Significance
Robust predictions of future changes in global biogeochemical cycling require an understanding of how microorganisms adapt to stressful and changing environments. In the ocean, rates of adaptation will be a function of both evolutionary timescales and physical dynamics. However, little is known about this interaction. We examined evolutionary dynamics of marine microbes by combining a model of microbial adaptation with varying selection pressures with a high-resolution ocean circulation model. A trade-off emerged between two evolutionary strategies: (i) ability to adapt plastically to short-term environmental fluctuations with delayed genetic adaptation and (ii) more rapid genetic adaptation with limited response to short-term environmental fluctuations. This trade-off determines evolutionary timescales and provides a foundation for understanding distributions of microbial traits and biogeochemistry.
Keywords: evolution, marine microbes, fluctuating environment, adaptation timescales, advection
Abstract
Marine microbes form the base of ocean food webs and drive ocean biogeochemical cycling. Yet little is known about the ability of microbial populations to adapt as they are advected through changing conditions. Here, we investigated the interplay between physical and biological timescales using a model of adaptation and an eddy-resolving ocean circulation climate model. Two criteria were identified that relate the timing and nature of adaptation to the ratio of physical to biological timescales. Genetic adaptation was impeded in highly variable regimes by nongenetic modifications but was promoted in more stable environments. An evolutionary trade-off emerged where greater short-term nongenetic transgenerational effects (low-γ strategy) enabled rapid responses to environmental fluctuations but delayed genetic adaptation, while fewer short-term transgenerational effects (high-γ strategy) allowed faster genetic adaptation but inhibited short-term responses. Our results demonstrate that the selective pressures for organisms within a single water mass vary based on differences in generation timescales resulting in different evolutionary strategies being favored. Organisms that experience more variable environments should favor a low-γ strategy. Furthermore, faster cell division rates should be a key factor in genetic adaptation in a changing ocean. Understanding and quantifying the relationship between evolutionary and physical timescales is critical for robust predictions of future microbial dynamics.
Planktonic microorganisms in the oceans are at the mercy of ocean circulation, which transports cells throughout the ocean basins and results in significant variations in the physical and chemical environment experienced by the cells (1–3). As a result, long-term shifts in the average ocean environment, such as a temperature increase from global warming, are experienced by phytoplankton as gradual changes overlain on top of a highly dynamic regime of environmental fluctuations. Previous work has shown that microbes have the potential to evolve faster through neutral genetic processes than their dispersal by large-scale currents, thereby creating biogeographic provinces even in the absence of selection (2). However, little is known about the interaction of ocean circulation with adaptive evolution of microbial populations to new environments. Constraining rates of adaptive evolution in the ocean presents a significant challenge because evolutionary timescales are a function of many factors including environmental fluctuations driven by physical dynamics, chemical cycling, microbial growth rates, population sizes, and the rate at which genetic variation can be generated—all of which are variable in the marine systems. Improving our understanding of these interactions is critical for accurately predicting future shifts in microbial diversity, ecosystem dynamics, and biogeochemical cycling as the oceans respond to global warming induced changes.
Microbial populations—defined as clusters of closely related organisms exhibiting population-specific gene flow—are acted upon by both natural selection and neutral evolutionary processes. Laboratory-based experimental evolution studies have demonstrated relatively fast timescales (<350 generations) of selective adaptation for marine microbes under constant conditions (4) and shown that fluctuations impact the outcome of evolution (5). These studies are consistent with theory (6) and laboratory experiments in nonmarine model systems (e.g., ref. 7). However, our understanding of how marine microbial evolution will proceed in situ in a fluctuating environment remains in its infancy. One reason for this is that models of microbial adaptation rarely include common nongenetic responses, which can affect adaptive outcomes (8–12). Second, until recently, we did not have the ability to model the dynamic environment experienced by pelagic microbes with high enough resolution to capture realistic environmental dynamics critical for driving evolution (1). Here, we develop two criteria that describe microbial adaptation strategies as a function of physical fluctuations and both nongenetic and genetic biological response timescales. These criteria identify constraints on different adaptive strategies and on rates of microbial adaptation to environmental change, which can be applied across vastly different oceanographic regions and to diverse microbial species. This insight into marine microbial adaptation will allow for an improved understanding of general patterns of trait distributions among marine microbial functional groups (13–15) and how these distributions might shift in a changing world.
Adaptation under Variable Selection Pressures
Correctly accounting for different biological response timescales is central to understanding adaptation in fluctuating environments. Adaptation to a new environment (defined as a heritable increase in fitness) can be generated through a range of processes from transgenerational plasticity (defined as any heritable, nongenetic change in phenotype) to genetic mutations. These processes for generating and transmitting trait variation can be classified on a spectrum from fast variation, low transmission (LT) to slow variation, high transmission (HT) modifications (16). HT modifications are relatively rare, and so generate variation in fitness in growing populations slowly but have a high probability of being transmitted to offspring through a large number of cell divisions. Classic examples of HT modifications are point mutations, genome rearrangement, horizontal gene transfer, and transposon insertions. In contrast, LT modifications are common relative to HT modifications, and so generate variation in fitness in growing populations quickly, but are nongenetic and so have a lower probability of being transmitted to offspring. LT modifications include—but aren’t limited to—transgenerational plastic effects and some changes to DNA methylation and acetylation patterns (i.e., epigenetics). Immediately following environmental change, LT modifications may allow for flexible and rapid diversification in phenotype within or over very few generations. This can result in different rates of adaptation to a new environment (increase in fitness) relative to what would be expected due to HT modifications alone (8–11). However, because LT modifications are reversible, the fitness benefits and trait changes from LT modifications will be lost from the population more quickly than would be expected from HT modifications alone, especially in a dynamic environment where selective pressure can fluctuate. Theoretical (8, 17) and empirical (9, 12) data suggest that HT and LT modifications acting together best explain patterns of microbial evolution on timescales of hundreds of generations. For example, experimental evolution studies in yeast have shown that the interaction of short-term epigenetic inheritance with genetic mutation modifies the rate and type of adaptation, thereby impacting long-term evolution (12).
Before tackling the complexities of adaptation in the ocean, we first quantified how the interplay between LT and HT modifications can affect both the timescale and outcome of marine microbial adaptation in an idealized fluctuating environment. When considering adaptation in a variable environment, it is necessary to clearly define the effects of selection pressure across different types of environments. We distinguish between two types of environments: the “new” environment where populations are under directional selection (i.e., the selective fixation of new beneficial alleles where the population is in the process of adapting); and the “ancestral” environment where the population is well adapted and assumed to be under stabilizing selection (i.e., the selective removal of new nonneutral alleles, which are deleterious). We used an individual-based model of adaptation modified from Fisher’s model (18) in which the simulated population moved between the “new” and “ancestral” environment following a step function with varying frequencies. In the model simulations, adaptation—increases in fitness in the “new” environment—could be driven by both LT and HT modifications. Critically, LT modifications were introduced at a higher frequency than HT modifications but were also associated with a transmission timescale or reversion rate (Methods). As a result, the model simulations captured both the high frequency occurrence of LT modifications (e.g., transgenerational plastic responses) in populations following an environmental change and the degradation of this signal over several generations once the environmental cue was removed (SI Appendix, Fig. S2). In contrast, HT modifications (e.g., genetic mutations) occurred at low frequencies in the population but were transmitted with high fidelity between generations.
An ensemble of model simulations was conducted, varying the time spent in each environment (τf) from short-duration fluctuations (τf =10 generations) to long-duration fluctuations (τf = 500 generations). Similarly, a large range of transmission timescales for LT modifications (τLT) was explored from no LT modifications (τLT = 1 generation) to maternal effects (τLT = 4 generations) to experimentally confirmed timescales (τLT = 10 and 20 generations; refs. 19 and 20), and to a proof-of-concept long-lasting LT effect (τLT = 150 generations). In addition to τf and τLT, the timescale required for a beneficial HT modification to fix in the population through a selective sweep once it occurred in an individual (τHT) emerged as a critical timescale in the model (Fig. 1). τHT is an emergent property of the model that varied as a function of HT modification supply and effect (SI Appendix, Fig. S1). τHT was systematically varied by running the model with varying strengths of stabilizing selection (SI Appendix, S1 and Figs. S2–S4) and a range of population sizes and mutation rates (SI Appendix, S2 and Fig. S7). These parameter ranges were sufficient to understand how model behavior varied as a function of τHT. Since our primary aim was to test the robustness of our predicted relationships between physical and biological timescales (described below), we examined ranges of physical and biological parameters around thresholds that determined evolutionary outcomes and showed that the overall patterns were robust (SI Appendix, S1 and S2).
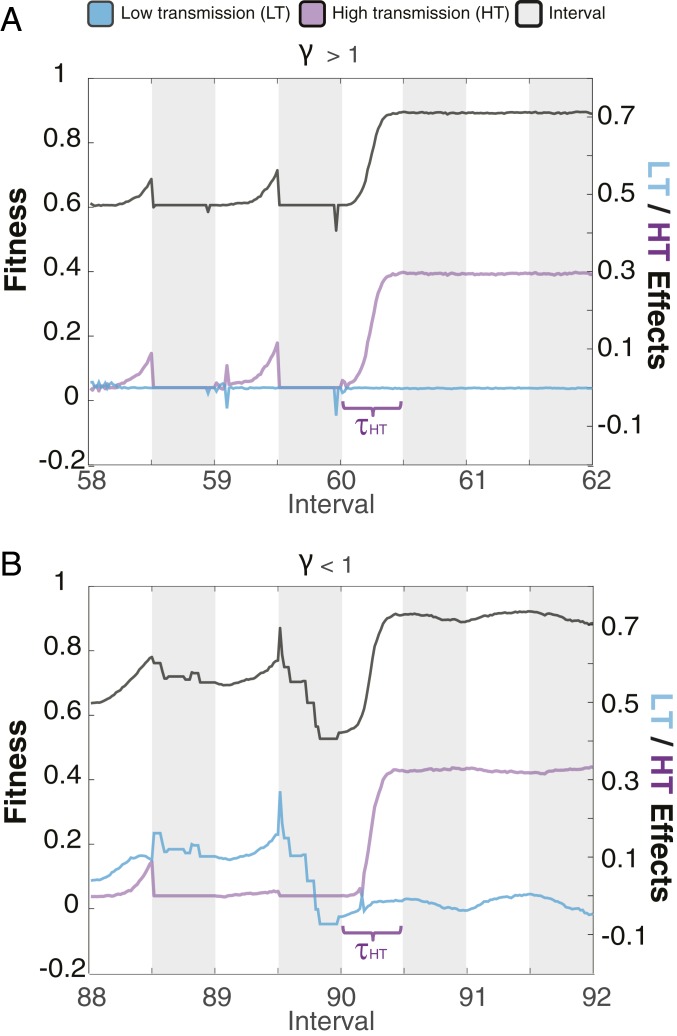
Fig. 1.
Illustrative example of model dynamics for a high-γ (A) and low-γ (B) simulation. Fitness changes (black line) are primarily driven by HT modifications (purple line) in the high-γ simulation and by both HT and LT (blue line) modifications in the low-γ simulation. The time-to-sweep (τsweep) is longer for the low-γ simulation (B) than the high-γ simulation (A). White shading denotes the “new” environment while gray shading denotes the “ancestral” environment.
In all model simulations, fitness increased rapidly with exposure to the “new” environment, consistent with laboratory experiments (5, 21–27). With stabilizing selection applied during the “ancestral” environment periods, selective sweeps driven by HT modifications emerged if the fluctuation intervals (τf) were long enough. We identified two dimensionless criteria of the relative timescales of fluctuations to the timescales of high-transmission (ε) and low-transmission (γ) modifications:
| [1] |
| [2] |
Together these criteria determined model behavior across the wide range of parameter values tested. When ε <1, the timescales of environmental variability (τf) were short relative to the fixation timescale for HT modifications (τHT) and so selective sweeps based on HT modifications were inhibited (Fig. 2A). Conversely, when ε > 1, HT selective sweeps always occurred and the time to sweep (τsweep) decreased as τf increased. In other words, longer exposure times to a new environment drove higher rates of genetic adaptation to that environment, consistent with previous results using a variety of different modeling approaches (e.g., refs. 28–30).
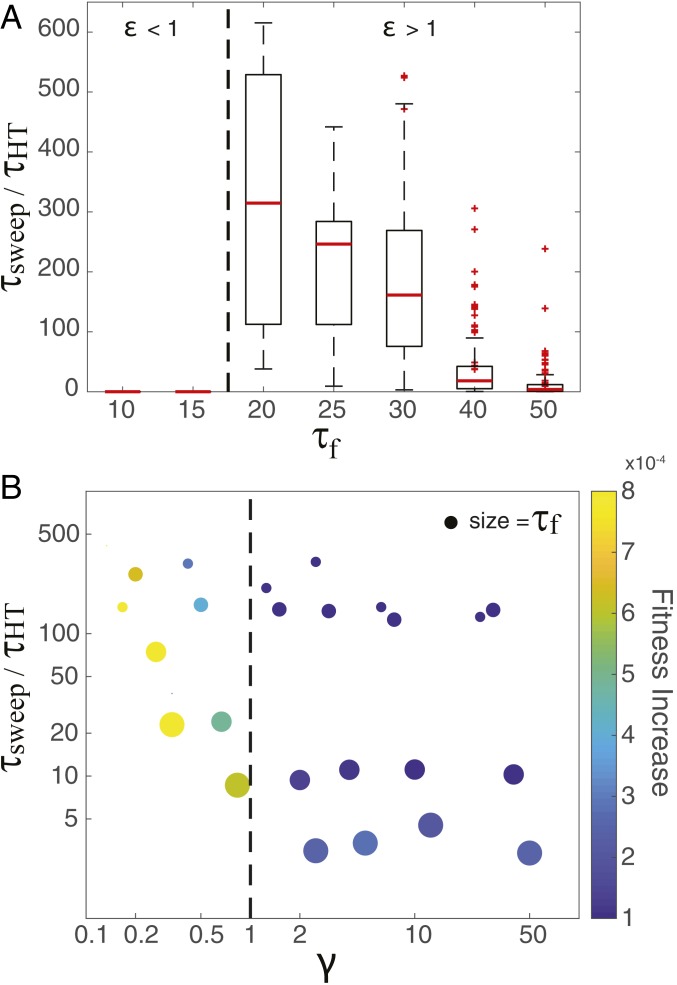
Fig. 2.
Timescales and outcomes of adaptation are determined by the values ε and γ. A illustrates the ε criteria by showing the impact of environmental fluctuations (τf) on τsweep normalized to τHT. Boxplots show the distribution of τsweep/τHT across all replicates with the median value indicated by the red central line. Outlier values are denoted with a red + sign and represent replicates with timescales more than 1.5 times the interquartile range (25th to 75th percentile). The dashed line indicates ε = 1. B illustrates the trade-off associated with a low-γ strategy by showing the relationship between the rate of fitness increase in a “new” environment (colorbar) with τsweep normalized to τHT. In B, τf is represented by the size of the symbol. The dashed line indicates γ = 1.
The second criteria, γ, identifies a key evolutionary trade-off for organisms in a fluctuating environment. When γ > 1, HT modifications drove adaptive fitness changes while LT modifications played a minor role, resulting in little or no short-term responses (i.e., fitness changes) to environmental fluctuations (Fig. 1A). However, when γ < 1, LT modifications enabled short-term fitness responses to environmental fluctuations both before and after a HT selective sweep, resembling previously observed short-term epigenetic dynamics (6) (Fig. 1B). Although simulations with γ < 1 had a more rapid response to environmental change (faster increase in fitness), it also took longer for a HT sweep to occur (larger τsweep) than simulations where γ > 1 (Fig. 2B).
These results provide a framework for understanding and predicting population-level rates of adaptation based on the relationship between environmental and microevolutionary (genetic and nongenetic) timescales. Defining the critical model timescales in terms of generations instead of days allows us to generate intuition about microbial adaptation that applies to microbes with very different growth rates and experience different environmental conditions. In a stable environment, it is advantageous to minimize adaptive timescales (smaller τsweep) and so instances where γ < 1 will be detrimental. However, in a fluctuating environment, longer adaptive timescales may be advantageous because they avoid a HT selective sweep that may be beneficial in one environment but deleterious in the other. This trade-off between short-term and long-term benefits can be framed in terms of two opposing evolutionary strategies: (i) a low-γ strategy with more persistent LT modifications which facilitates rapid environmental tracking with less heritability; and (ii) a high-γ strategy favoring more rapid selective sweeps of innovative HT modifications at the expense of shorter-term environmental fitness tracking. A low-γ strategy should be favored under enhanced environmental variability (6), while a high-γ strategy should be favored under stable conditions. In most oceanic regions, a range of strategies would be expected, since individual water masses experience different environmental fluctuation patterns before they arrive at a given location and, critically, the apparent timescale of the fluctuations will vary by species as a function of the generation time of the population (described in detail below). The ε and γ criteria provides a way to make strong hypotheses about the diversity of strategies expected in different oceanic regions.
Microevolution in a Dynamic Ocean
In the oceans, environmental fluctuations (τf) will be driven by advection into different ecoregions with distinct chemical and physical characteristics, seasonal variability, and other physical dynamics (e.g., eddies). Understanding the implications of these fluctuations on rates of microbial adaptation requires translating our understanding of the timescales of environmental variability into a microbially relevant timescale (i.e., generation times), which will be a function of cell division rates. Critically, two populations in a single parcel of water can experience the same changes in environmental conditions differently based on differences in cell division rates. The ε and γ criteria provide a framework for distilling these complex interactions between organismal and environmental timescales and generating predictions about differences in evolutionary strategies and rates of adaptation between taxa, ocean regions, and environmental drivers.
To demonstrate how the ε and γ criteria provide insight into marine microbial adaptation, we use temperature adaptation as a timely and important example. Warm temperature adaptation also provides a useful simplification in that the skewed nature of temperature tolerance curves means that the approximation of a rapid transition from “ancestral” to “new” environment is reasonable. However, the ε and γ criteria can be used to assess evolutionary strategies for any new environment with fluctuating selection. To quantify the relevant rates of environmental fluctuations, we focus on variability driven by Lagrangian movement in the ocean, as in ref. 2. This is consistent with our current understanding of the primary driver of environmental variability for marine microbes (1). The impact of more complicated physical dynamics, for example mixing of water masses, will be similar to increasing mutation rates (decreasing τHT) in our model, as these dynamics have the potential to add genetic variation to the population through immigration instead of mutation.
Using the output from the global eddy-resolving GFDL Coupled Climate CM2.6 Model (31) 2xCO2 simulation, we analyzed Lagrangian trajectories released at the surface every 1° x 1° (36,895 ocean trajectories per analysis), integrated using the OceanParcels code (32) (Methods and SI Appendix, Figs. S5 and S6). For illustrative purposes, we contrast two populations being advected along the same trajectories with environmentally relevant growth rates for marine phytoplankton (33): 0.1 d−1 (popA) and 1 d−1 (popB). We analyzed trajectories for 350 generations for each hypothetical population (2,426 d and 242 d) and calculated environmental fluctuations over each trajectory relative to both a temperature threshold (≥28 °C) and to the generation time (τf). This provides a quantitative comparison of how the same environmental variability (τf) can be experienced very differently by populations with different growth rates. For example, 30 d in waters ≥28 °C would translate into a τf = 4.3 generations for popA and τf = 43 generations for popB. In other words, for the same physical dynamics, a slower growing population (popA) would experience a more variable environment while a faster growing population (popB) would experience a more stable environment. Assuming constant growth rates is a simplification as growth rates in the real ocean will clearly vary in response to environmental fluctuations. However, one could reasonably expect that the adaptive dynamics of a population with variable growth rates would fall between the adaptive dynamics of the slow growth and fast growth populations presented here. The 350-generation timeframe was selected as experimental evolution studies have demonstrated that this is a sufficient period for adaptation to occur (25–27, 34, 35), although the conclusions of this study are not impacted by this choice. Finally, we conducted a sensitivity analysis of the 28 °C threshold and showed that the results were not a function of this specific temperature choice (SI Appendix, Figs. S9 and S10).
Differences in generation times of the two populations resulted in significantly different adaptive dynamics along the same trajectories. As a result of a shorter generation time, the exposure times of popB to ≥28 °C waters were long enough that adaptation through genetic modifications (HT) was predicted to occur. Specifically, based on the duration of physical fluctuations (τf) and a conservative estimate of τHT = 50 generations, we predict that ε >1 for 70–79% of the popB trajectories that experienced ≥28 °C (Fig. 3C). A faster τHT, due to higher genetic modification supply rates, would increase the fraction with ε >1. In contrast, because popA experienced a more variable environment due to its longer generation time, we estimate that selective sweeps (ε >1) would occur in only 2–11% of the popA trajectories (Fig. 3A). Critically, even when popA was exposed to the “new” environment every year (e.g., through seasonal fluctuations), we predict that the duration of the exposure was not sufficient to result in a selective sweep for the majority of trajectories. PopA trajectories that experienced selective sweeps were retained in warm waters for an extended period of time (>346 d). As growth rate increases and generation time decreases, the perceived environment will become less variable and seasonal fluctuations will become sufficient to drive selective sweeps. We confirmed our predictions using two representative trajectories (SI Appendix, S3). These results suggest that, within a given water parcel, directional selection is more effective for faster growing marine microbes than slower growing populations, making it more likely for HT selective sweeps to occur. This is because faster-growing populations experience the selective environment for a larger number of generations (τf).
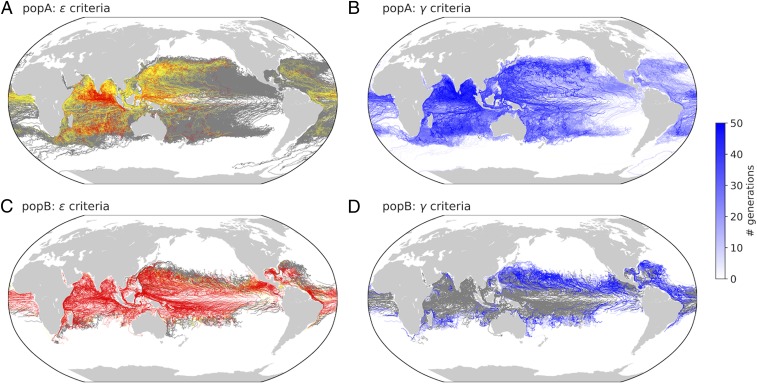
Fig. 3.
Differences in selective pressure for popA (A and B) versus popB (C and D). A and C show trajectories predicted to have ε > 1 and so experience a HT selective sweep. Here, we assume that τHT < 50 generations and so ε > 1 for trajectories with mean τf > 50 (red trajectories). This is a conservative estimate since the average model τHT = 15 ± 7 with max τHT = 60. Trajectories with the potential for a HT sweep (mean τf <50 but the maximum τf > 50) are shown in yellow, and trajectories where a sweep is unlikely (maximum τf < 50) are shown in gray. B and D show the estimated timescale of τLT necessary for a low-γ strategy. Trajectories with τLT < 50 generations are shown in shades of blue, while trajectories with τLT > 50 are shown in gray. Here, we plot a subset of the trajectories (2° x 2° grid) for clarity (see SI Appendix, Fig. S11 for all trajectories).
Consideration of the γ criteria allows us to identify the most effective strategy for each population and each trajectory. Slow-growing populations (popA) experienced fluctuation timescales that were short enough (in terms of generational time) that a low-γ strategy was beneficial based on reasonable LT transmission timescales (τLT = 10–50). Specifically, we find that 41% of the popA trajectories could employ a low-γ strategy to better track environmental fluctuations (Fig. 3B). This is in contrast to the popB trajectories where only 24% could employ a low-γ strategy (Fig. 3D); 76% of trajectories experienced environmental fluctuations that were either too fast (τf < 10) or too slow (τf > 50). Combining these results with the idealized simulations (Fig. 2) suggests that the average adaptation timescale for warm temperature adaptation (time to sweep, τsweep) could be less than 170 generations for the majority (70–79%) of popB trajectories and over 430 generations for the majority (89–98%) of popA trajectories.
This analysis identifies two contrasting strategies for marine microbes: (i) faster response to variable environments through a low-γ strategy where LT modifications provide a competitive advantage versus (ii) faster selective sweeps that provide an advantage based on HT modifications. We predict that the low-γ strategy with more persistent LT modifications will be favored by organisms that experience subjectively shorter timescale fluctuations. The above example contrasts two populations experiencing the same physical environment. However, the hypothesis also applies to organisms living in different regions. For example, relatively stable environments (e.g., oligotrophic) should favor a high-γ strategy (less LT mechanisms) while more variable environments (e.g., upwelling/coastal) should favor a low-γ strategy (more LT mechanisms). One condition needed for these dynamics to occur is that at least a subset of individuals in the population show adaptive plastic responses to the new environment before a beneficial genetic modification can occur and rise to a high frequency.
The results of our model are consistent with several recent environmental genomic studies that have attributed patterns in marine microbial diversity to local adaptation to environmental gradients driven by large-scale ocean circulation (36–38). Here, we propose an evolutionary mechanism for these biogeographical patterns and develop a mathematical framework for distilling the complexity of marine microbial adaptation into a testable hypothesis for future targeted sampling and experimental efforts. While we present a single case study for warm temperature adaptation, which is constrained to low latitudes, as the climate changes new combinations of environmental parameters (39) will drive microbial adaptation throughout the global ocean—the timescales of which can be understood in terms of the ε and γ criteria.
Untangling the interactions between the physical timescales of advection and the biological timescales of evolution is necessary to accurately predict how and where marine microbes will adapt to novel environments. Specifically, our results demonstrate that different evolutionary strategies (e.g., low-γ versus high-γ) are favored by different combinations of fluctuation patterns and cell growth rates and that these strategies can play key roles in shaping microbial fitness and underlying trait values. The importance of the interaction between physical and biological timescales in determining adaptation outcomes identifies the need to incorporate these dynamics into global carbon cycle models. Understanding these dynamics and constraining marine microbial adaptation timescales will require an improved mechanistic understanding of adaptation that includes variation from LT modifications and the quantification of critical biological timescales including τLT and τHT. This work suggests that marine microbial populations commonly experience dynamic ocean conditions that favor short-term adaptive strategies (i.e., low-γ). Expanding models of adaptive evolution to include both nongenetic processes and highly dynamic environments provides a foundation for understanding future shifts in microbial trait distributions and biogeochemical cycling in oceans.
Methods
EpiGen Model and Simulations.
To model an individual-based adaptive walk, we used a modified version of Fisher’s (18) geometric adaptation model from Kronholm and Collins (8)—the EpiGen model. LT and HT modifications drove changes in fitness where HT modifications were fixed and LT modifications reverted with probability µrev (LT reversion rate). The model was initialized with a population of N uniform individuals: here N was varied from N = 103 to N = 105. The modification supply (population size × modification rate) remained constant in each generation, and no more than one LT and one HT modification per generation was allowed to occur in a single individual. Simulations were run for 15,000 generations, and each simulation was done with 50 replicates.
We analyzed variable selection pressures through the introduction of intervals during the adaptive walk where the population moved between a “new” environment (Fig. 1, white shading) and the “ancestral” environment (Fig. 1, gray shading). Selection was based on fitness in the “new” environment such that the sampling probability of an individual was weighted by its fitness in the “new” environment until N offspring had been produced. In the “ancestral” environment, selection occurred through the stochastic removal of organisms with relatively more HT modifications (i.e., higher HT modification abundance), which corresponds to stabilizing selection. We assumed that all modifications had an equal chance of being conditionally deleterious (being neutral or adaptive in the “selection” or “new” environment, but deleterious in some other environment) so that individuals who had accumulated a high number of modifications in the selection environment had a higher probability of decreased fitness in the ancestral environment. Simulations were conducted with a range of population sizes, LT transmission timescales, and strength of stabilizing selection. A full description of the model framework and simulations are detailed in SI Appendix, Supplement Methods. The EpiGen model code is available on GitHub (https://github.com/LevineLab/EpiGen).
Global Trajectory Analysis.
Lagrangian trajectories were computed with surface velocity and sea surface temperature output from the eddy resolving, 0.1° x 0.1° horizontal resolution, GFDL Coupled Climate CM2.6 Model (31) with 2xCO2 forcing. For this study, we analyzed trajectories initialized on a 1° × 1° horizontal grid from 80°S to 70°N (resulting in 36,895 trajectories released in the ocean). Trajectories were integrated using OceanParcels code (32) version 1.0.3 with a timestep of 10 min. Location and temperature along the trajectories were recorded for illustration once per day. Two trajectory lengths were analyzed: 2,426 d (6.6 y) and 242 d of output both starting 60 y after the branch. A trajectory of length 2,426 d corresponds to 350 generations of a phytoplankton population growing at an average rate of 1 d−1. These growth rates were chosen for illustrative purposes as representative of typical growth rates for eukaryotic phytoplankton (33). Out of the 36,895 trajectories released, 27–29% experienced ≥28 °C at least once within 350 generations. Additional details on the global trajectory analysis can be found in SI Appendix, Supplemental Methods.
Supplementary Material
Acknowledgments
This work was supported by Simons Foundation Grants 509727 and 542389 (to N.M.L.), Moore Foundation Grant MMI 7397 (to N.M.L. and S.C.), National Science Foundation Grant OCE 1538525 (to N.M.L.), National Oceanic and Atmospheric Administration’s Office of Oceanic and Atmospheric Research, and a Royal Society University Research Fellowship (to S.C.). We thank L. Resplandy and J. Busecke for their assistance with the CM2.6 output and C. Thrash and three anonymous reviewers for comments on the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The model code has been deposited on GitHub (https://github.com/LevineLab/EpiGen).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919332117/-/DCSupplemental.
References
- 1.Doblin M. A., van Sebille E., Drift in ocean currents impacts intergenerational microbial exposure to temperature. Proc. Natl. Acad. Sci. U.S.A. 113, 5700–5705 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hellweger F. L., van Sebille E., Fredrick N. D., Biogeographic patterns in ocean microbes emerge in a neutral agent-based model. Science 345, 1346–1349 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Jönsson B. F., Watson J. R., The timescales of global surface-ocean connectivity. Nat. Commun. 7, 11239 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Donnell D. R., et al. , Rapid thermal adaptation in a marine diatom reveals constraints and trade-offs. Glob. Change Biol. 24, 4554–4565 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Schaum C.-E., Rost B., Collins S., Environmental stability affects phenotypic evolution in a globally distributed marine picoplankton. ISME J. 10, 75–84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veening J.-W., Smits W. K., Kuipers O. P., Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62, 193–210 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Saarinen K., Laakso J., Lindström L., Ketola T., Adaptation to fluctuations in temperature by nine species of bacteria. Ecol. Evol. 8, 2901–2910 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kronholm I., Collins S., Epigenetic mutations can both help and hinder adaptive evolution. Mol. Ecol. 25, 1856–1868 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Jablonka E., Raz G., Transgenerational epigenetic inheritance: Prevalence, mechanisms, and implications for the study of heredity and evolution. Q. Rev. Biol. 84, 131–176 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Kronholm I., Bassett A., Baulcombe D., Collins S., Epigenetic and genetic contributions to adaptation in Chlamydomonas. Mol. Biol. Evol. 34, 2285–2306 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Schmitz R. J., et al. , Transgenerational epigenetic instability is a source of novel methylation variants. Science 334, 369–373 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stajic D., Perfeito L., Jansen L. E. T., Epigenetic gene silencing alters the mechanisms and rate of evolutionary adaptation. Nat. Ecol. Evol. 3, 491–498 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Coles V. J., et al. , Ocean biogeochemistry modeled with emergent trait-based genomics. Science 358, 1149–1154 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Kiørboe T., Visser A., Andersen K. H., A trait-based approach to ocean ecology. ICES J. Mar. Sci. 75, 1849–1863 (2018). [Google Scholar]
- 15.Louca S., et al. , Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2, 936–943 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Klironomos F. D., Berg J., Collins S., How epigenetic mutations can affect genetic evolution: Model and mechanism. BioEssays 35, 571–578 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Denman K. L., A model simulation of the adaptive evolution through mutation of the Coccolithophore Emiliania huxleyi based on a published laboratory study. Front. Mar. Sci. 3, 487 (2017). [Google Scholar]
- 18.Fisher R. A., The Genetical Theory of Natural Selection (Oxford, 1930). [Google Scholar]
- 19.Kronholm I., “Adaptive evolution and epigenetics” in Handbook of Epigenetics, Tollefsbol T. O., Ed. (Academic Press, 2017), pp. 427–438. [Google Scholar]
- 20.Johannes F., Schmitz R. J., Spontaneous epimutations in plants. New Phytol. 221, 1253–1259 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Schaum C. E. , Collins S., Plasticity predicts evolution in a marine alga. Proc. Biol. Sci. 281, 20141486 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyd P. W., et al. , Experimental strategies to assess the biological ramifications of multiple drivers of global ocean change-A review. Glob. Change Biol. 24, 2239–2261 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Ward B. A., et al. , Considering the role of adaptive evolution in models of the ocean and climate system. J. Adv. Model. Earth Syst. 11, 3343–3361 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walworth N. G., et al. , Mechanisms of increased Trichodesmium fitness under iron and phosphorus co-limitation in the present and future ocean. Nat. Commun. 7, 12081 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walworth N. G., Lee M. D., Fu F.-X., Hutchins D. A., Webb E. A., Molecular and physiological evidence of genetic assimilation to high CO2 in the marine nitrogen fixer Trichodesmium. Proc. Natl. Acad. Sci. U.S.A. 113, E7367–E7374 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutchins D. A., et al. , Irreversibly increased nitrogen fixation in Trichodesmium experimentally adapted to elevated carbon dioxide. Nat. Commun. 6, 8155 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlüter L., Lohbeck K. T., Gröger J. P., Riebesell U., Reusch T. B., Long-term dynamics of adaptive evolution in a globally important phytoplankton species to ocean acidification. Sci. Adv. 2, e1501660 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiser M. J., Ribeck N., Lenski R. E., Long-term dynamics of adaptation in asexual populations. Science 342, 1364–1367 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Botero C. A., Weissing F. J., Wright J., Rubenstein D. R., Evolutionary tipping points in the capacity to adapt to environmental change. Proc. Natl. Acad. Sci. U.S.A. 112, 184–189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cvijović I., Good B. H., Jerison E. R., Desai M. M., Fate of a mutation in a fluctuating environment. Proc. Natl. Acad. Sci. U.S.A. 112, E5021–E5028 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffies S. M., et al. , Impacts on ocean heat from transient mesoscale eddies in a hierarchy of climate models. J. Clim. 28, 952–977 (2015). [Google Scholar]
- 32.Lange M., van Sebille E., Parcels v0.9: Prototyping a Lagrangian ocean analysis framework for the petascale age. Geosci. Model Dev. 10, 4175–4186 (2017). [Google Scholar]
- 33.Boyd P. W., et al. , Marine phytoplankton temperature versus growth responses from polar to tropical waters–outcome of a scientific community-wide study. PLoS One 8, e63091 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lenski R. E., Convergence and divergence in a long-term experiment with bacteria. Am. Nat. 190, S57–S68 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Collins S., Rost B., Rynearson T. A., Evolutionary potential of marine phytoplankton under ocean acidification. Evol. Appl. 7, 140–155 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delmont T. O., et al. , Single-amino acid variants reveal evolutionary processes that shape the biogeography of a global SAR11 subclade. eLife 8, e46497 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whittaker K. A., Rynearson T. A., Evidence for environmental and ecological selection in a microbe with no geographic limits to gene flow. Proc. Natl. Acad. Sci. U.S.A. 114, 2651–2656 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richter D. J., et al. , Genomic evidence for global ocean plankton biogeography shaped by large-scale current systems. bioRxiv:10.1101/867739 (6 December 2019). [DOI] [PMC free article] [PubMed]
- 39.Henson S. A., et al. , Rapid emergence of climate change in environmental drivers of marine ecosystems. Nat. Commun. 8, 14682–14689 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.