Significance
ESCC is a deadly disease with few prevention or treatment options. In a dietary Zn deficiency-promoted rat ESCC model with miR-31–controlled inflammation pathway up-regulation, systemic antimiR-31 reduces miR-31 level and inflammation, suppressing ESCC development. We identified Egln3 as a direct miR-31 target and developed a constitutive miR-31 knockout rat model. Interactions of oncogenic miR-31, EGLN3 down-regulation, and inflammation occur in rat and human ESCC tissue. Since miR-31 genetic knockout prevents Zn deficiency-associated ESCC by eliminating or greatly reducing the entire miR-31-EGLN3/STK40-NF-κB–controlled inflammatory process, we conclude that miR-31 and downstream signaling proteins should be tested as prognostic and therapeutic targets and inexpensive Zn supplementation should be implemented for ESCC prevention in deficient populations.
Keywords: zinc deficiency, esophageal squamous cell carcinoma, esophageal cancer rat model, constitutive miR-31 knockout rat, in vivo antimiR-31 delivery
Abstract
MicroRNA-31 (miR-31) is overexpressed in esophageal squamous cell carcinoma (ESCC), a deadly disease associated with dietary Zn deficiency and inflammation. In a Zn deficiency-promoted rat ESCC model with miR-31 up-regulation, cancer-associated inflammation, and a high ESCC burden following N-nitrosomethylbenzylamine (NMBA) exposure, systemic antimiR-31 delivery reduced ESCC incidence from 85 to 45% (P = 0.038) and miR-31 gene knockout abrogated development of ESCC (P = 1 × 10−6). Transcriptomics, genome sequencing, and metabolomics analyses in these Zn-deficient rats revealed the molecular basis of ESCC abrogation by miR-31 knockout. Our identification of EGLN3, a known negative regulator of nuclear factor κB (NF-κB), as a direct target of miR-31 establishes a functional link between oncomiR-31, tumor suppressor target EGLN3, and up-regulated NF-κB–controlled inflammation signaling. Interaction among oncogenic miR-31, EGLN3 down-regulation, and inflammation was also documented in human ESCCs. miR-31 deletion resulted in suppression of miR-31–associated EGLN3/NF-κB–controlled inflammatory pathways. ESCC-free, Zn-deficient miR-31−/− rat esophagus displayed no genome instability and limited metabolic activity changes vs. the pronounced mutational burden and ESCC-associated metabolic changes of Zn-deficient wild-type rats. These results provide conclusive evidence that miR-31 expression is necessary for ESCC development.
Despite recent advances in surgery, radiation, and chemotherapy, esophageal squamous cell carcinoma (ESCC) remains a deadly disease with only 10% of patients surviving for 5 y (1). Thus, a critical need exists to elucidate further the molecular pathogenesis of ESCC that could in turn provide the development of new therapeutic strategies.
Alcohol and tobacco use, nutritional deficiencies, and exposure to environmental carcinogens, including N-nitrosomethylbenzylamine (NMBA) (2), are among the known risk factors for ESCC. In many populations (3–5), and in individuals with chronic alcohol consumption (6), dietary zinc (Zn) deficiency is implicated in the etiology of ESCC. Abnet et al. (5) showed that high vs. low tissue Zn concentration is associated with a reduced risk of developing ESCC, providing evidence in humans of an association between Zn deficiency and ESCC. We have developed a Zn-deficient (ZD) rat esophageal cancer model that reproduces the Zn-deficiency and inflammation features of human ESCC (7) and is highly sensitive to esophageal tumorigenesis induced by NMBA (8). Rats on a ZD diet for 5 wk develop hyperplastic esophagi with a distinct gene signature that includes up-regulation of the proinflammation mediators S100a8/a9 (9) and overexpression of the oncogenic microRNA-31 (miR-31) (10). Prolonged Zn deficiency leads to an expanded cancer-associated inflammatory program and an oncogenic miRNA signature, with miR-31 as the top up-regulated species that fuels ESCC development (8, 11).
miRNAs are commonly dysregulated in human cancers and miR-31 is among the most frequently altered of the miRNAs in cancers (12). Depending on tumor type, miR-31 can exhibit oncogenic or tumor suppressive roles. miR-31 is overexpressed and oncogenic in colorectal cancer (13) and squamous cell carcinomas of the esophagus (14), tongue (15), head and neck (16), and skin (17), but the mechanisms whereby miR-31 up-regulation promotes development of these cancers remain unclear.
Locked nucleic acid (LNA)-modified RNA oligonucleotides exhibit high biostability, low toxicity, and adequate biodistribution in vivo (18). For example, in vivo LNA-antimiR-122 was shown to mediate effective inhibition of liver-expressed miR-122 function in mice and nonhuman primates (19). Using LNA-antimiR-31, we previously demonstrated in a ZD rat esophageal preneoplasia model overexpressing miR-31 that silencing miR-31 in vivo inhibits the development of esophageal preneoplasia by repressing the miR-31–associated STK40-NF-κB (nuclear factor κB)–based inflammation signaling (10). Since the end point was the preneoplastic inflammatory esophageal phenotype, we questioned whether miR-31 silencing could suppress progression to ESCC in NMBA-treated Zn-deficient rats.
Using a Zn deficiency-promoted rat ESCC model with prominent miR-31 overexpression (11) and a cancer-associated inflammatory gene signature (8), we investigated the effects of LNA-mediated antimiR-31 delivery, or constitutive knockout of miR-31, on NMBA-induced ESCC development in ZD rats. We found that in vivo LNA-antimiR-31 delivery moderately suppressed ESCC development, whereas genetic miR-31 knockout completely protected ZD:miR-31−/− rats from ESCC development. In parallel, we examined the molecular consequences of miR-31 silencing in ESCC suppression by analyzing gene expression and metabolomics profiles. Because genome instability is an important “hallmark” of cancer development, we were also interested, during this “omics” study, to include whole-genome sequencing (WGS) of esophageal tissue samples from NMBA-treated rat cohorts: ESCC-bearing Zn-deficient wild-type (WT) rats, Zn-sufficient (ZS) WT rats with undetectable tumors, and tumor-free ZD:miR-31−/− rats.
Results
AntimiR-31 Delivery Inhibits ESCC Development in Zn-Deficient Rats.
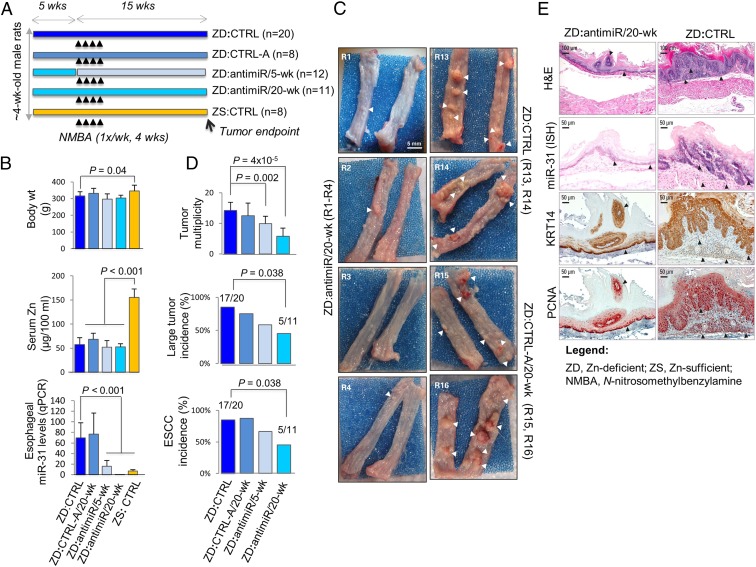
To determine if in vivo antimiR-31 delivery silences miR-31 and suppresses the development of ESCC, we performed a 20-wk anticancer study in the ZD rat ESCC model with miR-31 up-regulation (11) and a cancer-associated inflammatory gene signature (8). We used an LNA-mediated rno-miR-31a-5p inhibitor probe (designated antimiR) to inhibit esophageal miR-31 expression and an LNA-negative-control-A probe (designated CTRL-A) as a control oligo (10). As depicted in Fig. 1A (see treatment details in Materials and Methods), 4-wk-old rats were randomized into four Zn-deficient cohorts (ZD:antimiR/20-wk, ZD:antimiR/5-wk, ZD:CTRL-A, ZD:CTRL) and one Zn-sufficient group (ZS:CTRL). ZD:antimiR/20-wk and ZD:CTRL-A/20-wk rats received 30 i.v. doses of antimiR or CTRL-A over the 20-wk study, ZD:antimiR/5-wk rats received 10 doses over the first 5 wk, and ZD:CTRL and ZS:CTRL rats were untreated. At week 5, the animals received intragastric NMBA doses once a week for 4 wk and were killed 15 wk post first NMBA dose or 48 h after the final oligo delivery. At tumor end point, ZD cohorts displayed similar body weights, indicating the LNA-mediated oligo treatment was well-tolerated (Fig. 1B). Serum Zn level was similar in all four ZD cohorts (∼55 μg/100 mL) but significantly lower than in ZS rats (P < 0.001) (Fig. 1B). As expected (10), ZD:CTRL and ZD:CTRL-A rats were comparable in all parameters measured (Fig. 1 B–D). Thus, ZD:CTRL rats served as a control in the analysis.
Fig. 1.
Systemic delivery of locked nucleic acid-mediated antimiR-31 suppresses esophageal carcinogenesis. (A) Study design. (B) Body weights, serum Zn levels, and esophageal miR-31 levels (qPCR) (rat snoRNA as normalizer; Tukey-HSD post hoc unpaired t test for multiple comparisons; error bars represent SD; n = 8 rats per group). (C) Macroscopic view of whole esophagus. Representative photos of ZD:CTRL (R13, R14) and ZD:CTRL-A/20-wk (R15, R16) with multiple large/sessile esophageal tumors vs. ZD:antimiR/20-wk (R1 to R4) esophagi with small/isolated tumors (arrowheads). (D) Tumor multiplicity (number of tumors per esophagus; error bars represent mean ± SD) (two-tailed Welch t test; n = 8 to 20 rats per cohort); large tumor (size > 2 mm) and ESCC incidence (%) (two-tailed Fisher’s exact test; n = 8 to 20 rats per cohort). (E) Histopathologic changes (arrowheads highlight the most relevant histological findings) in ZD esophagus after antimiR-31 treatment. Representative photos of ZD:antimiR/20-wk vs. ZD:CTRL esophagus showing H&E staining, IHC staining for KRT14 (brown, 3,3′-diaminobenzidine tetrahydrochloride; DAB), and PCNA (red, 3-amino-9-ethylcarbazole substrate chromogen; AEC) and ISH localization of miR-31 (blue, BCIP/NBT; counterstain, nuclear fast red) (n = 8 rats per cohort).
As in previous studies (11), qPCR analysis showed that dietary Zn deficiency prominently up-regulated miR-31 expression in the esophagus (ZD:CTRL vs. ZS:CTRL, P < 0.001) (Fig. 1B). AntimiR treatment (20 wk) resulted in ∼90% reduction of esophageal miR-31 levels in ZD:antimiR/20-wk rats vs. ZD:CTRL (P < 0.001) (Fig. 1B). At the cellular level, miR-31 analysis by in situ hybridization (ISH) in formalin-fixed, paraffin-embedded (FFPE) esophageal tissues revealed marked reduction in intensity/extent of miR-31 signal in ZD:antimiR/20-wk vs. ZD:CTRL cohort (Fig. 1E; n = 10 rats per cohort), validating the qPCR data. Consistent with the long-lasting effect of antimiR (19), ZD:antimiR/5-wk esophagus, assessed 15 wk after antimiR dosing, showed an ∼70% decrease in miR-31 levels vs. ZD:CTRL tissues (P < 0.001) (Fig. 1B).
Independent of antimiR treatment, ZD cohorts had 100% overall esophageal tumor incidence vs. ZS:CTRL rats with 25% incidence. Macroscopically (Fig. 1C; n = 8 to 20 rats per group), ZD:antimiR/20-wk esophagus (rats R1 to R4) typically had smaller and fewer tumors vs. ZD:CTRL (R13, R14) or ZD:CTRL-A/20-wk (R15, R16) esophagus showing multiple large/sessile tumors. Thus, antimiR delivery (20 wk) led to a large decrease in tumor multiplicity (5.7 vs. 14, P = 4 × 10−5), in large tumor incidence (45 vs. 85%, P = 0.038; size >2 mm), and in ESCC incidence (45 vs. 85%, P = 0.038) (Fig. 1D), as assessed by histological examination of hematoxylin & eosin (H&E)-stained and KRT14-immunostained sections (Fig. 1E). Thus, despite sustained dietary ZD, prolonged antimiR-31 delivery (20 wk) significantly reduces ESCC incidence by 40%. In contrast, a short-term antimiR delivery (5 wk) led to a modest reduction in tumor multiplicity vs. ZD:CTRL rats (9.9 vs. 14, P = 0.002) (Fig. 1D).
miR-31 Knockout Abrogates ESCC Development in ZD:miR-31−/− Rats.
To understand the molecular interaction between miR-31 and dietary Zn deficiency in ESCC development, we generated a constitutive miR-31 knockout (KO) rat (Materials and Methods) with deletion of a 466-bp DNA fragment containing miR-31 in one allele by CRISPR/Cas9 technology (20, 21) in the same Sprague–Dawley rat strain as our previous Zn deficiency-associated esophageal tumor studies. miR-31–null rats were generated by breeding and selection of rats carrying two KO alleles. Because off-target effects are a concern of gene silencing technology, 10 potential off-targets for the upstream single-guide (sg)RNA and 10 sites for the downstream sgRNA were searched. Cas9-mediated mutations were not detected among those 20 potential off-target sites. The production of homozygous miR-31 KO rat pups in normal sex ratios showed that miR-31 deletion is well-tolerated in the rat and miR-31−/− rats exhibit no discernible abnormal phenotypes.
First, we assessed the effect of dietary Zn deficiency alone on esophageal cell proliferation and inflammation in miR-31−/− rats (SI Appendix, Fig. S1). Zn deficiency (20 wk) induced pronounced miR-31 up-regulation and a highly proliferative and inflammatory esophageal phenotype in WT ZD:miR-31+/+ vs. ZS:miR-31+/+ rats, with overexpression of the inflammation markers COX-2 and S100A8/S100A9 and the transcription factor NF-κB p65. In contrast, deficient ZD:miR-31−/− rats displayed a noninflammatory and nonproliferative esophageal phenotype similar to the ZS:miR-31−/− counterpart (SI Appendix, Fig. S1).
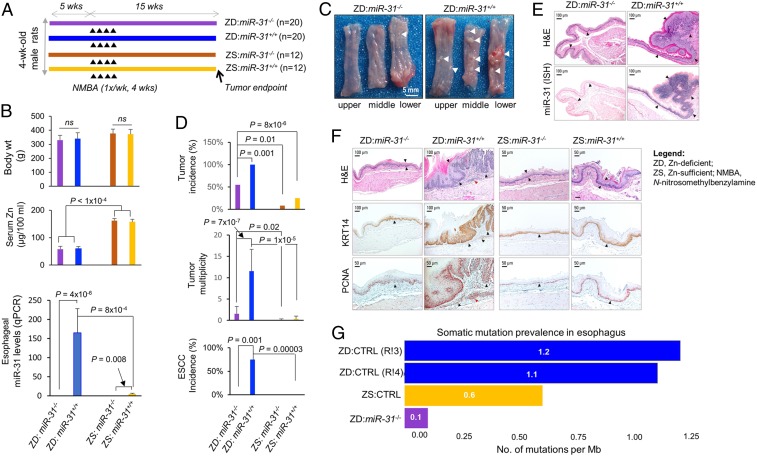
Next, we investigated the consequences of miR-31 genetic knockout on ESCC development by performing a 20-wk esophageal tumor bioassay by NMBA, as for in vivo antimiR-31 delivery. As shown in Fig. 2A, 4-wk-old miR-31 KO and WT littermates were fed a ZD or ZS diet to form four cohorts: ZD:miR-31−/−, ZD:miR-31+/+, ZS:miR-31−/−, and ZS:miR-31+/+. At week 5, the rats received intragastric NMBA doses, once a week for 4 wk. The study was concluded at 15 wk post first NMBA dose. At tumor end point, regardless of genotype, Zn-deficient cohorts had comparable body weights and lower serum Zn levels vs. the Zn-sufficient cohort (P < 1 × 10−4) (Fig. 2B). qPCR analysis showed that esophageal miR-31 expression was barely detectable in KO cohorts (0.12 ± 0.17 and 0.06 ± 0.06 in ZD:miR-31−/− and ZS:miR-31−/−). Conversely, WT cohorts showed the expected prominent miR-31 expression in ZD:miR-31+/+ vs. ZS:miR-31+/+ rats (164 ± 63 vs. 4.8 ± 1.6) (Fig. 1B). At the cellular level, in situ miR-31 analysis in FFPE esophageal tissues revealed no miR-31 signal in ZD:miR-31−/− or ZS:miR-31−/− (Fig. 3D) esophagus compared with abundant/intense miR-31 signal in esophageal mucosa and tumor in ZD:miR-31+/+ (Fig. 2E) (Fig. 3D, ZD:CTRL). Therefore, the qPCR and ISH data confirm that miR-31 is knocked out in miR-31−/− rats.
Fig. 2.
Genetic miR-31 knockout completely prevents ESCC development. (A) Study design. (B) Body weights, serum Zn levels, and qPCR analysis of esophageal miR-31 levels (snoRNA as normalizer; error bars represent mean ± SD; Welch’s t test; n = 12 rats per cohort; ns, not significant). (C) Macroscopic view of whole esophagus. Representative photos of ZD:miR-31−/− vs. ZD:miR-31+/+ esophagus (arrowheads indicate esophageal tumors). (D) Overall tumor incidence and ESCC incidence (%) (Fisher’s exact test) and tumor multiplicity (number of tumors per esophagus; error bars represent mean ± SD) (Welch t test). Statistical tests were two-sided (n = 20 rats per ZD cohort). (E) miR-31 localization in esophagus. Representative photos of ZD:miR-31−/− vs. ZD:miR-31+/+ tissues (miR-31 ISH signal; blue, BCIP/NBT) (n = 20 rats per cohort). Arrowheads indicate esophageal epithelium. (F) Histopathologic changes in miR-31−/− rat esophagus (arrowheads emphasize the difference in epithelia response to dietary ZD in miR-31−/− vs. miR-31+/+ esophagus and the similarity in the epithelia response in the ZS:miR-31−/− vs. ZS:miR-31+/+ esophagus). Representative photos of ZD:miR-31−/− vs. ZD:miR-31+/+ and ZS:miR-31−/− vs. ZS:miR-31+/+ esophagus showing histology (H&E), IHC staining for KRT14 (brown, DAB), and PCNA (red, AEC) (n = 20 rats per cohort). (G) Prevalence of somatic mutations in esophageal epithelia in ZD:CTRL (WT), ZS:CTRL (WT), and ZD:miR-31−/− rats with divergent esophageal tumor outcome. ZD:CTRL (R13) and ZD:CTRL (R14) esophagus, respectively, had high and low tumor burden; ZS:CTRL had no observable tumors, and nonproliferative ZD:miR-31−/− esophagus was tumor-free.
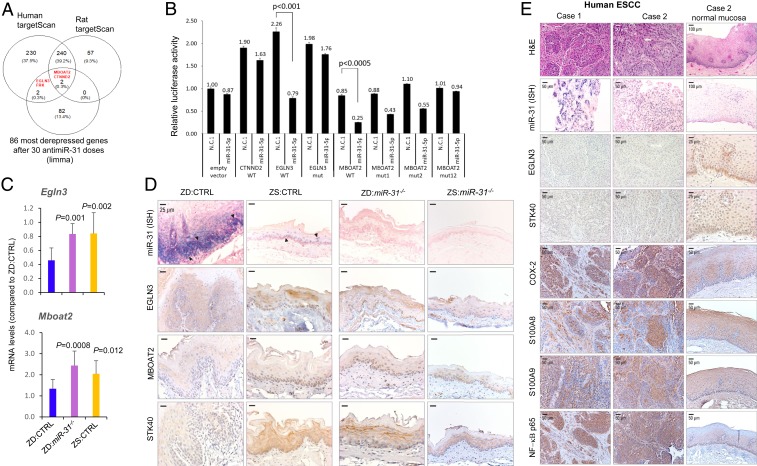
Fig. 3.
miR-31-5p targets EGLN3 and MBOAT2 expression. (A) Venn diagram showing the intersection between 86 genes that were the most derepressed after antimiR-31 treatment and predicted miR-31 target genes from TargetScan human/rat software. (B) Transfection experiments were performed in HEK-293FT cells using constructs for 3 predicted miR-31 targets, MBOAT2, EGLN3, and CTNND2. Renilla luciferase activity was normalized to firefly luciferase activity. The normalized luciferase activity in 293FT cells transfected with empty psiCHECK2 vector and scrambled negative control 1 (premiR-N.C.1) and relative luciferase activities of 293FT cells transfected with all other combinations of psiCHECK2 constructs and premiR oligos are shown. Experiments were repeated thrice in duplicate and data are presented as mean ± SD. Luciferase reporter assays indicate direct interactions between miR-31-5p and MBOAT2 (P < 0.005) and between miR-31-5p and EGLN3 (P < 0.001), two-tailed Student’s t test. (C) mRNA expression of Egln3 and Mboat2 in esophageal mucosa-derived RNA from ZD:CTRL, ZS:CTRL, and ZD:miR-31−/− rats (Psmb6 as normalizer; error bars represent mean ± SD; n = 7 to 11 rats). P values represent the significant difference between each individual group and ZD:CTRL rats; two-tailed, one-way ANOVA and Tukey post hoc t tests. (D) Protein expression of miR-31 targets EGLN3, MBOAT2, and STK40 (arrowheads emphasize the abundant/intense miR-31 overexpression in ZD:CTRL vs. ZS:CTRL esophagus). Representative photos of ZD:CTRL, ZS:CTRL, ZD:miR-31−/−, and ZS:miR-31−/− esophagus showing miR-31 ISH signal (blue, NBT/BCIP) (EGLN3, MBOAT2, and STK40 protein expression; brown, DAB; n = 10 rats per cohort). (E) Connection between miR-31 expression, protein expression of EGLN3 and STK40, and inflammation markers in human ESCC tissue. Representative photos of human ESCC tissue (FFPE sections): Case 1, case 2, and normal esophageal mucosa in case 2 showing miR-31 ISH signal (blue, NBT/BCIP) (EGLN3, STK40, COX-2, S100A8, S100A9, and NF-κB p65 protein expression; brown, DAB; n = 10 patient cases).
Macroscopically (Fig. 2C), we found small/isolated esophageal tumors (∼0.5 mm) in a few ZD:miR-31−/− rats compared with the omnipresent large/sessile tumors (>2 mm) in ZD:miR-31+/+ esophagus. Thus, ZD:miR-31−/− rats had significantly lower esophageal tumor incidence/multiplicity than ZD:miR-31+/+ rats (tumor incidence, 55 vs. 100%, P = 0.007; multiplicity, 1.6 ± 1.7 vs. 11.6 ± 5.1, P = 7 × 10−7) (Fig. 2D); 0% (0/20) of ZD:miR-31−/− rats developed ESCC compared with 75% (15/20) of ZD:miR-31+/+ rats (P = 0.00003). While ZD:miR-31−/− esophageal epithelia were mostly thin (Fig. 2F; H&E), with an occasional hyperplastic hyperkeratotic focus featuring basal cell proliferation (Fig. 2F; proliferating cell nuclear antigen [PCNA]), ESCC-bearing ZD:miR-31+/+ esophagus regularly showed uncontrolled cellular proliferation and inflammatory cell infiltrates in the stroma (Fig. 2 E and F). The data showed that Zn deficiency is unable to promote ESCC in ZD:miR-31−/− rats, providing conclusive genetic evidence that miR-31 is necessary for ESCC development.
Expression of Cancer-Related Inflammation Genes in miR-31–Silenced Esophagus.
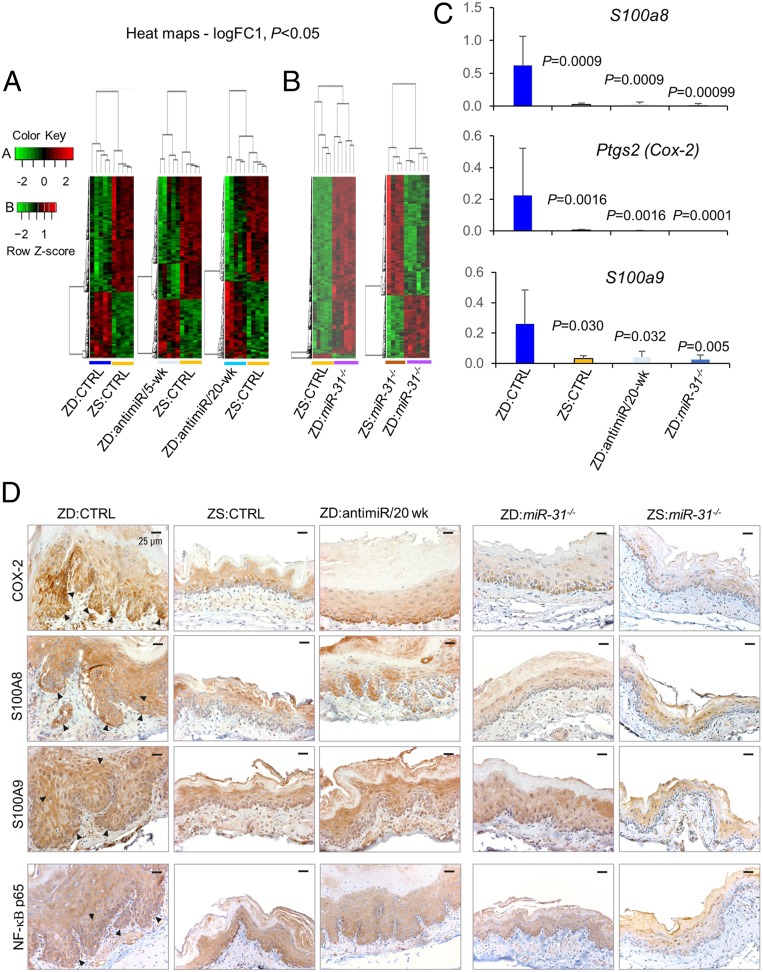
To identify genes critical in ESCC suppression after miR-31 knockdown or knockout, we performed comparative transcriptomics profiles of esophageal mucosa-derived RNA from six cohorts, ZD:CTRL, ZS:CTRL, ZD:antimiR/5-wk, ZD:antimiR/20-wk, ZD:miR-31−/−, and ZS:miR-31−/−, using the Affymetrix Rat 230 2.0 Genome GeneChip (n = 4 to 6 rats per cohort). Hierarchical clustering of messenger (m)RNA profiles using a cutoff of P < 0.05 and logFC (fold change) of 1 revealed that the ZD esophagi had distinct gene expression patterns compared with ZS:CTRL esophagus (Fig. 4 A and B). ZD:CTRL esophagus showed an inflammatory signature with prominent up-regulation of six cancer-related inflammation genes: S100a8, Ptgs2, S100a9, Ccl2, Cxcl14, and Cxcl1 (SI Appendix, Table S1 and Dataset S1). None of the six up-regulated inflammation genes was differentially expressed in ZD:antimiR/20-wk vs. ZS:CTRL (SI Appendix, Table S1 and Dataset S2), ZD:miR-31−/− vs. ZS:CTRL esophagus (SI Appendix, Table S1 and Dataset S4), or ZD:miR-31−/− vs. ZS:miR-31−/− esophagus (Dataset S3), although three inflammation genes (S100a8, Cxcl14, and Cxcl1) were moderately up-regulated in ZD:antimiR/5-wk esophagus (SI Appendix, Table S1). Additionally, the expression profile of ZD:miR-31−/− esophagus revealed prominent down-regulation of Ptgs2, S100a9, and S100a8 compared with ZD:CTRL esophagus (ranging from logFC −3.8 to −2.2) (Dataset S5). These gene expression profiles showed that the Zn deficiency-induced up-regulation of key inflammation genes was reversed by in vivo miR-31 silencing or by genetic miR-31 knockout. qPCR data (Fig. 4C) confirmed that these three genes that were markedly up-regulated by Zn deficiency were strikingly restored to normal ZS:CTRL levels in ZD:antimiR/20-wk and ZD:miR-31−/− esophagus. At the protein level, immunohistochemistry (IHC) analysis (Fig. 4D) showed strong/abundant cytoplasmic staining for all three inflammation markers COX2, S100A8, and S100A9 and their transcription factor NF-κB p65 (22, 23) in cancerous ZD:CTRL vs. ZS:CTRL esophagus. Expression of the same inflammation markers and NF-κB p65, a key mediator of inflammation (24), was reduced in ZD:antimiR/20-wk esophagus and became weak/diffuse in ZD:miR-31−/−, as well as in ZS:miR-31−/− esophagus, both featuring a noninflammatory and nonproliferative esophageal phenotype.
Fig. 4.
In vivo miR-31 silencing abolishes overexpression of cancer-related inflammation genes in esophagus. (A) Heatmaps (logFC 1, P < 0.05) showing distinct expression profiles of ZD:CTRL vs. ZS:CTRL, ZD:antimiR/5-wk vs. ZS:CTRL, and ZD:antimiR/20-wk vs. ZS:CTRL. (B) Heatmaps (logFC 1, P < 0.05) showing distinct expression profiles of ZD:miR-31−/− vs. ZS:CTRL and ZD:miR-31−/− vs. ZS:miR-31−/−. (C) mRNA expression (qPCR) of S100a8, Ptgs2, and S100a9 (top up-regulated inflammation genes; SI Appendix, Table S1) in ZD:CTRL, ZS:CTRL, ZD:antimiR/20-wk, and ZD:miR-31−/− esophagus (error bars represent mean ± SD; n = 7 to 11 rats per cohort; P values represent the significant difference between each individual group and the ZD:CTRL group; two-tailed, one-way ANOVA and Tukey post hoc t tests). (D) Protein expression (IHC) of COX-2, S100A8, S100A9, and NF-κB p65 (arrowheads emphasize the overexpression of the four inflammation markers in ZD:CTRL esophagus; brown, DAB; n = 10 rats per cohort). Representative photos of ZD:CTRL, ZS:CTRL, ZD:antimiR/20-wk, ZD:miR-31−/−, and ZS:miR-31−/− esophagus.
Egln3 and Mboat2 Are Tumor Suppressor Targets of miR-31 in ESCC.
We previously identified STK40 as a direct miR-31 target in esophageal preneoplasia (10). To identify physiological miR-31 targets in ESCC, we performed genome-wide mRNA expression profiling and analyzed genes with derepressed transcript levels in response to in vivo antimiR-31 delivery (20 wk). Among the 86 most derepressed mRNAs in ZD:antimiR/20-wk vs. ZD:CTRL esophagus (Fig. 4A, first and third heatmaps), the human/rat TargetScan tool predicts that miR-31-5p potentially targets MBOAT2, CTNND2, FRK, and EGLN3 (Fig. 3A). To determine if miR-31-5p is a negative regulator of MBOAT2, CTNND2, and EGLN3 genes, we performed luciferase assays. As shown in Fig. 3B, HEK-293FT cells were cotransfected with a vector expressing either the WT or the mutated versions of the 3′ untranslated regions (UTRs) of the MBOAT2, CTNND2, and EGLN3 genes, and either premiR-31-5p or scrambled negative control 1. Luciferase reporter assays (Fig. 3B) indicated direct interactions between miR-31-5p and both MBOAT2 (P < 0.0005) and EGLN3 (P < 0.001), confirming that they are direct miR-31 targets.
The comparative transcriptomics data revealed that genetic miR-31 knockout led to a marked up-regulation of all three miR-31 tumor suppressor targets Egln3 (logFC 1.04, P = 0.00059), Mboat2 (logFC 1.18, P = 1.51E-5), as well as Stk40 (logFC 1.38, P = 1. 51E-11) in ZD:miR-31−/− vs. ZD:CTRL esophagus (Dataset S5). This result was borne out in our CDF (cumulative distribution function) plot of Egln3, Stk40, and Mboat2 in ZD:CTRL (WT), ZS:CTRL (WT), and ZD:miR-31−/− rat esophagus showing that the expression of Egln3, Stk40, and Mboat2 was down-regulated in miR-31–overexpressing ZD:CTRL esophagus vs. the ZS:CTRL counterpart but was up-regulated in ZD:miR-31−/− esophagus compared with ZD:CTRL esophagus (SI Appendix, Fig. S2). Additionally, qPCR analysis confirmed that Egln3 (P = 0.001) and Mboat2 (P = 0.0008) levels were significantly higher in ZD:miR-31−/− vs. ZD:CTRL esophagus (Fig. 3C), validating the microarray result. Using ISH for miR-31 detection and IHC for protein expression analysis (Fig. 3D), ZD:miR-31−/− esophagus had moderate cytoplasmic immunostaining of EGLN3, MBOAT2, and STK40 protein in basal and suprabasal cells, whereas ZD:CTRL esophagus with intense/abundant miR-31 signal displayed weak/absent expression of the same three proteins compared with ZD:miR-31−/− and ZS:CTRL esophagus, validating EGLN3 and MBOAT2 as miR-31 direct targets at both the mRNA and protein levels.
EGLN3 (also known as PHD3) belongs to the Caenorhabditis elegans gene egl-9 (EGLN) family of prolyl hydroxylases. EGLN3 is down-regulated in colorectal cancer cells (25) and is a negative regulator of the NF-κB–controlled pathway (25, 26). The role of MBOAT2 (a member of the membrane-bound O-acyltransferase family) in pathological conditions is largely unknown. Because EGLN3 is a known negative regulator of NF-κB, a key mediator of inflammation, we focused our study on EGLN3.
Effects of Genetic miR-31 Knockout on the EGLN3-NF-κB–Based Inflammatory Pathway.
To determine if a functional connection occurs among oncomiR-31, EGLN3, STK40, and NF-κB–controlled signaling in ESCC, we examined the FFPE esophageal tissue findings on in situ detection of miR-31 and IHC analysis of its target EGLN3/STK40 protein (Fig. 3D) along with IHC analysis of the NF-κB–controlled COX-2/S100A8/S100A9 inflammation pathway (Fig. 4D). In ESCC-bearing ZD:CTRL esophagus, miR-31 overexpression was associated with down-regulation of EGLN3 and STK40 proteins (Fig. 3D) and accompanying up-regulation of the NF-κB–controlled COX-2/S100A8/S100A9 inflammation pathway (Fig. 4D). Strikingly, genetic knockout of miR-31 in ZD:miR-31−/− esophagus up-regulated EGLN3 and STK40 protein expression (Fig. 3D) and abolished this NF-κB p65-controlled inflammation network (Fig. 4D), giving rise to a nonproliferative and tumor-free esophageal phenotype. Similarly, in vivo antimiR-31 delivery derepressed miR-31 tumor suppressor targets (SI Appendix, Fig. S3) and attenuated the same NF-κB–controlled inflammatory pathway (Fig. 4D), decreasing ESCC development (Fig. 1 C–E). These in vivo data establish the functional connections between oncomiR-31, EGLN3, and STK40 and NF-κB p65-controlled signaling in ESCC, providing a mechanism for miR-31 promotion of ESCC.
miR-31 Overexpression, EGLN3 Down-Regulation, and Inflammation in Human ESCC Tissue.
To examine a similar mechanistic link between miR-31 and inflammation in human ESCC, we retrieved archived human FFPE ESCC tissues (n = 10 patient cases) and performed in situ miR-31 analysis by ISH, EGLN3 and STK40 protein expression, and the NF-κB–controlled COX-2/S100A8/S100A9 inflammation pathway by immunohistochemistry. Overexpression of the proinflammation markers S100A8 (9), S1009 (27), and COX-2 (28) has been reported in human ESCC. Consistent with our previous data (10, 11), miR-31 ISH signal was abundant and strong in all 10 cases of ESCC tumor tissue (Fig. 3E). This miR-31 overexpression was accompanied by weak to absent expression of its tumor suppressor EGLN3 and STK40 protein (Fig. 3E). Because EGLN3 (29) and STK40 (30) are negative regulators of NF-κB–mediated transcription, their down-regulation unleashed a robust inflammatory response, with strong expression of the inflammation mediator NF-κB p65 and its target proinflammation markers COX-2, S100A8, and S100A9 (Fig. 3E), establishing a mechanistic connection between oncogenic miR-31, its tumor suppressor targets EGLN3 and STK40, and inflammation in human ESCC.
ZD:CTRL Rat Esophagus Exhibits a Tumor-Associated Metabolome.
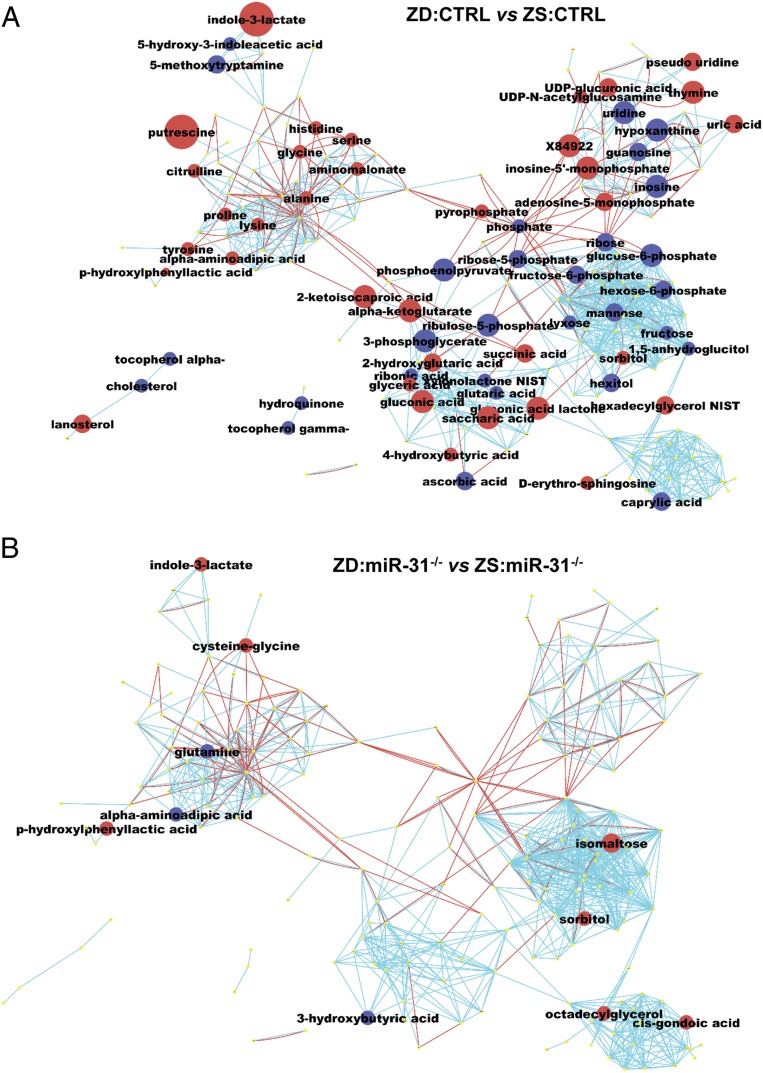
Cancer cells rewire cellular metabolism to adapt to increased demands for energy and cellular biosynthesis to sustain tumor growth (31). Changes in metabolite levels define the purpose of cellular regulation by genes and proteins (32). Metabolites are also key actors in biological networks by feedback inhibition or activation of enzymes. Additionally, miRNAs have emerged as key regulators of cellular metabolism in normal and pathological conditions (33). To determine if the diverse esophageal tumor outcomes in ZD:CTRL (WT) vs. ZD:miR-31−/− are reflected in their metabolomes, we performed nontargeted metabolomics profiling on esophageal mucosa in five cohorts (ZD:CTRL, ZD:antimiR/20-wk, ZS:CTRL, ZD:miR-31−/−, and ZS:miR-31−/−) using gas chromatography time-of-flight mass spectrometry (GC-TOF MS), a method previously used in metabolomics analyses in ovarian cancer (34).
Most metabolomics analyses in ESCC had been performed on serum/plasma from patients, with limited ESCC tissue analyses (35, 36). Based on P value < 0.05, our differential analysis identified 69 significantly altered metabolites (38 up- and 31 down-regulated) in the high ESCC-burden ZD:CTRL esophagus vs. ESCC-free ZS:CTRL. To obtain a biochemical overview of classic univariate statistical differences between ZD:CTRL and ZS:CTRL esophagi (SI Appendix, Table S2), we constructed a metabolomics network (37) among the structurally identified metabolites using the Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (38) and PubChem compound ID number (CID) identifiers (39). This network analysis (Fig. 5A) showed that of the 38 up-regulated metabolites in ZD:CTRL esophagus, about 1/3 (32%) were associated with metabolic alteration in amino acid (AA) metabolism. Differences in AA levels have been reported in many human cancers, including ESCC (35, 36, 40). Six AAs (lysine, histidine, proline, serine, tyrosine, glycine) are similarly up-regulated in human ESCC (36). The remaining up-regulated metabolites are involved in anabolic/biosynthetic pathways, including purine/pyrimidine metabolism (adenosine-5-monophosphage, thymine). In particular, putrescine (an intermediate in polyamine biosynthesis) was up-regulated 6.4-fold (P = 0.0001) in ZD:CTRL esophagus. Polyamines are indispensable for cell growth and malignant transformation (41).
Fig. 5.
Metabolomics profiling by GC-TOF MS reveals divergent esophageal metabolic phenotypes in ZD:CTRL (WT) and ZD:miR-31−/− rats. (A) ESCC-bearing ZD:CTRL vs. ZS:CTRL esophagus showing 38 up- and 31 down-regulated metabolites. (B) Tumor-free ZD:miR-31−/− vs. ZS:miR-31−/− esophagus showing limited metabolic changes (eight up- and three down-regulated metabolites). Each node denotes an identified metabolite (red, up-regulated; blue, down-regulated; yellow, insignificant change; Mann–Whitney U test, P < 0.05). Metabolite size reflects median fold change. Metabolites are connected based on biochemical relationships (red lines) or structural similarity (blue lines). Molecules not directly participating in biochemical transformations but sharing structural properties were connected at a threshold of Tanimoto similarity coefficient ≥0.7 (n = 9 rats per group).
Of the 31 down-regulated metabolites in ZD:CTRL esophagi (Fig. 5A and SI Appendix, Table S2), 16 (52%) were carbohydrates, including the glycolytic intermediates (glucose-6-phosphate, phosphoenolpyruvate). Increased glycolysis that produces glycolytic intermediates to furnish subsidiary pathways for sustaining tumor growth is a characteristic of cancer cell metabolism.
For insights into biological mechanisms underlying the effect of dietary ZD on ESCC development, we performed analysis of chemical similarity enrichments (ChemRICH) (42) that detects sets of metabolites and uses Kolmogorov–Smirnov statistics to test the significance of differential regulation. ChemRICH analysis showed that a total of 13 metabolite clusters differed significantly (FDR < 0.05) in ZD:CTRL vs. ZS:CTRL esophagus (SI Appendix, Fig. S4A and Table S5). Three clusters were found at decreased levels (hexoses, hexosephosphates, purine nucleosides), three clusters at increased levels (AAs, dicarboxylic acids, purine nucleotides), and seven clusters contained up-regulated and down-regulated compounds. Thus, ChemRICH showed that levels of glycolytic intermediates were decreased and anabolic/biosynthetic compounds increased, pointing to a classic cancer cell remodeling of energy and nucleoside metabolism.
Few studies have investigated whether metabolite biomarkers can be used to predict the efficacy of cancer treatment (43). Our analysis of ZD:antimiR/20-wk vs. ZS:CTRL esophagus yielded 95 significantly altered metabolites (SI Appendix, Table S3). Thirty up- and 28 down-regulated metabolites were common to ZD:antimiR/20-wk vs. ZD:CTRL esophagus (SI Appendix, Fig. S4C). Because polyamine biosynthesis is altered in human ESCC (35), the result that the level of putrescine was reduced by 48% with a P value approaching significance (P = 0.0625) (SI Appendix, Fig. S3D) suggests that putrescine could be a metabolite biomarker in ESCC treatment.
ZD:miR-31−/− Rat Esophageal Metabolome Shows Limited Perturbation.
Finally, we determined the influence of genetic miR-31 knockout on the esophageal metabolome in ZD:miR-31−/− rats that exhibited an ESCC-free esophageal phenotype. In sharp contrast to the ZD:CTRL metabolome, metabolomics analyses of ZD:miR-31−/− vs. ZS:miR-31−/− rat esophagus yielded only 11 significantly altered metabolites (Fig. 5B and SI Appendix, Table S4) and two significantly altered metabolite clusters (SI Appendix, Fig. S3B and Table S6).
ZD:miR-31−/− Rat Esophagus Exhibits a Stable Genome.
Cancer is a genetic disease and the genome instability observed, a “hallmark of cancer,” occurs in virtually all cancers, though there has been less focus on genome instability in rodent tumors. Our nutrigenomics profiling of the rat ZD esophageal models for following hyperplasia and neoplasia development has shown that the miRNA and metabolomics data for these rat lesions mirror the miRNA and metabolome alterations in human ESCC, prompting us to determine, by WGS, if there were differential genomic alterations in the NMBA-induced cancerous tissues of rat models.
After quality checks and filtering of the WGS data and analysis of sequences for altered genes in four rat tissue DNAs, we observed very few genetic alterations in the ZS:CTRL vs. ZD:CTRL (rats R13 and R14, Fig. 1C). In WT genomes, the number of variants per Mb of genome was computed (Fig. 2G). The ZD:miR-31−/− mucosa, without evident tumors, showed only two in-frame deletions and a single nucleotide insertion in a different gene on a different chromosome, all with possible moderate effect on protein products. The ZS:CTRL rat (WT) tissue without evident tumors showed only four genome alterations at three loci, each with a possible moderate effect on protein products (SI Appendix, Table S7). The ZD:CTRL (R14) rat with low tumor burden and ZD:CTRL (R13) rat with high tumor burden showed 7 and 14 mostly in-frame deletions/insertions, with moderate effects on specific genes (SI Appendix, Table S7). These sequences, with relatively high mutational effect, showed significant mutations in genes that might have affected tumor development, nine genes in the R14 sample and 22 genes in the ZD:CTRL R13 mucosal DNA. A gene ontology analysis was performed for the gene alterations with “HIGH” and “MODERATE” effects to find most enriched gene processes (SI Appendix, Tables S8 and S9). The most commonly affected processes in both HIGH and MODERATE effect mutations were biological regulation, metabolic process, and response to stimulus.
Discussion
In a Zn deficiency-promoted rat ESCC model, we used omics tools to examine the molecular consequences of miR-31 silencing by antimiR-31 delivery and by genetic miR-31 knockout. This model that mimics features of human ESCC, including dietary ZD, miR-31 overexpression, and inflammation, exhibits a high ESCC burden, prominent miR-31 up-regulation, and a cancer-associated inflammatory signature (5, 7, 8, 10, 11, 14). We find that prolonged antimiR-31 delivery knocks down esophageal miR-31 levels in ZD:antimiR/20-wk vs. ZD:CTRL rats and reduces ESCC incidence by 40%, counteracting the ESCC-promoting effects of Zn deficiency. This result extends our previous antimiR-31 delivery study in esophageal preneoplasia (10) and affirms the oncogenic role of miR-31 in human ESCC (14). Remarkably, with genetic miR-31 knockout, Zn deficiency per se does not create a precancerous esophageal phenotype in ZD:miR-31−/− rats as it does in WT ZD:miR-31+/+ rats. At tumor end point, nonmalignant small/isolated esophageal tumors were found in a few ZD:miR-31−/− rats in contrast to large/sessile tumors in most ZD:miR-31+/+ esophagi. Strikingly, none of the 20 ZD:miR-31−/− rats developed ESCC compared with 15 of the 20 ZD:miR-31+/+ rats. While ZD:miR-31−/− esophageal epithelia were thin with occasional hyperplastic foci, ESCC-bearing ZD:miR-31+/+ esophagi showed unrestrained cellular proliferation and inflammatory cell infiltrates in the stroma. These data show that Zn deficiency does not promote ESCC in ZD:miR-31−/− rats, providing conclusive genetic evidence that miR-31 expression is required for ESCC development.
Our comparative transcriptomics data showed that the high ESCC-burden ZD:CTRL esophagus displayed a Zn deficiency-induced inflammation gene signature with up-regulation of six human ESCC-associated inflammation genes (8), including S100A8 (9), S100A9 (27), and COX-2 (28). In contrast, expression of proinflammatory genes is absent in ESCC-free ZD:miR-31−/− esophagus or in ZD:antimiR/20-wk esophagus, with a 40% reduction in ESCC incidence. Because inflammation, a hallmark of cancer (31), is central to the pathogenesis of ESCC (7, 44), the data suggest that suppression of inflammation is a mechanism by which genetic miR-31 knockout abrogates ESCC development.
We also identified and validated EGLN3, a known colorectal cancer tumor suppressor and negative regulator of the NF-κB–controlled pathway, as a miR-31 direct target. Loss of EGLN3 allows tumors to overcome hypoxic growth inhibition and sustain proliferation through EGFR (45), though its regulation in cancer remains not well-understood. Our in situ miR-31 analysis by ISH and EGLN3/STK40-NF-κB–controlled COX-2/S100A8/S100A9 protein expression analysis revealed that genetic miR-31 deletion leads to suppression of the EGLN3/STK40-NF-κB–controlled inflammation pathway, opposite of results with ESCC-bearing ZD:CTRL esophagus and of human ESCC tissue, in which miR-31 overexpression suppresses EGLN3, setting free the NF-κB p65-controlled inflammation signal. These data establish a connection between oncomiR-31 and EGLN3/STK40-NF-κB–controlled signaling in ESCC and establish the mechanism by which miR-31 promotes human ESCC. We predict this could be a mechanism for oncogenic miR-31 in inflammation-associated human cancers such as colorectal cancer, where miR-31 overexpression (13), EGLN3 down-regulation (25), and NF-κB activation (46) have been reported.
Because Egln3 is a direct target of miR-31 and miR-31 KO rats do not develop Zn deficiency-associated ESCC even after exposure to NMBA, a limitation of the study is whether overexpressing the EGLN3-negative regulator of NF-κB will eliminate ESCC development as does the miR-31 knockout, assuming that the NF-κB inflammatory signal pathway is the main inducer of ESCC in this rat model. Studies are in progress to specifically address this issue.
Metabolomics analysis showed that the ESCC-bearing ZD:CTRL esophagus has a highly dysregulated metabolome, with 69 significantly altered metabolites. In particular, ∼32% of the up-regulated metabolites are implicated in active amino acid biosynthesis, a common feature of human ESCC (35, 36, 40). Phosphoenolpyruvate, the top down-regulated species in ZD:CTRL vs. ZS:CTRL esophagus (−5.83-fold, P = 0.010), is a metabolic checkpoint of antitumor T cell responses (47). Uridine, involved in the regulation of carbohydrate and pyrimidine nucleotide synthesis, is decreased 3.58-fold (P = 0.0008) in ZD:CTRL esophagus. Uridine levels were also down-regulated in human ESCC (35). In combination, these data establish that dietary Zn deficiency causes a tumor-associated esophageal metabolome profile resembling that of human ESCC.
Remarkably, despite sustained Zn deficiency, genetic miR-31 knockout produced an esophageal metabolome with limited metabolic changes in ZD:miR-31−/− esophagus compared with its ZS:miR-31−/− counterpart. This nontumor metabolome is reflected by the ESCC-free esophageal phenotype in ZD:miR-31−/− animals.
The results of the genome sequence analysis of the rat esophageal tissues at tumor end point did not uncover large numbers of mutations but found alterations with high or moderate effect on the proteins mutated in the ZD:CTRL WT rats, while there were virtually no mutations in the ZD:miR-31−/− rat or the ZS:CTRL WT rat. This genomic instability could be due to exposure of the rat GI tract to the nitrosamine carcinogen NMBA, and the accumulated mutations would likely show the mutational signature recently described for the environmental carcinogen NMBA (48). It is interesting that without expression of miR-31, an NMBA mutational signature appears to be suppressed, suggesting that the inflammation signal pathway might contribute to production of the carcinogen-induced mutations. But we emphasize that the suppression of epithelial proliferation and associated cellular signal pathways are also necessary for NMBA induction of mutations. The most interesting finding from the WGS study was that the miR-31 KO rat, even with ZD and carcinogen treatment, did not show genome alterations, confirming that the miR-31 signaling pathway is required for development of ESCC in this rat model, as in the counterpart human cancers.
In summary, in a Zn deficiency-promoted rat ESCC model, with miR-31 overexpression and inflammation pathway up-regulation, we reveal that in vivo antimiR-31 delivery suppresses, and miR-31 genetic knockout abrogates, ESCC development. Our identification of EGLN3, a known negative regulator of NF-κB, as a direct target of miR-31 establishes a functional link between the oncomiR-31 tumor suppressor gene target, EGLN3, and the NF-κB p65-controlled inflammation signaling, which in turn promotes ESCC in both human and rat tissue. Additionally, our study shows that counteracting the ESCC-promoting effect of dietary Zn-deficiency genetic miR-31 knockout leads to the eliminating of entire biological processes, including the miR-31-EGLN3/STK40-NF-κB–controlled inflammatory pathway, esophageal cancer metabolism, and an unstable genome. Thus, our findings provide insight into the mechanisms whereby Zn deficiency or miR-31 promotes ESCC. Our data also provide a mechanistic rationale for development of therapeutic strategies to target miR-31 in ESCC and for Zn replenishment in ESCC prevention.
Materials and Methods
Rats, Diets, and Carcinogen.
Weanling male Sprague–Dawley rats (55 ± 5 g) were from Taconic Laboratory. Custom-formulated ZD and ZS diets (Harlan Teklad) were identical except for the amount of Zn, which was 3 to 4 ppm for the ZD and ∼60 ppm for the ZS diet. NMBA was from the Midwest Research Institute. Animal protocols were approved by the Thomas Jefferson University Animal Care and Use Committee.
Locked Nucleic Acid AntimiR-31 Inhibitor.
An in vivo grade of phosphorothioated miRCURY LNA-enhanced rno-miR-31a-5p inhibitor probe (SI Appendix, Table S10) was dissolved in sterile phosphate-buffered saline (PBS) and stored at −80 °C.
Generation of an miR-31−/− Rat Model.
Sage Labs was commissioned to custom generate a constitutive miR-31−/− rat model on the Sprague–Dawley genetic background by CRISPR/Cas9 technology (20, 21).
Anticancer Studies.
AntimiR-31 delivery in the ZD rat ESCC model.
Four-week-old male Sprague–Dawley rats were divided into four ZD groups and a ZS group. ZD rats were fed ad libitum and ZS rats were pair fed to deficient animals to match their decreased food consumption (10). The animals received PBS-formulated LNA-antimiR or LNA-CTRL-A oligonucleotides or were untreated, forming five cohorts: ZD:antimiR/5-wk (n = 12), ZD:antimiR/20-wk (n = 11), ZD:CTRL-A/20-wk (n = 8), ZD:CTRL (untreated, n = 20), and ZS:CTRL (untreated, n = 8). Over a period of the first 5 wk (10), ZD:antimiR/5-wk, ZD:antimiR/20-wk, and ZD:CTRL-A/20-wk cohorts received via tail vein a loading dose of antimiR or CTRL-A (20 mg⋅kg−1), followed by nine maintenance doses (6 mg⋅kg−1, twice weekly). Starting at week 5 (11), the animals were given four intragastric doses of NMBA (2 mg⋅kg−1, once per week for 4 wk). During and following NMBA administration (15 wk), ZD:antimiR/20-wk and ZD:CTRL-A/20-wk rats received 20 additional antimiR or CTRL-A doses (6 mg⋅kg−1, twice per week for 5 wk and then once per wk for the remaining 10 wk). The animals were weighed weekly and killed 48 h after the final dose of antimiR or CTRL-A or 15 wk after the first NMBA dose. Blood was obtained from the retroorbital venous plexus for serum preparation. Whole esophagus was excised, longitudinally slit open, and photographed. Tumors >0.5 mm in diameter were mapped. Esophagi were cut into two equal portions. Esophageal epithelium was isolated, snap-frozen in liquid nitrogen, and stored at −80 °C. The remaining portion was fixed in 10% buffered formalin.
Esophageal tumorigenesis in miR-31−/− rats.
Four-week-old male miR-31−/− and miR-31+/+ littermates were randomly divided into a ZD or ZS dietary group. The animals were divided into NMBA-untreated groups (10 rats per group) and NMBA-treated groups (20 rats per group), using the same NMBA-induced tumor assay as described above.
RNA Isolation.
Total RNA was extracted from the pulverized esophageal mucosal samples using an RNA extraction kit (Norgen Biotek; 25700). The integrity of the RNA was analyzed by an Agilent 2100 Bioanalyzer and RNA integrity number was ≥8 for all samples.
TaqMan miRNA Assay.
Reverse transcription (Applied Biosystems) of miRNAs was performed. qRT-PCR was performed using TaqMan miRNA assays, mmu-miR-31 (ID 000185), and endogenous controls small nucleolar (sno)RNA (ID 001718) and U87 (ID 001712). As an overall quality control, threshold cycle values above 35 were excluded from analysis.
ISH.
ISH was performed (10) using miRCURY LNA microRNA detection probes rno-miR-31 and negative control (rno-miR-31 with mismatches at two positions) double DIG-labeled at the 5′ and 3′ ends (Exiqon). miRNA was localized by incubation with 4-nitro-blue tetrazolium (NBT) and 5-bromo-4-chloro-3′-indolylphosphate (BCIP) (Roche). Nuclear fast red (Vector Labs) was used as a counterstain.
qRT-PCR.
cDNA was reverse-transcribed using the High-Capacity cDNA Archive Kit (Applied Biosystems). qPCR was performed using the ΔΔCt method of qPCR data analysis and single-tube TaqMan gene expression assays with S100A8 (Rn00587579_g1), S100A9 (Rn00585879_m1), Ptgs2 (Rn01483828_m1), Egln3 (Rn00571341_m1), and Mboat2 (Rn01472966_m1) and normalizers Psmb6 (Rn02377140_g1) and Oaz1 (Rn01408148_g1).
Gene Expression Profiling.
Expression profiling of esophageal epithelia was performed at the Ohio State University Comprehensive Cancer Center Genomics Facility using the Affymetrix Rat Genome 230 2.0 GeneChip (10).
Expression Data Analysis.
The processing of raw files was performed using the Expression Console (Affymetrix). The normalization was performed using robust multiarray normalization implemented in the Expression Console and Rat_230.CDF. Differentially expressed genes were identified using the R package (limma). The heatmaps were created using custom R scripts and the function heatmap.2 of gplots.
DNA Constructs.
For psiCHECK2-MBOAT2-WT, a 1,494-bp-long fragment of the MBOAT2 3′ UTR (corresponding to positions 1733 to 3226 of National Center for Biotechnology Information [NCBI] reference sequence NM_138799.4) containing two predicted target sites for miR-31-5p was cloned downstream of the Renilla luciferase gene in the psiCHECK2 vector (Promega). For psiCHECK2-CTNND2-WT, an 864-bp-long fragment of the CTNND2 3′ UTR (corresponding to positions 4298 to 5161 of NCBI reference sequence NM_001332.4) was cloned in the psiCHECK2 vector. For psiCHECK2-EGLN3-WT, a 368-bp-long fragment of the EGLN3 3′ UTR (corresponding to positions 1047 to 1414 of NCBI reference sequence NM_022073.3) was cloned in the psiCHECK2 vector. The predicted miR-31-5p site in EGLN3 is located at position 159 to 166 of the EGLN3 3′ UTR. Two predicted miR-31-5p sites in MBOAT2 are located at positions 376 to 382 and 1268 to 1274 of the MBOAT2 3′ UTR. Substitutions in predicted miR-31-5p binding sites to produce mutated sites unable to bind miR-31-5p were introduced by using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene) following the manufacturer’s instructions (see oligonucleotides used in SI Appendix, Table S10).
Cell Cultures and Luciferase Assay.
HEK-293FT cells were cultured in RPMI medium 1640 (Sigma-Aldrich) supplemented with 10% FBS, 25 μg/mL gentamicin, and 1% glutamine (Sigma-Aldrich) at 37 °C in a 5% CO2 incubator. Transfections were carried out with Lipofectamine 2000 following the standard protocol (Life Technologies). HEK-293FT cells were seeded in 12-well plates the day before transfection. Cells were cotransfected with 250 ng of various psiCHECK2 vector constructs and 100 nM premiR-31-5p or scrambled negative control 1 (Thermo Fisher Scientific). Twenty-four hours after transfection, firefly and Renilla luciferase activities were measured using the Dual-Luciferase Report Assay (Promega).
IHC.
IHC was performed (10) using primary antibodies for PCNA (clone PC-10; Ab-1; Thermo Scientific), KRT14 (NCL-LL002; Novocastra), COX-2 (12282; Cell Signaling), S100A8 (T-1032; BMA), S100A9 (NB110-89726; Novus Biologicals), NF-κB p65 (ab7970; Abcam), STK40 (orb101780; Biorbyt), EGLN3 (orb443107; Biorbyt), and MBOAT2 (orb185503; Biorbyt). Protein was localized by incubation with the 3-amino-9-ethylcarbazole substrate chromogen (Dako) or 3,3′-diaminobenzidine tetrahydrochloride (Sigma-Aldrich).
Metabolomics Profiling by GC-TOF MS.
Frozen rat esophageal mucosa were shipped to the NIH West Coast Metabolomics Center (University of California, Davis). Esophageal tissue was extracted, derivatized, and processed as described (34). Primary metabolite analysis was performed by GC-TOF MS using cold injection/automatic liner exchange (CIS-ALEX GC-TOF MS; Leco Pegasus IV) (49).
ChemRICH.
Chemical set enrichment statistical analysis was performed on metabolomics data as described (42, 49).
Metabolome Network Visualization.
Using KEGG and PubChem CIDs, a biochemical and chemical similarity network (37, 49) was calculated for all measured metabolites.
Genomic DNA Isolation.
High-quality nondegraded genomic DNA was extracted from esophageal epithelia isolated using the DNeasy Blood & Tissue Kit (Qiagen).
WGS.
DNA library preparation, library quality control, and nonhuman WGS to yield ∼30× coverage were performed on the Illumina HiSeq X at HudsonAlpha Genomic Services Laboratory.
WGS Analysis.
WGS reads were mapped against the Rnor6.0 (Rattus norvegicus) reference using BWA (50). The read postprocessing analysis was performed after the alignment using Picard (http://broadinstitute.github.io/picard/). The read duplicate marking was performed using MarkDuplicate. GATK was used for the step of calling variants. Single-nucleotide variants were selected considering QD (QualByDepth) <2.0, FS (FisherStrand) >60, MQ (Mapping Quality) <40, MQRankSum <−12.5, and ReadPosRankSum <−8.0. Only the indels with QD <2.0, FS >200, and ReadPosRankSum <20 were selected for downstream analysis. The resulting VCF files were annotated with the variant effect predictor (VEP) (https://www.biorxiv.org/content/10.1101/501817v2) using the following options: –species Rattus_norvegicus–offline–pick–ccds–hgvs–symbol–protein. VEP allows the annotation of the variants considering the different effects on the proteins. Only the variants with MODERATE or HIGH effect were selected for downstream analysis. Additional filters were applied to maintain only the most deleterious variants at the level of the sequence (e.g., frameshift_variants, splice_acceptor_variant, etc.).
Microscopy.
IHC and ISH analyses were performed by light microscopy (Olympus BX51 microscope), and photographs were taken with a Spot RT3 camera and Spot software version 5.2.
Zn Measurement.
Serum Zn content was determined using Atomic Absorption Spectrometer Analyst 400 (PerkinElmer).
Statistical Analysis.
ANOVA and Tukey post hoc t tests (or Kruskal–Wallis and pairwise Wilcoxon rank-sum tests for groups with nonnormally distributed data) were used to determine the differences among the groups. Tumor and ESCC incidence data among the groups were evaluated using the χ2 test. Individual differences in incidence data were then assessed using Fisher’s exact test. The Mann–Whitney U test was used to detect significant compounds in metabolomics. All statistical tests were two-sided and considered significant at P < 0.05. Statistical analysis was performed by R (http://www.R-project.org).
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (R01CA118560 to L.Y.F.; R35CA197706 to C.M.C.) and the NIH West Coast Metabolomics Center (U24 DK097154 to O.F.). We thank Professor Stephan Peiper and the Department of Pathology, Anatomy, and Cell Biology, Thomas Jefferson University, for funds to generate the miR-31 KO rat model. We also thank Timothy Flanagan of Medical Media Services, Thomas Jefferson University, for assistance with figure preparation.
Footnotes
The authors declare no competing interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE140190). Whole-genome sequencing data have been deposited in the the Sequence Read Archive (SRA code PRJNA607070).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920333117/-/DCSupplemental.
References
- 1.Ferlay J., et al. , Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359–E386 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Magee P. N., The experimental basis for the role of nitroso compounds in human cancer. Cancer Surv. 8, 207–239 (1989). [PubMed] [Google Scholar]
- 3.Yang C. S., Research on esophageal cancer in China: A review. Cancer Res. 40, 2633–2644 (1980). [PubMed] [Google Scholar]
- 4.Hashemian M., et al. , Dietary intake of minerals and risk of esophageal squamous cell carcinoma: Results from the Golestan Cohort Study. Am. J. Clin. Nutr. 102, 102–108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abnet C. C., et al. , Zinc concentration in esophageal biopsy specimens measured by X-ray fluorescence and esophageal cancer risk. J. Natl. Cancer Inst. 97, 301–306 (2005). [DOI] [PubMed] [Google Scholar]
- 6.McClain C. J., Su L. C., Zinc deficiency in the alcoholic: A review. Alcohol. Clin. Exp. Res. 7, 5–10 (1983). [DOI] [PubMed] [Google Scholar]
- 7.Mandard A. M., Hainaut P., Hollstein M., Genetic steps in the development of squamous cell carcinoma of the esophagus. Mutat. Res. 462, 335–342 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Taccioli C., et al. , Dietary zinc deficiency fuels esophageal cancer development by inducing a distinct inflammatory signature. Oncogene 31, 4550–4558 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taccioli C., et al. , Zinc replenishment reverses overexpression of the proinflammatory mediator S100A8 and esophageal preneoplasia in the rat. Gastroenterology 136, 953–966 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taccioli C., et al. , Repression of esophageal neoplasia and inflammatory signaling by anti-miR-31 delivery in vivo. J. Natl. Cancer Inst. 107, djv220 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fong L. Y., et al. , MicroRNA dysregulation and esophageal cancer development depend on the extent of zinc dietary deficiency. Oncotarget 7, 10723–10738 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laurila E. M., Kallioniemi A., The diverse role of miR-31 in regulating cancer associated phenotypes. Genes Chromosomes Cancer 52, 1103–1113 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Bandrés E., et al. , Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol. Cancer 5, 29 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang T., et al. , The oncogenetic role of microRNA-31 as a potential biomarker in oesophageal squamous cell carcinoma. Clin. Sci. (Lond.) 121, 437–447 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Wong T. S., et al. , Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin. Cancer Res. 14, 2588–2592 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Lajer C. B., et al. , Different miRNA signatures of oral and pharyngeal squamous cell carcinomas: A prospective translational study. Br. J. Cancer 104, 830–840 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruegger C., et al. , MicroRNA expression differs in cutaneous squamous cell carcinomas and healthy skin of immunocompetent individuals. Exp. Dermatol. 22, 426–428 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Fluiter K., et al. , In vivo tumor growth inhibition and biodistribution studies of locked nucleic acid (LNA) antisense oligonucleotides. Nucleic Acids Res. 31, 953–962 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elmén J., et al. , LNA-mediated microRNA silencing in non-human primates. Nature 452, 896–899 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Bassett A. R., Tibbit C., Ponting C. P., Liu J. L., Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 4, 220–228 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H., et al. , One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamamoto K., et al. , Suppressive effects of a selective cyclooxygenase-2 inhibitor, etodolac, on 4-nitroquinoline 1-oxide-induced rat tongue carcinogenesis. Exp. Toxicol. Pathol. 56, 145–151 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Németh J., et al. , S100A8 and S100A9 are novel nuclear factor kappa B target genes during malignant progression of murine and human liver carcinogenesis. Hepatology 50, 1251–1262 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Barnes P. J., Karin M., Nuclear factor-kappaB: A pivotal transcription factor in chronic inflammatory diseases. N. Engl. J. Med. 336, 1066–1071 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Xue J., et al. , Prolyl hydroxylase-3 is down-regulated in colorectal cancer cells and inhibits IKKbeta independent of hydroxylase activity. Gastroenterology 138, 606–615 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Fu J., Taubman M. B., EGLN3 inhibition of NF-κB is mediated by prolyl hydroxylase-independent inhibition of IκB kinase γ ubiquitination. Mol. Cell. Biol. 33, 3050–3061 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan N. J., et al. , Identification of the up-regulation of TP-alpha, collagen alpha-1(VI) chain, and S100A9 in esophageal squamous cell carcinoma by a proteomic method. J. Proteomics 75, 3977–3986 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Zimmermann K. C., et al. , Cyclooxygenase-2 expression in human esophageal carcinoma. Cancer Res. 59, 198–204 (1999). [PubMed] [Google Scholar]
- 29.Fu J., Taubman M. B., Prolyl hydroxylase EGLN3 regulates skeletal myoblast differentiation through an NF-kappaB-dependent pathway. J. Biol. Chem. 285, 8927–8935 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang J., et al. , Identification of a novel serine/threonine kinase that inhibits TNF-induced NF-kappaB activation and p53-induced transcription. Biochem. Biophys. Res. Commun. 309, 774–778 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Ward P. S., Thompson C. B., Metabolic reprogramming: A cancer hallmark even Warburg did not anticipate. Cancer Cell 21, 297–308 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rottiers V., Näär A. M., MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 13, 239–250 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denkert C., et al. , Mass spectrometry-based metabolic profiling reveals different metabolite patterns in invasive ovarian carcinomas and ovarian borderline tumors. Cancer Res. 66, 10795–10804 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Sun C., et al. , Spatially resolved metabolomics to discover tumor-associated metabolic alterations. Proc. Natl. Acad. Sci. U.S.A. 116, 52–57 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tokunaga M., et al. , Metabolome analysis of esophageal cancer tissues using capillary electrophoresis-time-of-flight mass spectrometry. Int. J. Oncol. 52, 1947–1958 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Barupal D. K., et al. , MetaMapp: Mapping and visualizing metabolomic data by integrating information from biochemical pathways and chemical and mass spectral similarity. BMC Bioinformatics 13, 99 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kotera M., Hirakawa M., Tokimatsu T., Goto S., Kanehisa M., “The KEGG databases and tools facilitating omics analysis: Latest developments involving human diseases and pharmaceuticals” in Next Generation Microarray Bioinformatics: Methods and Protocols, Wang J., Tan A., Tian T., Eds. (Methods in Molecular Biology, Humana Press, New York, 2012), vol. 802, pp. 19–39. [DOI] [PubMed] [Google Scholar]
- 39.Bolton E. E., Wang Y., Thiessen P. A., Bryant S. H., PubChem: Integrated platform of small molecules and biological activities. Annu. Rep. Comput. Chem. 4, 217–241 (2008). [Google Scholar]
- 40.Zhu X., et al. , Metabolic perturbation and potential markers in patients with esophageal cancer. Gastroenterol. Res. Pract. 2017, 5469597 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pegg A. E., Casero R. A. Jr, Current status of the polyamine research field. Methods Mol. Biol. 720, 3–35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barupal D. K., Fiehn O., Chemical similarity enrichment analysis (ChemRICH) as alternative to biochemical pathway mapping for metabolomic datasets. Sci. Rep. 7, 14567 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iemoto T., et al. , Serum level of octanoic acid predicts the efficacy of chemotherapy for colorectal cancer. Oncol. Lett. 17, 831–842 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balkwill F., Charles K. A., Mantovani A., Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer Cell 7, 211–217 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Henze A. T., et al. , Loss of PHD3 allows tumours to overcome hypoxic growth inhibition and sustain proliferation through EGFR. Nat. Commun. 5, 5582 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lind D. S., et al. , Nuclear factor-kappa B is upregulated in colorectal cancer. Surgery 130, 363–369 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Ho P. C., et al. , Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell 162, 1217–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kucab J. E., et al. , A compendium of mutational signatures of environmental agents. Cell 177, 821–836.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fong L. Y., et al. , Human-like hyperplastic prostate with low ZIP1 induced solely by Zn deficiency in rats. Proc. Natl. Acad. Sci. U.S.A. 115, E11091–E11100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.