Abstract
Mulberry sclerotiniose caused by Ciboria shiraiana is a devastating disease of mulberry (Morus alba L.) fruit in Northwest China. At present, no disease-resistant varieties are used in production, as the molecular mechanisms of this disease are not well understood. In this study, to explore new prevention methods and provide direction for molecular breeding, transcriptomic sequencing and un-targeted metabolomics were performed on healthy (CK), early-stage diseased (HB1), and middle-stage diseased (HB2) mulberry fruits. Functional annotation, gene ontology, a Kyoto encyclopedia of genes and genomes (KEGG) analysis, and a Mapman analysis of the differentially expressed genes revealed differential regulation of genes related to plant hormone signal transduction, transcription factors, and phenylpropanoid biosynthesis. A correspondence between the transcript pattern and metabolite profile was observed in the phenylpropanoid biosynthesis pathway. It should be noted that the log2 ratio of eugenol (isoeugenol) in HB1 and HB2 are 85 times and 23 times higher than CK, respectively. Our study shows that phenylpropanoid biosynthesis may play an essential role in response to sclerotiniose pathogen infection and eugenol(isoeugenol) enrichment in mulberry fruit, which may provide a novel method for mulberry sclerotiniose control.
Keywords: mulberry fruit, sclerotiniose, Ciboria shiraiana, transcriptome, metabolite, eugenol/isoeugenol
1. Introduction
Mulberry fruit is a valuable food because of its high nutritional value, pleasant taste, and large amount of biologically active components that might be associated with some potential pharmacological functions that are beneficial for human health [1,2]. In recent years, with both ecological and economic benefits, large-scale mulberry orchards have been set up and the mulberry fruit industry has developed rapidly in China, becoming a model for the diversified development of mulberry in Northwest China. However, incidences of disease in the fruit have also increased. Mulberry sclerotiniose is a devastating fungal disease of mulberry fruit; it not only causes mulberry fruit to lose their color and turn white, but also heavily affects mulberry fruit quality and yield.
Mulberry sclerotiniose can be classified into hypertrophy sorosis sclerotiniose, reduced sorosis sclerotiniose, and small particles sorosis sclerotiniose, according to its morphology and symptoms. Hypertrophy sorosis sclerotiniose is a common and major disease in Northwest China, and its pathogen is Ciboria shiraiana or Sclerotinia sclerotinia. The diseased fruit fall onto the ground and the pathogen overwinter in the soil as sclerotium. During the mulberry flowering time of the following spring, the sclerotium germinates and the apothecium forms. The ascospore infects the stigma of mulberry pistillate, specifically when the apothecium has matured, and the hyphae multiply, eventually forming sclerotia, and thus completing the infection cycle.
Understanding plant responses to pathogen infection is essential to elucidate the mechanisms of plant–microbe interactions and to develop novel strategies for therapy [3]. Unfortunately, the pathogenesis and molecular mechanism of mulberry sclerotiniose have not been well investigated. Of the two pathogens of hypertrophy sorosis sclerotiniose, S. sclerotinia was only discovered recently [4]. At present, few genes, such as CsCelA [5] and CsPG1 [6], have been discovered piecemeal in the study of C. shiraiana.
Recently, transcriptome, metabolome, and proteome analyses have been applied as powerful tools to explain the plant–pathogen interactions in many plants, such as Zymoseptoria tritici [7], Fusarium culmorum [8], Botrytis cinerea [9], Bradyrhizobium diazoefficiens [10], Candidatus Liberibacter asiaticus [11], Xanthomonas oryzae [12], and Cucumber mosaic virus [13].
With the success of Morus notabilis genome sequencing [14], multi-omics technology has also been applied in mulberry sclerotiniose. A transcriptomic and proteomic study of Ciboria carunculoides, the pathogen of small particles sorosis sclerotinioseis, which has occurred often in east China, was carried out this year [15,16]. In order to provide a global view of C. shiraiana infection regulatory mechanisms in mulberry fruit, in this study, the influence of diseased fruit was investigated using transcriptome and metabolite profiling analysis. The differential gene expression during fungal infection was screened, and changes in selected secondary metabolites were recorded. Our research provides foundational information which may lead to a better understanding of the molecular mechanisms of sclerotiniose in mulberry fruit.
2. Results
2.1. Symptoms of Infected Mulberry Fruits and Fungal Detection
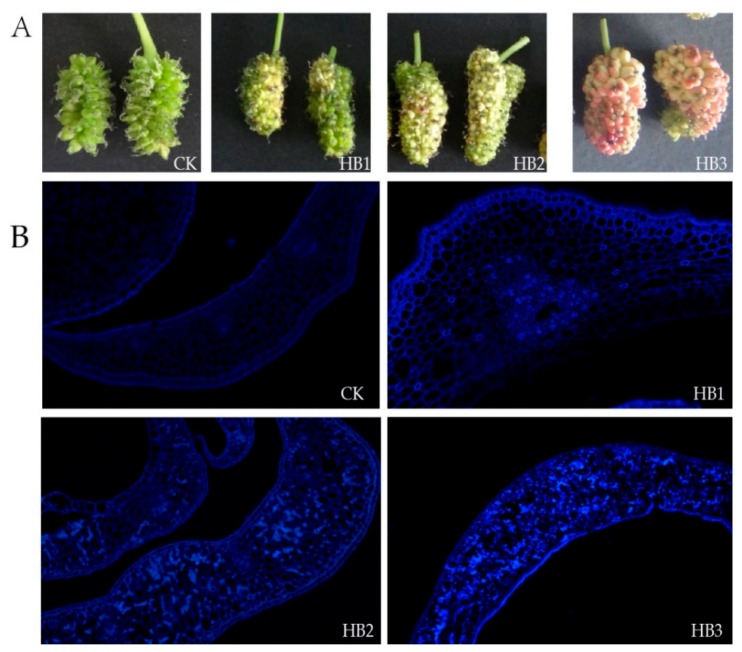
Healthy mulberry fruits were collected from a normal mulberry orchard and used as the control (CK). Diseased mulberry fruits were divided into three classes: HB1, HB2, and HB3. In the early stage of infection, only a few achenes turned white or gray at stage HB1. The ovary of the mulberry flower swelled due to hyphal proliferation, and the whole fruit was malformed at stage HB2. At stage HB3, the ovary swelled further, some of the diseased fruit turned pink, and the sclerotium was forming (Figure 1A). This was the typical morphological characteristics of hypertrophy sorosis sclerotiniose. In addition, a lot of different levels of diseased mulberry fruits could be found on infected mulberry trees (Figure S1A) and a large number of fungal fruiting bodies were discovered on the ground (Figure S1B,C). Plenty of matured ascospores existed in the apothecium of fruiting bodies (Figure S1D). Moreover, the pathogen in diseased fruits was identified as C. shiraiana by fungal ITS primers and a phylogenetic tree analysis, while no pathogens were detected in mulberry fruit from a healthy mulberry orchard (Figure S2A,B). The hyphal of C. Shiraiana that had proliferated could be observed by Calcofluor White fluorescent staining (Figure 1B) and pectinase activity increased in diseased fruits (Figure S2C). The plant tissue degraded rapidly, in keeping with hyphal proliferation, and mulberry fruit DNA was barely detected at stage HB3. Therefore, HB1 and HB2 were chosen for subsequent transcriptomic and metabolomic analyses.
Figure 1.
Physical and fluorescent staining of healthy and diseased mulberry fruits infection by Ciboria shiraiana. (A) Physical appearance of healthy mulberry fruits (CK) and progressive stage of diseased mulberry fruits (HB1, HB2, HB3). (B) Fluorescent staining of healthy and diseased mulberry fruits by Calcofluor White Stain.
2.2. Identified Differentially Expressed Genes (DEGs), Their Gene Ontology (GO) Enrichments, and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichments by Transcriptome Analysis
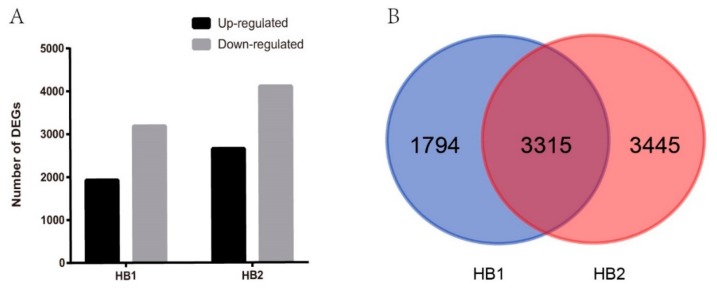
In order to compare the difference in the transcriptome of mulberry fruit after fungal infection, we performed RNA-seq profiling with the samples CK, HB1, and HB2. A total of 5109 (with 1926 up- and 3183 down-regulated) and 6760 (with 2655 up- and 4105 down-regulated) differentially expressed genes (DEGs) were identified in infection stages HB1 and HB2, respectively, as compared with CK (Figure 2A). As shown in the Venn diagram in Figure 2B, 3315 genes of all DEGs were regulated by C. shiraiana at both HB1 and HB2. These results show the differences at the molecular level between healthy fruits and diseased fruits, and also between different disease levels at HB1 and HB2.
Figure 2.
Differential expressed genes (DEGs) in HB1 and HB2 after infection by Ciboria shiraiana against CK. (A) Up-regulated and down-regulated genes in HB1 and HB2 compared with CK. (B) Venn diagrams showing the number of DEGs of infection stage HB1 and HB2.
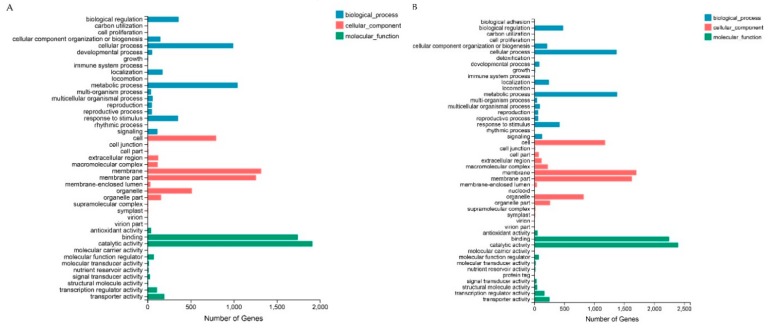
A GO analysis was carried out to classify the functions of the DEGs of HB1 and HB2 compared with healthy mulberry fruit CK. Within the biological process categories, The DEGs from the comparisons of HB1 and HB2 were significantly enriched in the terms of cellular processes, metabolic processes, biological regulation, and response to stimuli. Within the cellular component category, the membrane, membrane parts, cells, and organelles were significantly enriched. Within the molecular function category, the DEGs reveal significant enrichments in categories for binding and catalytic activity. The results indicate that the DEGs were functionally enriched in the same GO terms for the top 10 categories (Figure 3, Table S1). However, further study of GO enrichment showed that the DEGs of HB1 and HB2 were different (Figure S3).
Figure 3.
GO analysis of differentially expressed genes (DEGs) in HB1 (A) and HB2 (B) against CK.
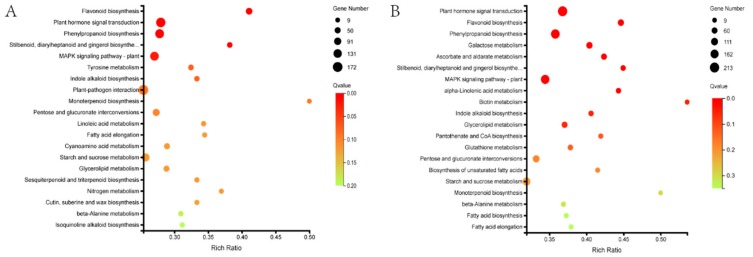
KEGG pathway analysis of the DEGs identified the terms global and overview maps, carbohydrate metabolism, biosynthesis of other secondary metabolites, signal transduction, amino acid metabolism, and lipid metabolism as being significantly enriched as compared with the whole genomic background in both HB1 and HB2 (Figure S4). KEGG function annotations were used to analyze the KEGG enrichment results as a scatter plot. In both HB1 and HB2, the top three highest significantly enriched KEGG pathways were flavonoid biosynthesis, plant hormone signal transduction, and phenylpropanoid biosynthesis (Figure 4).
Figure 4.
KEGG pathway enrichment analysis of DEGs in HB1 (A) andHB2 (B).
To further validate the expression profiles of the RNA-Seq data, eight transcripts from top three of the highest significant terms were selected for analysis using quantification real-time PCR (qRT-PCR). The results from the qRT-PCR analysis are generally in agreement with the expression profiles obtained using the RNA-Seq data (Figure S5, Table S2).
2.3. Transcriptomic Response of Mulberry Fruits Infected by C. shiraiana
According to the GO and KEGG enrichment results, plant hormone signal transduction, phenylpropanoid biosynthesis, and transcription factors were analyzed to help understand the response of mulberry fruits to fungal pathogen C. shiraiana infection.
2.3.1. Plant Hormone Signal Transduction Pathway
A group of genes changed involved in hormone biosynthesis, mobilization, and signal transduction in response to C. shiraiana infection. Transcripts related to plant hormone signal transduction pathway showed that the most were involved in auxin, brassinosteroid, cytokinine, jasmonic acid, gibberellin, abscisic acid, ethylene, salicylic acid, and so on. Log fold ratios for differentially expression genes in HB1 and HB2 compared with CK were shown (Table 1). The transcripts’ up-regulated or down-regulated trends in HB1 and HB2 were similar. Transcripts for abscisic acid receptor (PY1 and PY4), involved in the abscisic acid pathway, were more abundant in HB1 and HB2. Several transcripts related to jasmonic acid and brassinosteroid pathway were down-regulated in HB1 and HB2, including a large number of TIFY protein and protein kinase. Of auxin biosynthesis and signal transduction, transcripts for auxin-responsive protein IAA21 and SAUR36 were down-regulated, while IAA4, IAA29, SAUR24 and SAUR50 were up-regulated. Their detailed roles involved in the responses to C. shiraiana infection remain to be explored.
Table 1.
Differential expressed genes (DEGs) related to the plant hormone signal transduction pathway. Log-fold ratio is shown as a gradient between red (up-regulated) and green (down-regulated).
| Gene ID | Predicted Gene Description | Gene Expression (log2 Ratio) | |
|---|---|---|---|
| HB1 | HB2 | ||
| Auxin | |||
| LOC21401961 | Auxin-responsive protein SAUR36 | −2.51 | −1.61 |
| LOC21401600 | Probable WRKY transcription factor protein 1 | −1.80 | −2.52 |
| LOC21388680 | Auxin response factor 2B | −1.55 | −1.85 |
| LOC21408902 | Auxin-responsive protein IAA21 | −1.39 | −1.45 |
| LOC21386679 | Auxin response factor 9 | −1.29 | −2.27 |
| LOC21403994 | Indole-3-acetic acid-amido synthetase GH3.6 | −1.27 | −1.59 |
| LOC21391765 | Auxin-responsive protein SAUR15 | −1.08 | 1.86 |
| LOC21395462 | Auxin-induced protein 6B | 1.01 | 1.25 |
| LOC21392112 | Auxin-responsive protein SAUR24 | 1.38 | 1.60 |
| LOC21402398 | Auxin-induced protein AUX22 | 1.46 | 1.74 |
| LOC21394746 | Probable indole-3-acetic acid-amido synthetase GH3.1 | 1.51 | 2.24 |
| LOC21392317 | Auxin-responsive protein IAA29 | 1.66 | 2.86 |
| LOC21407314 | Auxin-responsive protein IAA4 | 1.74 | 2.34 |
| LOC21409344 | Auxin-responsive protein SAUR50 | 2.24 | 3.32 |
| LOC21399379 | Auxin-responsive protein IAA4 | 2.93 | 4.86 |
| Cytokinine | |||
| LOC21385677 | Histidine-containing phosphotransfer protein 1 | −2.01 | −1.49 |
| LOC21407993 | Transcription factor BOA | 1.15 | 1.14 |
| LOC21410451 | Two-component response regulator ARR5 | 1.23 | 0.46 |
| LOC21406708 | Two-component response regulator ORR9 | 1.59 | 1.27 |
| LOC21406707 | Two-component response regulator ARR9 | 1.61 | 2.77 |
| LOC21394517 | Two-component response regulator ARR17 | 1.93 | 2.05 |
| LOC21388228 | Two-component response regulator ARR5 | 2.17 | 2.89 |
| LOC21384736 | Two-component response regulator ARR9 | 2.43 | 3.10 |
| Gibberellin | |||
| LOC21405471 | Transcription factor PIF3 | −1.37 | −1.51 |
| LOC21398268 | Transcription factor PIF1 | −1.30 | −1.21 |
| LOC21392291 | Transcription factor bHLH79 | −1.10 | −0.94 |
| LOC21402833 | Transcription factor bHLH62 | 1.03 | 1.16 |
| LOC21399703 | Transcription factor BIM1 | 1.32 | 0.94 |
| LOC21405713 | Transcription factor mef2A | 1.41 | 2.47 |
| LOC21392198 | Transcription factor bHLH137 | 2.48 | 3.16 |
| LOC21409470 | SCARECROW-LIKE protein 7 | 2.58 | 2.10 |
| Abscisic acid | |||
| LOC21396136 | Probable protein phosphatase 2C 75 | −2.00 | −3.45 |
| LOC21394160 | Probable protein phosphatase 2C 6 | −1.02 | −1.52 |
| LOC21406025 | Abscisic acid receptor PYL1 | 1.01 | 0.54 |
| LOC21402805 | Basic leucine zipper 9 | 1.36 | 2.17 |
| LOC21396186 | Light-inducible protein CPRF2 | 2.35 | 2.54 |
| LOC21390361 | Abscisic acid receptor PYL4 | 2.37 | 1.69 |
| Ethylene | |||
| LOC21398777 | Phosphate transporter PHO1 homolog 3 | −3.17 | −3.55 |
| LOC21393854 | Probable serine/threonine-protein kinase DDB_G0271682 | −2.79 | −2.66 |
| LOC21383879 | Serine/threonine-protein kinase CTR1 | −1.30 | −1.17 |
| LOC21387330 | Protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 10 | 1.16 | 2.17 |
| LOC21396016 | Ethylene-responsive transcription factor ERF096 | 1.55 | 2.96 |
| LOC21385827 | EIN3-binding F-box protein 1 | 1.68 | 1.49 |
| LOC21394948 | Phosphate transporter PHO1 | 1.99 | 3.13 |
| LOC21408627 | Protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 4 | 2.18 | 2.73 |
| Brassinosteroid | |||
| LOC21404629 | Squamosa promoter-binding-like protein 8 | −2.52 | −4.69 |
| LOC21400451 | Teosinte glume architecture 1 | −2.48 | −1.83 |
| LOC21383985 | Probable serine/threonine-protein kinase At4g35230 | −1.78 | 0.19 |
| LOC21397624 | Squamosa promoter-binding protein 1 | −1.41 | −2.00 |
| LOC21396408 | Probable disease resistance protein At5g66900 | −1.27 | −1.67 |
| LOC21407610 | Probable serine/threonine-protein kinase At4g35230 | −1.24 | −1.37 |
| LOC21400300 | Probable serine/threonine-protein kinase At5g41260 | −1.17 | −1.01 |
| LOC21394077 | Squamosa promoter-binding-like protein 9 | −1.01 | −0.88 |
| LOC21393228 | LRR receptor-like serine/threonine-protein kinase At2g24230 | 1.35 | 0.24 |
| LOC21400957 | Tyrosine-sulfated glycopeptide receptor 1 | 1.58 | 1.32 |
| LOC21406401 | Probable serine/threonine-protein kinase BSK3 | 1.80 | 1.87 |
| LOC21405696 | Xyloglucan endotransglucosylase/hydrolase protein 23 | 1.99 | 2.03 |
| Jasmonic acid | |||
| LOC21398450 | Transcription factor MYC2 | −4.37 | −4.89 |
| LOC21386546 | Transcription factor bHLH35 | −2.63 | −2.53 |
| LOC21404490 | Protein TIFY 5A | −2.43 | −1.15 |
| LOC21386105 | Protein TIFY 10A | −2.26 | −2.37 |
| LOC21406254 | Transcription factor bHLH13 | −1.73 | −1.88 |
| LOC21387505 | Protein TIFY 6B | −1.72 | −2.38 |
| LOC21406020 | Transcription factor MYC2 | −1.46 | −1.99 |
| LOC21396351 | Protein TIFY 9 | −1.42 | −1.70 |
| LOC21401260 | Transcription factor ILR3 | −1.21 | −1.50 |
| LOC21384725 | Transcription factor bHLH18 | 1.89 | 2.90 |
| Salicylic acid | |||
| LOC21396732 | Pathogenesis-related protein 1 | −1.08 | −0.76 |
| LOC21403015 | Transcription factor TGA1 | 1.05 | 1.71 |
| LOC21409923 | BTB/POZ domain and ankyrin repeat-containing protein NPR1 | 1.05 | 0.69 |
| LOC21387250 | Pathogenesis-related protein 1 | 3.04 | 2.26 |
2.3.2. Phenylpropanoid Biosynthesis Pathway
The phenylpropanoid biosynthesis pathway plays an important role in the regulation of secondary metabolites in plants. In this study, 40 DEGs were detected in both HB1 and HB2 (Table 2). Most of the DEGs were up-regulated in HB1 and HB2 compared with CK, such as phenylalanine ammonia-lyase, eugenol synthase, cinnamyl alcohol dehydrogenase, and cinnamoyl-CoA reductase. However, β-glucosidase was down-regulated. It should be noted that both up-regulated and down-regulated genes existed in peroxidase, caffeic acid 3-O-methyltransferase, and 4-coumarate-CoA ligase.
Table 2.
Classification of the differentially expressed genes identified from HB1 and HB2 compared with CK in the phenylpropanoid biosynthesis pathway. Log-fold ratio is shown as a gradient between red (up-regulated) and green (down-regulated).
| Gene ID | Predicted Gene Description | Gene Expression (log2 Ratio) | |
|---|---|---|---|
| HB1 | HB2 | ||
| LOC21407114 | Phenylalanine ammonia-lyase | 0.52 | 0.34 |
| LOC21409963 | Phenylalanine ammonia-lyase | 1.95 | 1.20 |
| LOC21407113 | Phenylalanine ammonia-lyase 1 | 1.18 | 0.00 |
| LOC21407115 | Phenylalanine ammonia-lyase 1 | 0.82 | 0.21 |
| LOC21391685 | Eugenol synthase 1 | 0.91 | −0.49 |
| LOC21391683 | Eugenol synthase 1 | −0.69 | −0.49 |
| LOC21391684 | Eugenol synthase 1 | 0.74 | 0.82 |
| LOC21409656 | Cinnamyl alcohol dehydrogenase 1 | 1.03 | 0.24 |
| LOC21409658 | Cinnamyl alcohol dehydrogenase 1 | 0.32 | 0.54 |
| LOC21396683 | Cinnamoyl-CoA reductase 1 | 0.12 | −0.65 |
| LOC21398414 | Cinnamoyl-CoA reductase 1 | 1.11 | 1.43 |
| LOC21398415 | Cinnamoyl-CoA reductase 1 | 0.96 | 1.39 |
| LOC21398416 | Cinnamoyl-CoA reductase 1 | 0.55 | 1.04 |
| LOC21398417 | Cinnamoyl-CoA reductase 1 | 0.25 | 0.36 |
| LOC21399536 | Cinnamoyl-CoA reductase 1 | 0.10 | 0.54 |
| LOC21401355 | β-glucosidase 12 | −1.08 | −0.69 |
| LOC21385981 | β-glucosidase 17 | −2.54 | −3.96 |
| LOC21402010 | β-glucosidase 3 | −1.34 | −1.58 |
| LOC21385542 | Peroxidase 12 | −0.63 | −1.91 |
| LOC21402709 | Peroxidase 17 | −1.44 | −0.65 |
| LOC21407160 | Peroxidase 21 | −1.01 | −0.26 |
| LOC21387029 | Peroxidase 31 | 1.01 | −1.02 |
| LOC21385792 | Peroxidase 4 | −0.35 | 0.50 |
| LOC21396696 | Peroxidase 42 | 0.89 | 0.09 |
| LOC21385871 | Peroxidase 45 | 0.94 | 0.30 |
| LOC21394066 | Peroxidase 55 | 1.10 | 1.28 |
| LOC21396430 | Peroxidase 72 | −1.81 | −4.39 |
| LOC21401444 | Peroxidase A2 | 1.59 | 2.17 |
| LOC21387006 | Caffeic acid 3-O-methyltransferase | −0.86 | −1.80 |
| LOC21396224 | Caffeic acid 3-O-methyltransferase | 1.00 | 2.31 |
| LOC21405422 | Caffeic acid 3-O-methyltransferase | 0.34 | −0.21 |
| LOC21406195 | Caffeic acid 3-O-methyltransferase | 0.04 | 0.70 |
| LOC21384962 | 4-coumarate--CoA ligase 1 | −1.06 | −2.32 |
| LOC21389140 | 4-coumarate--CoA ligase 2 | 0.38 | −0.32 |
| LOC21402620 | 4-coumarate--CoA ligase 2 | 0.44 | 0.46 |
| LOC21386277 | 4-coumarate--CoA ligase-like 7 | 1.65 | 0.60 |
| LOC21403500 | 4-coumarate--CoA ligase-like 7 | 0.75 | 0.46 |
| LOC21407615 | 4-coumarate--CoA ligase-like 5 | −1.95 | −1.97 |
| LOC21384166 | 4-coumarate--CoA ligase-like 9 | −0.79 | −1.34 |
| LOC21409371 | Cytochrome P450 84A1 | 2.11 | 2.35 |
2.3.3. Transcription Factors
In the present study, 1345 transcription factors (TFs) genes were detected, 306 and 443 TFs were differentially expressed in HB1 and HB2, respectively. There were 122 up-regulated and 184 down-regulated TFs in HB1, and 164 up-regulated and 279 down-regulated TFs in HB2 (Table 3). Of all the differentially expressed TFs, the MYB cluster showed the largest percentage, followed by AP2-EREBP, bHLH, NAC, WRKY, C2H2, MADS, etc. Most of the TFs in HB2, both up- and down- regulated, were increased compared with HB1. Specially, some TFs were down-regulated only in HB2, such as SBP, GRF, C3H, TCP and Tify.
Table 3.
Numbers of up- and down-regulated transcription factors in HB1 and HB2 compared with CK.
| Transcription Factors | HB1 | HB2 | ||
|---|---|---|---|---|
| Up-Regulated | Down-Regulated | Up-Regulated | Down-Regulated | |
| MYB | 17 | 35 | 22 | 45 |
| AP2-EREBP | 18 | 17 | 23 | 30 |
| bHLH | 11 | 26 | 16 | 25 |
| NAC | 12 | 14 | 15 | 14 |
| WRKY | 4 | 11 | 10 | 11 |
| C2H2 | 10 | 5 | 12 | 6 |
| MADS | 2 | 10 | 5 | 12 |
| ABI3VP1 | 4 | 4 | 4 | 10 |
| G2-like | 2 | 3 | 6 | 6 |
| SBP | 0 | 11 | 0 | 11 |
| GRAS | 3 | 6 | 4 | 7 |
| LOB | 5 | 1 | 5 | 5 |
| GRF | 0 | 6 | 0 | 10 |
| C2C2-Dof | 6 | 0 | 6 | 2 |
| HSF | 4 | 2 | 5 | 3 |
| Trihelix | 1 | 0 | 2 | 6 |
| C3H | 0 | 1 | 0 | 8 |
| OFP | 4 | 5 | 2 | 5 |
| mTERF | 4 | 2 | 3 | 4 |
| zf-HD | 2 | 4 | 1 | 6 |
| TCP | 0 | 4 | 0 | 7 |
| bZIP | 1 | 1 | 4 | 3 |
| ARF | 0 | 2 | 1 | 6 |
| Tify | 0 | 5 | 0 | 5 |
| C2C2-CO-like | 2 | 0 | 4 | 1 |
| C2C2-GATA | 0 | 2 | 1 | 4 |
| CPP | 1 | 1 | 0 | 4 |
| PLATZ | 1 | 0 | 2 | 2 |
| FAR1 | 0 | 1 | 2 | 2 |
| FHA | 0 | 0 | 0 | 4 |
| ARR-B | 2 | 0 | 1 | 2 |
| SRS | 0 | 0 | 2 | 1 |
| E2F-DP | 0 | 0 | 0 | 3 |
| LIM | 0 | 3 | 0 | 2 |
| HB | 1 | 0 | 1 | 1 |
| C2C2-YABBY | 2 | 0 | 1 | 0 |
| BSD | 1 | 0 | 1 | 0 |
| DBP | 1 | 0 | 0 | 1 |
| Sigma70-like | 0 | 1 | 1 | 0 |
| TIG | 0 | 1 | 0 | 1 |
| S1Fa-like | 0 | 0 | 1 | 0 |
| TAZ | 0 | 0 | 1 | 0 |
| BES1 | 0 | 0 | 0 | 1 |
| GeBP | 0 | 0 | 0 | 1 |
| SAP | 0 | 0 | 0 | 1 |
| TUB | 0 | 0 | 0 | 1 |
| EIL | 1 | 0 | 0 | 0 |
2.4. Overview of the Metabolites in the Phenylpropanoid Pathway by Metabolimics Data
In order to obtain an overview of metabolic changes in response to C. shiraiana of mulberry fruits, comparative metabolite analyses were performed at CK, HB1, and HB2 using an untargeted metabolomics approach. A principal component analysis (PCA), and a partial least squares discriminant analysis (PLS-DA) were used to analyze these data (Figures S6 and S7). Evaluation parameters of PLS-DA are shown in Table S3. The results show that the three samples were easy to distinguish from the data and the model was stable and reliable.
A total of 1799, 2238, 1642, and 2322 different expression ion numbers were detected from metabolite profiling by negative or positive ionization mode (Table S4). In the phenylpropanoid pathway, 16 metabolites were detected; the m/z values, retention time, and putative identification of the detected molecular formula are reported in Table 4. The results indicate that phenylalanine, sinapic acid, cinnamaldehyde, p-coumaraldehyde, coniferyl acetate, chavicol/isochavicol, methylchavicol/anethole, eugenol/isoeugenol, and methyleugenol/isomethyleugenol were up-regulated, whereas cinnamic acid, p-coumaric acid, caffeic acid, caffeyl aldehyde, coniferyl aldehyde, β-d-glucosyl-2-coumarinate, and coumarine were down-regulated in the phenylpropanoid pathway. It should be noted that ratio of eugenol/isoeugenol in HB1 and HB2 were 85-fold and 22-fold higher than that of CK, respectively.
Table 4.
Annotation of distinguishable metabolites belonging to phenylpropanoid pathway and related to Ciboria shiraiana-induced metabolic reprogramming in mulberry fruits. The reported ratio for HB1 and HB2 were obtained from a PLS-DA model of HB1, HB2 vs. CK, respectively.
| Metabolites | m/z | Retention Time (min) | Adduct | Ion Mode | Molecular Formula | HB1 | HB2 | Regulated | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Ratio | p-Value | Ratio | p-Value | |||||||
| Phenylalanine | 164.0707 | 5.026367 | M − H | neg | C9H11NO2 | 2.1720 | 1.29 × 10−2 | - | - | up |
| Cinnamic acid | 149.0596 | 7.963617 | M + H | pos | C9H8O2 | 11.4905 | 2.32 × 10−3 | 4.6224 | 1.84 × 10−3 | down |
| p-Coumaric acid | 163.039 | 4.4665 | M − H | neg | C9H8O3 | 0.3405 | 1.27 × 10−2 | 0.3196 | 3.00 × 10−4 | down |
| Caffeic acid | 179.0339 | 4.141967 | M − H | neg | C9H8O4 | 0.1961 | 1.64 × 10−2 | 0.1146 | 1.76 × 10−3 | down |
| Sinapic acid | 223.0598 | 4.5345 | M − H | neg | C11H12O5 | 2.3948 | 9.14 × 10−3 | 2.9807 | 1.97 × 10−3 | up |
| Cinnamaldehyde | 150.0909 | 1.136233 | M + NH4 | pos | C9H8O | 8.4712 | 1.47 × 10−4 | - | - | up |
| p-Coumaraldehyde | 149.0596 | 7.963617 | M + H | pos | C9H8O2 | 11.4905 | 2.32 × 10−3 | 4.6224 | 1.84 × 10−3 | up |
| Caffeyl aldehyde | 147.0439 | 4.702383 | M + H − H2O | pos | C9H8O3 | 0.3493 | 9.79 × 10−3 | 0.1463 | 5.00 × 10−5 | down |
| Coniferyl aldehyde | 179.0694 | 4.716667 | M + H | pos | C10H10O3 | 0.5148 | 1.83 × 10−4 | 0.4065 | 1.91 × 10−4 | down |
| β-d-Glucosyl-2-coumarinate | 325.092 | 4.4665 | M − H | neg | C15H18O8 | 0.3771 | 7.64 × 10−3 | 0.3288 | 2.31 × 10−4 | down |
| Coumarine | 147.0438 | 4.1807 | M + H | pos | C9H6O2 | 0.4280 | 3.80 × 10−3 | 0.3453 | 1.02 × 10−5 | down |
| Coniferyl acetate | 205.086 | 7.407367 | M + H − H2O | pos | C12H14O4 | 5.0356 | 1.44 × 10−3 | 4.4807 | 6.96 × 10−6 | up |
| Chavicol; Isochavicol | 135.0802 | 3.767783 | M + H | pos | C9H10O | 4.1766 | 6.69 × 10−4 | 5.2547 | 1.35 × 10−6 | up |
| Methylchavicol; Anethole | 131.0853 | 8.6643 | M + H − H2O | pos | C10H12O | 3.4420 | 7.26 × 10−5 | 3.1045 | 1.28 × 10−5 | up |
| Eugenol; Isoeugenol | 182.1174 | 4.759533 | M + NH4 | pos | C10H12O2 | 85.0298 | 3.30 × 10−6 | 22.8484 | 5.22 × 10−4 | up |
| Methyleugenol; Isomethyleugenol | 177.0911 | 7.402767 | M − H | neg | C11H14O2 | 9.5141 | 2.26 × 10−2 | 7.4019 | 1.73 × 10−2 | up |
3. Discussion
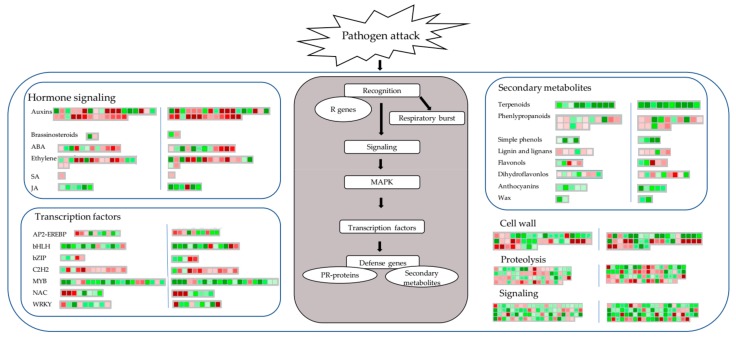
In order to maintain predominance, plants are engaged in a co-evolutionary fight against their pathogens [17]. The widespread changes in plants during pathogen infection make understanding the interactions a huge task [18]. To have a global view of all DEGs in mulberry fruits at HB1 and HB2, a large number of genes involved in metabolism were assessed by a Mapman analysis. Mapman is a common software for displaying large data sets onto pictorial diagrams that symbolically depict areas of biological function [19]. In this study, mainly DEGs of plant hormone signal transduction, phenylpropanoid pathway and transcription factors were discussed.
Figure 5 summarizes the global view of DEGs in response to C. shiraiana infection in both HB1 and HB2 using Mapman analysis. Plant hormones play key roles in the regulation of immune responses to microbial pathogens [20,21,22,23]. Jasmonic acid and salicylic acid coordinate growth and defense response upon fungal infection in poplar [24]. Jasmonic acid and terpenoids play roles in resistance against Phytophthora cinnamomic in Zea mays roots [25]. Ethylene plays a positive role in adapting mulberry to cope with the drought and salinity condition [26]. Transcription factors are also an important factor of gene expression change [27]. PbMYB10b and PbMYB9 regulate anthocyanins and flavonols of the flavonoid biosynthesis in pear fruit [28]. The bHLH factor VvMYC1 regulates anthocyanin and/or proanthocyanidin (PA) synthesis of the flavonoid pathway in grapevine [29]. In this study, the down-regulation of most MYB and bHLH clusters in HB1 and HB2, which indicates that they may affect the mulberry flavonoid biosynthesis.
Figure 5.
Schematic diagram of biotic stress categories using the Mapman visualization platform. The gene expression ratios of HB1 (left of blue vertical line) and HB2 (right of blue vertical line) compared with CK were used in a Mapman analysis. The gradient of red squares and green squares indicates up- or down-regulated genes involved, respectively.
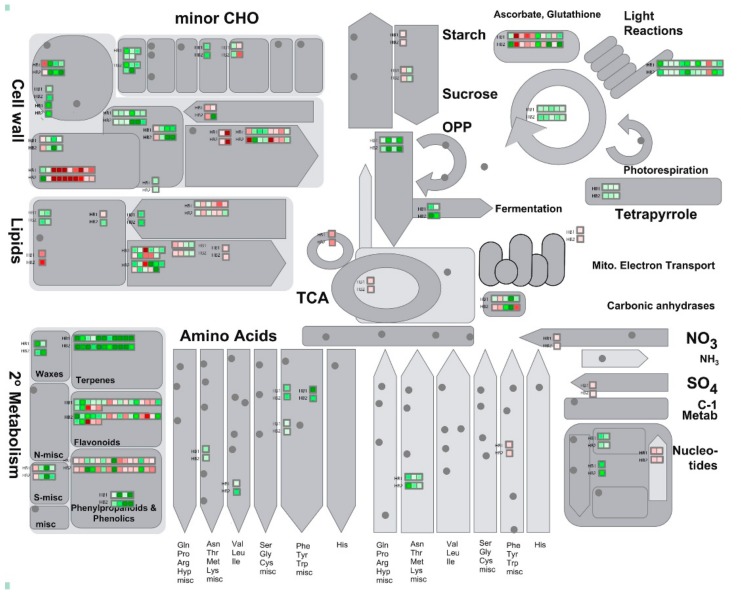
As shown in Figure 6, in the case of pathogen infection, primary metabolism, such as raffinose synthesis, lipid synthesis, hemicellulose synthesis, cellulose synthesis, and cell wall proteins were decreased, while cell wall modification and degradation, and lipid degradation increased in plants. Light reactions, the Calvin cycle, and glycolysis were also indicated to be suppressed in plants. Changes in plant secondary metabolites were more obvious. Terpenes, phenols, and waxy compounds were suppressed, and phenylpropanoids and flavonoids were increased (Figure 5 and Figure 6). The Mapman analysis results show that most of genes related to secondary metabolism had decreased expression levels, which is similar to the results of the GO and KEGG enrichment analyses. With the infection of pathogens, the infection hyphae rapidly expanded, which took away a lot of plant nutrients, resulting in the inhibition of plant metabolism and the destruction of plant tissue. These also appear during infection by other pathogens. In rice blast disease, the photoassimilates were shifted to mannitol and glycerol for mycelial growth [30]. The bean metabolites that varied in leaves included amines/amino acids, organic acids, phytoalexins, and ureides during Sclerotinia sclerotiorum infection [31]. Terpene down-regulation triggers pathogen defense responses in transgenic orange [32].
Figure 6.
Metabolism overview in Mapman depicting differential regulated genes in HB1 and HB2. Log-fold ratio is shown as a gradient between red (up-regulated) and green (down-regulated).
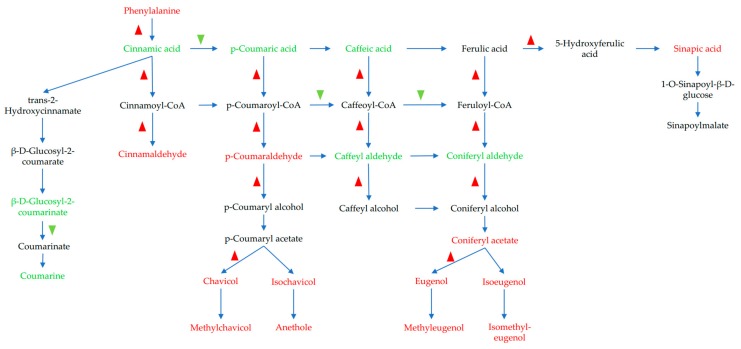
Today, numerous questions about the mechanism of pathogen infection still need to be addressed. Metabolomics is an important platform for studying stress in plant–fungal pathogen interactions [25,33]. We can qualify and quantify the metabolites of plants with biotic stress using modern analytical techniques. When pathogens infect, a large number of plant secondary metabolites are found to be up-regulated. In barley, higher levels of flavonoids, phenylpropanoids, and metabolites of fatty acids and terpenoid pathways were found in the resistant barley lines compared with the susceptible lines during infection with Fusarium [34]. Phenylpropanoid pathway intermediates, like 4-hydroxybenzoate, cinnamic acid, ferulic acid, and caffeic acid, were highly accumulated in the resistant soybean line against Sclerotinia sclerotiorum infection [35]. Moreover, in trees displaying symptoms of wood decay, large amounts of lignans were observed [36]. Mulberry trees contain large quantities of flavonoids and other secondary metabolites, such as phenylpropanoids and alkaloids, which have wide roles in defenses against biotic and abiotic stresses [26] and have long been used in Chinese traditional medicine for human disease treatment [37]. A large number of flavonoid metabolites from mulberry fruit have been evaluated both in vitro and in vivo for their antioxidant activity [38], such as morcin N [39], anthocyanins [40], and so on. Combined with transcriptome and metabolite profiling data, the significant genes and metabolites were found by co-expression analysis of the phenylpropanoid biosynthesis pathway of mulberry fruit in response to C. shiraiana (Figure 7, Table S5). We found that a large number of phenylpropanoid pathway intermediates, cinnamic acid, coumaric acid, caffeic acid, caffeyl aldehyde, coniferyl aldehyde were decreased. However, the final metabolites, like chavicol, isochavicol, methychavicol, anethole, eugenol, isoeugenol, methyleugenol, isomethyleugenol, and sinapic acid were increased. In particular, the large increase in eugenol/isoeugenol levels indicates that this compound may play an important role in response to pathogen infection. Eugenol is widely present in plants and is mainly used for antibacterial purposes, and can also be used in various cosmetic flavors and soap flavor formulations. There is no clear evidence that eugenol is carcinogenic. Eugenol exhibits an antimicrobial effect against carbapenem-resistant Klebsiella pneumoniae (CRKP) strains and could be potentially used to control CRKP-related infections [41]. The antimicrobial sachet containing the synergistic inhibitory effect of eugenol and citral (SEC) was developed for bread preservation [42]. Eugenol-casein nanoparticles (EC-NPs) exhibited a great antifungal activity against spore germination of fungus and have shown potential as an environmentally friendly preservative in the food industry [43]. Our data reveal important roles of eugenol/isoeugenol enrichment in mulberry fruit in response to C. shiraiana, which may provide a novel method for mulberry sclerotiniose control.
Figure 7.
Model summarizing metabolism network in mulberry fruit in response to C. shiraiana. The red font indicates significantly up-regulated metabolites (ratio > 1.5); the green font indicates significantly down-regulated metabolites (ratio < 0.5); the black font indicates a low cutoff value (ratio between 0.5 and 1.5) or not detected in this study; the red upward triangle arrow indicates up-regulated genes; the green downward triangle arrow indicates down-regulated genes.
4. Materials and Methods
4.1. Plant Growth Conditions and Fungus Identification
Mulberry cultivar Hongguo2 (Morus atropurpurea), which is a widely cultivated mulberry fruit tree in Northwest China, was bred by the Institute of Sericulture and Silk, Northwest A&F University.
A four-year high-yielding fruit mulberry orchard with area of about 3000 m2, and a space of 3 m between the rows and 1.5 m between the plants, was located in Guangji Town, Zhouzhi County, Shannxi Province, China. The climate and soil conditions of the orchard were consistent. A total of 20 mulberry trees suffered a serious outbreak of mulberry sclerotiniose on one side of the mulberry orchard near the low slope. A large number of diseased fruits dropped into the ground and formed sclerotia later. About 30 sclerotia per tree were discovered in winter, and PCR amplification was done using ITS sequences as primers, C. shiraiana was identified by morphological characters and phylogenetic tree. The diseased fruit could be divided into three stages according to the degree of the disease. Only a few achenes turning light white from green were marked as HB1, and a few appearing white and swelling slightly marked as HB2, while most achenes of mulberry fruit turned white and the whole fruit showed malformation, being marked as HB3. On the other side of mulberry orchard, about 200 m from the diseased area, mulberry fruits grew healthy and no pathogens in the fruit were detected by ITS sequencing, and marked as CK.
HB1, HB2, HB3, CK, fungal fruiting bodies, and sclerotia were collected at the same time. The samples were frozen in liquid nitrogen for storage at −80 °C. Three biological replicates were analyzed using transcriptome sequencing, qRT-PCR validation, and the enzyme activity test. Six biological replicates were used for metabolite profiling. Each replicate consisted of three single fruit, and a single fruit collected from an independent tree.
4.2. Pathogen Detection and Enzyme Activity Analysis
Genome DNA of CK, HB1, HB2, and HB3 were extracted from 0.1 g tissue using a rapid plant genomic DNA isolation kit (Sangon, Shanghai, China). Fungal genome DNA were extracted with a rapid fungi genomic DNA isolation kit (Sangon, Shanghai, China), and PCR amplification using fungal universal ITS1 and ITS4 primers.
Pectinase activity was measured by a pectinase assay kit (Jiancheng Biotechnology, Nanjing, China). The activity of the pectinase enzyme was expressed as units per milligrams of protein. Statistical differences were based on one-way ANOVA analysis followed by Tukey’s HSD t-test using SPSS 19.
4.3. Tissue Paraffin Section and Fluorescent Staning
Several samples were fixed in 4% buffered formalin, embedded in paraffin, and sectioned. For toluidine blue staining, the sections were cut and stained with toluidine blue solution (0.5% toluidine blue, 1% borax). For lactophenol-cotton blue staining, the staining included 0.05 g cotton blue, 20 g phenol crystals, 40 mL glycerol, 20 mL lactic acid, and 20 mL distilled water. For Calcofluor White Staining, one drop of Calcofluor White Stain (18909, Sigma-Aldrich, St. Louis, MO, USA) and one drop of 10% potassium hydroxide were added to the sections, a coverslip was placed over the specimen, let stand for 1 min, and finally examined under UV light (355 nm) [44].
4.4. RNA Extraction, RNA Sequencing, and Functional Annotations
Total RNA in each sample was extracted using the MiniBEST Plant RNA Extraction Kit (Takara Bio Inc., Dalian, China) by following the manufacturer’s procedure. An ethanol precipitation protocol and CTAB-PBIOZOL reagent was used for the purification of total RNA. The total RNA quantity and purity were analyzed by a Nano Drop and Agilent 2100 bioanalyzer (Thermo Fisher Scientific, Massachusetts, USA). Oligo (dT)-attached magnetic beads were used to purify mRNA. Sequencing was carried out using the BGISEQ-500 platform (BGI, Shenzhen, China).
The raw reads were filtered by removing adaptor sequences and low-quality sequences which contain more than 5% unknown nucleotides (N), and low-quality reads (those with >20% of bases with a quality value of <15) with SOAPnuke v1.5.2 [45]. The clean reads were aligned to the reference index by HISAT2 v2.0.4 [46]. String Tie (http://ccb.jhu.edu/software/stringtie) v1.0.4 was used to assemble the transcripts depending on the mulberry (Morus notabilis) annotation. The gene expression level was calculated with fragments per kilobase of transcript per million (FPKM) mapped reads. FPKM > 1, Fold Change > 2, and Adjusted p-value < 0.001 were used as the thresholds for the differential gene expression (DEGs) for the comparisons of HB1 vs. CK and HB2 vs. CK, and enrichment analyses of the gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of all the DEGs were analyzed.
4.5. Validation of Gene Expression by Quantitative Real-Time PCR (qRT-PCR)
To confirm the RNA-Seq results, 10 genes were selected to examine the consistency of their expression using qRT-PCR assays. All of the gene-specific primers were designed by Primer-BLAST (https://blast.ncbi.nlm.nih.gov/blast.cgi), and the primers are listed in Table S2. PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa, Dalian, China) was used for cDNA synthesis. The ABI 7500 Fast Real-Time PCR Systems (Applied Biosystems, Waltham, Massachusetts, USA) and the abm®EvaGreen qPCR MasterMix-No dye kit (Applied Biological Materials Inc., Vancouver, BC, Canada) were used for all of the qRT-PCR assays. The 2−ΔΔCt method was used to calculate the relative gene expression based on the qRT-PCR data [47].
4.6. Metabolite Profiling Assay by Ultra-Performance Liquid Chromatography (UPLC) and Mass Spectrometry (MS)
All samples were prepared according to the LC-MS system machine orders. All chromatographic contents were performed using an ultra-performance liquid chromatography (UPLC) system (Waters, Manchester, UK), and ACQUITY UPLC HSS T3 column (100 mm × 2.1 mm, 1.8 μm, Waters, UK) was used for the reversed phase separation. The mobile phase composed of solvent A (water, 0.1% formic acid) and solvent B (acetonitrile, 0.1% formic acid), and the flow rate of the mobile phases was 0.4 mL/min. Gradient elution conditions were set as follows: 0~2 min, 100% A; 2~11 min, 0% to 100% B; 11~13 min, 100% B; 13~15 min, 0% to 100% A. The column oven was maintained at 50 °C and the injection volume for each sample was 10 μL.
A high-resolution tandem mass spectrometer Xevo G2 XS QTOF (Waters, Manchester, UK) was used to test metabolites eluted from the column and operated in both positive and negative ion modes. The capillary and sampling cone voltages were set at 3.0 kV and 40.0 V, respectively, in positive ion mode, while 2.0 kV and 40.0 V were set in negative ion mode. The TOF mass range was between 50 and 1200 Da, and the scan time was set as 0.2 s. For the MS/MS detection, all precursors were fragmented using 20–40 eV, and the scan time was 0.2 s. In the data acquisition process, real-time quality correction was performed on the LE signal every 3 s. At the same time, quality control samples were collected every 10 samples to evaluate the stability of the instrument state during sample collection.
4.7. Metabolite Data Processing and Analysis
The raw data, both ESI negative and positive, were pretreated by Progenesis QI v 2.2, (Waters, UK) and metaX [48] (BGI, Shenzhen, China). Principal component analysis (PCA) and partial least square discriminant analysis (PLS-DA) were used for comparing the groups among CK, HB1, and HB2. The metabolites were initially identified based on their retention time, molecular weight, fragmentation patterns, and ultraviolet absorption, compared to standards and published references. The variable importance in the projection (VIP) value and ANOVA methods, under the condition of VIP > 1.0, p < 0.05, and fold change < 0.5 or > 1.5, were used for preliminary assessment of significantly different metabolites identified from UPLC–QTOF-MS analysis.
4.8. Mapman Analysis
Protein functions were categorized using MapMan software (available online: http://mapman.gabipd.org/). Annotations were transferred to the Arabidopsis genome with consideration for orthologous genes to predict functions of identified proteins from M. notabilis.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/5/1789/s1.
Author Contributions
L.B., C.S. and Y.Q. initiated and designed the research. L.B., H.G., Z.Z., and X.Z. performed the experiments. L.B. analyzed the data. F.J. and M.Z. prepared materials. L.B. and Y.Q. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by Modern Agricultural Industry Technology System (CARS-18), Shannxi Provincial Agriculture Department Research Project (NYKJ-2019-YL64), Promotion Team Project of Northwest A & F University (TGTD2017-03), Shannxi Provincial Education Department Research Project (No. 17JS002).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Mahboubi M. Morus alba (mulberry), a natural potent compound in management of obesity. Pharmacol. Res. 2019;146:104341. doi: 10.1016/j.phrs.2019.104341. [DOI] [PubMed] [Google Scholar]
- 2.Zhang H., Ma Z.F., Luo X., Li X.-L. Effects of Mulberry Fruit (Morus alba L.) Consumption on Health Outcomes: A Mini-Review. Antioxidants. 2018;7:69. doi: 10.3390/antiox7050069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinelli F., Uratsu S.L., Albrecht U., Reagan R.L., Phu M.L., Britton M., Buffalo V., Fass J., Leicht E., Zhao W., et al. Transcriptome Profiling of Citrus Fruit Response to Huanglongbing Disease. PLoS ONE. 2012;7:e38039. doi: 10.1371/journal.pone.0038039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu R., Zhao A., Li J., Wang X., Yu Y., Lu C., Yu M. Biological study of hypertrophy sorosis scleroteniosis and its molecular characterization based on LSU rRNA. Afr. J. Microbiol. Res. 2013;7:3405–3411. [Google Scholar]
- 5.Lu R., Zhao A., Li J., Liu C., Wang C., Wang X., Wang X., Pei R., Lu C., Yu M. Screening, cloning and expression analysis of a cellulase derived from the causative agent of hypertrophy sorosis scleroteniosis, Ciboria shiraiana. Gene. 2015;565:221–227. doi: 10.1016/j.gene.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 6.Li M., Lu R., Yu J., Cai Y., Wang C., Zhao A., Lu C., Yu M. Cloning and Functional Analysis of Polygalacturonase Genes from Ciboria shiraiana. Acta Agron. Sin. 2016;42:190. doi: 10.3724/SP.J.1006.2016.00190. [DOI] [Google Scholar]
- 7.Rudd J.J., Kanyuka K., Hassani-Pak K., Derbyshire M.C., Andongabo A., Devonshire J., Lysenko A., Saqi M., Desai N.M., Powers S.J., et al. Transcriptome and metabolite profiling of the infection cycle of Zymoseptoria tritici on wheat reveals a biphasic interaction with plant immunity involving differential pathogen chromosomal contributions and a variation on the hemibiotrophic lifestyle definition. Plant Physiol. 2015;167:1158–1185. doi: 10.1104/pp.114.255927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt R., Durling M.B., De Jager V., Menezes R.C., Nordkvist E., Svatos A., Dubey M., Lauterbach L., Dickschat J.S., Karlsson M., et al. Deciphering the genome and secondary metabolome of the plant pathogen Fusarium culmorum. FEMS Microbiol. Ecol. 2018;94 doi: 10.1093/femsec/fiy078. [DOI] [PubMed] [Google Scholar]
- 9.Liu X., Cao X., Shi S., Zhao N., Li D., Fang P., Chen X., Qi W., Zhang Z. Comparative RNA-Seq analysis reveals a critical role for brassinosteroids in rose (Rosa hybrida) petal defense against Botrytis cinerea infection. BMC Genet. 2018;19:62. doi: 10.1186/s12863-018-0668-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lardi M., Murset V., Fischer H.-M., Mesa S., Ahrens C.H., Zamboni N., Pessi G. Metabolomic Profiling of Bradyrhizobium diazoefficiens-Induced Root Nodules Reveals Both Host Plant-Specific and Developmental Signatures. Int. J. Mol. Sci. 2016;17:815. doi: 10.3390/ijms17060815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao L., Yu Q., Huang M., Hung W., Grosser J., Chen S., Wang Y., Gmitter F.G. Proteomic and metabolomic analyses provide insight into the off-flavour of fruits from citrus trees infected with ‘Candidatus Liberibacter asiaticus’. Hortic. Res. 2019;6:31. doi: 10.1038/s41438-018-0109-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C., Tariq R., Ji Z., Wei Z., Zheng K., Mishra R., Zhao K. Transcriptome analysis of a rice cultivar reveals the differentially expressed genes in response to wild and mutant strains of Xanthomonas oryzae pv. oryzae. Sci. Rep. 2019;9:3757. doi: 10.1038/s41598-019-39928-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu D., Zhao Q., Cheng Y., Li D., Jiang C., Cheng L., Wang Y., Yang A. Transcriptome analysis of two cultivars of tobacco in response to Cucumber mosaic virus infection. Sci. Rep. 2019;9:3124. doi: 10.1038/s41598-019-39734-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He N., Zhang C., Qi X., Zhao S., Tao Y., Yang G., Lee T.-H., Wang X., Cai Q., Li N., et al. Draft genome sequence of the mulberry tree Morus notabilis. Nat. Commun. 2013;4:2445. doi: 10.1038/ncomms3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai F., Wang Z., Li Z., Luo G., Wang Y., Tang C. Transcriptomic and proteomic analyses of mulberry (Morus atropurpurea) fruit response to Ciboria carunculoides. J. Proteom. 2019;193:142–153. doi: 10.1016/j.jprot.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Yin P., Li X., Zeng Q., Shen C., Gao L., Ton J. Effect of Popcorn Disease Infected Leaves on Silkworm Performance and Differential Proteome Analysis of Mulberry Popcorn Disease. Pak. J. Zool. 2017;50 doi: 10.17582/journal.pjz/2018.50.1.15.27. [DOI] [Google Scholar]
- 17.Dodds P.N., Rathjen J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 18.Windram O.P.F., Penfold C., Denby K. Network Modeling to Understand Plant Immunity. Annu. Rev. Phytopathol. 2014;52:93–111. doi: 10.1146/annurev-phyto-102313-050103. [DOI] [PubMed] [Google Scholar]
- 19.Thimm O., Nagel A., Kruger P., Selbig J., Müller L.A., Bläsing O., Gibon Y., Meyer S., Rhee S.Y., Stitt M. mapman: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313X.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 20.Pieterse C.M., Van Der Does D., Zamioudis C., Leon-Reyes A., Van Wees S.C.M. Hormonal Modulation of Plant Immunity. Annu. Rev. Cell Dev. Boil. 2012;28:489–521. doi: 10.1146/annurev-cellbio-092910-154055. [DOI] [PubMed] [Google Scholar]
- 21.Bari R., Jones A. Role of plant hormones in plant defence responses. Plant Mol. Boil. 2008;69:473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- 22.Blázquez M.A., Nelson D.C., Weijers D. Evolution of Plant Hormone Response Pathways. Annu. Rev. Plant Boil. 2020;71 doi: 10.1146/annurev-arplant-050718-100309. [DOI] [PubMed] [Google Scholar]
- 23.Chanclud E., Morel J.-B. Plant hormones: A fungal point of view. Mol. Plant Pathol. 2016;17:1289–1297. doi: 10.1111/mpp.12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo J., Xia W., Cao P., Xiao Z., Zhang Y., Liu M., Zhan C., Wang N. Integrated Transcriptome Analysis Reveals Plant Hormones Jasmonic Acid and Salicylic Acid Coordinate Growth and Defense Responses upon Fungal Infection in Poplar. Biomolecules. 2019;9:12. doi: 10.3390/biom9010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castro-Moretti F.R., Gentzel I.N., Mackey D.M., Alonso A.P. Metabolomics as an Emerging Tool for the Study of Plant–Pathogen Interactions. Metabolites. 2020;10:52. doi: 10.3390/metabo10020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y., Ji D., Turgeon R., Chen J., Lin T., Huang J., Luo J., Zhu Y., Zhang C., Lv Z. Physiological and Proteomic Responses of Mulberry Trees (Morus alba. L.) to Combined Salt and Drought Stress. Int. J. Mol. Sci. 2019;20:2486. doi: 10.3390/ijms20102486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng Y., Wang P., Chen X., Sun Y., Yue C., Ye N. Transcriptome and Metabolite Profiling Reveal Novel Insights into Volatile Heterosis in the Tea Plant (Camellia sinensis) Molecules. 2019;24:3380. doi: 10.3390/molecules24183380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhai R., Wang Z., Zhang S., Meng G., Song L., Wang Z., Li P., Ma F., Xu L. Two MYB transcription factors regulate flavonoid biosynthesis in pear fruit (Pyrus bretschneideri Rehd.) J. Exp. Bot. 2015;67:1275–1284. doi: 10.1093/jxb/erv524. [DOI] [PubMed] [Google Scholar]
- 29.Hichri I., Heppel S.C., Pillet J., Léon C., Czemmel S., Delrot S., Lauvergeat V., Bogs J. The Basic Helix-Loop-Helix Transcription Factor MYC1 Is Involved in the Regulation of the Flavonoid Biosynthesis Pathway in Grapevine. Mol. Plant. 2010;3:509–523. doi: 10.1093/mp/ssp118. [DOI] [PubMed] [Google Scholar]
- 30.Parker D., Beckmann M., Zubair H., Enot D.P., Overy D.P., Snowdon S., Talbot N.J., Draper J., Caracuel-Rios Z. Metabolomic analysis reveals a common pattern of metabolic re-programming during invasion of three host plant species by Magnaporthe grisea. Plant J. 2009;59:723–737. doi: 10.1111/j.1365-313X.2009.03912.x. [DOI] [PubMed] [Google Scholar]
- 31.Robison F.M., Turner M.F., Jahn C., Schwartz H.F., Prenni J., Brick M., Heuberger A.L. Common bean varieties demonstrate differential physiological and metabolic responses to the pathogenic fungus Sclerotinia sclerotiorum. Plant Cell Environ. 2018;41:2141–2154. doi: 10.1111/pce.13176. [DOI] [PubMed] [Google Scholar]
- 32.Rodríguez A., Shimada T., Cervera M., Alquezar B., Gadea J., Gómez-Cadenas A., De Ollas C., Rodrigo M.J., Zacarias L., Peña L. Terpene Down-Regulation Triggers Defense Responses in Transgenic Orange Leading to Resistance against Fungal Pathogens1[W] Plant Physiol. 2013;164:321–339. doi: 10.1104/pp.113.224279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen F., Ma R., Chen X.-L. Advances of Metabolomics in Fungal Pathogen-Plant Interactions. Metabolites. 2019;9:169. doi: 10.3390/metabo9080169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choo T.M. Breeding Barley for Resistance to Fusarium Head Blight and Mycotoxin Accumulation. Plant Breed. Rev. 2010:125–169. [Google Scholar]
- 35.Ranjan A., Westrick N.M., Jain S., Piotrowski J.S., Ranjan M., Kessens R., Stiegman L., Grau C.R., Conley S.P., Smith D., et al. Resistance against Sclerotinia sclerotiorum in soybean involves a reprogramming of the phenylpropanoid pathway and up-regulation of antifungal activity targeting ergosterol biosynthesis. Plant Biotechnol. J. 2019;17:1567–1581. doi: 10.1111/pbi.13082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kovalchuk A., Zeng Z., Ghimire R.P., Kivimäenpää M., Raffaello T., Liu M., Mukrimin M., Kasanen R., Sun H., Julkunen-Tiitto R., et al. Dual RNA-seq analysis provides new insights into interactions between Norway spruce and necrotrophic pathogen Heterobasidion annosum s.l. BMC Plant Boil. 2019;19:2. doi: 10.1186/s12870-018-1602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodrigues E.L., Marcelino G., Silva G.T., Figueiredo P.S., Garcez W.S., Corsino J., Guimarães R.D.C.A., Gielow K.D.C.F. Nutraceutical and Medicinal Potential of the Morus Species in Metabolic Dysfunctions. Int. J. Mol. Sci. 2019;20:301. doi: 10.3390/ijms20020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gui Z., Raman S.T., Ganeshan A.K.P.G., Chen C., Jin C., Li S.-H., Chen H.-J. In vitro and In vivo antioxidant activity of flavonoid extracted from mulberry fruit (Morus alba L.) Pharmacogn. Mag. 2016;12:128–133. doi: 10.4103/0973-1296.177910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tu J., Shi D., Wen L., Jiang Y., Zhao Y., Yang J., Liu H., Liu G., Yang B. Identification of moracin N in mulberry leaf and evaluation of antioxidant activity. Food Chem. Toxicol. 2019;132:110730. doi: 10.1016/j.fct.2019.110730. [DOI] [PubMed] [Google Scholar]
- 40.Jiang Y. Effects of anthocyanins derived from Xinjiang black mulberry fruit on delaying aging. J. Hyg. Res. 2010:39. [PubMed] [Google Scholar]
- 41.Qian W., Sun Z., Wang T., Yang M., Liu M., Zhang J., Li Y. Antimicrobial activity of eugenol against carbapenem-resistant Klebsiella pneumoniae and its effect on biofilms. Microb. Pathog. 2020;139:103924. doi: 10.1016/j.micpath.2019.103924. [DOI] [PubMed] [Google Scholar]
- 42.Ju J., Xie Y., Yu H., Guo Y., Cheng Y., Zhang R., Yao W. Synergistic inhibition effect of citral and eugenol against Aspergillus niger and their application in bread preservation. Food Chem. 2019;310:125974. doi: 10.1016/j.foodchem.2019.125974. [DOI] [PubMed] [Google Scholar]
- 43.Xue Y., Zhou S., Fan C., Du Q., Jin P. Enhanced Antifungal Activities of Eugenol-Entrapped Casein Nanoparticles against Anthracnose in Postharvest Fruits. Nanomaterials. 2019;9:1777. doi: 10.3390/nano9121777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasconi S., Jobard M., Jouve L., Sime-Ngando T. Use of Calcofluor White for Detection, Identification, and Quantification of Phytoplanktonic Fungal Parasites. Appl. Environ. Microbiol. 2009;75:2545–2553. doi: 10.1128/AEM.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cock P.J., Fields C.J., Goto N., Heuer M.L., Rice P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2009;38:1767–1771. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim D., Langmead B., Salzberg S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miao L., Di Q., Sun T., Li Y., Duan Y., Wang J., Yan Y., He C., Wang C., Yu X. Integrated Metabolome and Transcriptome Analysis Provide Insights into the Effects of Grafting on Fruit Flavor of Cucumber with Different Rootstocks. Int. J. Mol. Sci. 2019;20:3592. doi: 10.3390/ijms20143592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wen B., Mei Z., Zeng C., Liu S. metaX: A flexible and comprehensive software for processing metabolomics data. BMC Bioinform. 2017;18:183. doi: 10.1186/s12859-017-1579-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.