Significance
The lancet fluke Dicrocoelium dendriticum can precisely manipulate its ant host to facilitate transmission to its ungulate host. However, the sole parasite manipulator in the ant brain is presumed not infective to its next host whereas the parasites in the abdomen of the same ant are infective. We conducted a test of genetic relatedness between the brain fluke to those flukes in the abdomen. Our data show that clonemates (i.e. genetically identical individuals) are cotransmitted into ants and that the brain fluke is often a clonemate of flukes in the abdomen. Hence, our study provides empirical evidence that supports a role for kin selection in the evolution of a host-manipulating behavior.
Keywords: Trematoda, host manipulation, host behavior, brain fluke, genetic relatedness
Abstract
Host manipulation by parasites is a fascinating evolutionary outcome, but adaptive scenarios that often accompany even iconic examples in this popular field of study are speculative. Kin selection has been invoked as a means of explaining the evolution of an altruistic-based, host-manipulating behavior caused by larvae of the lancet fluke Dicrocoelium dendriticum in ants. Specifically, cotransmission of larval clonemates from a snail first host to an ant second host is presumed to lead to a puppeteer parasite in the ant’s brain that has clonemates in the ant abdomen. Clonal relatedness between the actor (brain fluke) and recipients (abdomen flukes) enables kin selection of the parasite’s host-manipulating trait, which facilitates transmission of the recipients to the final host. However, the hypothesis that asexual reproduction in the snail leads to a high abundance of clonemates in the same ant is untested. Clonal relationships between the manipulator in the brain and the nonmanipulators in the abdomen are also untested. We provide empirical data on the lancet fluke’s clonal diversity within its ant host. In stark contrast to other trematodes, which do not exhibit the same host-manipulating behavioral trait, the lancet fluke has a high abundance of clonemates. Moreover, our data support existing theory that indicates that the altruistic behavior can evolve even in the presence of multiple clones within the same ant host. Importantly, our analyses conclusively show clonemate cotransmission into ants, and, as such, we find support for kin selection to drive the evolution and maintenance of this iconic host manipulation.
“Is it true that Dicrocoelium dendriticum actually consists of a mix of altruists and nonaltruists?…Wickler (1) called attention to the brain worm as an example of altruism, Wilson (2) predicted the likelihood of a polymorphism, and no relevant studies have appeared since then. No one has estimated the basic parameters of the model, such as the numbers of eggs ingested by snails and the number of mucus masses ingested by ants. The brain worm remains a fascinating prima facie example of altruism from the field of natural history, but the conceptually relevant details have only been guessed.” –Sober and Wilson (3)
Research on host manipulation by parasites has gained popularity (4–6), as evidenced, in part, by the steady increase of empirical studies on trophically transmitted helminths since the early 1970s (7). Moreover, theoretical/conceptual studies involving host manipulation have spiked since the mid-2000s (7). Despite these encouraging trends for evolutionary ecology research on parasites, Poulin and Maure (7) provided a reality check by showing that there has been a proportionate decrease (relative to the identification of new host–parasite systems) in experimental studies that demonstrate adaptive benefits to host manipulation (e.g., ref. 8) or that elucidate proximate mechanisms for host modifications (see also ref. 9). More broadly, they suggested that an increased publication rate of synthesis/theoretical studies relative to empirical studies may create an imbalance between facts and ideas. Indeed, a persisting imbalance between an adaptive idea and a perceived ecological fact already exists in one of the most iconic examples of a host-manipulating parasite, larvae of the lancet fluke Dicrocoelium dendriticum in ants.
The lancet fluke has an obligate, three-host life cycle (10, 11). Adult flukes reside within the bile ducts of ungulate and lagomorph definitive hosts wherein they have obligate sexual reproduction. Eggs pass into the external environment via host feces. Terrestrial snails consume eggs encountered on pasture. Repeated bouts of obligate asexual reproduction occur within the snail, leading to the release of packets of larvae (= cercariae) within discrete, fluid-filled packages known as “slime balls” (reviewed by ref. 12). The fate of individual slime balls is not known, but slime balls are either ingested by the worker ant that encounters them or are transported to the nest for potential ingestion by nestmates. Following ingestion, cercariae penetrate the lining of the ant crop. One cercaria, rarely two to three (13), makes its way to the anterioventral-most region of the subesophageal ganglion of the brain where it resides, unencysted. The remaining cercariae migrate to the gaster of the ant where they encyst as metacercariae within a double-layered cyst wall (13).
Two remarkable aspects of the life cycle have elevated D. dendriticum to “textbook” status. First, the single unencysted metacercaria located in the ant’s subesophageal ganglion (i.e., the brain fluke) alters the ant’s behavior in a precise temperature-dependent manner (14) such that the ant attaches with its mandibles onto vegetation, particularly inflorescences, growing adjacent to their nests (15, 16). Second, the unencysted brain fluke is considered to have reduced fitness relative to the encysted flukes in the abdomen (13, 16, 17); thus, the brain fluke apparently displays an altruistic behavior. Given that an ant may consume a slime ball with potentially many clonemates (i.e., individuals of the same clone [= progenitor egg]), it is presumed that the brain fluke has encysted clonemates in the abdomen of the same ant. This clonemate cotransmission into ants enables kin selection of the host-manipulating behavior. Specifically, the ultimate cost experienced by the altruistic brain fluke is thought to be mitigated because it facilitates the transmission of its encysted clonemates (i.e., the highest form of genetic relatedness) to the final host (1, 2, 18). The adaptive story that emerges from the above ecology and life history is one where selection for increased transmission to a final host (grazing mammals) has led to the evolution of a host-manipulating mechanism, which itself is the result of an evolved behavior that reduces the fitness of the parasite. This altruistic behavior is possible via kin selection because the life cycle is conducive to cotransmission of clonemates into ants.
Although a fantastical narrative, there exist no data on clonemate cotransmission within ant hosts to support the idea that kin selection may be at play. Indeed, supportive evidence for the “clonemate cotransmission hypothesis” into ants is absent, despite coverage in theoretical and conceptual works discussing the evolution of host manipulation in D. dendriticum (e.g., refs. 1, 18, and 19). Furthermore, theoretical work by Wilson (2) has shown that the altruistic behavior could evolve even in the presence of multiple clones within an ant host and that a balanced polymorphism could exist between altruistic and nonaltruistic behavioral variants in the parasite’s population (3). Regardless of the number of clones, studies examining the perception that trematodes should have an abundance of clonemates in hosts subsequent to the mollusc first host did not become common until the 2000s (20). A current summary of the available data suggests that clonemates in second or third hosts are not common in many trematode systems (20, 21). Hence, it is necessary to revisit the iconic host-manipulating lancet fluke to test the clonemate cotransmission hypothesis with empirical data from ants.
The “the basic parameters” requested by Sober and Wilson (3) call for data on how many eggs are ingested by snails and how many slime balls are eaten by ants. These two factors combine to shape the cotransmission of clonemates to an ant and how many clones cooccur in an ant. Because kin selection on the host-manipulation trait would be manifested in the transmission from ant to final host, the parameters of primary importance are the clone and clonemate distributions within and among ant hosts. Specifically, for kin selection to promote host manipulation, parasite clonemates must be unevenly distributed in the population such that parasite relatedness within hosts is higher than between hosts. This uneven distribution of clonemates is defined as cotransmission. We therefore asked three questions that are necessary to assess a possible role for kin selection in the evolution of host manipulation by the lancet fluke. 1) Is there evidence of cotransmission of clonemates into ants? Kin selection could also be made possible via sibling relationships so we took advantage of newly developed methods based on pedigree reconstruction data to also ask if there was evidence of sibling cotransmission into ants (22). 2) Does a brain fluke have clonemates or siblings encysted in the same host? 3) If there is clonemate cotransmission, can multiple clones coexist in an ant host?
Materials and Methods
Study Site and Ant Collections.
Our study included samples of infected ants, Formica aserva, from two sites in Cypress Hills Park in southeastern Alberta, Canada. The lancet fluke was introduced into the park from Europe prior to the 1980s and then emerged in the mid 1990s (23). The two collection sites were selected from ∼32 known locations in the park where ants have been observed attached to vegetation (24). We selected one of the sites (Staff Camp [SC]) because we had observed attached ants adjacent to a well-defined nest each year since 2009 (25) and because we had background information on patterns of D. dendriticum infection in populations of ant (26) and snail (27) intermediate hosts. Our ongoing studies that involve monitoring the behavior of marked ants as they leave and enter their nest also occur at this site (16). We have also observed attached ants on plants at the second site (Trans-Canada Trail [TC]) each year since 2009 (25). This site is 12 km southeast of the SC site. The morphological characteristics of brain flukes and the nature of the brain fluke–brain interface have been well-characterized for ants sampled from this site (13). At each site, a 3-m2 area surrounding the nest was demarcated with flagging tape, and then the first nine ants observed attached to a plant were manually detached and then fixed whole in 90% ethanol. Samples of individual ants were collected on 8 June 2013 between 6:00 and 9:00 AM from the two sites.
DNA Extraction, PCR, and Genotyping.
For consistency, we refer to encysted metacercariae from the abdomen as “body flukes” and the single nonencysted metacercaria in the subesophageal ganglion as the “brain fluke.” Individual body flukes were isolated from ants by opening the abdomen into a Petri dish containing distilled water. The numbers of body flukes (= intensity) (28) were assessed in each ant following methods in van Paridon et al. (26). Individual body flukes were selected for DNA extraction and placed in 200-μL PCR tubes containing 50 μL of lysis buffer (29) and Proteinase K (10 mg/mL; New England BioLabs). To extract DNA from the brain fluke, the head of each ant was removed from each body and placed in a 1.5-mL Eppendorf tube containing 50 μL of lysis buffer and Proteinase K. Heads were crushed using a sterile plastic pestle within the tube to increase exposure of the metacercaria to the lysis solution. All metacercariae were scored at five microsatellite loci (DdMs21, DdMs28, DdMs60, DdMs70, and DdMs95) as per the PCR and genotyping methods in van Paridon et al. (29). When present, up to 22 larvae per ant were used in DNA extractions (except one ant where we did 28). In total, we attempted DNA extractions on 376 larvae.
Clonemate Identification.
We first note that about 27% of the larvae failed to produce a quality DNA template as evidenced by failed PCRs across multiple attempts and loci. We suspect that preservation of whole ants in ethanol may have decreased extraction efficiency of the tiny larval flukes. We only retained metacercariae that were successfully genotyped at all five loci to allow for appropriate clonality tests and to avoid low quality samples. The final dataset included a total of 272 metacercariae across 18 hosts. F-statistic results (see below) did not show any evidence of null alleles, and, thereby, the obtained multilocus genotypes enabled unbiased estimates of clonal metrics. GENCLONE 2.0 was used to identify individuals with identical multilocus genotypes (MLGs) and to test whether identical MLGs arose from asexual reproduction: i.e., test if identical MLGs were clonemates (30). The clonemate testing was done by calculating Psex, which is the probability of observing n copies of an MLG in a sample size of N given sexual reproduction. If Psex < 0.05 at n = 2, then all copies of that MLG can be considered to be the product of asexual reproduction (31). Prior work on a sample of adult flukes indicated that the D. dendriticum at our sampling locations represented a panmictic population (29). Thus, we looked for identical MLGs and calculated Psex across the entire sample of n = 272 metacercariae from n = 18 ant hosts. We also did Psex calculations for each sampling site separately and obtained the same clone/clonemate identification results.
We note the Psex can only evaluate if identical MLGs are part of the same clone. As indicated by Arnaud-Haond et al. (32), however, mutation, PCR errors, or scoring errors may lead to small allelic differences within a single clone. Following the recommendations of Arnaud-Haond et al. (32), we plotted the number of allelic differences between pairs of unique MLGs and found a bimodal distribution where six pairs of unique MLGs differed at a single allele. We assessed the significance of each of these pairs separately by removing the one locus at which the pair differed and then recalculated Psex. In each of these cases, the six pairs were found to be significant (Psex < 0.01 at n = 2): i.e., there were six unique clones and not 12. Again, Psex calculations by sampling site led to the same results. Therefore, we modified the dataset such that the genotype that will be used to represent the unique clone of these pairs will be the more frequent genotype. This carries the assumption that the less frequent genotype was due to mutation, PCR error (such as allele dropout), or scoring error. We refer to this dataset as n = 272mod. After identifying all clones and determining the significance of all clonemates, the index of clonal diversity was calculated as the number of clones per number genotyped (32).
Clonal Structure Analyses.
To broadly test for evidence of cotransmission of clonemates, we analyzed average within-host FIS and FST among hosts (multilocus estimators) in two datasets: the total dataset (n = 272mod) and a dataset where the number of unique clones was reduced to one representative within each host (n = 59). FIS and FST measure the proportional change in heterozygosity (expected under Hardy–Weinberg equilibrium) that is due to nonrandom union of gametes in a subpopulation or subdivision of a population into subpopulations, respectively. According to the theoretical work of Prugnolle et al. (33), the expectation is that a high variance in clonal reproductive success will drive within-host FIS negative and increase among-host FST. Significance was tested in FSTAT (34) using 10,000 randomizations of alleles among individual flukes within hosts and of genotypes among hosts, respectively. For two-tailed tests of FIS across the entire component population (i.e., ignoring among-host delineations) (28), we used SPAGeDI with 10,000 randomizations of alleles among individuals (35). The presence of clonemates should also increase linkage disequilibrium. Across the entire component population, we tested genotypic equilibrium (GD) between pairs of loci in GENEPOP 4.2 (Markov chain parameters: 5,000 dememorizations; 5,000 batches; 5,000 iterations) (36) on the total dataset (n = 272mod) and in a reduced dataset of one representative of each clone (n = 54). Note the reduced dataset of the component population (n = 54) is smaller than the among-host reduced dataset (n = 59) because a few clones occurred in more than one host (Results).
Pedigree Reconstruction.
Pedigree reconstruction analyses were conducted to determine if sibling cotransmission, as well as clonemate cotransmission, could also be a factor in driving kin selection. In order to carry out the pedigree reconstruction analyses, only individuals that are the product of sexual reproduction need to be included. Thus, at this point, clonal inference is assumed fixed without error, and the dataset was reduced to one individual per clone (n = 54) for the sibship reconstruction analysis. We used the full-likelihood method of sibship reconstruction implemented in the software COLONY v2.0.6.4 (37). Details of the method are given in Wang and Santure (38) and Wang et al. (39). The following settings were specified in COLONY: female and male polygamy with inbreeding for a monoecious species, length of run was very long under the full-likelihood method with very high precision, three runs, allele frequencies estimated from the dataset and were updated as the analysis was run, and sibship scaling was set to yes. We did not use the sibship prior option as individual adult flukes have very high fecundity (24). Allelic dropout rate was set to 0.005, and mutation/error rate was set to 0.005. We also did two additional analyses: 1) error rates set to 0, and 2) error rates set to 0 and a strong sibship prior with maternal and paternal family sizes set to 3.6. The latter was done to determine if sib inference was sensitive to this prior. Qualitatively, these additional analyses were nearly identical to the results from the initial analysis so we only report on the main analysis.
Of specific interest were the sibling relationships of brain flukes to body flukes within the same ant. Sibling relationships between pairs of clones (= dyads) were based on the best configuration of the sibship analysis, and the probability of the sibling relationship was based on that given in the FullSibDyad or HalfSibDyad output files in COLONY.
Assessing Clonemate and Sibling Cotransmission.
As described in Detwiler and Criscione (22), the percentage of sibling dyads (full- and half-sib dyads summed) within hosts (PS) compared to the percentage of sibling dyads over the entire component population (PES) (i.e., the expectation by random chance alone) provides a direct test of sibling cotransmission. Note, in Detwiler and Criscione (22), these statistics were referred to as kin dyads (PK and PEK, respectively), but, here, we use sibling dyads to avoid confusion in the fact that clonemates also represent a kin relationship. We adopt their approach to analyze clonemate cotransmission. In particular, the percentage of clonemate dyads within hosts (PC) compared to the percentage of clonemate dyads over the entire component population (PEC) can be used to test clonemate cotransmission. The following was done to estimate the above percentages. After the sibship analysis was conducted on the clones (n = 54), clonemates were included back into the dataset (n = 272mod) and assigned the same parent identifications as that of their clonemate used in the sibship analysis. Parent identifications are based on the best configuration of the sibship analysis (22). Based on the clonemate and parent identifications, clonemate, full-sib, half-sib, and unrelated relationships could be assigned to all dyads across the component population and within ant hosts.
The percentages of clonemate dyads and sibling dyads (full and half sibs summed) within hosts (PC and PS, respectively) are calculated as weighted averages over hosts where the weights are based on the sample sizes of genotyped metacercariae of each host (i.e., the percentage of genotyped metacercariae in a host relative to the total genotyped). Specific details on significance testing are given in Detwiler and Criscione (22). In short, resampling procedures are used to assess error and generate confidence intervals (CIs). Clonemate inference is assumed without error so only sampling error, evaluated by bootstrapping over hosts, is estimated for PC and PEC. For PS and PES, pedigree estimation error is based on a resampling procedure using the plausible sibship configurations provided in the plausible configuration archive by COLONY whereas sampling error is assessed by bootstrapping over host individuals (22). CIs of PC and PS are compared to CIs of PEC and PES, respectively. Nonoverlapping 84% CIs are considered to approximate a significant difference at a P = 0.05 (40). Odds ratios were calculated to assess the magnitude of difference between PC (PS) and PEC (PES). The odds ratio of clonemate dyads within hosts compared to the component population was ORC = [(PC)/(1 − PC)]/[(PEC)/(1 − PEC)], and the odds ratio for sibling dyads was ORS = [(PS)/(1 − PS)]/[(PES)/(1 − PES)]. Odds ratios were calculated within each bootstrap replicate, and the resulting simulated distributions were used to determine if the observed ORC or ORS differed from 1 (the value expected if PC and PS did not differ from PEC and PES, respectively).
Mean Number of Clones per Host.
We calculated the mean number of clones per ant host in two ways. First, we used a weighted mean based on the raw number of clones per host where the weighting was based on the sample size of genotyped flukes of each host. The second was to calculate the Shannon–Weiner diversity index (H′) per host and then convert the value to N1 = eH′ where N1 represents the number of equally common clones that would produce the same clonal diversity H′ in a host (41). A weighted mean of N1 was calculated among hosts where, as above, the weight was the genotyped sample size. The reason for using N1 is that Wilson (2) used several simplifying parameters in his model, one of which was that clones were equally frequent within a host. The variable N1 would be analogous to this latter assumption. To test if the mean number of clones per host or mean N1 per host was greater than 1, we bootstrapped over hosts (10,000 times) to determine the proportion of simulated values (i.e., the means) >1. Randomization tests were conducted with POPTOOLS V3.2.5 (42).
Clonal Metrics by Location.
The clonal metrics described above (index of clonal diversity, mean number of clones per host, N1 per host, PEC, PC, and ORC) were analyzed across the entire dataset (n = 272mod). However, clonal transmission could be influenced by local scale factors (Discussion). Thus, we analyzed clonal metrics for samples from each location separately to determine if there were any differences in clonal patterns between SC and TC.
Data Availability.
All data discussed in the paper are available in SI Appendix, Tables S1 and S2.
Results
Sampling and Genotyping.
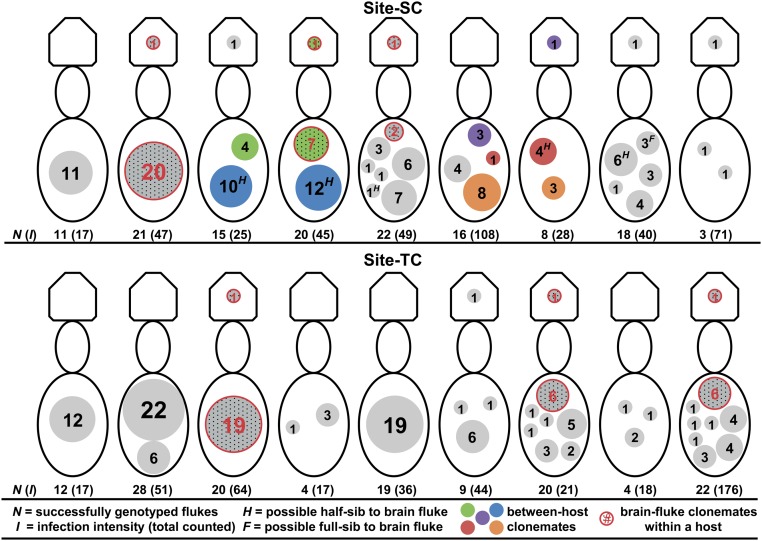
The total number of flukes collected from the 18 ants was 874 with a range of 17–176 per ant (Fig. 1). Including 11 brain flukes, a total of 272 metacercariae were successfully genotyped at all 5 loci (Fig. 1). The full genotype dataset (n = 272) of all metacercariae is available in SI Appendix, Table S1.
Fig. 1.
Distribution of larval D. dendriticum clones and clonemates among infected ants collected from two sampling locations (SC, top; and TC, bottom) in Cypress Hills Park, Alberta. Each circle represents a unique clone, except for the following two criteria: 1) Solid colors denote clonemates that occur between hosts, and 2) red-outlined/patterned circles denote a brain fluke and its abdomen clonemates within a host. The area of the circle is proportional to the number (shown in the circle) of individuals of the respective circle. For example, the fourth ant in site SC has eight individuals of the green clone, one of which is a brain fluke, and 12 individuals of the blue clone. The green and blue clones are also found in the third ant in site SC. The third ant has a brain fluke that does not have a clonemate in the abdomen.
Clonemate Identification.
A total of 60 unique MLGs was identified among the 272 metacercariae that were successfully genotyped. In all cases where there was more than one copy of an MLG, the Psex < 0.01 at n = 2. Thus, all repeated copies of an MLG are clonemates. In addition, the six pairs of MLGs that differed by a single allele also had Psex < 0.01 at n = 2 after removing the discrepant locus. In five of the six pairs, allele dropout may explain the single allele difference as the less frequent MLG was homozygous at the locus that differed by 1 allele. Furthermore, the paired MLGs were always found cooccurring within the same host individuals, a result that seems unlikely if pairs truly represented different clones. Hence, we are confident that the modified dataset (Methods) more appropriately reflects clone identifications. After making the modified dataset (n = 272mod) (SI Appendix, Table S2), there were 54 unique MLGs, and, once again, all repeated copies of an MLG were found to be clonemates (i.e., Psex < 0.01 at n = 2). Therefore, the overall index of clonal diversity expressed as a percentage was 19.9% (54 of 272). We note that the five loci we used had very high discriminatory power to identify the maximal clonal diversity in the sample, as evidenced by the resampling of all loci combinations from one to five loci (option in GENCLONE 2.0). In fact, at just three loci, the mean number of unique MLGs was 53.2 (range: 51 to 54), and, with four loci, all 54 MLGs were identified.
None of the 54 clones or clonemates were shared between the two sites (Fig. 1). Thus, clonemates only occurred within sampling locations (i.e., within ant nests) and not between them. Although up to eight clones were detected in an individual ant (ant 9 at TC), 66% of the ants contained one to three clones. The summary distribution data also showed that five clones had clonemates shared between different ants collected at the same site (all at site SC) (Fig. 1). Ants 3 and 4 shared the “green” and “blue” clones, and ants 6 and 7 shared the “orange,” “magenta,” and “purple” clones (Fig. 1). For example, the “purple” clone present in the head of ant 7 was found in the body of ant 6. In 11 of the ants where both a brain and body fluke were genotyped, there were six cases where the brain fluke had clonemates in the abdomen of the same host (red outlined circles, Fig. 1).
Clonal Structure.
In the complete dataset (n = 272mod), average within-host FIS was significantly negative (−0.407; one-tailed P < 0.001), and there was significant among-host genetic structure (FST = 0.275; one-tailed P < 0.001). Ignoring among-host partitioning, FIS of the component parasite population was significantly negative (−0.039, two-tailed P = 0.003), and all 10 pairwise combinations of loci had significant GD (all P values < 0.001). When the dataset was reduced to only unique clones within each host (n = 59), average within-host FIS was positive and not significant (0.031; one-tailed P = 0.15), and among-host FST was no longer significant (0.015; one-tailed P = 0.1). When only unique clones were analyzed as a single population (n = 54), FIS was no longer negative (= 0.052; two-tailed P = 0.076). SI Appendix, Table S3 provides the per locus number of alleles, gene diversity, and FIS values for the unique clone data (n = 54) as this would reflect the product of sexual reproduction in the parental generation of the sample. In addition, there was no significant GD for any pair of loci (all P values > 0.17). These results provide strong evidence for clonal genetic structure at the metacercarial stage of D. dendriticum. Moreover, the lack of genetic structure in the reduced datasets indicates flukes from both sites belong to the same underlying genetic population, a result consistent with the panmixia we observed using adult flukes in a prior study (29).
Pedigree Reconstruction.
With n = 54 unique individuals (i.e., clones), there were 1,431 dyads of which there was a single full sib dyad and 157 half-sib dyads inferred in the best configuration. Hence, 11% of the dyads among clones were inferred to be siblings. Specific sibling and clonemate relationships among brain fluke and body fluke clones are shown in Fig. 1. There were five ants in which the brain fluke had a half-sib relationship to another clone within the same host. In general, the probabilities of these half-sib relationships were low (P values = 0.26, 0.38, 0.48, 0.69, and 0.73) (Fig. 1). Thus, caution is advised as these half-sib relationships may be false positives. In contrast, the one full-sib dyad, which was between a brain fluke and body fluke in the same host (Fig. 1), had strong support (P = 0.92) and was found in all 1,000 plausible sibship configurations provided in the COLONY archive file.
Assessing Clonemate and Sibling Cotransmission.
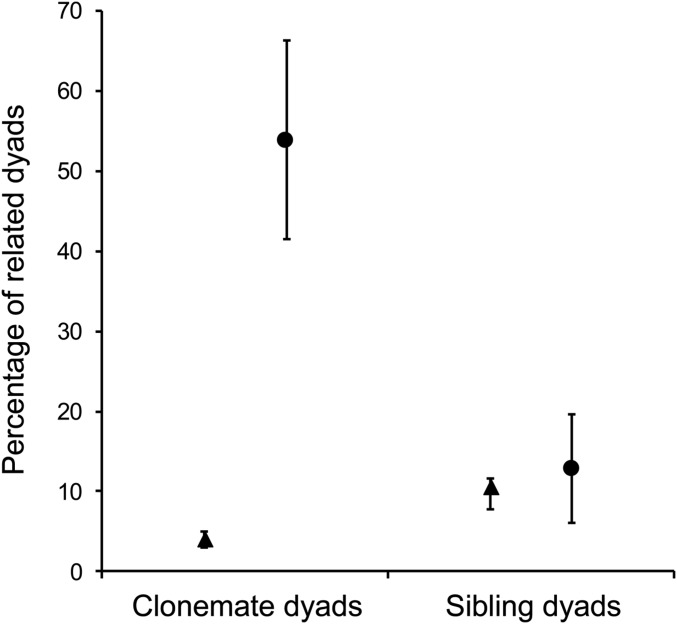
Across all individuals (n = 272), there were 36,856 possible dyads of which 1,471 were clonemate dyads (PEC = 4%). There were three full-sib dyads and 3,872 half-sib dyads (PES = 10.5%). Within ant hosts, there were 2,369 possible dyads; PC = 54% and PS = 12.8%. There was no overlap in the 84% CI of PC and PEC (Fig. 2), and the odds of being a clonemate dyad within hosts was 28 times higher than that expected by chance (ORC = 28.1; 95% CI: [18.9, 48.4]; P>1 = 0.0001). In contrast, the 84% CI of PS and PES completely overlapped (Fig. 2), and ORS was not significantly greater than 1 (ORS = 1.3; 95% CI: [0.4, 2.6]; P>1 = 0.28). Thus, there was significant clonemate cotransmission, but no significant sibling cotransmission greater than that expected by chance alone.
Fig. 2.
Percentages of clonemate and sibling dyads within hosts (PC and PS, respectively; circles) compared to the percentages of clonemate and sibling dyads across the whole component population (PEC and PES, respectively; triangles). The 84% CIs are shown.
Mean Number of Clones per Host.
Using the raw number of clones, the weighted mean number of clones per ant host was 3.5 (95% CI: 2.2 to 4.9), which was significantly greater than 1 (P < 0.001). The weighted mean N1, number of equally common clones per ant host, was 2.9 (95% CI: 1.9 to 3.9), which was also significantly greater than 1 (P < 0.001).
Clonal Metrics by Location.
There was no statistical difference in the index of clonal diversity between SC and TC (Fisher’s exact test, P = 0.65). Moreover, there was overlap in 84% CI for all other clonal metrics (Table 1). Therefore, resulting clonal transmission patterns are nearly identical at both locations, and the results based on analyses of the whole dataset (n = 272mod) are the main focus of the discussion.
Table 1.
Parasite clonal patterns in D. dendriticum-infected ants from two sampled locations in Cypress Hills Park, Alberta
| Site-SC | Site-TC | |
| No. of individuals genotyped | 134 | 138 |
| Total no. of clones | 25 | 29 |
| Index of clonal diversity* | 18.7% | 21.0% |
| Clones per host† | 3.5 (2.4, 4.7) | 3.4 (1.9, 5) |
| N1 per host‡ | 2.9 (2.1, 3.8) | 2.7 (1.6, 3.9) |
| PEC§ | 0.08 (0.05, 0.11) | 0.08 (0.05, 0.11) |
| PC¶ | 0.48 (0.32, 0.65) | 0.60 (0.41, 0.78) |
| ORC# | 10.5 (7.0, 17.5) | 17.1 (11.6, 33.4) |
Overlap in 84% CI (in parentheses) shows no differences between sites for any of the clonal metrics.
The percentage of clones among genotyped individuals.
Weighted mean of the raw number of clones per host.
Weighted mean of the number of equally common clones per ant host.
Percentage of clonemate dyads over the entire component population (see text for details).
Percentage of clonemate dyads within hosts (see text for details).
Odds ratio of clonemate dyads within hosts to the component population (see text for details).
Discussion
Natural History Inference on Clonemate Abundance.
A striking feature we observed for D. dendriticum metacercariae is that its index of clonal diversity among ants, 19.9% (54 of 272), is the lowest reported to date for any trematode species. Given the asexual reproduction and high numbers of cercariae often produced in molluscs by trematodes, it may seem intuitive that numerous clonemates would be present in subsequent hosts. However, existing data among trematodes indicate that an abundance of clonemates and, hence, a low index of clonal diversity are not an inevitable outcome. The environments of transmission, peculiarities of life cycle patterns, and sampling scale will drive estimates of clonal diversity (21). Among trematodes with three-host, fully aquatic or aquatic-to-bird life cycles, and no matter if sampling was done at the definitive host or second intermediate host stage, most reported clonal diversities are from 95 to 100% (43–50). Aquatic life cycles are favorable for cercarial dispersal, and mobile second intermediate hosts may acquire cercariae from multiple first hosts. Therefore, even if sampling is completed on a local scale, the chance of finding an abundance of clonemates will be low.
The lowest clonal diversities have been reported from semiterrestrial trematode/host systems. For example, in Schistosoma mansoni, where cercariae directly penetrate the final host, clonal diversities were 80 to 85% among rat final hosts (51, 52). Among sheep hosts on a single farm, Vilas et al. (53) reported a clonal diversity of 58% for Fasciola hepatica, a species where cercariae encyst on vegetation. In these semiterrestrial systems, there is less opportunity for dispersal of clonemates prior to infection of the next host. We do note that, when sampling was conducted on larger scales (e.g., across multiple farms), high clonal diversities have been reported for F. hepatica: 92% from cattle in Spain (53), and 89% and 92% from sheep and cattle, respectively, in the United Kingdom (54).
Why does D. dendriticum have a high abundance of clonemates among ant hosts? Certain features of the lancet fluke’s terrestrial life cycle likely promote localized transmission from snail to ant. Terrestrial snails have low vagility, lack a dispersive larval stage, and rates of passive transportation by other animals are low (55). The results of marking studies indicate that individuals move less than 2 m over the course of 1 y (56) and only 350 m over their lifetime (57). Likewise, the movement of the ants that are used as second intermediate host for the lancet fluke is probably restricted to a few meters within the vicinity of the nest. Lastly, the cercariae within exuded slime balls cannot disperse, and the slime balls themselves tend to adhere to the surface of the substrate.
The clonemate distributions we observed among ant hosts are consistent with the restricted movement of snails, ants, and slime balls across the landscape. First, clonemates only occurred within, and not between, the two sampling locations that are separated by 12 km (Fig. 1). Given that asexual reproduction occurs in the snail host, this distribution of clonemates makes sense because of the limited dispersal ability of both the snail and ant hosts. If clonemates were to be found among relatively distant locations, the expectation would be that it would occur among the more mobile ungulate definitive hosts. Second, because ants likely ingest slime balls at or near the nest, any sharing of fluke clonemates among ants would occur among nestmates. Consistent with the latter idea, we found two pairs of ants that shared either two or three clones (Fig. 1). These ants that shared clonemates were likely nestmates because they were sampled in close proximity to a nest at the same site. Overall, the low index of clonal diversity (i.e., high clonemate abundance) in our local samples is concordant with the biology of the lancet fluke and sets the stage for the potential of kin selection in D. dendriticum.
Kin Selection Inferences.
Is there evidence of clonemate or sibling cotransmission? The answers are yes and no, respectively. In the dataset retaining clonemates (n = 272mod), there was significant structure among hosts (FST = 0.275), but, upon reducing to one unique clone within each host, there was no longer significant among-host structure (FST = 0.015). Although the former test shows a clear nonrandom distribution of clonemates among ant hosts, comparing FST values among systems is problematic due to the dependency of FST on high marker gene diversity (i.e., high mutation rates) (58). Building upon the methods of Detwiler and Criscione (22), the proportion of clonemate dyads within hosts provides a comparable quantitative measure that will be useful for comparisons among systems in future studies. We found that 54% of all dyad relationships within hosts were clonemates, and this value was significantly greater than the 4% expectation by chance. Indeed, clonemate cotransmisison was common as 17 of 18 hosts had at least one pair of clonemates and five of the 18 contained only a single clone (Fig. 1). In contrast, we found little to no support that sibling cotransmission would play a role in potential kin selection; the 12.8% of sibling dyads within hosts did not deviate from the expectation by chance alone of 10.5% (Fig. 2). These results confirm the original hypothesis by Wickler (1) that the life cycle of D. dendriticum is conducive to clonemate cotransmission. Hence, we find support for the potential of kin selection to drive the host-manipulation behavior in the parasite.
Do brain flukes have clonemates or siblings encysted in the same host? While clonemate cotransmission was high, the brain fluke still needs to have clonemates in the same host for kin selection to be possible. Our data support this second condition for kin selection to be possible as, in six of the 11 (55%) ants where a brain fluke was genotyped, there were clonemates of the brain fluke encysted in the abdomen. In the other five ants with genotyped brain flukes, it is possible there could still be a clonemate as we did not genotype all body flukes. Therefore, the 55% frequency of clonemates to a brain fluke should be considered a lower bound. Although we did not have strong evidence for sibling cotransmission, it is interesting to note that the single full sib relationship that was identified was between a brain fluke and a coinfecting metacercarial clone with three copies (Fig. 1). This result raises another interesting clonal dynamic in that the presence of clonemates could amplify the number of sibling relationships present in a host even though the overall chance of sibling cotransmission is low. In this particular case, instead of just one pair of full sibs, there are now three in relation to the brain fluke.
Wilson’s (2) model showed that larval D. dendriticum could evolve the altruistic behavior even if ants are infected with multiple clones. Our results support this prediction of the model. Overall, a minimum (as we did not genotype all individuals) of 54 progenitor eggs seeded the metacercariae collected from the 18 ants. With regard to the number of progenitor eggs that can coinfect an ant, we found the mean number of clones per ant host was 3.5 and that the mean number of equally frequent clones per ant host was 2.9. Both values were significantly greater than 1. It is interesting to compare these values to figure 2 in Wilson (2). For a fixed value of where a brain fluke doubles the probability of an ant being ingested compared to an infected ant with no brain fluke, the figure shows that, when three equally frequent clones (which is the value we observed) infect ants, the frequency of parasitized ants that have a brain flukes is ∼98 to 99%. In accordance with this prediction, Romig et al. (59) reported 97.7% (43 of 44 via dissection) of ants collected from vegetation contained a brain fluke. Each of the 12 infected ants examined by micro-CT in Martin-Vega et al. (13) had a brain fluke. Further, in the hundreds of ants collected in our annual monitoring studies (e.g., ref. 26), each individual contained a single brain fluke.
It is possible that metacercarial clonal distribution patterns within and among ant hosts could vary across sampling locations. As discussed above, the scale of clonemate transmission is likely to be highly localized. Thus, factors that affect cercarial clonemate reproduction in snails (e.g., local adaptations of host–parasite compatibility or intraspecific competition within snails) and/or contact and consumption rates of slime balls by ants (e.g., habitat heterogeneity, ant colony size, or behavioral differences in ant species) may impact the number of clones, abundance of clonemates, and how both are partitioned among individual ants on a local scale. In our study system, we did not detect differences in clonemate abundance or differences in various clonal metrics within or among ants between the two sampled locations (Table 1). Thus, despite being 12 km apart, there appears to be consistency in the clonal transmission process from snail to ant in the Cypress Hills system. For future studies, it will be of interest to test if the mean number of clones per ant varies among locations and, if so, if the frequency of parasitized ants with brain flukes declines as a function of within-ant clonal diversity as predicted in Wilson (2).
Conclusion
Kin selection was invoked over 40 y ago as a means of driving the evolution of the lancet fluke’s altruistic-based, host-manipulating behavior (1). However, data on the level of genetic relatedness between the brain fluke and abdomen metacercariae have remained lacking. Our results provide empirical evidence that conclusively shows the conditions necessary for kin selection to operate in this system. Specifically, we find a high degree of parasite clonemate cotransmission into ant hosts and that the brain fluke is often a clonemate of its coinfecting abdomen metacercariae. Moreover, our results are in agreement with an existing model that shows the altruistic behavior can evolve in the presence of multiple parasite clones within the same ant host. Hence, our clonal data from ants fill an important empirical gap in the long-perpetuated adaptive story of the lancet fluke in that we find support for the potential of kin selection to drive the evolution and maintenance of this iconic host manipulation.
Recent, high-resolution imaging data of the larval flukes inside ants (13, 16) provide support to another part of the adaptive story: i.e., the host-manipulation trait is the result of an evolved altruistic behavior in the parasite. In the original experimental infections of final hosts (17), the lack of explicit methodologies precludes conclusive assessment of the reported noninfectivity of the brain fluke. Nonetheless, structural differences between the brain fluke and abdomen flukes provide inferential support for fitness costs associated with the host-manipulation behavior displayed by the brain fluke (13, 16). Whereas the abdomen flukes are enveloped by a thick, double-layered cyst wall, the brain flukes are unencysted. It follows that brain flukes likely have reduced survivorship during the harsh ingestion processes in a final host (e.g., mastication, digestive enzymes). Moreover, in contrast to the abdomen flukes, the teguments of brain flukes are dominated by mitochondria-rich secretory tissue, which is consistent with its active energy-expending role of manipulating the ant. Hence, even if a brain fluke established in a final host, its reduced energy stores would lead to lower fecundity than an abdomen fluke. While additional experimental exposures into final hosts are warranted, the structural differences along with the original experimental infection report (17) collectively support the hypothesis of reduced fitness of the brain fluke as a consequence of its behavior in manipulating the ant host. Hence, the brain fluke’s behavior fits the definition of an altruistic trait.
D. dendriticum is already distinctive among parasite puppeteers in that its induced ant “zombie-ism” involves discrete and daily “on–off” phases of host manipulation. Our clonemate cotransmission results highlight additional unique features of this parasite. For example, kin selection has played a role in the evolution of a diverse array of organismal traits from warning behaviors and cooperative breeding to the formation of microbial multicellular bodies and plant root growth (60). The lancet fluke provides the only known example where a kin-selected, altruistic trait facilitates parasite transmission. In addition, in other animal systems where kin selection operates, the genetic relatedness is largely determined by sibling and/or parent–offspring relationships. The obligate asexual reproductive stage of D. dendriticum in its snail first host has enabled the highest degree of genetic relatedness: i.e., clonality. Given the ubiquity of obligate asexual reproduction among digeneans, it will be of interest to quantify clonemates and explore the role of kin selection in the evolution of other trematode traits: e.g., larval-stage division of labor within mollusc hosts (61, 62) or competition outcomes for coinfecting metacercariae (e.g., ref. 63).
Supplementary Material
Acknowledgments
C.D.C.’s studies on the population genetics and evolution of parasite life cycles are supported by National Science Foundation Grant DEB-1655147. C.P.G.’s studies on the ecology and evolution of host/parasite interactions are supported by Natural Sciences and Engineering Research Council of Canada (NSERC) Grant 05067. J.S.G.’s studies on parasite genetics are supported by NSERC Discovery Grant 03976 and a grant from the National Centre for Veterinary Parasitology. B.J.v.P. was supported from a Grant in Biodiversity from the Alberta Conservation Association and the NSERC-Collaborative Research and Training Experience (CREATE) Host–Parasite Interactions Program.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922272117/-/DCSupplemental.
References
- 1.Wickler W., Evolution-oriented ethology, kin selection, and altruistic parasites. Z. Tierpsychol. 42, 206–214 (1976). [DOI] [PubMed] [Google Scholar]
- 2.Wilson D. S., How nepotistic is the brain worm. Behav. Ecol. Sociobiol. 2, 421–425 (1977). [Google Scholar]
- 3.Sober E., Wilson D. S., Unto Others: The Evolution of Psychology and Unselfish Behavior (Harvard University Press, Cambridge, MA, 1998). [Google Scholar]
- 4.Moore J., Parasites and the Behavior of Animals (Oxford University Press, Oxford, UK, 2002). [Google Scholar]
- 5.Libersat F., Delago A., Gal R., Manipulation of host behavior by parasitic insects and insect parasites. Annu. Rev. Entomol. 54, 189–207 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Hughes D. P., Brodeur J., Thomas F., Host Manipulation by Parasites (Oxford University Press, Oxford, UK, 2012). [Google Scholar]
- 7.Poulin R., Maure F., Host manipulation by parasites: A look back before moving forward. Trends Parasitol. 31, 563–570 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Lafferty K. D., Morris K., Altered behavior of parasitized killifish increases susceptibility to predation by bird final hosts. Ecology 77, 1390–1397 (1996). [Google Scholar]
- 9.Herbison R. E. H., Lessons in mind control: Trends in research on the molecular mechanisms behind parasite-host behavioral manipulation. Front. Ecol. Evol. 5, 102 (2017). [Google Scholar]
- 10.Krull W. H., Mapes C. R., Studies on the biology of Dicrocoelium dendriticum (Rudolphi, 1819) looss, 1899 (Trematoda: Dicrocoeliidae), including its relation to the intermediate host, Cionella lubrica (Müller). VII. The second intermediate host of Dicrocoelium dendriticum. Cornell Vet. 42, 603–604 (1952). [PubMed] [Google Scholar]
- 11.Krull W. H., Mapes C. R., Studies on the biology of Dicrocoelium dendriticum (Rudolphi, 1819) Looss, 1899 (Trematoda: Dicrocoeliidae), including its relation to the intermediate host Cionella lubrica (Müller). IX. Notes on the cyst, metacercaria, and infection in the ant, Formica fusca. Cornell Vet. 43, 389–410 (1953). [PubMed] [Google Scholar]
- 12.Goater T. M., Goater C. P., Esch G. W., Parasitism: The Diversity and Ecology of Animal Parasites (Cambridge University Press, New York, NY, ed. 2, 2014). [Google Scholar]
- 13.Martín-Vega D., et al. , 3D virtual histology at the host/parasite interface: Visualisation of the master manipulator, Dicrocoelium dendriticum, in the brain of its ant host. Sci. Rep. 8, 8587 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Botnevik C. F., Malagocka J., Jensen A. B., Fredensborg B. L., Relative effects of temperature, light, and humidity on clinging behavior of metacercariae-infected ants. J. Parasitol. 102, 495–500 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Spindler E. M., Zahler M., Loos-Frank B., Behavioural aspects of ants as second intermediate hosts of Dicrocoelium dendriticum. Z. Parasitenkd. 72, 689–692 (1986). [Google Scholar]
- 16.Unrau S., “Interactions between ants and larvae of the host-manipulating parasite, dicrocoelium dendriticum,” MSc thesis, University of Lethbridge, Lethbridge, Alberta, Canada (2019).
- 17.Hohorst W., Graefe G., Ameisen–obligatorische zwischenwirte des lanzettegels (Dicrocoelium dendriticum). Naturwissenschaften 48, 229–230 (1961). [Google Scholar]
- 18.Poulin R., Fredensborg B. L., Hansen E., Leung T. L. F., The true cost of host manipulation by parasites. Behav. Processes 68, 241–244 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Vickery W. L., Poulin R., The evolution of host manipulation by parasites: A game theory analysis. Evol. Ecol. 24, 773–788 (2010). [Google Scholar]
- 20.Criscione C. D., “History of microevolutionary thought in parasitology: The integration of molecular population genetics” in A Century of Parasitology: Discoveries, Ideas and Lessons Learned by Scientists Who Published in the Journal of Parastiology, 1914–2014, Janovy J. Jr, Esch G. W., Eds. (Wiley, Chichester, UK, 2016), pp. 93–109. [Google Scholar]
- 21.Gorton M. J., Kasl E. L., Detwiler J. T., Criscione C. D., Testing local-scale panmixia provides insights into the cryptic ecology, evolution, and epidemiology of metazoan animal parasites. Parasitology 139, 981–997 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Detwiler J. T., Criscione C. D., Role of parasite transmission in promoting inbreeding: II. Pedigree reconstruction reveals sib-transmission and consequent kin-mating. Mol. Ecol. 26, 4405–4417 (2017). [DOI] [PubMed] [Google Scholar]
- 23.van Paridon B. J., Colwell D. D., Goater C. P., Gilleard J. S., Population genetic analysis informs the invasion history of the emerging trematode Dicrocoelium dendriticum into Canada. Int. J. Parasitol. 47, 845–856 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Beck M. A., Goater C. P., Colwell D. D., Comparative recruitment, morphology and reproduction of a generalist trematode, Dicrocoelium dendriticum, in three species of host. Parasitology 142, 1297–1305 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beck M. A., “Ecological epidemiology of an invasive host generalist parasite, dicrocoelium dendriticum, in cypress hills interprovincial park, Alberta,” PhD thesis, University of Lethbridge, Lethbridge, Alberta, Canada (2015).
- 26.van Paridon B. J., Gilleard J. S., Colwell D. D., Goater C. P., Life cycle, host utilization, and ecological fitting for invasive lancet liver fluke, Dicrocoelium dendriticum, emerging in southern Alberta, Canada. J. Parasitol. 103, 207–212 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Dempsey Z. W., Burg T. M., Goater C. P., Spatio-temporal patterns of infection for emerging larval liver fluke (Dicrocoelium dendriticum) in three species of land snail in southern Alberta, Canada. J. Parasitol. 105, 155–161 (2019). [PubMed] [Google Scholar]
- 28.Bush A. O., Lafferty K. D., Lotz J. M., Shostak A. W., Parasitology meets ecology on its own terms: Margolis et al. revisited. J. Parasitol. 83, 575–583 (1997). [PubMed] [Google Scholar]
- 29.van Paridon B. J., Goater C. P., Gilleard J. S., Criscione C. D., Characterization of nine microsatellite loci for Dicrocoelium dendriticum, an emerging liver fluke of ungulates in North America, and their use to detect clonemates and random mating. Mol. Biochem. Parasitol. 207, 19–22 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Arnaud-Haond S., Belkhir K., Genclone: A computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Mol. Ecol. Notes 7, 15–17 (2007). [Google Scholar]
- 31.Gregorius H. R., Testing for clonal propagation. Heredity 94, 173–179 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Arnaud-Haond S., Duarte C. M., Alberto F., Serrão E. A., Standardizing methods to address clonality in population studies. Mol. Ecol. 16, 5115–5139 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Prugnolle F., Liu H., de Meeûs T., Balloux F., Population genetics of complex life-cycle parasites: An illustration with trematodes. Int. J. Parasitol. 35, 255–263 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Goudet J., FSTAT (Version 1.2): A computer program to calculate F-statistics. J. Hered. 86, 485–486 (1995). [Google Scholar]
- 35.Hardy O., Vekemans X., SPAGeDI: A versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2, 618–620 (2002). [Google Scholar]
- 36.Rousset F., genepop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 8, 103–106 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Jones O. R., Wang J., Colony: A program for parentage and sibship inference from multilocus genotype data. Mol. Ecol. Resour. 10, 551–555 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Wang J., Santure A. W., Parentage and sibship inference from multilocus genotype data under polygamy. Genetics 181, 1579–1594 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J., El-Kassaby Y. A., Ritland K., Estimating selfing rates from reconstructed pedigrees using multilocus genotype data. Mol. Ecol. 21, 100–116 (2012). [DOI] [PubMed] [Google Scholar]
- 40.MacGregor-Fors I., Payton M. E., Contrasting diversity values: Statistical inferences based on overlapping confidence intervals. PLoS One 8, e56794 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krebs C., Ecological Methodology (Benjamin/Cummings, Menlo Park, CA, ed. 2, 1999). [Google Scholar]
- 42.Hood G. M., PopTools, Version 3.2.5. http://www.poptools.org. Accessed 31 January 2017.
- 43.Rauch G., Kalbe M., Reusch T. B. H., How a complex life cycle can improve a parasite’s sex life. J. Evol. Biol. 18, 1069–1075 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Criscione C. D., Blouin M. S., Minimal selfing, few clones, and no among-host genetic structure in a hermaphroditic parasite with asexual larval propagation. Evolution 60, 553–562 (2006). [PubMed] [Google Scholar]
- 45.Keeney D. B., Waters J. M., Poulin R., Clonal diversity of the marine trematode Maritrema novaezealandensis within intermediate hosts: The molecular ecology of parasite life cycles. Mol. Ecol. 16, 431–439 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Keeney D. B., Waters J. M., Poulin R., Diversity of trematode genetic clones within amphipods and the timing of same-clone infections. Int. J. Parasitol. 37, 351–357 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Lagrue C., Poulin R., Keeney D. B., Effects of clonality in multiple infections on the life-history strategy of the trematode Coitocaecum parvum in its amphipod intermediate host. Evolution 63, 1417–1426 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Leung T. L. F., Poulin R., Keeney D. B., Accumulation of diverse parasite genotypes within the bivalve second intermediate host of the digenean Gymnophallus sp. Int. J. Parasitol. 39, 327–331 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Criscione C. D., Vilas R., Paniagua E., Blouin M. S., More than meets the eye: Detecting cryptic microgeographic population structure in a parasite with a complex life cycle. Mol. Ecol. 20, 2510–2524 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Valdivia I. M., Criscione C. D., Cárdenas L., Durán C. P., Oliva M. E., Does a facultative precocious life cycle predispose the marine trematode Proctoeces cf. lintoni to inbreeding and genetic differentiation among host species? Int. J. Parasitol. 44, 183–188 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Prugnolle F., De Meeûs T., Durand P., Sire C., Théron A., Sex-specific genetic structure in Schistosoma mansoni: Evolutionary and epidemiological implications. Mol. Ecol. 11, 1231–1238 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Prugnolle F., Choisy M., Théron A., Durand P., De Meeûs T., Sex-specific correlation between heterozygosity and clone size in the trematode Schistosoma mansoni. Mol. Ecol. 13, 2859–2864 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Vilas R., Vázquez-Prieto S., Paniagua E., Contrasting patterns of population genetic structure of Fasciola hepatica from cattle and sheep: Implications for the evolution of anthelmintic resistance. Infect. Genet. Evol. 12, 45–52 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Beesley N. J., Williams D. J. L., Paterson S., Hodgkinson J., Fasciola hepatica demonstrates high levels of genetic diversity, a lack of population structure and high gene flow: Possible implications for drug resistance. Int. J. Parasitol. 47, 11–20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hausdorf B., Hennig C., Nestedness of north-west European land snail ranges as a consequence of differential immigration from Pleistocene glacial refuges. Oecologia 135, 102–109 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Schilthuizen M., Lombaerts M., Population structure and levels of gene flow in the Mediterranean land snail Albinaria corrugata (Pulmonata: Clausiliidae). Evolution 48, 577–586 (1994). [DOI] [PubMed] [Google Scholar]
- 57.Clark S. A., Richardson B. J., Spatial analysis of genetic variation as a rapid assessment tool in the conservation management of narrow-range endemics. Invertebr. Syst. 16, 583–587 (2002). [Google Scholar]
- 58.Wang J., Does GST underestimate genetic differentiation from marker data? Mol. Ecol. 24, 3546–3558 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Romig T., Lucius R., Frank W., Cerebral larvae in the second intermediate host of Dicrocoelium dendriticum (Rudolphi, 1819) and Dicrocoelium hospes Looss, 1907 (Trematodes, Dicrocoeliidae). Z. Parasitenkd. 63, 277–286 (1980). [DOI] [PubMed] [Google Scholar]
- 60.Kay T., Lehmann L., Keller L., Kin selection and altruism. Curr. Biol. 29, R438–R442 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Garcia-Vedrenne A. E., et al. , Trematodes with a reproductive division of labour: Heterophyids also have a soldier caste and early infections reveal how colonies become structured. Int. J. Parasitol. 47, 41–50 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Poulin R., Kamiya T., Lagrue C., Evolution, phylogenetic distribution and functional ecology of division of labour in trematodes. Parasit. Vectors 12, 5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weinersmith K. L., et al. , A lack of crowding? Body size does not decrease with density for two behavior-manipulating parasites. Integr. Comp. Biol. 54, 184–192 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in the paper are available in SI Appendix, Tables S1 and S2.