Significance
Current approaches for producing T cells for adoptive immunotherapy for cancer rely on interleukin (IL)-2–based strategies that generate large numbers of tumor-reactive T cells but also drive the cells toward terminal differentiation and exhaustion, thereby diminishing their effectiveness. By characterizing the metabolic effects of IL-2 versus IL-21, a closely related cytokine that expands cells but lacks the detrimental effects of IL-2, we identified a pivotal role of lactate dehydrogenase (LDH) in regulating CD8+ T cell effector differentiation. Remarkably, LDH inhibition combined with IL-21 enhanced the formation of T memory stem cells and mitochondrial fitness while suppressing programs of exhaustion and senescence and markedly enhancing antitumor responses. These findings have potential therapeutic implications.
Keywords: IL-2, IL-21, LDH, adoptive immunotherapy, immunometabolism
Abstract
Interleukin (IL)-2 and IL-21 dichotomously shape CD8+ T cell differentiation. IL-2 drives terminal differentiation, generating cells that are poorly effective against tumors, whereas IL-21 promotes stem cell memory T cells (TSCM) and antitumor responses. Here we investigated the role of metabolic programming in the developmental differences induced by these cytokines. IL-2 promoted effector-like metabolism and aerobic glycolysis, robustly inducing lactate dehydrogenase (LDH) and lactate production, whereas IL-21 maintained a metabolically quiescent state dependent on oxidative phosphorylation. LDH inhibition rewired IL-2–induced effects, promoting pyruvate entry into the tricarboxylic acid cycle and inhibiting terminal effector and exhaustion programs, including mRNA expression of members of the NR4A family of nuclear receptors, as well as Prdm1 and Xbp1. While deletion of Ldha prevented development of cells with antitumor effector function, transient LDH inhibition enhanced the generation of memory cells capable of triggering robust antitumor responses after adoptive transfer. LDH inhibition did not significantly affect IL-21–induced metabolism but caused major transcriptomic changes, including the suppression of IL-21–induced exhaustion markers LAG3, PD1, 2B4, and TIM3. LDH inhibition combined with IL-21 increased the formation of TSCM cells, resulting in more profound antitumor responses and prolonged host survival. These findings indicate a pivotal role for LDH in modulating cytokine-mediated T cell differentiation and underscore the therapeutic potential of transiently inhibiting LDH during adoptive T cell-based immunotherapy, with an unanticipated cooperative antitumor effect of LDH inhibition and IL-21.
Immune responses are initiated by engagement of the T cell receptor (TCR) and then critically controlled by cytokines, which influence differentiation, proliferation, and survival. Altering metabolic pathways can affect the actions of immune cells (1–4), and different cellular subtypes vary in how they produce and expend energy (5). For example, naïve T cells are quiescent, with a low energy demand that is met primarily via oxidative phosphorylation, but after TCR activation, T cells markedly increase their metabolic activity, acutely engaging in aerobic glycolysis and later also up-regulating oxidative ATP production (6), with production of a range of cytokines.
Interleukin (IL)-2 is a type I cytokine with pleiotropic actions and therapeutic effects that has been approved for the treatment of melanoma and renal cell carcinoma and is also used to expand cells for adoptive cell therapy (7). However, besides its beneficial effects, IL-2 can induce T cell differentiation and diminish antitumor efficacy (8). In contrast, IL-21, which like IL-2 uses the common cytokine receptor γ chain (γc) as a receptor component (9, 10), has greater antitumor activity in adoptive transfer experiments (8, 11) and also exhibits antitumor activity in other settings (12). IL-2 primarily activates STAT5 and mediates T cell expansion following antigen activation, but it also induces a CD8+ T cell effector phenotype and promotes regulatory T cell (Treg) differentiation (13). IL-21 primarily activates STAT3 (12), cooperatively expands CD8+ T cells with IL-7 and IL-15 (14), and promotes T follicular helper cell development (12). These cytokines can exhibit opposing actions for Th9 and Treg cells (promoted by IL-2 but inhibited by IL-21) or Th17 and Tfh cells (promoted by IL-21 but inhibited by IL-2) (11, 15). Moreover, they also have different metabolic effects: IL-2 induces aerobic glycolysis, characteristic of effector-like metabolism in CD8+ T cells (16–22), whereas IL-21 maintains a naïve-like metabolically quiescent state dependent on oxidative phosphorylation (23). Here we further explored the metabolic differences between IL-2 and IL-21 and investigated how these phenotypes relate to their differential antitumor activity.
Results
IL-21 Sustains Metabolic Quiescence and Mitochondrial Morphology.
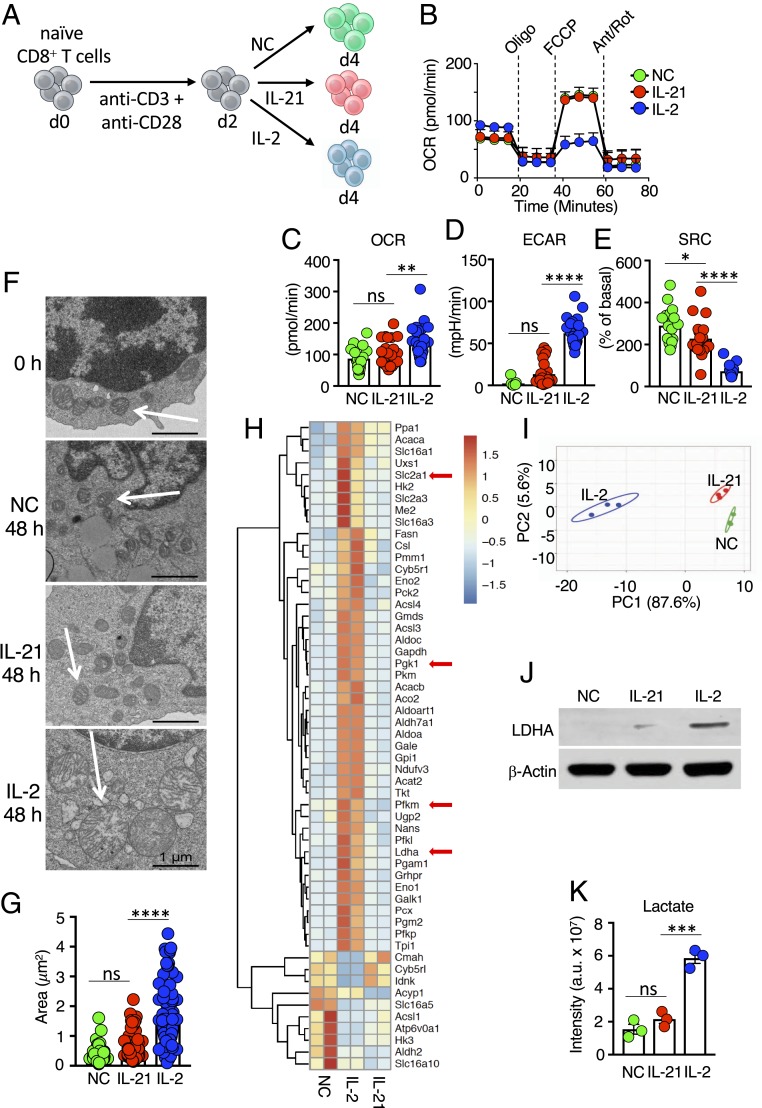
We initially compared the metabolic effects of IL-2 and IL-21 on mouse splenic CD8+ T cells activated with anti-CD3 and anti-CD28 for 48 h and then cultured for 48 h with no cytokine (NC), IL-2, or IL-21 (Fig. 1A). As reported previously (23), IL-2–treated cells exhibited higher basal metabolism than IL-21–treated cells (Fig. 1B), with a higher basal oxygen consumption rate (OCR) (Fig. 1C) and extracellular acidification rate (ECAR) (Fig. 1D). IL-21–treated cells had limited metabolic differences compared with cells cultured without exogenous cytokines (Fig. 1 C and D). This more quiescent state was consistent with preservation of mitochondrial spare respiratory capacity (SRC) (Fig. 1E), a feature of memory T cells correlating with the potential for expansion on restimulation (19). In contrast, SRC was minimal in IL-2–treated cells (Fig. 1E). A major difference in ECAR between IL-2 and IL-21 was not evident at 4 or 12 h but appeared by 24 h and increased at 48 h, with differences in OCR also seen at 48 h (SI Appendix, Fig. S1A). Importantly, IL-2 also induced more energetic basal metabolism in human CD8+ T cells (SI Appendix, Fig. S1B), with higher OCR and ECAR, whereas IL-21 sustained a higher SRC (SI Appendix, Fig. S1C).
Fig. 1.
Distinctive metabolic effects of IL-2 versus IL-21. (A) Schematic of protocol for stimulating CD8+ T cells. Naive T cells were isolated, activated with anti-CD3 and anti-CD28 for 2 d, and cultured with NC, IL-21, or IL-2 for another 2 d. (B) Seahorse experiment measuring the OCR for CD8+ T cells treated with NC, IL-21, or IL-2 following treatment with oligomycin, FCCP, and antimycin A/rotenone. Data are representative of 11 independent experiments. (C–E) Bar graphs indicating basal OCR (C), ECAR (D), and SRC (E), as measured from Seahorse assays. (F) Electron micrographs are representative images from two independent experiments. (Scale bar: 1 μm.) (G) Calculated area of mitochondria from electron micrographs from cells treated with IL-2, IL-21, or NC. (H) RNA-Seq heatmap of differentially expressed metabolic genes from cells treated with NC, IL-2, or IL-21. Shown is the scale for fold induction or repression. (I) Principal component analysis plot based on metabolomics data of cells treated with NC, IL-2, or IL-21. (J) Western blotting of LDHA protein. (K) LC-MS–based analysis of intracellular lactate. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
Metabolic reprogramming is accompanied by mitochondrial remodeling, and differentiation in effector T cells is associated with punctate mitochondria, loosely structured cristae, disrupted oxidative phosphorylation, and Warburg metabolism (21). Therefore, we used electron microscopy to examine the impact of IL-2 and IL-21 on mitochondrial morphology. After 48 h of anti-CD3 and anti-CD28 activation, cells had mitochondria with tight cristae (Fig. 1 F, Top), indicating maintenance of redox chemistry and efficient oxidative phosphorylation (21, 24). After 48 h in medium or IL-21, cells retained tight mitochondrial cristae (Fig. 1 F, Middle), but IL-2–treated cells had large dysmorphic mitochondria with loosely structured cristae (Fig. 1 F, Bottom and Fig. 1G).
Differential Effects of IL-2 versus IL-21 on Metabolic Gene Expression and Metabolomic Profiles in T Cells.
We next assessed the effects of IL-2 and IL-21 by RNA-Seq. Compared with cells cultured without exogenous cytokines, IL-2 induced the expression of key glycolytic genes, including Slc2a1, encoding the glucose transporter GLUT1; Pgk1, encoding the enzyme in the first ATP-generating step of glycolysis; Pfkm, a regulator of glycolytic flux; and Ldha, encoding the major isoform of lactate dehydrogenase (LDH) (Fig. 1H, arrows). We confirmed the induction of these genes by RT-qPCR (SI Appendix, Fig. S2A). Overall, many more metabolic genes were differentially expressed after stimulation with IL-2 compared with IL-21 (Fig. 1H). Despite phenotypic similarities between NC cells and IL-21–treated cells, the latter proliferated faster, as assessed by fold expansion (SI Appendix, Fig. S2B) and carboxyfluorescein succinimidyl ester dilution (SI Appendix, Fig. S2C), and had increased viability compared with NC cells (SI Appendix, Fig. S2 D and E). IL-2 induced even greater expansion than IL-21 (SI Appendix, Fig. S2 B and C), and like IL-21, it also increased viability compared with NC cells (SI Appendix, Fig. S2 D and E). Consistent with the differences in expression of metabolic genes by RNA-Seq, metabolomics analysis showed that IL-2–treated cells had the most distinctive metabolite changes compared with the NC control, whereas the effect of IL-21 was more modest (Fig. 1I and SI Appendix, Fig. S2F). Together with bioenergetic and transcriptomic data, this reveals that IL-2 and IL-21 differ in their regulation of key genes and metabolites involved in glycolysis.
Correspondingly, studies using a fluorescently-labeled glucose derivative revealed that IL-2–treated CD8+ T cells had higher glucose uptake than cells treated with NC or IL-21 (SI Appendix, Fig. S2G); however, IL-2–treated cells had lower intracellular glucose (SI Appendix, Fig. S2H) and higher intracellular levels of pyruvate (SI Appendix, Fig. S2I), consistent with elevated glycolysis. Moreover, consistent with the higher Ldha mRNA expression (Fig. 1H and SI Appendix, Fig. S2 A, Right), IL-2–treated cells expressed higher levels of LDHA than IL-21–treated cells (Fig. 1J) and correspondingly generated more lactate (Fig. 1K). Interestingly, IL-2 treatment increased the level of the inactive phosphorylated form of pyruvate dehydrogenase compared with IL-21– and NC-treated cells (SI Appendix, Fig. S2J), showing that while IL-2 induces glycolysis, it restricts pyruvate incorporation into the tricarboxylic acid (TCA) cycle. Compared with IL-2, IL-21 is metabolically more inert.
Inhibiting LDH Rewires T Cell Metabolism to Promote Oxidative Phosphorylation.
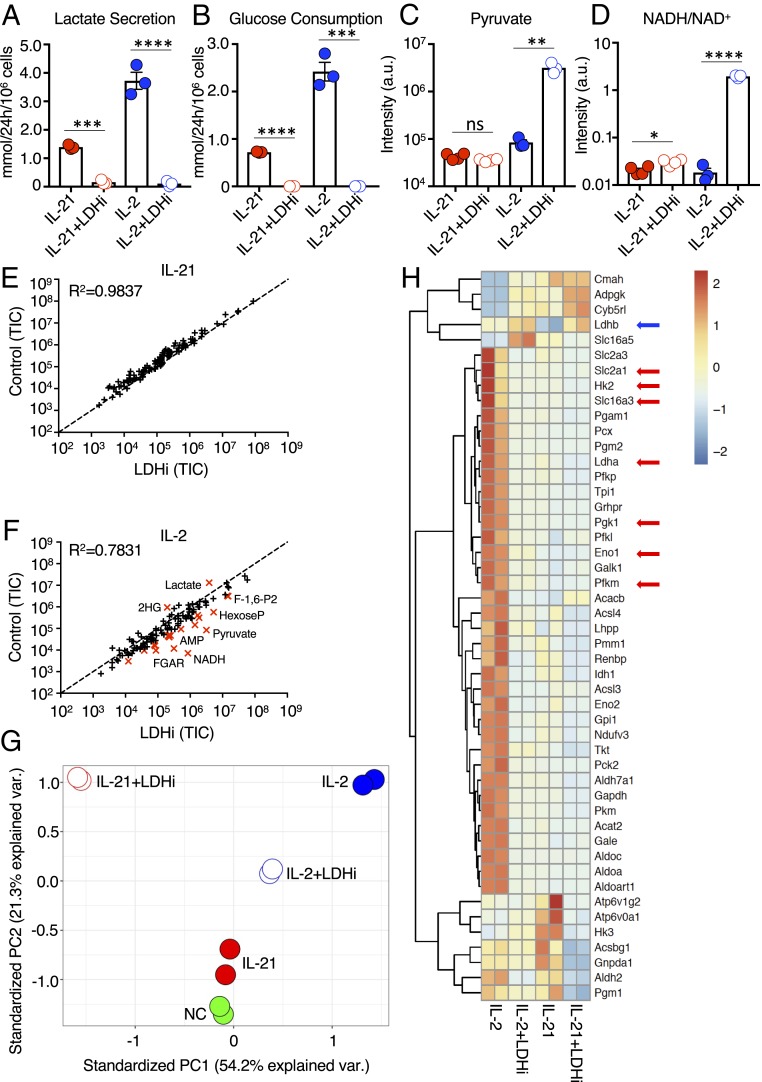
We next investigated whether the proglycolytic effects of IL-2 in CD8+ T cells are important for the developmental and functional programs induced by this cytokine. We tested the capacity of a small molecule inhibitor of LDH, herein denoted LDHi (SI Appendix, Fig. S3A), that reversibly inhibits the activity of the LDHA and LDHB isoforms of LDH (SI Appendix, Fig. S3B) (25, 26) to suppress aerobic glycolysis and reprogram T cell metabolism. Along with the enzymatic conversion of pyruvate to lactate, LDH activity maintained cytosolic levels of the oxidized form of nicotinamide adenine dinucleotide (NAD+) and the NADH/NAD+ ratio to sustain glycolytic flux. LDHi treatment of IL-2– or IL-21–stimulated cells indeed reduced lactate secretion (Fig. 2A), with a corresponding color change of the pH indicator dye in the culture media, particularly for IL-2 (SI Appendix, Fig. S3C), and also diminished glucose consumption (Fig. 2B). LDHi significantly inhibited IL-2–induced proliferation, whereas its effect on IL-21 proliferation was not statistically significant (SI Appendix, Fig. S3D). Moreover, for IL-21–stimulated cells, LDHi treatment did not alter intracellular pyruvate (Fig. 2C), raised the NADH/NAD+ ratio only slightly (Fig. 2D), and had little effect on metabolite levels (Fig. 2E). In contrast, in IL-2–treated cells, LDHi increased intracellular pyruvate (Fig. 2C) and caused an ∼100-fold increase in the NADH/NAD+ ratio (Fig. 2D). Furthermore, LDHi treatment lowered intracellular lactate in cells stimulated with IL-2 or IL-21 (SI Appendix, Fig. S3E). This large decrease in lactate coupled with the large increase in pyruvate in cells treated with LDHi+IL-2 resulted in a marked decrease in the lactate/pyruvate ratio (SI Appendix, Fig. S3F). Thus, overall, in the IL-2 setting, LDHi treatment had the greatest effect on levels of lactate, pyruvate, and NADH levels (Fig. 2F), which are direct substrates/products of LDH.
Fig. 2.
LDH inhibition alters glycolytic flux and transcriptional programming. (A and B) Using a YSI 2900 Series Biochemical Analyzer, media measurements for lactate secretion (A), and glucose consumption (B) were determined for cells treated with IL-2 or IL-21 without or with LDHi. (C and D) Pyruvate levels (C) and intracellular NADH/NAD+ ratio (D) were assessed by LC-MS for cells treated with IL-2 or IL-21 in the absence or presence of LDHi. (E and F) Truth plot showing correlation of intracellular metabolites for cells treated with IL-21 versus those treated with IL-21+LDHi (E) or IL-2 versus IL-2+LDHi (F); in F, the metabolites in red had a fold change >4. (G) Principal component analysis generated from RNA-Seq data of cells treated with NC, IL-2, IL-21, IL-2+LDHi, or IL-21+LDHi. (H) RNA-Seq heatmap displaying differentially expressed metabolic genes between cells treated with IL-2, IL-2+LDHi, IL-21, or IL-21+LDHi. Each stimulation was performed in duplicate. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
A >4-fold increase was also observed in glycolytic intermediates, presumably due to downstream inhibition of glycolysis, as well as higher levels of FGAR (Fig. 2F), an intermediate in purine biosynthesis. As expected, 2-hydroxyglutarate, which plays a role in T cell differentiation and is produced in CD8+ T cells substantially via LDH (27), was decreased by LDHi (Fig. 2F). LDHi also globally affected gene expression induced by IL-21 and IL-2 (see the principal component analysis of the RNA-Seq data; Fig. 2G), with a greater effect on metabolism-associated genes in IL-2–treated cells compared with IL-21–treated cells (Fig. 2H). Interestingly, most genes differentially expressed by IL-21 versus IL-21+LDHi were not metabolism-associated genes, and many were cell cycle-related (SI Appendix, Fig. S3G).
Strikingly, cells treated with IL-2+LDHi had a metabolic transcriptomic pattern resembling that of cells treated with IL-21 rather than cells treated with IL-2 (Fig. 2H and SI Appendix, Table S1). Compared with IL-21, in the IL-2 context LDHi had a greater effect on genes related to glycolysis and gluconeogenesis, with lower induction of Slc2a1, Hk2, Slc16a3, Pgk1, Eno1, and Pfkm, as well as of Ldha (Fig. 2H, red arrows), although Ldhb levels were increased (Fig. 2H, blue arrow).
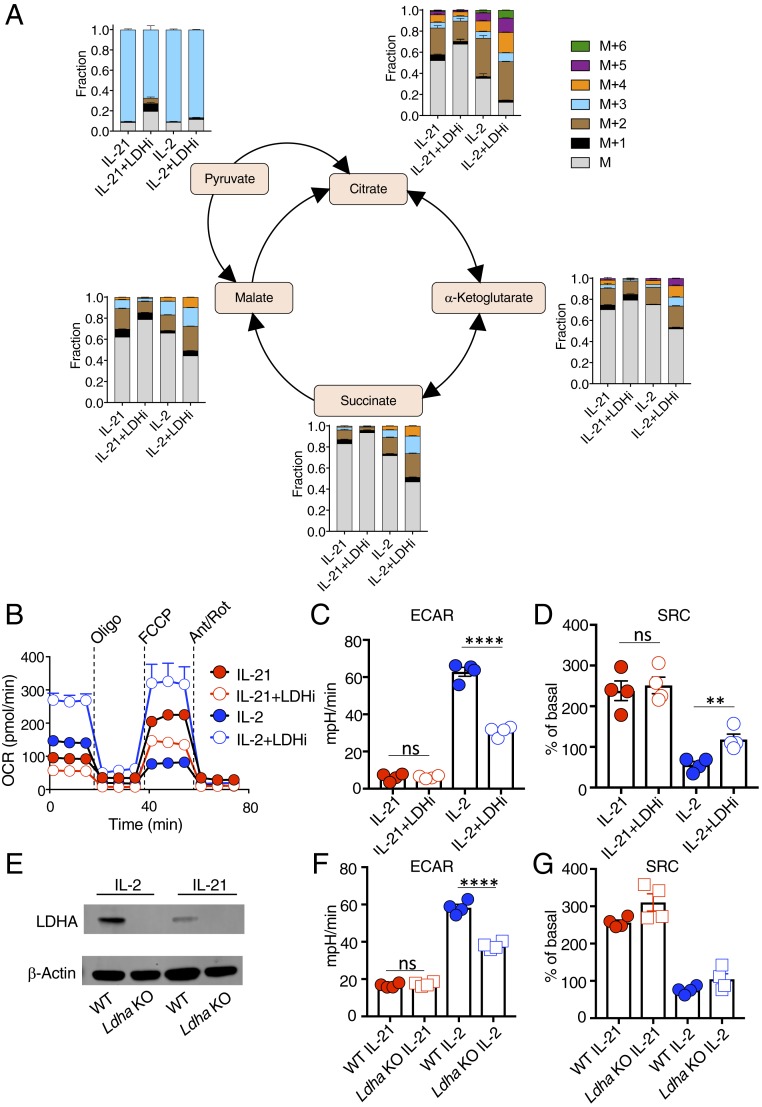
To elucidate the metabolomic effects of LDH inhibition, we performed a U-13C-glucose tracer study. LDHi treatment had little effect on isotopomer distribution in IL-21–stimulated cells but caused a greater redirection of glucose carbons into the TCA cycle in IL-2–stimulated cells (Fig. 3A). Correspondingly, LDHi did not substantially alter basal OCR (Fig. 3B), ECAR (Fig. 3C), or SRC (Fig. 3D) in IL-21–treated cells but significantly increased OCR and SRC and decreased ECAR in IL-2–treated cells (Fig. 3 B–D). Thus, LDHi limits pyruvate conversion to lactate while favoring pyruvate oxidation and entry into the TCA cycle, promoting mitochondrial metabolism in IL-2–stimulated cells. In Seahorse experiments using human CD8+ T cells (SI Appendix, Fig. S4 A and B), LDHi had similar effects to those in mouse cells, but with a critical difference: it decreased ECAR (SI Appendix, Fig. S4C) and raised SRC (SI Appendix, Fig. S4D) in cells treated with either IL-21 or IL-2. Thus, inhibiting LDH rewires metabolism in CD8+ T cells and promotes pyruvate oxidation and entry into the TCA cycle, thereby making IL-2 act more like IL-21 and, especially in human T cells, enhancing oxidative capacity even in IL-21–treated cells.
Fig. 3.
LDHi increases the contribution of glucose to the TCA cycle in IL-2–treated cells and alters mitochondrial metabolism. (A) Corrected 13C-labeled fraction of TCA cycle intermediates from 13C-glucose in IL-2– and IL-21–treated cells in the absence or presence of LDHi, assessed by LC-MS. Measurements are displayed as total distribution of labeled fractions ranging from the unlabeled mass of the metabolite, M, to the mass plus the addition of six 13C-labeled atoms (M+6). (B) OCR of cells treated with IL-2, IL-2+LDHi, IL-21, or IL-21+LDHi following injection with oligomycin, FCCP, and antimycin A/rotenone. (C and D) Quantitative measurements of ECAR (C) and SRC (D) for cells treated with IL-21, IL-21+LDHi, IL-2, or IL-2+LDHi, as indicated. (E) Ldha-deletion in T cells. Western blot demonstrating the absence of LDHA following tamoxifen-mediated deletion of Ldha (Materials and Methods). (F and G) Graphs showing ECAR (F) and SRC (G) measurements for WT and Ldha KO cells. **P < 0.01; ****P < 0.0001; ns, not significant.
We also compared T cells from WT mice to Ldha-deficient T cells, which we confirmed lack expression of LDHA (Fig. 3E), and found that Ldha-deficient cells behaved similarly to LDHi-treated wild-type T cells, in that IL-21 had no effect on ECAR or SRC (compare Ldha KO data in Fig. 3 F and G with LDHi data in Fig. 3 C and D), and ECAR was lower in Ldha-deficient cells treated with IL-2 (Fig. 3F; compare with Fig. 3C). However, unlike LDHi treatment, Ldha deletion was not sufficient to significantly raise SRC in IL-2–treated cells (Fig. 3G; compare with Fig. 3D), suggesting that inhibition of both LDHA and LDHB activity may be required to substantially raise SRC in this setting. Alternatively, Ldha KO cells permanently lack LDHA, whereas LDHi treatment of WT cells represents acute inhibition, which also might be a contributing factor.
Inhibiting LDH In Vitro Enhances Stem Cell Memory T Cell Formation, with Increased Antitumor Activity In Vivo following Adoptive Transfer.
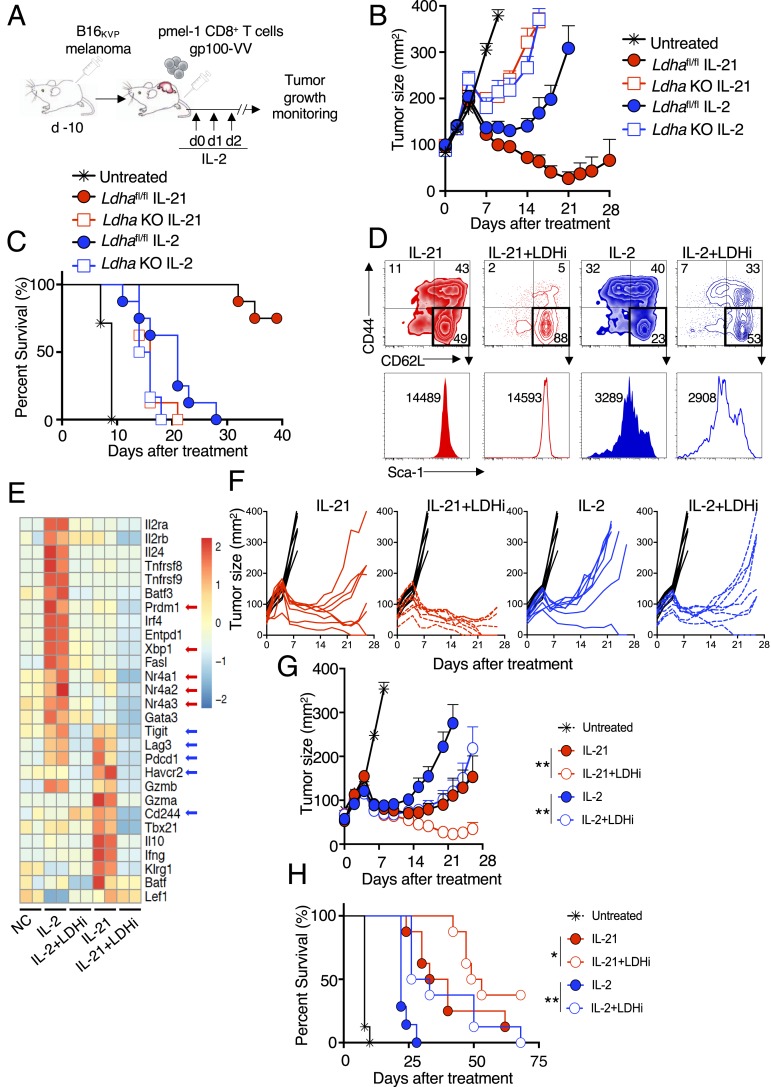
To investigate whether lowering LDH activity could reprogram CD8+ T cell function, we used a mouse model of adoptive cancer immunotherapy in which pmel-1 CD8+ T cells recognizing the melanoma-melanocyte differentiation antigen gp100 were transferred into C57BL/6 mice bearing subcutaneous B16 melanoma expressing the mutated gp100 antigen B16KVP (Fig. 4A) (28, 29). Strikingly, adoptively transferred Ldha-deficient T cells poorly eliminated tumors (Fig. 4B), with lower host survival compared with WT controls (Fig. 4C) irrespective of treatment with IL-2 or IL-21, indicating that LDHA is required for the development of effector CD8+ T cells with antitumor activity. However, we hypothesized that selectively inhibiting LDH only during the in vitro culture phase might be therapeutically beneficial, as this might maintain cells in a stem cell memory T cell (TSCM) state, with enhanced antitumor activity following adoptive transfer. When pmel-1 CD8+ T cells were activated with anti-CD3 and anti-CD28, IL-2 was more potent than IL-21 in driving differentiation of CD44hiCD62Llo effector memory cells, whereas IL-21 resulted in more CD44loCD62LhiSca1hi cells (Fig. 4D), typical of TSCM cells (30). Combining IL-2 and LDHi resulted in more CD44loCD62Lhi cells than seen in IL-2–treated cells, and LDHi synergized with IL-21 to augment TSCM cell numbers (Fig. 4D). Moreover, LDHi repressed IL-2– and IL-21–specific programs of exhaustion/senescence; for example, transcripts for IL-2–induced transcription factors associated with exhaustion, including Nr4a1, Nr4a2, Nrfa3, and Prdm1 (31, 32), were decreased by LDHi to levels similar to those observed with IL-21, and treatment with IL-21+LDHi resulted in even lower expression (Fig. 4E, red arrows and SI Appendix, Table S1). Transcripts for XBP1, which plays a role in the unfolded protein response and can diminish antitumor efficacy of T cells in human ovarian cancer by regulating mitochondrial activity (33), were also induced by IL-2 but not by IL-21, and Xbp1 expression was inhibited by LDHi (Fig. 4E). Furthermore, LDHi repressed transcripts for exhaustion markers Lag3, Pdcd1 (encoding PD1), Cd244 (encoding 2B4), and Havcr2 (encoding TIM3), which were induced by IL-21 (Fig. 4E, blue arrows).
Fig. 4.
Inhibiting LDH in vitro enhances T cell priming and antitumor activity in vivo. (A) Schematic of the adoptive cell transfer protocol. pmel-1+ CD8+ T cells were cultured in the presence of anti-CD3 and anti-CD28 for 4 d, with concomitant stimulation with IL-2 or IL-21 in the absence or presence of LDHi. Cells were then adoptively transferred into animals that had been injected with B16 melanoma 10 d earlier. IL-2 was administered at the time of adoptive cell transfer and on each of the next 2 d. (B) Average tumor curves for transferred WT or Ldha KO CD8+ T cells treated with IL-2 or IL-21 or left untreated, as indicated. (C) Percent survival for each treatment group in B. (D) pmel-1 CD8 T cells were treated with IL-2 or IL-21 in the absence of presence of LDHi, and CD44 vs. CD62L expression was assessed on live cells (Upper). Gating was done on the CD62LhiCD44lo cells from the upper panels to assess Sca-1+ cells (histograms, Lower). (E) Heatmap of RNA-Seq data for cells treated with NC, IL-2, IL-2+LDHi, IL-21, or IL-21+LDHi. (F) Graphs showing individual tumor size over time for each mouse in the experiment. (G) Average tumor curves from data in F for untreated (black) or treated with IL-21 (solid red circles), IL-21+LDHi (open red circles), IL-2 (solid blue circles), or IL-2+LDHi (open blue circles). Statistics comparing tumor curves were performed using the Wilcoxon rank-sum test. (H) Percent survival for each treatment group in the tumor experiment shown in F and G. Statistics comparing percent survival data were performed using a log-rank (Mantel–Cox) test. All tumor data are from one of two similar experiments. *P < 0.05; **P < 0.01.
Although the basis for the transcriptomic changes is not known, nuclear LDHA can affect H3K79 methylation (34), and histone lactylation might also contribute (35), an area for further investigation. These phenotypic and transcriptomic data suggest that LDHi might be capable of enhancing the antitumor activity of CD8+ T cells. Following adoptive transfer, as expected (8, 11), IL-21 conferred a therapeutic advantage compared with IL-2 (individual tumor curves in Fig. 4F; summarized in Fig. 4G). Remarkably, culturing cells with LDHi and IL-2 before adoptive transfer augmented the antitumor effect to the level of IL-21–treated cells (Fig. 4G), and combining IL-21 with LDHi further enhanced antitumor activity (Fig. 4 F and G). These antitumor effects corresponded to prolonged animal survival (Fig. 4H), revealing a role for LDH in regulating CD8+ T cell fate decisions in response to IL-2 and IL-21 and demonstrating that temporally-specific LDH inhibition has therapeutic potential in adoptive T cell-based immunotherapy.
Discussion
Our findings underscore distinctive metabolic programs of IL-2 and IL-21 in CD8+ T cells. IL-2 promotes metabolic reprogramming, leading to energetic metabolism, with higher OCR and ECAR but essentially absent SRC and enlarged dysmorphic mitochondria, whereas IL-21 sustains metabolic quiescence yet induces expression of cellular fitness genes. By combining RNA-Seq, Seahorse, metabolomics, 13C-glucose carbon tracing, and Western blot analysis, we found that favoring lactate instead of the TCA cycle as the end product of glucose energy metabolism is a major distinction between IL-2 and IL-21, with dramatic differences between these cytokines on CD8+ T cell differentiation and antitumor responses in vivo.
Previously, 2-deoxyglucose, an inhibitor of glucose conversion to glucose-6-phosphate, the first step in glycolysis, was shown to enhance the generation of memory cells and antitumor functionality (36), but that study did not investigate the fate of glucose-generated pyruvate, which here we specifically interrogated using an inhibitor of LDH (which acts on pyruvate, the product of glycolysis) to elucidate the relative role of lactic acid fermentation versus pyruvate decarboxylation and entry into the TCA cycle. Strikingly, adding LDHi to IL-2–treated cells during the in vitro priming phase markedly improved antitumor activity in vivo following adoptive cell transfer.
These data reveal that transiently inhibiting LDH activity only before adoptive transfer is associated with improved tumor clearance, helping clarify the augmented antitumor response by pmel-1 CD8+ T cells that are primed with IL-21 as opposed to IL-2. However, the greater antitumor activity of cells treated with IL-21+LDHi versus IL-21 alone or IL-2+LDHi indicates that IL-21 has beneficial effects beyond sustaining low LDH activity, potentially involving the repression of prosenescence genes such as Prdm1, Xbp1, and Nr4a family members. Importantly, our data also show that while genomic deletion of Ldha (i.e., permanent lack of LDHA) resulted in a similar metabolic phenotype (albeit with a difference in IL-2–induced SRC) compared with that observed with LDH inhibition in vitro, a permanent lack of LDHA prevented the development of effector CD8+ T cells with potent antitumor activity. These data for CD8+ T cells indicate that LDHA activity is necessary in vivo for effector function and complement a previous report showing that LDHA is important for CD4+ T cell differentiation into IFNγ-producing Th1 cells (37), underscoring the broad role of LDHA in effector T cell differentiation. Moreover, we show that inhibiting LDH during the in vitro culture phase, thereby inhibiting lactic acid fermentation and favoring pyruvate entry into the TCA cycle before adoptive transfer of the cells, is beneficial and a mechanism for increasing the number of pmel-1 TSCM cells, which are known to have antitumor potential (38, 39). Thus, the timing of LDH inhibition is critical, and sustaining LDH activity following adoptive transfer is necessary for the development of effector T cells with antitumor activity. Similar in vitro metabolic effects of LDHi were observed for human CD8+ T cells, underscoring the potential therapeutic benefits of LDH inhibition before cell transfer in adoptive cell transfer-based immunotherapy.
In summary, we have elucidated the metabolic basis for the differential actions of IL-2 versus IL-21, and moreover, by comparing LDH inhibition with Ldha gene deletion, we have identified LDH as a negative regulator of formation of TSCM cells but a positive regulator of effector T cell formation, with striking antitumor effects of combining LDHi with IL-2 and unanticipated cooperative effects of combining LDHi with IL-21, findings with important therapeutic implications.
Materials and Methods
Cell Preparation and Culture.
Except when specified otherwise, all experiments were performed with murine splenic CD8+ T cells isolated with the STEMCELL Technologies EasySep Mouse CD8+ T Cell Isolation Kit (catalog no. 19853). Following isolation, cells were resuspended at 1 × 106 cells/mL in T cell culture medium: RPMI medium 1640 (Thermo 11875093) containing 10% FBS (FBS), 1 x Glutamax (Life Technologies #35050–061), 1 mM sodium pyruvate, 0.1% β-mercaptoethanol, and penicillin/streptomycin. Cells were plated in 24-well tissue culture plates precoated with mouse anti-CD3 (2 μg/mL) and cultured with soluble mouse anti-CD28 (2 μg/mL) for 48 h. After incubation, cells were pooled, washed twice in media, and replated at 1 × 106 cells/mL Cells were stimulated without cytokine or with 100 IU/mL human IL-2 or 100 ng/mL mouse IL-21. Unless specified otherwise, cells were stimulated for 48 h before being harvested. Where noted, cells were treated (1 μM) with a novel pyrazole-based LDH inhibitor (NCI-737; herein denoted LDHi) that was developed under the auspices of the National Cancer Institute’s Experimental Therapeutics Program (25, 26). LDHi was added at the time of anti-CD3 and anti-CD28 treatment and replenished after cells were washed and replated in either IL-2 or IL-21, as indicated.
Western Blot Analysis.
After 3 d of stimulation with medium, IL-2, or IL-21, cells were pelleted at 1,500 rpm for 5 min, washed with ice-cold PBS, resuspended in radioimmunoprecipitation assay buffer, and frozen on dry ice. Frozen lysates were thawed on ice, cell debris was removed by centrifuging at 13,000 rpm for 15 min at 4 °C, and supernatant was collected. Protein was quantified using the DC Protein Assay Kit (Bio-Rad; 5000111) and 20 μg of supernatant protein was loaded into the wells of a NuPAGE 4 to 12% Bis-Tris Gel (Invitrogen; NP0335BOX) and electrophoresed at 100 V in NuPAGE Mops SDS Running Buffer (Life Technologies; 1908733). Proteins were transferred onto a 0.45-μm polyvinylidene difluoride membrane using an XCell II Blot Module (Invitrogen; EI9051) run at 30 V for 1 h in NuPAGE Transfer Buffer (Life Technologies; 1904537). Following transfer, membranes were blocked by shaking for 1 h at room temperature with 5% nonfat dry milk in TBS. Blots were probed overnight at 4 °C with primary antibodies dissolved in 5% nonfat dry milk in TBS + 0.1% Tween-20 (TBSt). The following primary antibodies were used: anti-human or anti-mouse recombinant LDHA purified sheep IgG (R&D Systems; AF7304), anti-pyruvate dehydrogenase E1-alpha subunit (phospho S293) rabbit polyclonal antibody (Abcam; ab92696) and anti–β-actin mouse monoclonal antibody clone AC-15 (Sigma-Aldrich; A1978). Following incubation with primary antibodies, blots were washed four times in TBSt and then incubated for 1 h at room temperature with secondary antibodies dissolved in TBSt. The secondary antibodies used were rabbit anti-sheep IgG DyLight 800 (Thermo Fisher Scientific; SA5-10060), infrared dye 680RD goat anti-rabbit (LI-COR; 926-68071), or infrared dye 680RD goat anti-mouse (LI-COR; 926-68070). Following secondary antibody incubations, blots were washed with TBSt and imaged using the LI-COR Odyssey CLx Imaging System.
Metabolomics and 13C Labeling.
CD8+ T cells were isolated from 6-wk-old C57BL/6 mouse spleens (STEMCELL Technologies; 19853), activated for 48 h with anti-CD3 and anti-CD28, washed, and replated for 48 h in medium alone or medium supplemented with human IL-2 or mouse IL-21. Cells were then washed with medium supplemented with dialyzed FBS (Thermo Fisher Scientific; A3382001); replated in RPMI medium 1640 containing 10% dialyzed FBS, 1× Glutamax, 1 mM sodium pyruvate, 0.1% β-mercaptoethanol, and penicillin-streptomycin; and restimulated with same cytokine treatment for 5 h. For 13C labeling, cells were instead replated in medium containing 2 g/L U-13C-glucose (Cambridge Isotope Laboratories; CLM-1396) for 6 h. Then 3 × 106 cells per replicate (three or four replicates per treatment) were transferred to microcentrifuge tubes and centrifuged for 1 min at 6,000 × g. Room temperature PBS was added to wash cell pellets, followed by another round of centrifugation for 1 min. Pellets were then immediately resuspended for 5 min in 150 μL of ice-cold extraction solvent (40:40:20 acetonitrile:methanol:water plus 0.5% formic acid; the inclusion of acetonitrile and acid in the extraction solvent is important to prevent interconversion of NAD+ and NADH). After incubation, 13.2 μL of 15% ammonium bicarbonate was added, and samples were centrifuged for 15 min at maximum speed at 4 °C. Supernatant containing cellular metabolites was transferred to a fresh tube, frozen in dry ice, and stored at -80 °C. Cell extracts were analyzed using a quadrupole-orbitrap mass spectrometer (Q Exactive; Thermo Fisher Scientific) coupled to hydrophilic interaction chromatography via electrospray ionization.
LC separation was done on a XBridge BEH Amide column (2.1 mm × 150 mm, 2.5 μm particle size; Waters) using a gradient of solvent A (20 mM ammonium acetate, 20 mM ammonium hydroxide in 95:5 water:acetonitrile, pH 9.45) and solvent B (acetonitrile). For untargeted metabolomics, the flow rate was 150 μL/min, column temperature was 25 °C, autosampler temperature was 5 °C, and injection volume was 10 µL. The LC gradient was as follows: 0 min, 90% B; 2 min, 85% B; 3 min, 75% B; 7 min, 75% B; 8 min, 70% B; 9 min, 70% B; 10 min, 50% B; 12 min, 50% B; 13 min, 25% B; 14 min, 25% B; 16 min, 0% B; 21 min, 0% B; 22 min, 90% B; 25 min, 90% B. The autosampler temperature was 5 °C, and the injection volume was 10 μL. The mass spectrometer was operated in negative ion mode to scan from m/z 70 to 1,000 at 1 Hz and a resolving power of 140,000 (40).
For the analysis of NAD+ and NADH, the flow rate was 200 μL/min, column temperature was 25 °C, autosampler temperature was 5 °C, and injection volume was 10 µL. The LC gradient was as follows: 0 min, 85% B; 2 min, 85% B; 3 min, 60% B; 9 min, 60% B; 9.5 min, 35% B; 13 min, 5% B; 15.5 min, 5% B; 16 min, 85% B; 20 min, 85% B. The mass spectrometer was operated in negative ion mode to scan from 640 to 765 at a resolving power of 140,000 at m/z 200 (41). Data were analyzed using El-MAVEN software (Elucidata). Isotope labeling was corrected for natural 13C abundance (42). Media measurements were acquired using a YSI 2900 Series Biochemical Analyzer (Xylem).
Mitochondrial Stress Test.
OCR and ECAR were recorded using a Seahorse XFe96 Analyzer (Agilent). Cells were washed and resuspended in Agilent XF Assay Medium supplemented with 25 mM glucose, 1 mM sodium pyruvate, and 2 mM l-Glutamine. Cells were plated on Seahorse assay plates at 0.2 × 106 cells/well and during the assay were exposed to 1 μM oligomycin, 1.5 μM FCCP, 100 nM rotenone, and 1 μM antimycin A, as indicated.
RNA-Seq Analysis Preparation and Bioinformatics Analysis.
Total RNA was extracted using an RNA Miniprep Kit (Zymo Research), and 500 ng RNA was used for library preparation. RNA-Seq libraries were prepared using mRNA HyperPrep Kits (KK8580; Kapa Biosystems) and indexed with NEXTflex DNA Barcodes-24 as reported previously (43). After final amplification, samples were loaded onto 2% E-Gel precast gels (Thermo Fisher Scientific), and 250- to 400-bp DNA fragments were recovered and purified with the Zymoclean Gel DNA Recovery Kit (Thermo Fisher Scientific). After quantification by Qubit (Invitrogen), barcoded samples were mixed and sequenced on the Illumina HiSeq3000 platform. Sequenced reads (50 bp, single end) were obtained with the Illumina CASAVA pipeline and mapped to the mouse genome mm10 (GRCm38; December 2011) using Bowtie 2.2.6 (44) and Tophat 2.2.1 (44). Only uniquely mapped reads were retained. Raw counts that fell on exons were calculated and normalized using reads per kilobase per million mapped reads. Differentially expressed genes were identified with the R Bioconductor package edgeR (45), and expression heat maps were generated with the R package “pheatmap”. Metabolic genes were extracted from the KEGG pathway database, including those corresponding to fatty acid biosynthesis/degradation, ox/phos, pyruvate metabolism, amino sugar and nucleotide sugar metabolism, PPP, TCA, and glycolysis/gluconeogenesis. Metabolic genes that overlapped with RNA-Seq data were subsequently shown in heat maps.
Real-Time PCR Assay.
Total RNA was isolated as specified by the Direct-zoI RNA MiniPrep Kit (Zymo Research, R2052) and cDNA was synthesized with the 5X All-In-One RT MasterMix Kit (abm, G590). TaqMan probes were used for the RT-qPCR measurements of Pfkm (Mm01309576_m1), Pgk1 (Mm00435617_m1), and Slc2a1 (Mm00441480_m1). The SYBR method was used for amplifying Ldha using forward primer 5′TATCTTAATGAAGGACTTGGCGGATGAG3′ and reverse primer 5′GGAGTTCGCAGTTACACAGTAGTC3′. All measurements were performed on a Bio-Rad CFX96 Real-Time System.
Flow Cytometric Analysis.
Live cells were stained in FACS buffer containing 0.5% BSA and 0.02% sodium azide in PBS. Cells were stained at 4 °C for 30 min in darkness with anti-CD8 (53-6.7, BioLegend), anti-CD62L (MEL-14, BioLegend), anti-CD44 (IM7, BD Pharmingen), anti-Sca-1 (D7, BioLegend), and LIVE/DEAD Aqua (Invitrogen). After staining, cells were washed and analyzed on an LSRFortessa X-20 cytometer (BD Biosciences). For glucose uptake experiments, cultured cells were starved for 7 h in glucose-free media, then treated with a fluorescently-labeled glucose derivative (Cayman, 600471) for 10 min and assessed by flow cytometry. All analyses were done using FlowJo software (Tree Star).
Electron Microscopy.
Cells were stimulated as indicated and chemically fixed for 1 h with 2.5% glutaraldehyde and 1% formaldehyde in 0.12 M sodium cacodylate buffer, pH 7.4, for conventional transmission electron microscopy. Specimens were then postfixed in 1% osmium tetroxide in cacodylate buffer, en bloc stained with 1% uranyl acetate, dehydrated in an ethanol series/propylene oxide, and embedded in EMbed 812 resin (Electron Microscopy Sciences). Ultrathin sections (50 to 60 nm) were obtained using a Leica EM UC7 ultramicrotome. Sections were examined with a JEOL JEM-1200EX transmission electron microscope (accelerating voltage 80 keV) equipped with a bottom-mounted AMT 6-megapixel digital camera (Advanced Microscopy Techniques). Mitochondrial area measurements were done using Fiji imaging software (ImageJ).
Mouse Tumor Model and Adoptive Transfer.
For tumor experiments, 3.5 × 105 hpg100-transduced B16 melanoma (B-16KVP) cells were subdermally injected into WT C57BL/6 mice. Tumors were allowed to grow for 10 d before antigen-specific CD8+ T cell transfer. Splenic pmel-1 CD8+ T cells were harvested from C57BL/6 mice (STEMCELL Technologies, 19853) and cultured for 4 d in anti-CD3 and anti-CD28 with either 100 IU/mL IL-2 or 100 ng/mL IL-21 with or without LDHi (1 μM). After the first 2 d of the 4-d culture, media, cytokine, and LDHi (where appropriate) were replenished. To delete Ldha, pmel-1+ Ldhafl/fl mice (46) were bred to Cre-ERT2 mice to generate Ldhafl/fl mice containing Cre-ERT2 and were then injected i.p. with 2 mg of tamoxifen each day for 4 d. T cells were prepared as described for WT mice, and then after 4 d of culture, 1 × 106 T cells were injected into the tail vein of tumor-bearing mice in conjunction with hgp100 vaccinia virus. Subsequently, mice were injected i.p. with six doses of IL-2 (1 μg/mouse twice daily) on days 0, 1, and 2 after adoptive transfer. Tumor growth was monitored until tumors exceeded 20 mm in size. All animal studies were performed under protocols approved by the National Heart, Lung, and Blood Institute or National Cancer Institute Animal Care and Use Committees and followed NIH guidelines for the use of animals in intramural research.
Statistics.
Datasets were analyzed using Prism 7.0 software; mean ± SEM values are shown. Unless stated otherwise, datasets comparing IL-2, IL-21, and NC were analyzed for significance using one-way ANOVA and Tukey’s multiple comparison test, and statistics comparing IL-2 vs. IL-2+LDHi and IL-21 vs. IL-21+LDHi were performed using a two-tailed unpaired Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; n.s., not significant. The metabolomics heatmap and principal component analysis were done using ClustVis (47).
Data Deposition.
All next-generation sequencing data have been deposited in the Gene Expression Omnibus (GEO) database. All analyzed data are available in the main text or SI Appendix, Supplementary Materials.
Supplementary Material
Acknowledgments
This work was supported by the Division of Intramural Research, National Heart, Lung, and Blood Institute (NHLBI; D.H., D.G., S.M., R.S., P.L., S.C., J.-X.L., J.O., N.D., R.E.-S., C.B., and W.J.L.) and the Division of Intramural Research, National Cancer Institute (S.G., L.M.N., J.F., and L.G.), and NIH grants R01 CA163591 and DP1DK113643 (to J.D.R.). J.C.G.-C. was supported by funding from the European Union’s Horizon 2020 research and innovation program under Marie Sklodowska-Curie Grant Agreement 751423. D.G. was supported by the NIH Medical Research Scholars Program. R.E.-S. was supported by a Public Interest Incorporated Foundation Scholarship from the MSD Life Science Foundation. All next generation DNA sequencing was performed in the NHLBI DNA sequencing core.
Footnotes
Competing interest statement: W.J.L. is an inventor on patents or patent applications related to IL-2 and IL-21. D.H., S.G., L.M.N., L.G., and W.J.L. are inventors on patent application(s) related to the LDH inhibitor described herein. L.G. is an inventor on a patent related to methods for generating TSCM cells.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE143903).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920413117/-/DCSupplemental.
References
- 1.Mathis D., Shoelson S. E., Immunometabolism: An emerging frontier. Nat. Rev. Immunol. 11, 81 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Neill L. A., Kishton R. J., Rathmell J., A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 16, 553–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee Y. S., Wollam J., Olefsky J. M., An integrated view of immunometabolism. Cell 172, 22–40 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim J., Regulation of immune cell functions by metabolic reprogramming. J. Immunol. Res. 2018, 8605471 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buck M. D., O’Sullivan D., Pearce E. L., T cell metabolism drives immunity. J. Exp. Med. 212, 1345–1360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang R., Green D. R., Metabolic reprogramming and metabolic dependency in T cells. Immunol. Rev. 249, 14–26 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tran E., Robbins P. F., Rosenberg S. A., “Final common pathway” of human cancer immunotherapy: Targeting random somatic mutations. Nat. Immunol. 18, 255–262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinrichs C. S., et al. , IL-2 and IL-21 confer opposing differentiation programs to CD8+ T cells for adoptive immunotherapy. Blood 111, 5326–5333 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin J. X., Leonard W. J., The common cytokine receptor γ chain family of cytokines. Cold Spring Harb. Perspect. Biol. 10, a028449 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leonard W. J., Lin J. X., O’Shea J. J., The γc family of cytokines: Basic biology to therapeutic ramifications. Immunity 50, 832–850 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Spolski R., Gromer D., Leonard W. J., The γ c family of cytokines: Fine-tuning signals from IL-2 and IL-21 in the regulation of the immune response. F1000 Res. 6, 1872 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spolski R., Leonard W. J., Interleukin-21: A double-edged sword with therapeutic potential. Nat. Rev. Drug Discov. 13, 379–395 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Liao W., Lin J. X., Leonard W. J., Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity 38, 13–25 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng R., et al. , The molecular basis of IL-21–mediated proliferation. Blood 109, 4135–4142 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao W., et al. , Opposing actions of IL-2 and IL-21 on Th9 differentiation correlate with their differential regulation of BCL6 expression. Proc. Natl. Acad. Sci. U.S.A. 111, 3508–3513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markley J. C., Sadelain M., IL-7 and IL-21 are superior to IL-2 and IL-15 in promoting human T cell-mediated rejection of systemic lymphoma in immunodeficient mice. Blood 115, 3508–3519 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng H., et al. , mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature 499, 485–490 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finlay D. K., et al. , PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J. Exp. Med. 209, 2441–2453 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Windt G. J., et al. , Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Sullivan D., et al. , Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity 41, 75–88 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buck M. D., et al. , Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166, 63–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L., et al. , Mammalian target of rapamycin complex 2 controls CD8 T cell memory differentiation in a Foxo1-dependent manner. Cell Rep. 14, 1206–1217 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Loschinski R., et al. , IL-21 modulates memory and exhaustion phenotype of T-cells in a fatty acid oxidation-dependent manner. Oncotarget 9, 13125–13138 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patten D. A., et al. , OPA1-dependent cristae modulation is essential for cellular adaptation to metabolic demand. EMBO J. 33, 2676–2691 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rai G., et al. , Discovery and optimization of potent, cell-active pyrazole-based inhibitors of lactate dehydrogenase (LDH). J. Med. Chem. 60, 9184–9204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeung C., et al. , Targeting glycolysis through inhibition of lactate dehydrogenase impairs tumor growth in preclinical models of Ewing sarcoma. Cancer Res. 79, 5060–5073 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tyrakis P. A., et al. , S-2-hydroxyglutarate regulates CD8+ T-lymphocyte fate. Nature 540, 236–241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanada K. I., Yu Z., Chappell G. R., Park A. S., Restifo N. P., An effective mouse model for adoptive cancer immunotherapy targeting neoantigens. JCI Insight 4, 124405 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gautam S., et al. , The transcription factor c-Myb regulates CD8+ T cell stemness and antitumor immunity. Nat. Immunol. 20, 337–349 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gattinoni L., et al. , Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat. Med. 15, 808–813 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mognol G. P., et al. , Exhaustion-associated regulatory regions in CD8+ tumor-infiltrating T cells. Proc. Natl. Acad. Sci. U.S.A. 114, E2776–E2785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welsh R. M., Blimp hovers over T cell immunity. Immunity 31, 178–180 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song M., et al. , IRE1α-XBP1 controls T cell function in ovarian cancer by regulating mitochondrial activity. Nature 562, 423–428 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y., et al. , Nuclear lactate dehydrogenase A senses ROS to produce α-hydroxybutyrate for HPV-induced cervical tumor growth. Nat. Commun. 9, 4429 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang D., et al. , Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sukumar M., et al. , Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J. Clin. Invest. 123, 4479–4488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng M., et al. , Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 354, 481–484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sabatino M., et al. , Generation of clinical-grade CD19-specific CAR-modified CD8+ memory stem cells for the treatment of human B-cell malignancies. Blood 128, 519–528 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gattinoni L., Speiser D. E., Lichterfeld M., Bonini C., T memory stem cells in health and disease. Nat. Med. 23, 18–27 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailis W., et al. , Distinct modes of mitochondrial metabolism uncouple T cell differentiation and function. Nature 571, 403–407 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu W., Wang L., Chen L., Hui S., Rabinowitz J. D., Extraction and quantitation of nicotinamide adenine dinucleotide redox cofactors. Antioxid. Redox Signal. 28, 167–179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su X., Lu W., Rabinowitz J. D., Metabolite spectral accuracy on orbitraps. Anal. Chem. 89, 5940–5948 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin J. X., et al. , Critical functions for STAT5 tetramers in the maturation and survival of natural killer cells. Nat. Commun. 8, 1320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y. H., et al. , Cell-state-specific metabolic dependency in hematopoiesis and leukemogenesis. Cell 158, 1309–1323 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Metsalu T., Vilo J., ClustVis: A web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 43, W566-70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.