abundant than CD63 (in MCF-7 cells), such effect on syntenin degradation seems to be specific rather than by default or gain of global TSPN mass. Thus, the fate of syntenin is clearly differentially regulated by cognate tetraspanins. We presume the negative effect of TSPN6 on exosome production is reflecting this syntenin degradation. TSPN6 would direct syntenin-loaded endosomal ILVs toward lysosomal degradation instead of exosomal secretion. By inference, to account for a decrease in the global cellular levels of syntenin, the rate of TSPN6-mediated syntenin lysosomal degradation must be more rapid than the rate at which syntenin is cleared from cells by exosomal secretion.
The observation that the lysosomal degradation of syntenin does not rely on the presence of the PDZ-binding motif of TSPN6 but specifically on the presence of SDC4, another protein engaging its PDZ domains (ref. 27 and Fig. 3 E and G), is truly surprising. Intriguingly, while SDC1 similarly engages the PDZ domains of syntenin and is also highly expressed in MCF-7 cells (SI Appendix, Fig. S3A), SDC1 has no impact on TSPN6-syntenin functional relationships (Fig. 3 E, G, and I). Consistent with this specific TSPN6-SDC4 functional interaction, in coimmunoprecipitation experiments, using detergents with different strengths, we observed a quite robust molecular association between TSPN6 and SDC4 (Fig. 3 B–D). An unexplored possibility is that the association between SDC4 and TSPN6 might depend on the GxxxG motif. Indeed, the transmembrane domain of all syndecans contains a GxxxG motif that promotes formation of sodium dodecyl sulfate (SDS)-resistant dimers (33) and TSPN6 has two (overlapping) GxxxG motifs in its fourth transmembrane domain. Yet the presence of such motif cannot explain the preferential association of TSPN6 with SDC4 (and not SDC1). Possibly, specific associations of the complex with lipids and/or differential states of TSPN and SDC oligomerization also play a role. Noteworthy, syntenin and SDC4-intracellular domain (ICD), but not SDC1-ICD interact with phosphatidylinositol 4,5-bisphosphate (34).
This work is establishing an intimate and specific crosstalk between members of the tetraspanin and syndecan families. This may have wider implications. On one hand, syndecans are well-known coreceptors for a plethora of growth factors and adhesion molecules (29, 35, 36). On the other hand, it is extensively documented that tetraspanins impact on the trafficking and activity of a plethora of signaling complexes and this property was so far mainly attributed to their ability to organize membrane webs. In this respect, with TSPN6 having a preference for SDC4 rather than for SDC1, it is interesting to consider that SDC4 and SDC1 core proteins directly associate with different growth factor receptors (29, 37, 38). In other words, specific SDC-TSPN networks might contribute to signaling specificity by engaging different molecular determinants and might also differentially control the dynamics of signaling, by impacting on the trafficking and degradation of signaling complexes. According to the high abundance of syndecans in cells and the extensive versatility of their binding properties [including those that engage their sugar moieties (39–41)], deciphering the functional impact of tetraspanin-syndecan crosstalk promises to be quite challenging, but also extremely interesting. Such is illustrated by the impact of TSPN6 on the fate of SDC4. At steady-state, cellular SDC4 exists in two main protein forms; i.e., full-length forms of SDC4 (SDC4-FL; observable in Western Blot as a discrete band of 35 kDa, but only after digestion of the glycosaminoglycan chains) and so-called C-terminal fragments (CTFs), of around 15 kDa (SI Appendix, Figs. S3C and S4D), corresponding to membrane-integrated remnant pieces of SDC4 produced by proteolytic cleavage. TSPN6 supports the lysosomal degradation of the latter (Fig. 4C), similar to its effect on syntenin (Fig. 2E). TSPN6 supports also the degradation of potentially SDC4-associated specific cargo, as illustrated by EGFR, which has been shown to specifically associate with SDC4 (37). It is tempting to speculate that after SDC4 proteolytic cleavage, SDC4-CTF:TSPN6:syntenin complexes and cognate cargos are processed for degradation (Fig. 6, Left). Intriguingly, in the absence of SDC4, almost half of the syntenin disappears from the lysate (Fig. 3E) and this is not accompanied by an increase of the exosomal pool of syntenin (16). Similar effects on cellular syntenin are not observed in the absence of SDC1. These observations await further investigations, but imply that syntenin might also be addressed to degradation by the lack of SDC4, independently of TSPN6.
A discrete SDC4-ectodomain fragment that represents the part complementary to the CTF (SI Appendix, Figs. S3C and S4E) is never observed in cell lysates (also not in this study where TSPN6 levels were modulated). Yet, such fragment (of around 25 kDa after heparan sulfate digestion) is easily observed after immunoprecipitation from the cell media, suggesting that the SDC4 proteolytic cleavage that is yielding a discrete part of the ectodomain primarily takes place at the plasma membrane or—if occurring in endosomes—is nearly immediately followed by the release (via plasma membrane recycling) or degradation of that fragment (SI Appendix, Fig. S3C). TSPN6 clearly has negative impact on such SDC4 cleavage and concomitant shedding (Fig. 5C), and the significant effect is lost in cells that are treated with TMI-1 and that by themselves show markedly enhanced levels of SDC4-FL (Fig. 5E). This suggests that TSPN6 segregates SDC4 away from MMPs and ADAM17, either by inhibiting these enzymes or by preventing SDC access to cellular compartments where these enzymes operate, including the cell surface and early endosomes. The major colocalization of (overexpressed) TSPN6 with (endogenous) SDC4 occurring in early endosomes is compatible with this contention (Fig. 5D). The interpretation of the data must also accommodate the impact of TSPN6 on the subcellular distribution of full-length SDC4. Both the loss and the gain of TSPN6 result in increased cellular levels of SDC4-FL (Fig. 5 A and B), and in diminished SDC4 plasma membrane localization (Fig. 5 D and G). RUSH experiments are not directly suggestive of a negative effect of TSPN6 on SDC4 secretory pathway (SI Appendix, Fig. S4F). Similar effects of gain- and loss-of-expression suggest that TSPN6 overexpression might limit/compete with important components and in fine function as loss-of-function or, more appealingly, that different endosomal pools of SDC4-FL might exist, differentially affected by TSPN6. In the presence of TSPN6, more SDC4-FL may transiently accumulate in early endosomes because of increased net endocytosis, from where it progresses to be rapidly degraded by lysosomal hydrolases. In the absence of TSPN6, more SDC4-FL may transiently accumulate in early endosomes, as it is cleared from there by discrete cleavage, recycling and shedding, and not by the more rapid lysosomal route. In that sense, TSPN6 could be qualified as a rheostat, setting the amplitude of the secretion of SDC4, of the SDC4 ectodomain as soluble fragment and of the SDC4-CTF as part of exosomes, while stabilizing the cell surface levels of this SDC, keeping these levels low. This relationship of TSPN6 with SDC4 proteolytic processing and the negative effect of TSPN6 on exosome formation would also seem consistent with the notion that SDC-mediated ILV budding and exosome formation require the endosomal processing of SDCs in CTFs (16). The precise nature of this relationship and the mechanisms and proteases involved remain to be identified.
Obviously, networks might be cell type and context dependent. Differences in wiring, supported by different relative abundance in tetraspanins, syndecans, cognate cargo, and syntenin might possibly explain the discrepancy between the present study and the work in 2017 of Guix et al. (22), the latter claiming a positive role for TSPN6 in exosome formation, but performed with other cells (HEK293) and with focus on overexpressed APP (amyloid precursor protein).
In conclusion, our study shows that tetraspanins should not solely be appreciated for their positive role on exosome formation or cargo composition as previously suspected (19, 42). Our work is suggestive of TSPN6 sustaining/stimulating the production of degradative rather than secretory late endosomes. How that occurs precisely remains to be determined. Complex membrane networks under the control of interconnected scaffolds composed of the tetraspanins, syndecans, syntenin, and possibly other PDZ proteins might regulate the production and the molecular composition of (a specific subpopulation of) ILVs and exosomes. Our observations highlight a dimension of complexity that questions our views of how intracellular but also extracellular signaling specificity can be achieved.
Materials and Methods
Further details are provided in SI Appendix, SI Materials and Methods.
Exosomes and Total Cell Lysates.
Preliminary note: In this study, we used the term exosomes for small extracellular vesicles pelleting at 100,000 × g and microvesicles for those pelleting at 10,000 × g. This is an approximation and we recommend readers that might be confused by methodology to read the MISEV guidelines 2014 and 2018 (43, 44). For comparative analyses, in gain- and loss-of-function studies, exosomes were collected from equivalent amounts of culture medium, conditioned by equivalent amounts of cells. After the required time of complementary DNA (cDNA) or RNA interference (RNAi) treatments, the cell layers were washed twice with phosphate-buffered saline (PBS) and refreshed with DMEM/F12 (Dulbecco’s modified eagle medium: nutrient mixture F-12) containing 10% exosome-depleted FCS (fetal calf serum). Cell conditioned media were collected 16 h later. Exosomes were isolated from these media by three sequential centrifugation steps at 4 °C: 10 min at 500 × g, to remove cells; 30 min at 10,000 × g, to remove microvesicles; and 3 h at 100,000 × g, to pellet exosomes, followed by one wash in PBS. For lysates, cells were scraped and were pelleted by centrifugation at 300 × g for 5 min at 4 °C and then mixed directly with 1× loading buffer (250 mM Tris⋅HCl pH 6.8, 25% glycerol, 10% SDS) or lysis buffer (Tris 30 mM pH 7.4, NaCl 150 mM supplemented with 1% detergent [NP-40 or Brij97] and protease inhibitor mixture dilution 1/1000 reference P8340-5ML from Sigma-Aldrich).
GFP-Trap.
MCF-7 cells overexpressing GFP-TSPN6 or GFP alone as control for 24 h or 48 h were resuspended in lysis buffer supplemented with 1% detergent (NP-40 or Brij97) for 30 min at 4 °C. Extracts were then centrifuged for 30 min at 10,000 g at 4 °C. Immunoprecipitation was performed for 1 h at 4 °C by incubating GFP-Trap_A beads (Chromtek) with the cellular extracts. After immunoprecipitation, the beads were washed three times in PBS. Proteins coimmunoprecipitated with GFP-TSPN6 were detected with corresponding antibodies by Western blot analysis.
Mass Spectrometry Analysis and Protein Quantification.
Proteins associated to GFP-TSPN6 versus GFP alone were analyzed using label-free liquid chromatography (LC) mass spectrometry (MS/MS) relative quantitation. Briefly, immunoprecipitated complexes were submitted to an in-gel trypsin digestion. Peptides were extracted and analyzed by LC-tandem MS/MS using an Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Thermo Electron) online with an Ultimate3000 RSLCnano chromatography system (Thermo Fisher Scientific). Protein identification and quantification were processed using the MaxQuant computational proteomics platform, version 1.6.3.4 using the human subset of the UniProt Knowledgebase (date 2018.09; 20394 entries) (45, 46). The iBAQ intensities, roughly proportional to the molar quantities of the proteins, were processed (47). The statistical analysis was done with Perseus program (version 1.6.1.3). Differential proteins were detected using a two-sample t test at 0.01 permutation-based false discovery rate. The mass spectrometry proteomics data, including search results, have been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org/) via the PRIDE (48) partner repository with the dataset identifier PXD014559.
Statistical analysis was performed using the standard two-tailed Student t test, and *P < 0.05, **P < 0.01, ***P < 0.001 were considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by grants from the French National Research Agency (ANR-18-CE13-0017, Project SynTEV), the Fund for Scientific Research–Flanders (Fonds Wetenschappelijk Onderzoek—Vlaanderen Grants G.0846.15 and G0C5718N), and the Institut National du Cancer (Projets Libres de Recherche “Biologie et Sciences du Cancer” INCa 9474). This project has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement No 747025. We thank Etienne Morel for his feedback on autophagy analysis. Proteomic analyses were done using the mass spectrometry facility of Marseille Proteomics (http://marseille-proteomique.univ-amu.fr/) supported by IBISA (Infrastructures Biologie Santé et Agronomie), Plateforme Technologique Aix-Marseille, the Cancéropôle Provence-Alpes-Côte d’Azur, Région Sud-Alpes-Côte d’Azur, the Institut Paoli-Calmettes, the Centre de Recherche en Cancérologie de Marseille, Fonds Européen de Developpement Régional, and Plan Cancer. We thank Emilie Baudelet for technical assistance in mass spectrometry analysis. The J.-P.B. lab is also funded by La Ligue Nationale Contre le Cancer (Label Ligue J.-P.B.). J.-P.B. is a scholar of Institut Universitaire de France.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The mass spectrometry proteomics data, including search results, have been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org/) via the PRIDE partner repository with the dataset identifier PXD014559.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922447117/-/DCSupplemental.
References
- 1.Colombo M., Raposo G., Théry C., Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30, 255–289 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Yáñez-Mó M., et al. , Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 4, 27066 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maas S. L. N., Breakefield X. O., Weaver A. M., Extracellular vesicles: Unique intercellular delivery vehicles. Trends Cell Biol. 27, 172–188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bobrie A., et al. , Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 72, 4920–4930 (2012). [DOI] [PubMed] [Google Scholar]
- 5.van Niel G., D’Angelo G., Raposo G., Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19, 213–228 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Mathieu M., Martin-Jaular L., Lavieu G., Théry C., Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 21, 9–17 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Polo S., Di Fiore P. P., Sigismund S., Keeping EGFR signaling in check: Ubiquitin is the guardian. Cell Cycle 13, 681–682 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh B., Coffey R. J., Trafficking of epidermal growth factor receptor ligands in polarized epithelial cells. Annu. Rev. Physiol. 76, 275–300 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henne W. M., Buchkovich N. J., Emr S. D., The ESCRT pathway. Dev. Cell 21, 77–91 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Hurley J. H., Hanson P. I., Membrane budding and scission by the ESCRT machinery: It’s all in the neck. Nat. Rev. Mol. Cell Biol. 11, 556–566 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanson P. I., Cashikar A., Multivesicular body morphogenesis. Annu. Rev. Cell Dev. Biol. 28, 337–362 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Raiborg C., Stenmark H., The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458, 445–452 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Hurley J. H., Boura E., Carlson L.-A., Różycki B., Membrane budding. Cell 143, 875–887 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurley J. H., ESCRTs are everywhere. EMBO J. 34, 2398–2407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trajkovic K., et al. , Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244–1247 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Baietti M. F., et al. , Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 14, 677–685 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Imjeti N. S., et al. , Syntenin mediates SRC function in exosomal cell-to-cell communication. Proc. Natl. Acad. Sci. U.S.A. 114, 12495–12500 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charrin S., Jouannet S., Boucheix C., Rubinstein E., Tetraspanins at a glance. J. Cell Sci. 127, 3641–3648 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Andreu Z., Yáñez-Mó M., Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 5, 442 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Latysheva N., et al. , Syntenin-1 is a new component of tetraspanin-enriched microdomains: Mechanisms and consequences of the interaction of syntenin-1 with CD63. Mol. Cell. Biol. 26, 7707–7718 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roucourt B., Meeussen S., Bao J., Zimmermann P., David G., Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. 25, 412–428 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guix F. X., et al. , Tetraspanin 6: A pivotal protein of the multiple vesicular body determining exosome release and lysosomal degradation of amyloid precursor protein fragments. Mol. Neurodegener. 12, 25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Razi M., Futter C. E., Distinct roles for Tsg101 and Hrs in multivesicular body formation and inward vesiculation. Mol. Biol. Cell 17, 3469–3483 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klijn C., et al. , A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 33, 306–312 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Takino T., et al. , Tetraspanin CD63 promotes targeting and lysosomal proteolysis of membrane-type 1 matrix metalloproteinase. Biochem. Biophys. Res. Commun. 304, 160–166 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Talukdar S., et al. , MDA-9/Syntenin regulates protective autophagy in anoikis-resistant glioma stem cells. Proc. Natl. Acad. Sci. U.S.A. 115, 5768–5773 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grootjans J. J., et al. , Syntenin, a PDZ protein that binds syndecan cytoplasmic domains. Proc. Natl. Acad. Sci. U.S.A. 94, 13683–13688 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boucheix C., Rubinstein E., Tetraspanins. Cell. Mol. Life Sci. 58, 1189–1205 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H., Jin H., Beauvais D. M., Rapraeger A. C., Cytoplasmic domain interactions of syndecan-1 and syndecan-4 with α6β4 integrin mediate human epidermal growth factor receptor (HER1 and HER2)-dependent motility and survival. J. Biol. Chem. 289, 30318–30332 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kowal J., Tkach M., Théry C., Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 29, 116–125 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Kowal J., et al. , Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. U.S.A. 113, E968–E977 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grootjans J. J., Reekmans G., Ceulemans H., David G., Syntenin-syndecan binding requires syndecan-synteny and the co-operation of both PDZ domains of syntenin. J. Biol. Chem. 275, 19933–19941 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Dews I. C., Mackenzie K. R., Transmembrane domains of the syndecan family of growth factor coreceptors display a hierarchy of homotypic and heterotypic interactions. Proc. Natl. Acad. Sci. U.S.A. 104, 20782–20787 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh E. S., Woods A., Lim S. T., Theibert A. W., Couchman J. R., Syndecan-4 proteoglycan cytoplasmic domain and phosphatidylinositol 4,5-bisphosphate coordinately regulate protein kinase C activity. J. Biol. Chem. 273, 10624–10629 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Couchman J. R., Syndecans: Proteoglycan regulators of cell-surface microdomains? Nat. Rev. Mol. Cell Biol. 4, 926–937 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Sarrazin S., Lamanna W. C., Esko J. D., Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 3, a004952 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H., Jin H., Rapraeger A. C., Syndecan-1 and syndecan-4 capture epidermal growth factor receptor family members and the α3β1 integrin via binding sites in their ectodomains: Novel synstatins prevent kinase capture and inhibit α6β4-INTEGRIN-dependent epithelial cell motility. J. Biol. Chem. 290, 26103–26113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H., Leavitt L., Ramaswamy R., Rapraeger A. C., Interaction of syndecan and alpha6beta4 integrin cytoplasmic domains: Regulation of ErbB2-mediated integrin activation. J. Biol. Chem. 285, 13569–13579 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christianson H. C., Belting M., Heparan sulfate proteoglycan as a cell-surface endocytosis receptor. Matrix Biol. 35, 51–55 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Friand V., David G., Zimmermann P., - and syndecan in the biogenesis of exosomes. Biol. Cell 107, 331–341 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Lambaerts K., Wilcox-Adelman S. A., Zimmermann P., The signaling mechanisms of syndecan heparan sulfate proteoglycans. Curr. Opin. Cell Biol. 21, 662–669 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Y., et al. , Tetraspanin CD9 stabilizes gp130 by preventing its ubiquitin-dependent lysosomal degradation to promote STAT3 activation in glioma stem cells. Cell Death Differ. 24, 167–180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lötvall J., et al. , Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for extracellular vesicles. J. Extracell. Vesicles 3, 26913 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Théry C., et al. , Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for extracellular vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7, 1535750 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cox J., et al. , Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox J., Mann M., MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Schwanhäusser B., et al. , Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Vizcaíno J. A., et al. , ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.