Significance
Transfer RNAs (tRNAs) are essential for mRNA translation and protein synthesis. Their expression is tightly associated with cell states and is broadly controlled by the recruitment of Pol III through intragenic elements. Whether and how tRNA genes could also be individually regulated is not known. In this study, our results reveal that a polymerase II-dependent DNA-binding transcription factor can also regulate the expression of specific tRNAs through direct binding to DNA elements surrounding the tRNA genes. Such a regulatory mode may have broader implications into understanding how cells coordinate mRNA translation and codon usage with cell state and behavior, such as proliferation and reprogramming of human glioblastoma cells.
Keywords: tRNAiMet, tRNA expression, SOX4, glioblastoma, mRNA translation
Abstract
Transfer RNAs (tRNAs) are products of RNA polymerase III (Pol III) and essential for mRNA translation and ultimately cell growth and proliferation. Whether and how individual tRNA genes are specifically regulated is not clear. Here, we report that SOX4, a well-known Pol II-dependent transcription factor that is critical for neurogenesis and reprogramming of somatic cells, also directly controls, unexpectedly, the expression of a subset of tRNA genes and therefore protein synthesis and proliferation of human glioblastoma cells. Genome-wide location analysis through chromatin immunoprecipitation-sequencing uncovers specific targeting of SOX4 to a subset of tRNA genes, including those for tRNAiMet. Mechanistically, sequence-specific SOX4-binding impedes the recruitment of TATA box binding protein and Pol III to tRNA genes and thereby represses their expression. CRISPR/Cas9-mediated down-regulation of tRNAiMet greatly inhibits growth and proliferation of human glioblastoma cells. Conversely, ectopic tRNAiMet partially rescues SOX4-mediated repression of cell proliferation. Together, these results uncover a regulatory mode of individual tRNA genes to control cell behavior. Such regulation may coordinate codon usage and translation efficiency to meet the demands of diverse tissues and cell types, including cancer cells.
Human glioblastoma represents the most aggressive and deadly type of brain tumor, characterized by uncontrolled growth and proliferation. Recently, we and others have shown that glioblastoma cells can be reprogrammed into terminally differentiated neuron-like cells through ectopic expression of fate-determining factors (1–3). These include NGN2 (also known as NEUROG2) in combination with SOX4 or SOX11 (1, 2). NGN2 is a basic helix–loop–helix transcription factor that specifies neuronal fate during development (4). SOX4 and SOX11 belong to the Sry-related high mobility group (HMG) box (SOX) family and both are essential for development and neurogenesis (5, 6). NGN2 serves as a pioneer factor to induce a neurogenic programs but itself is not sufficient for robust cell-fate reprogramming (1, 2). On the other hand, SOX4 promotes chromatin remodeling and dramatically enhances reprogramming of both human fibroblasts and glioblastoma cells (1). Cell cycle exit is a key feature of this reprogramming process, but it is not clear how it is regulated.
Transfer RNAs (tRNAs) are essential for mRNA translation and protein synthesis (7–9). They perform housekeeping functions for all cell types under physiological and pathological conditions. Their expression is dependent on TFIIIC, TFIIIB, and RNA polymerase III (Pol III). Transcription of tRNA genes is initiated by binding of TFIIIC to two intragenic control sequence blocks, the A and B boxes. TFIIIC then guides and positions TFIIIB to the upstream-of-transcription start site. TFIIIB finally recruits Pol III to start tRNA transcription. Approximately 500 tRNA genes are dispersed throughout the human genome (10, 11). Although they generally serve as housekeeping genes, emerging evidence indicates that tRNA expression may also be under cell state-dependent regulations (12–16).
In this study, we performed a systematic analysis on how NGN2/SOX4-mediated cell-fate reprogramming leads to cell cycle exit of human glioblastoma cells. We found that SOX4, but not NGN2, quickly inhibits proliferation of these tumor cells. Unexpectedly, our chromatin immunoprecipitation sequencing (ChIP-seq) analysis revealed that a large fraction of SOX4 targets are tRNA genes. Binding of SOX4 to these genes down-regulates their expression by blocking recruitment of TATA box binding protein (TBP) and Pol III. Most importantly, knocking down one of the SOX4 targets, tRNAiMet, could mimic the inhibitory effect of SOX4 on human glioblastoma cells. Such an inhibitory effect of SOX4 could be partially rescued by the ectopic expression of a SOX4-irresponsive tRNAiMet. These results provide insights into how expression of certain tRNA genes can be specifically regulated and how such regulation plays a critical biological role, including proliferation of human glioblastoma cells.
Results
SOX4 Inhibits Proliferation of Human Glioblastoma Cells.
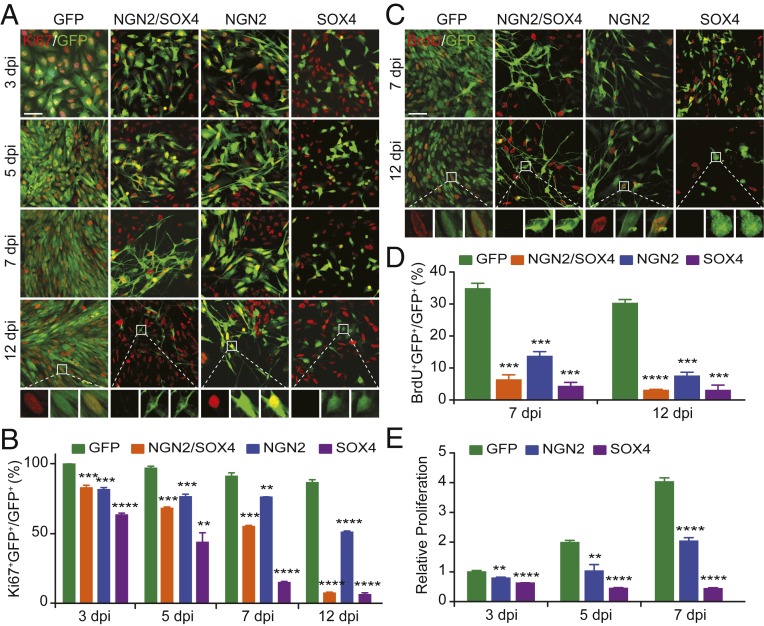
Neuronal reprogramming of human glioblastoma cells is accompanied by cell cycle exit (1, 2). We examined individual contribution of the reprogramming factors to this process. Consistent with previous findings, ectopic expression of NGN2 and SOX4 progressively but nearly completely reprogrammed U251 glioblastoma cells into neuronal morphology by 12 d postviral infection (dpi), whereas neither NGN2 nor SOX4 alone had such an effect (Fig. 1A). When cell proliferation was analyzed by expression of Ki67, an endogenous marker for all active cell cycles, coexpression of NGN2 and SOX4 also gradually induced cell cycle exit during this 12-d time-course analysis (Fig. 1 A and B). Unexpectedly, SOX4 but not NGN2 dramatically and rapidly enabled U251 cells to exit the cell cycle. Approximately 40% and 90% of SOX4-expressing cells stained negative for Ki67 at 3 and 12 dpi, respectively (Fig. 1B). This is in sharp contrast to NGN2-expressing cells, about 50% of which were still Ki67+ at 12 dpi.
Fig. 1.
SOX4 potently inhibits glioblastoma cell proliferation and growth. (A) Immunocytochemistry of proliferating glioblastoma cells indicated by Ki67-staining through a time-course analysis. Virus-transduced U251 glioblastoma cells are indicated by the coexpressed GFP. (Scale bar for lower magnification images, 50 µm.) (B) Quantification of Ki67+ glioblastoma cells at the indicated time points (mean ± SEM; n = 3; **P < 0.01, ***P < 0.001, and ****P < 0.0001). (C) Confocal images of glioblastoma cells undergoing DNA replication. BrdU was applied 2 h before immunocytochemistry. GFP expression indicates virus-transduced U251 glioblastoma cells. (Scale bar for lower magnification images, 50 µm.) (D) Quantification of BrdU+ glioblastoma cells at the indicated time points (mean ± SEM; n = 3; ***P < 0.001 and ****P < 0.0001). (E) A time-course analysis of relative cell proliferation by measuring ATP-dependent luminescence (mean ± SEM; n = 6; **P < 0.01 and ****P < 0.0001).
Cell proliferation was further analyzed by incorporation of BrdU, a synthetic analog of thymidine that can only be inserted into newly synthesized DNA during the S phase of the cell cycle. Virus-transduced U251 cells were pulse-labeled with BrdU for 2 h before immunocytochemistry at 7 and 12 dpi, respectively. Approximately 30% of the control GFP-expressing cells stained positive for BrdU at both 7 and 12 dpi (Fig. 1 C and D). Consistently, less than 5% of SOX4-expressing cells were found to be in S phase at both time points. NGN2 expression also inhibited U251 cells, but at a slower pace. The number of viable cells was then measured with luminescent assays for total cellular ATP, which serves as a proxy for metabolically active cells. A time-course analysis showed that SOX4 expression led to a significant reduction of cells as early as 3 dpi; an additional decrease was observed at 7 dpi (Fig. 1E). In contrast, the viable cells steadily increased for U251 cells expressing either NGN2 or the control GFP during these time periods.
In addition to U251 glioblastoma cells, we also examined primary glioblastoma stem cells (GSCs) directly isolated from human brains with glioblastoma. These GSCs were sorted based on expression of CD133, a marker enriched in tumor stem cells, and were maintained as self-renewable neurospheres in culture. When orthotopically transplanted, each of these GSC isolates could give rise to infiltrative and malignant brain tumors in nude mice (17). GSC proliferation was directly examined by Ki67 staining and BrdU incorporation assays (SI Appendix, Fig. S1). Expression of SOX4 remarkably decreased the number of GSCs that stained positive for Ki67 and BrdU at all of the analyzed time points (SI Appendix, Fig. S1 A–D). Measuring total cellular ATP contents of viable cells also showed completely halted increases by SOX4 expression in all three independent GSC isolates (SI Appendix, Fig. S1E). These were in sharp contrast to the control GFP-expressing GSCs, which in general doubled viable cells from 3 to 7 dpi. Together, the above results clearly indicate that SOX4 potently inhibits growth and proliferation of human glioblastoma cells and may serve as a tumor suppressor for this type of malignancy.
SOX4 Targets a Subset of tRNA Genes Revealed by ChIP-Seq.
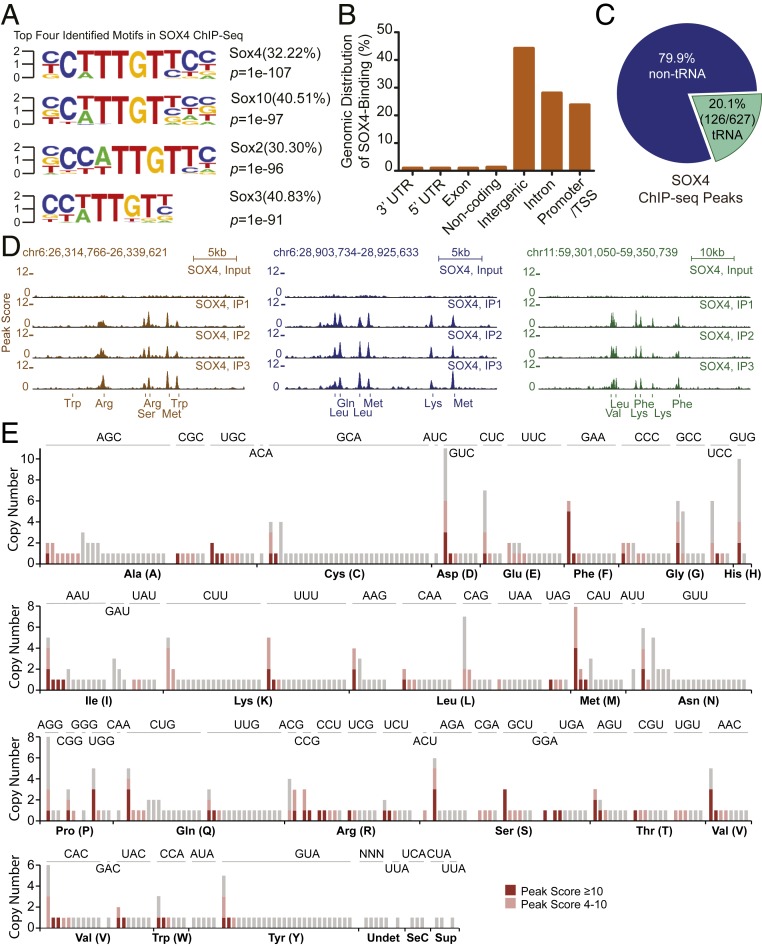
SOX4 belongs to the Sry-related high-mobility group (HMG) box (Sox) transcription factors that directly bind to consensus DNA sequences to control RNA polymerase II (Pol II)-dependent gene expression (5, 6). To understand the molecular mechanisms underlying SOX4-mediated inhibition of human glioblastoma cells, we examined genome-wide DNA-binding events of SOX4 in U251 cells through ChIP-seq in triplicates. ChIP-seq data were validated by ChIP-qPCR for candidate genes (SI Appendix, Fig. S2). A scan for transcription factor-binding motifs revealed that the top four motifs were all related to HMG-box (Fig. 2A). More than 30% of all peaks contained a consensus DNA-binding motif for SOX4, clearly showing ChIP-seq specificity. Nearly 50% of these peaks fell into intergenic regions, with introns and promoter/transcription start sites as the remaining major locations (Fig. 2B). Unexpectedly, a detailed examination of these intergenic or intronic peaks discovered that ∼20% of those ChIP-seq peaks were associated with tRNA genes (Fig. 2C and Dataset S1). Visualization of these peaks on the University of California Santa Cruz Genome Brower clearly showed that they were directly located on each respective tRNA genes and many of them are far away from neighboring Pol II-dependent genes (Fig. 2D and SI Appendix, Fig. S3).
Fig. 2.
SOX4 targets a subset of tRNA genes. (A) The top enriched DNA motifs identified through SOX4 ChIP-seq. The SOX4-binding motif represents 32.22% of all targets. (B) Genome-wide distribution of SOX4-binding sites. (C) The percentage of tRNA genes among all genes targeted by SOX4. (D) Representative ChIP-seq signal tracks surrounding SOX4-targeted tRNA genes. ChIP-seq was performed in biological triplicates (IP1, IP2, IP3). The input-DNA track was used as a control. (E) A genome-wide map of SOX4-targeted tRNA isodecoder genes indicated by pink and dark pink color.
We then mapped all SOX4-targeted human tRNA genes according to the tRNA database (GtRNAdb; http://gtrnadb.ucsc.edu/). tRNAs are well known for their diverse isodecoders, which have the same anticodons but different body sequences (18, 19). These tRNA genes, some of which have multiple genomic copies in different chromosome locations, are distributed throughout the human genome (Dataset S2). Examination of individual tRNA genes revealed that more than 200 of them were found to be bound by SOX4 from the ChIP-seq assay (Fig. 2E and Dataset S2). This number is slightly greater than the number of annotated peaks covering tRNA genes (Fig. 2C), since some of these peaks are closely located and were annotated as one.
Interestingly, SOX4 can target all copies of a single tRNA isodecoder gene on the same chromosome or even on different chromosomes (Chr). For example, both copies of a tRNAAla gene on Chr6 over 25 kb apart (tRNA-Ala-AGC-2-1 and tRNA-Ala-AGC-2-2) are targeted (Dataset S2). Each of the identical copies of another two tRNAAla genes on different chromosomes also has a SOX4-binding peak, such as tRNA-Ala-AGC-8-1 on Chr2 and tRNA-Ala-AGC-8-2 on Chr8, tRNA-Ala-TGC-3-1 on Chr5, and tRNA-Ala-TGC-3-2 on Chr12. In some cases, all copies of the same tRNA gene can be targeted by SOX4, such as eight genomic copies of one initiator tRNAiMet (tRNA-iMet-CAT-1-1 to tRNA-iMet-CAT-1-8, located in Chr1, Chr6, and Chr17) and six genomic copies of one special isodecoder gene of tRNAPhe (tRNA-Phe-GAA-1-1 to tRNA-Phe-GAA-1-6, located in Chr6, Chr11, Chr12, Chr13, and Chr19) (Fig. 2E and Dataset S2). On the other hand, none of the 23 genes for nuclear-encoded mitochondrial tRNAs (nmt-tRNA) harbors a SOX4-binding site (Dataset S2). Together, these results clearly indicate that SOX4 broadly but specifically targets many tRNA isodecoder genes in human glioblastoma cells.
SOX4 Represses Expression of Specific tRNAs.
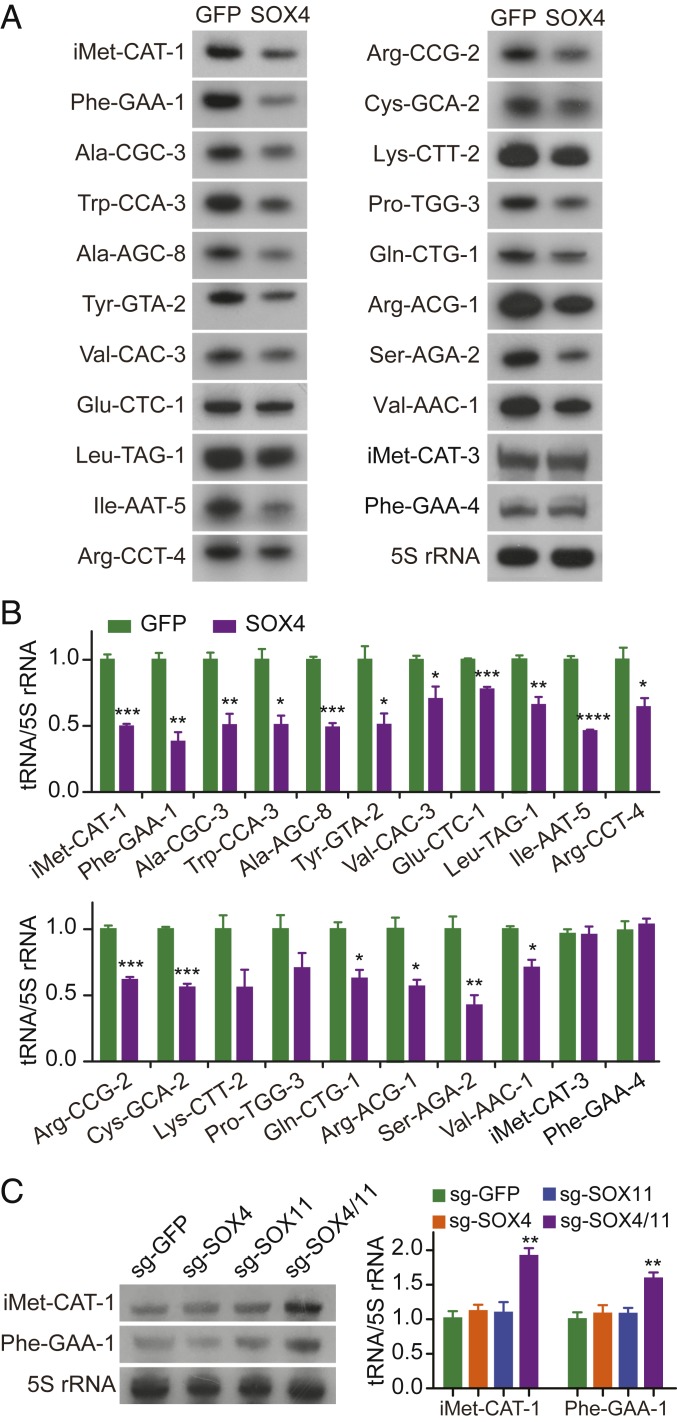
These above results are unanticipated, because SOX4 is a well-established DNA-binding transcription factor for Pol II-dependent gene expression and is not known for any role in regulating tRNA genes. To examine whether SOX4 binding to tRNA genes has any effect on their expression, we performed Northern blotting. We selected 19 of the SOX4-targeted tRNA isodecoders for analysis, based on ChIP-seq peak scores and covered gene copy numbers, and used 5S rRNA as a loading control (Dataset S2). The expression of all of those SOX4-targeted tRNA isodecoders was much reduced but to different degrees (Fig. 3 A and B). On the other hand, the expression of nontargeted and sequence divergent tRNA-iMet-CAT-3 and tRNA-Phe-GAA-4 was not affected.
Fig. 3.
SOX4 inhibits expression of specific tRNA genes. (A) Representative images of Northern blots for the indicated tRNAs at 3 dpi. 5S rRNA, which is not a SOX4 target, was used as a loading control. (B) Relative expression of SOX4-targeted and nontargeted tRNA genes after normalization to that of 5S rRNA (mean ± SEM; n = 3; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001). (C) Northern blotting analysis of the indicated tRNAs after sgRNA-mediated knockdown of SOX4 and/or SOX11 at 3 dpi. A sgRNA targeting GFP was used as a control. The expression of 5S rRNA was used as a loading control (mean ± SEM; n = 3; **P < 0.01).
We took a gene knockdown approach to further examine the effect of SOX4 on tRNA expression. Endogenous SOX4 was targeted through the CRISPR/Cas9 gene-editing technology. When compared to the GFP control, Western blotting showed a more than 60% reduction of the endogenous SOX4 protein in targeted U251 cells (SI Appendix, Fig. S4A). Nonetheless, it had no effect on the expression of the SOX4-target tRNA-iMet-CAT-1 and tRNA-Phe-GAA-1 (Fig. 3C). We then examined SOX11, since SOX4 and SOX11 share 97% similarity in the DNA-binding HMG-box (5) and exhibit redundant functions in neural development and reprogramming (1, 6). Although down-regulation of SOX11 alone also failed to show an effect, simultaneous knockdown of SOX4 and SOX11 resulted in a significant increase of the expression of tRNA-iMet-CAT-1 and tRNA-Phe-GAA-1 (Fig. 3C and SI Appendix, Fig. S4B). Together, these results indicate that the expression of specific tRNA genes is controlled by SOX4 and redundantly by SOX11 in human U251 cells.
SOX4 Reduces Protein Synthesis.
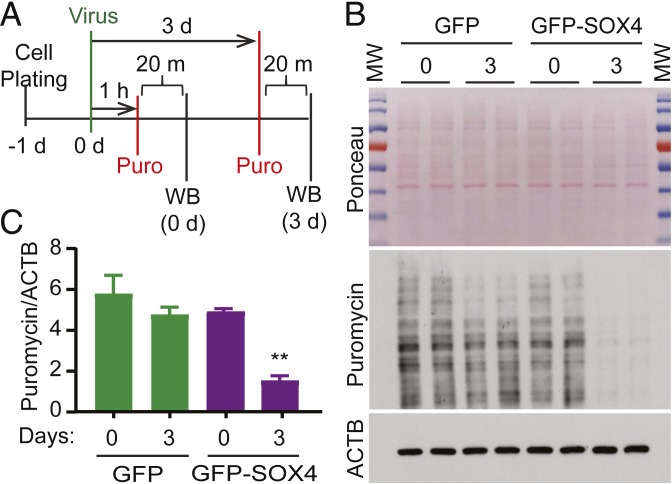
Since tRNAs are essential for mRNA translation (7–9), their down-regulation by SOX4 may have direct impacts on protein synthesis. This idea was examined by the puromycin-labeling method (20, 21). Incorporation of puromycin, an analog of aminoacyl tRNAs, into the elongating peptides results in the termination of mRNA translation and the release of the truncated peptides that are covalently linked with puromycin. The puromycin-labeled neosynthesized peptides can then be determined by Western blotting with a puromycin-specific monoclonal antibody. The amount of puromycin-labeled peptides correlates directly with the rate of mRNA translation (20, 21). We examined two time points after viral transduction of U251 cells (Fig. 4A). Western blotting revealed ∼70% reduction of puromycin-labeled peptides in cells transduced with the SOX4 but not the control GFP virus when examined at 3 dpi (Fig. 4 B and C). Equal loading of total cellular proteins for Western blotting was confirmed by both Ponceau S staining and using an antibody for β-actin (ACTB). Such a result clearly indicates that the rate of mRNA translation and protein synthesis is reduced by ectopic SOX4 presumably through its down-regulation of tRNA expression.
Fig. 4.
SOX4 represses mRNA translation. (A) A schematic diagram of the experimental design. Cells were incubated with puromycin (Puro) for 20 min (m) before lysis for Western blotting (WB). d, day; h, hour. (B) Western blotting analysis of the puromycin-labeled neosynthesized peptides. Ponceau staining and ACTB were used as loading controls. 0, 0 d; 3, 3 d; MW, molecular weight. (C) Quantification of puromycin-labeled neosynthesized peptides after normalization to ACTB expression (mean ± SEM; n = 5; **P < 0.01).
SOX4 Regulates Pol III Recruitment to Specific tRNA Genes.
We next examined in detail how SOX4 controls the expression of specific tRNA genes. Gene ontology analysis of the ChIP-seq data revealed that SOX4 targeted many genes involved in signal transduction and DNA-templated transcription (SI Appendix, Fig. S5), consistent with its well-known role in Pol II-dependent transcription (6). We checked by qRT-PCR the expression of critical genes required for tRNA transcription. Transcription of tRNA genes is initiated by binding of TFIIIC to two intragenic control sequence blocks, the A-box and B-box (7, 8). TFIIIC functions as an assembly factor by positioning TFIIIB upstream of the transcription start site. TFIIIB, which is composed of the subunits BRF1, BDP1, and TBP, then recruits Pol III to start the transcription. Our qRT-PCR data showed that the expression of all these genes was not significantly affected by SOX4 (SI Appendix, Fig. S6).
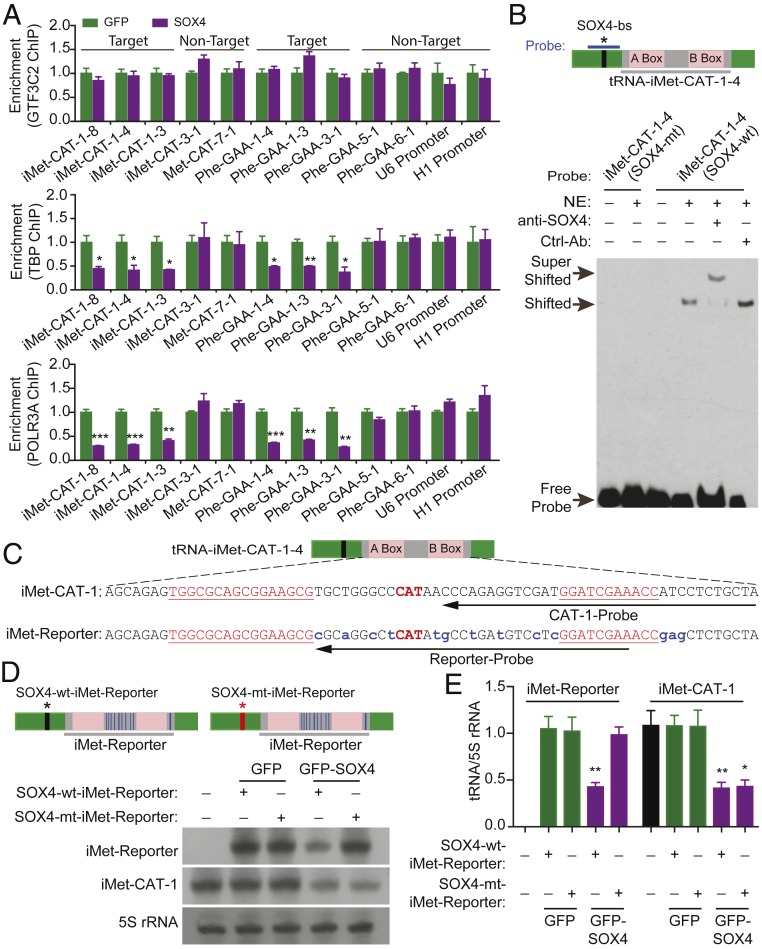
Since our ChIP-seq data clearly indicate that SOX4 binds specific tRNA genes, we then examined by ChIP-qPCR whether SOX4 had an effect on the recruitment of Pol III complexes to tRNA genes. We scanned several SOX4-targeted and nontargeted tRNA genes, and included U6 and H1 promoter regions as controls. The binding of GTF3C2 (a subunit of TFIIIC) was not influenced by the presence of SOX4 in U251 cells (Fig. 5A and SI Appendix, Fig. S7), suggesting that SOX4-binding had no effect on the initial step of tRNA transcription. In sharp contrast, the recruitments of TBP and POLR3A (the catalytic subunit of Pol III) were significantly reduced only to those SOX4-targeted tRNA genes—including iMet-CAT-1-3, iMet-CAT-1-4, iMet-CAT-1-8, Phe-GAA-1-3, Phe-GAA-1-4, and Phe-GAA-3-1—but not the nontargeted iMet-CAT-3-1, Met-CAT-7-1, Phe-GAA-5-1, and Phe-GAA-6-1 (Fig. 5A, SI Appendix, Fig. S7 and Dataset S2). In additional control tests, SOX4 had no any effect on the recruitments of Pol III transcription complexes to the U6 or H1 promoter.
Fig. 5.
SOX4 binds tRNA genes and blocks Pol III recruitment. (A) ChIP-qPCR assays for the indicated factors and genes. U6, H1, and some of the tRNA genes that are not targeted by SOX4 were used as controls (mean ± SEM; n = 3; *P < 0.05, **P < 0.01, and ***P < 0.001). (B) Gel-shift assays showing direct SOX4 binding to the indicated tRNAiMet gene. A SOX4-binding site (SOX4-bs, indicated by an asterisk)-containing DNA probe is located just upstream of the transcription initiation site. The relative positions of the intragenic control sequence blocks, A box and B box, are also shown. A DNA probe containing point mutations in the SOX4-binding site (SOX4-mt) was used as a control. Ctrl-Ab, control antibody; mt, mutant; NE, nuclear extracts from SOX4-expressing U251 cells; wt, wild-type. (C) DNA sequences of iMet-CAT-1 and a synthetic reporter. The divergent nucleotides in the reporter are marked with lowercase letters. The conserved intragenic control sequences are underlined. Probes for Northern blotting are also shown. (D) Requirement of SOX4-binding to repress tRNA expression. The synthetic reporter is under the regulation of genomic elements from tRNA-iMet-CAT-1-4. The same reporter with point mutations to the SOX4-binding site (indicated by an asterisk) was used as a control. Representative blots are shown. (E) Quantifications of reporter expression determined by Northern blotting (mean ± SEM; n = 3; *P < 0.05 and **P < 0.01).
SOX4 Directly Binds and Controls Specific tRNA Genes.
To further confirm that SOX4 indeed binds the identified tRNA genomic locations, we conducted gel-shift assays using a DNA probe located near to the 5′ region of the gene body of tRNAiMet-CAT-1-4 or tRNAThr-AGT-2-1. As a positive control, we used a 2HMG probe consisting of two tandem copies of a CD3E minimal enhancer, which harbors a core HMG box protein-binding sequence (5, 22). Incubation with nuclear extracts from SOX4-expressing U251 cells resulted in retarded migration of the probes, which could be further shifted by the addition of an antibody for SOX4 but not a control antibody (Fig. 5B and SI Appendix, Fig. S8). Importantly, such gel-shift was completely abolished when point mutations were introduced to the core SOX4-binding site of the tRNAiMet-CAT-1-4 probe (Fig. 5B).
To determine whether SOX4-binding is critical for tRNA expression, we created a tRNAiMet reporter. According to sequence comparisons between human and yeast tRNAiMet (23, 24), we made point mutations in the gene body of human tRNAiMet-CAT-1-4 (Fig. 5C). These mutations were not in the conserved regions thought to be required for transcription by Pol III and were meant to keep the cloverleaf structure of tRNAiMet. The large sequence differences between the reporter and the endogenous tRNAiMet enabled us to clearly distinguish them through Northern blotting. This reporter along with the 5′ and 3′ flanking genomic regions of tRNAiMet-CAT-1-4 was subcloned into a lentiviral vector. As a control, we also made point mutations to the SOX4-binding site in the 5′ genomic region of tRNAiMet-CAT-1-4 (Fig. 5D). Reporter activities in virus-transduced U251 cells were examined through Northern blotting. As expected, ectopic SOX4 greatly repressed the expression of both the endogenous tRNAiMet and the reporter with an intact SOX4-binding site (Fig. 5 D and E). Mutations to the SOX4-binding site rendered the reporter completely irresponsive to SOX4 expression, indicating that direct genomic binding of SOX4 is critical for its repressive activity on tRNA expression.
Repressing tRNAiMet Expression Inhibits Glioblastoma Cell Proliferation.
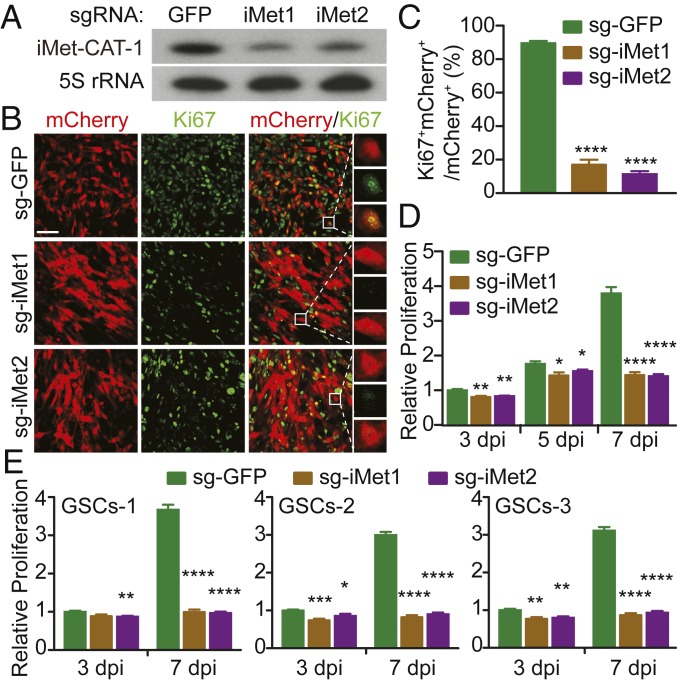
As shown above, one of the most significant SOX4 targets is tRNAiMet. tRNAiMet is encoded by three different isodecoder genes with one gene having eight copies and the remaining two being single-copy genes (Dataset S2). These genes are located in 10 separate genomic loci on 4 different chromosomes. All eight copies of one tRNAiMet isodecoder gene are targeted by SOX4 (tRNA-iMet-CAT-1-1 to tRNA-iMet-CAT-1-8) (Dataset S2), suggesting a critical role of this gene in mediating SOX4 function in human glioblastoma cells. Using CRISPR/Cas9 gene-editing technology, we designed two similar single-guide RNAs (sgRNAs) targeting all copies of this tRNAiMet gene (Dataset S2). A GFP-targeting sgRNA was used as a control. Both Cas9 enzyme and the sgRNA were delivered through lentivirus into glioblastoma cells. The coexpressed mCherry was used for monitoring viral transduction efficiency and identifying sgRNA-expressing cells. Northern blotting analysis confirmed that the two specific sgRNAs in U251 cells markedly reduced expression of tRNAiMet (Fig. 6A). When examined at 7 dpi, knockdown of tRNAiMet led to a significant reduction of cells stained positive for Ki67 (Fig. 6 B and C). A time-course analysis of viable cells also confirmed a significant inhibitory effect of knocking down tRNAiMet on U251 cell growth and proliferation (Fig. 6D). We extended this analysis to three primary human GSC isolates. The viable cells for all control sgRNA-expressing GSCs increased more than twofold at 7 dpi compared to 3 dpi (Fig. 6E). In contrast, knockdown of tRNAiMet nearly completely blocked this increase. These results clearly indicate that tRNAiMet expression is closely linked to growth and proliferation of human glioblastoma cells.
Fig. 6.
Expression of tRNAiMet controls glioblastoma cell growth. (A) Northern blotting analysis showing down-regulated expression of tRNAiMet by two separate targeting sgRNAs in U251 glioblastoma cells. A sgRNA targeting GFP was used as a control. The expression of 5S rRNA was used as loading controls. (B) Confocal images of proliferating U251 glioblastoma cells after transduction of virus expressing the indicated sgRNAs. Virus-transduced cells are identified by the coexpressed mCherry at 7 dpi. (Scale bar for lower magnification images, 50 µm.) (C) Quantification of the effect of knockdown of tRNAiMet on cell proliferation at 7 dpi (mean ± SEM; n = 3; ****P < 0.0001). (D) A time-course analysis of U251 cell proliferation by measuring ATP-dependent luminescence (mean ± SEM; n = 6; *P < 0.05, **P < 0.01, and ****P < 0.0001). (E) Knockdown of tRNAiMet greatly reduces proliferation of human GSCs. Cell proliferation was measured by ATP-dependent luminescence for three individual human GSC isolates (GSCs-1, GSCs-2, and GSCs-3; mean ± SEM; n = 6; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001).
Ectopic tRNAiMet Partially Rescues SOX4-Mediated Proliferation Inhibition.
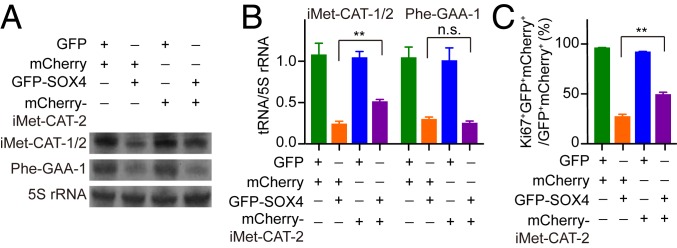
We finally examined through ectopic expression the role of tRNAiMet on SOX4-mediated proliferation inhibition of U251 cells. Although tRNA-iMet-CAT-1 and tRNA-iMet-CAT-2 differs only by one nucleotide in the gene body, the former but not the latter is targeted by SOX4 (Dataset S2). We made a lentivirus to express tRNA-iMet-CAT-2 under its endogenous regulatory elements, including the 5′ and 3′ genomic regions. U251 cells transduced with this virus could be identified by the coexpressed human PGK promoter-driven mCherry. Using a probe recognizing both iMet-CAT-1 and iMet-CAT-2 for Northern blotting, the result showed that ectopic iMet-CAT-2 moderately but significantly increased the overall tRNAiMet level in SOX4-overexpressing U251 cells. As a control, ectopic iMet-CAT-2 had no effect on the expression of endogenous Phe-GAA-1 (Fig. 7 A and B). Cell proliferation was determined by Ki67 staining at 7 dpi in virus-transduced U251 cells, which were identified by the coexpressed GFP and mCherry. A quantitative analysis revealed that ectopic iMet-CAT-2 partially but significantly rescued the proliferation defect of SOX4-overexpressing U251 cells (Fig. 7 B and C and SI Appendix, Fig. S9). On the other hand, both tRNAiMet expression and cell proliferation were not obviously altered by ectopic iMet-CAT-2 in the control GFP-expressing U251 cells, implicating a possible feedback mechanism in these cells under optimal growth conditions.
Fig. 7.
Rescue of SOX4-mediated inhibition of proliferation by ectopic tRNAiMet. (A) Representative Northern blots for tRNA expression. The probe for tRNAiMet recognizes both iMet-CAT-1 and iMet-CAT-2. (B) Quantification of tRNA expression through Northern blotting analysis (mean ± SEM; n = 3; **P < 0.01; n.s., not significant). (C) Quantification of cell proliferation indicated by Ki67-staining of U251 cells at 7 dpi (mean ± SEM; n = 3; **P < 0.01).
Discussion
Although it has long been observed that individual tRNA expression exhibits tissue- and cell-type–specific differences (25, 26), how such specificity is achieved is not clear (27). To our knowledge, our results are unique in revealing that a Pol II-dependent DNA-binding transcription factor can also regulate Pol III-transcribed individual tRNA genes through binding to genomic DNA sequences. Such a regulatory mode may have broader implications into understanding on how cells coordinate mRNA translation and codon usage with cell state and behavior. An example shown here is human glioblastoma cells, which can be efficiently reprogrammed into neurons by ectopic expression of NGN2 and SOX4. A key role of SOX4 is to promote cell cycle exit through direct suppression of a subset of tRNAs, thereby reducing protein translation (SI Appendix, Fig. S10).
Transcription of tRNA genes is generally believed to be globally regulated at the level of Pol III recruitment and activity controls (7, 8, 27). As limiting factors for recruitment of Pol III, both TFIIIB and TFIIIC are subject to transcriptional and posttranslational regulation, including phosphorylation (27). Through physical interactions with Pol III but not direct binding to tRNA genes, the evolutionarily conserved protein MAF1 is a converging point from multiple signaling pathways and broadly represses Pol III activity and tRNA expression (27–30). Another mode of regulation is through direct protein–protein interactions with TFIIIB to either prevent or stimulate promoter recruitment of Pol III and subsequent tRNA expression, such as p53 and MYC (31–33). Such regulation is not believed to be mediated through DNA-binding activity of the transcription factors, because no consensus DNA sequences are present in these tRNA genes. Studies also further demonstrate that DNA-binding of MYC is not required for it to stimulate Pol III transcription (34). Our genome-wide location analysis showed that SOX4-binding site can be specifically identified in a subset of tRNA genes. This is a regulatory mode in which the expression of individual tRNA genes can be finely tuned through DNA sequence-specific binding of a transcription factor. This work also raises the possibility that individual tRNA genes may potentially be regulated by many other DNA-binding transcription factors in a tissue and cell-type–specific manner.
The binding of SOX4 to multiple copies, or even all copies, of a tRNA gene is very intriguing and indicates the critical importance of conservation of SOX4-dependent regulation during tRNA gene duplication. For example, all eight copies of one tRNAiMet gene are targeted by SOX4, and its expression is greatly repressed by ectopic SOX4 expression. As an essential component for initiation of mRNA translation and protein synthesis (35), tRNAiMet abundance plays a critical role in protein translation rate (36). Overexpression of tRNAiMet is detected in multiple cancer cell lines (37) and it can increase metabolic activity, cell proliferation, and metastatic potential (38, 39). Consistent with these data, mRNA translation rate was greatly reduced in SOX4-overexpressing cells. Our CRISPR/Cas9-mediated knockdown of tRNAiMet also showed markedly reduced growth and proliferation of human glioblastoma cells. Conversely, SOX4-mediated proliferation inhibition could be partially rescued by the ectopic expression of a tRNAiMet. It should be noted that our data do not exclude the role of SOX4-targeted Pol II-dependent genes in controlling behaviors of U251 cells. As a transcription factor and shown by our ChIP-seq, SOX4 can regulate many genes in addition to tRNAs.
The role of SOX4 seems to be cell context-dependent. It is proposed as either an oncogene or a tumor suppressor depending on the cancer cell types (40). Even in gliomas, its role is also less clear. SOX4 expression is positively correlated with good prognosis of human patients with primary glioblastoma (41). It inhibits cell growth and promotes cell cycle arrests in a p53-dependent manner (41). On the other hand, SOX4 induction by TGF-β is proposed to be critical for stemness maintenance of glioma-initiating cells (42, 43). Our results that SOX4 represses specific tRNA expression and thereby cell proliferation add further complexity to its function. In addition to regulation at the level of gene expression, SOX4 function is also modulated by posttranslational modification and the binding of cofactors (40). Such regulations may be critical for conferring a cell context-dependent role of SOX4 by controlling its DNA-binding specificity, transcriptional activity, or cofactor function.
In conclusion, our results reveal a regulatory mode of tRNA expression, by which individual tRNA genes can be specifically regulated by a DNA-binding transcription factor. Such regulation may coordinate codon usage and translation efficiency to meet the demands of diverse tissues and cell types, including cancer cells. SOX4-dependent regulation of a subset of tRNA genes and its role in controlling human glioblastoma cells provides a rationale to further explore this regulatory mode for other transcription factors in a broad range of physiological and pathological conditions.
Materials and Methods
Cell Culture.
HEK293 (CRL-1573, ATCC) and U251 glioma cells (human U-251 MG, Sigma) were maintained in DMEM supplemented with 10% (vol/vol) FBS. GSC isolates were previously described (17). For neural differentiation, virus-transduced cells were switched 48 h later to differentiation medium consisting of DMEM/F12/neurobasal (2:2:1 dilutions), 0.8% (vol/vol) N2/0.4% (vol/vol) B27, 10 µM forskolin (Sigma), and 1 µM dorsomorphin (Millipore). The differentiation medium was half-changed every 2 d until analysis. For all other assays, cells were kept in DMEM supplemented with 10% FBS. All cells were maintained at 37 °C in a humidified incubator with 5% CO2.
Plasmid Construction and Lentivirus Production.
The CS-CDF-CG-PRE lentiviral vector was used for subcloning of NGN2-IRES-GFP, NGN2-IRES-GFP-T2A-SOX4, SOX4-IRES-GFP, or HA-SOX4-IRES-GFP. The pRRLSIN.cPPT.PGK-GFP.WPRE vector was used for expression of tRNA iMet-CAT-2 or synthetic tRNA iMet-Reporter, both of which were subcloned by PCR with corresponding flanking genomic sequences of iMet-CAT-2-1 or iMet-CAT-1-4 and were placed upstream of the PGK promoter. For the tRNA iMet-CAT-2 construct, the GFP cDNA was further replaced with the IRES-mCherry cDNA. The lentiCRISPRv2 vector (Addgene) was used for expression of sgRNAs. The selection marker puromycin in the lentiCRISPRv2 vector was replaced with mCherry for identification of virus-transduced cells. All plasmids were sequenced for confirmation. Lentivirus was prepared essentially as previously described (1, 2, 44, 45).
Immunocytochemistry and Cell Proliferation.
Cells for immunochemistry were cultured on glass coverslips in 24-well plates. Cells were gently washed with ice-cold 1×PBS, fixed with 4% paraformaldehyde for 15 min at room temperature, and then permeabilized with two washes of PBST (1×PBS containing 0.2% [vol/vol] Triton X-100) for 5 min each at room temperature. For BrdU staining, cells were further treated with 2 M HCl for 30 min at 37 °C and 0.1 M borate buffer for 10 min at room temperature. Cells were then blocked with 3% BSA-containing PBST solution and sequentially incubated with primary and secondary antibodies (Dataset S3). Cells were imaged by using the Zeiss LSM 700 confocal microscope. For quantifications, cells were processed in triplicates and about 100 to 400 cells were counted in three random areas for each replicate. Cell growth and proliferation was also measured by using the ATP-dependent CellTiter-Glo 2.0 Luminescent Assay kit (Promega). Briefly, cells were seeded in 96-well plates (1,000 cells per well) and were then transduced with the indicated lentivirus 16 h later. Six wells were used for each condition. Cells were monitored by the coexpressed GFP marker to confirm high transduction efficiency (∼95%). ATP-dependent luminescence for each well was measured by using a plate reader (BioTek) and was then normalized to the control GFP at 3 dpi.
ChIP-Seq and ChIP-qPCR.
ChIP-seq was performed as previously described (1). ChIP-qPCR was conducted by using the SYBR Green chemistry. See SI Appendix for more details.
qRT-PCR.
Total cellular RNAs were isolated by using the TRIzol method from cells at 3 dpi. One microgram total RNA from each sample were used for reserve transcription by using SuperScript III Reverse Transcriptase and random primers. Quantitative real-time PCR was then performed using parameters as above described. Relative gene expression was determined by the ΔΔCt method (where Ct is the threshold cycle) after normalizing to the expression of HPRT. Primer sequences are listed in Dataset S3.
tRNA Isodecoder Map.
The SOX4 ChIP-seq dataset was used to determine all SOX4-targeted tRNAs based on peak score. The tRNA database (GtRNAdb; http://gtrnadb.ucsc.edu/) was used to map tRNA copy numbers and isodecoder distributions. The detailed data on SOX4-targeted tRNA isodecoders and peak scores are listed in Dataset S2.
Gel-Shift Assays.
Gel-shift assays were conducted by using dsDNA probes as previously described (46). See SI Appendix for more details.
tRNAiMet Reporter Assays.
U251 cells were cotransduced with respective combinations of lentiviruses expressing GFP, GFP-SOX4, tRNAiMet reporter with a wild-type SOX4-binding site, or a tRNAiMet reporter with point-mutations in the SOX4-binding site. Total cellular RNAs were isolated by using the TRIzol method from virus transduced cells at 3 dpi. Gene expression was analyzed by Northern blotting with probes listed in Dataset S3.
Northern Blotting.
Total cellular RNAs were isolated from lentivirus transduced U251 cells at 3 dpi. A total of 10 µg RNAs were separated on a 15% denaturing acrylamide gel and blotted onto Zeta-Probe GT Genomic Tested Blotting Membranes (Bio-Rad). After UV cross-linking, membranes were hybridized to 32P-labeled oligonucleotide probes in SES solution (0.5 M sodium phosphate [pH7.2], 7% SDS, and 1 mM EDTA). The probes were designed to target the selected tRNA isodecoders, synthetic reporter, or the control 5S rRNA (Dataset S3). After overnight incubation with gentle shaking at 37 °C, the membranes were extensively washed with 6×SSPE buffer (20×SSPE: 3.6 M NaCl, 0.2 M NaH2PO4, 0.02 M EDTA [pH7.2]) twice at 37 °C and twice at 42 °C and then exposed to film for autoradiography. Band density (intensity) was analyzed by ImageJ and the Gel Analysis plugin.
Gene Ontology Analysis.
SOX4-targeted genes from ChIP-seq data were manually categorized into tRNA genes and nontRNA genes based on their detailed annotations (Dataset S1). The nontRNA genes were subjected to gene ontology analysis for biological processes or cell signaling using the DAVID Functional Annotation Tool (v6.7).
mRNA Translation Rate and Western Blotting.
For mRNA translation analysis, U251 cells were transduced with lentivirus expressing GFP or GFP-SOX4. At the indicated time points, puromycin (10 µg/mL, final concentration) was added into the medium and incubated for another 20 min. After a brief wash with ice-cold 1×PBS, cells were lysed in RIPA buffer (150 mM NaCl, 1% [vol/vol] Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris [pH 8.0] and 1% Protease Inhibitor Cocktail). For confirmation of sgRNA-mediated gene knockdown, U251 cells were transduced with the respective lentivirus and lysed in RIPA buffer at 3 dpi. Protein concentration was measured with the Pierce BCA Protein Assay Kit (Thermo Scientific, Cat #23227). Protein samples (10 µg each) were then separated by 10% SDS–PAGE and transferred onto PVDF (polyvinylidene difluoride) membranes (Millipore). Protein loading was also checked with Ponceau S staining, for which the PVDF membrane was incubated for 5 min with Ponceau S solution (0.1% [wt/vol] in 5% [vol/vol] acetic acid, Sigma, P7170) followed by rinsing with distilled water. For Western blotting, the PVDF membranes were sequentially blotted with the corresponding primary and secondary antibodies (Dataset S3) and processed for enhanced chemiluminescence detection.
Statistics.
Experiments were performed in triplicates unless otherwise indicated. Data are presented as mean ± SEM. Statistical analysis was performed by homoscedastic two-tailed Student’s t-test. Significant differences are indicated by *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Materials and Data Availability.
The accession number for the bioinformatics datasets is Gene Expression Omnibus GSE138328.
Supplementary Material
Acknowledgments
We thank members of the C.-L.Z. laboratory for discussions and reagents; Yuhua Zou for laboratory management; Robert Bachoo for glioblastoma stem cell isolates; and the Genomics & Microarray Core at University of Texas Southwestern for HiSeq and Bioinformatics. C.-L.Z. is a W. W. Caruth, Jr. Scholar in Biomedical Research and was supported by the Welch Foundation (I-1724), the Decherd Foundation, the Pape Adams Foundation, Texas Institute for Brain Injury and Repair, Kent Waldrep Foundation Center for Basic Research on Nerve Growth and Regeneration, and NIH Grants NS099073, NS092616, NS088095, and NS093502. Q.Z. was supported by the National Natural Science Foundation of China (No. 81771262), Zhejiang Health Science and Technology Project (2016RCA022), and Zhejiang Key Research and Development Project 2017C03027.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE138328).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920200117/-/DCSupplemental.
References
- 1.Smith D. K., Yang J., Liu M. L., Zhang C. L., Small molecules modulate chromatin accessibility to promote NEUROG2-mediated fibroblast-to-neuron reprogramming. Stem Cell Reports 7, 955–969 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su Z., et al. , Reprogramming the fate of human glioma cells to impede brain tumor development. Cell Death Dis. 5, e1463 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guichet P. O., et al. , Cell death and neuronal differentiation of glioblastoma stem-like cells induced by neurogenic transcription factors. Glia 61, 225–239 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Ma Q., Kintner C., Anderson D. J., Identification of neurogenin, a vertebrate neuronal determination gene. Cell 87, 43–52 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Dy P., et al. , The three SoxC proteins—Sox4, Sox11 and Sox12—exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 36, 3101–3117 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kavyanifar A., Turan S., Lie D. C., SoxC transcription factors: Multifunctional regulators of neurodevelopment. Cell Tissue Res. 371, 91–103 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Arimbasseri A. G., Maraia R. J., RNA polymerase III advances: Structural and tRNA functional views. Trends Biochem. Sci. 41, 546–559 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White R. J., Transcription by RNA polymerase III: More complex than we thought. Nat. Rev. Genet. 12, 459–463 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Schramm L., Hernandez N., Recruitment of RNA polymerase III to its target promoters. Genes Dev. 16, 2593–2620 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Parisien M., Wang X., Pan T., Diversity of human tRNA genes from the 1000-genomes project. RNA Biol. 10, 1853–1867 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan P. P., Lowe T. M., GtRNAdb 2.0: An expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44, D184–D189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kantidakis T., Ramsbottom B. A., Birch J. L., Dowding S. N., White R. J., mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc. Natl. Acad. Sci. U.S.A. 107, 11823–11828 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsang C. K., Liu H., Zheng X. F., mTOR binds to the promoters of RNA polymerase I- and III-transcribed genes. Cell Cycle 9, 953–957 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson S. A., Dubeau L., Johnson D. L., Enhanced RNA polymerase III-dependent transcription is required for oncogenic transformation. J. Biol. Chem. 283, 19184–19191 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felton-Edkins Z. A., et al. , The mitogen-activated protein (MAP) kinase ERK induces tRNA synthesis by phosphorylating TFIIIB. EMBO J. 22, 2422–2432 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cairns C. A., White R. J., p53 is a general repressor of RNA polymerase III transcription. EMBO J. 17, 3112–3123 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marian C. O., et al. , The telomerase antagonist, imetelstat, efficiently targets glioblastoma tumor-initiating cells leading to decreased proliferation and tumor growth. Clin. Cancer Res. 16, 154–163 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schimmel P., The emerging complexity of the tRNA world: Mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 19, 45–58 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Pan T., Modifications and functional genomics of human transfer RNA. Cell Res. 28, 395–404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marciano R., Leprivier G., Rotblat B., Puromycin labeling does not allow protein synthesis to be measured in energy-starved cells. Cell Death Dis. 9, 39 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt E. K., Clavarino G., Ceppi M., Pierre P., SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Harley V. R., et al. , DNA binding activity of recombinant SRY from normal males and XY females. Science 255, 453–456 (1992). [DOI] [PubMed] [Google Scholar]
- 23.Drabkin H. J., RajBhandary U. L., Attempted expression of a human initiator tRNA gene in Saccharomyces cerevisiae. J. Biol. Chem. 260, 5596–5602 (1985). [PubMed] [Google Scholar]
- 24.Drabkin H. J., Estrella M., Rajbhandary U. L., Initiator-elongator discrimination in vertebrate tRNAs for protein synthesis. Mol. Cell Biol. 18, 1459–1466 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dittmar K. A., Goodenbour J. M., Pan T., Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2, e221 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gingold H., et al. , A dual program for translation regulation in cellular proliferation and differentiation. Cell 158, 1281–1292 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Graczyk D., Cieśla M., Boguta M., Regulation of tRNA synthesis by the general transcription factors of RNA polymerase III: TFIIIB and TFIIIC, and by the MAF1 protein. Biochim. Biophys. Acta. Gene Regul. Mech. 1861, 320–329 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Moir R. D., Willis I. M., Regulation of pol III transcription by nutrient and stress signaling pathways. Biochim. Biophys. Acta 1829, 361–375 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oficjalska-Pham D., et al. , General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol. Cell 22, 623–632 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Roberts D. N., Wilson B., Huff J. T., Stewart A. J., Cairns B. R., Dephosphorylation and genome-wide association of Maf1 with Pol III-transcribed genes during repression. Mol. Cell 22, 633–644 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crighton D., et al. , p53 represses RNA polymerase III transcription by targeting TBP and inhibiting promoter occupancy by TFIIIB. EMBO J. 22, 2810–2820 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomez-Roman N., Grandori C., Eisenman R. N., White R. J., Direct activation of RNA polymerase III transcription by c-Myc. Nature 421, 290–294 (2003). [DOI] [PubMed] [Google Scholar]
- 33.White R. J., RNA polymerase III transcription and cancer. Oncogene 23, 3208–3216 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Steiger D., Furrer M., Schwinkendorf D., Gallant P., Max-independent functions of Myc in Drosophila melanogaster. Nat. Genet. 40, 1084–1091 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Kolitz S. E., Lorsch J. R., Eukaryotic initiator tRNA: Finely tuned and ready for action. FEBS Lett. 584, 396–404 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gingold H., Pilpel Y., Determinants of translation efficiency and accuracy. Mol. Syst. Biol. 7, 481 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pavon-Eternod M., et al. , tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 37, 7268–7280 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pavon-Eternod M., Gomes S., Rosner M. R., Pan T., Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. RNA 19, 461–466 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Birch J., et al. , The initiator methionine tRNA drives cell migration and invasion leading to increased metastatic potential in melanoma. Biol. Open 5, 1371–1379 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vervoort S. J., van Boxtel R., Coffer P. J., The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: Friend or foe? Oncogene 32, 3397–3409 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Zhang J., et al. , SOX4 inhibits GBM cell growth and induces G0/G1 cell cycle arrest through Akt-p53 axis. BMC Neurol. 14, 207 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ikushima H., et al. , Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell 5, 504–514 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Lin B., et al. , Massively parallel signature sequencing and bioinformatics analysis identifies up-regulation of TGFBI and SOX4 in human glioblastoma. PLoS One 5, e10210 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu M. L., et al. , Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat. Commun. 4, 2183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu M. L., Zang T., Zhang C. L., Direct lineage reprogramming reveals disease-specific phenotypes of motor neurons from human ALS patients. Cell Rep. 14, 115–128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang C. L., Zou Y., Yu R. T., Gage F. H., Evans R. M., Nuclear receptor TLX prevents retinal dystrophy and recruits the corepressor atrophin1. Genes Dev. 20, 1308–1320 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.