Abstract
Background
Many patients are not responsive or tolerant to medical therapies for carotid atherosclerosis. Thus, elucidating the molecular mechanism for the pathogenesis and progression of carotid atherosclerosis and identifying new potential molecular targets for medical therapies that can slow progression of carotid atherosclerosis and prevent ischemic events are quite important.
Material/Methods
We downloaded the expression profiling data of PBMC in Biobank of Karolinska Endarterectomy (BiKE, GSE21545) for GEO. The WGCNA and DEG screening were conducted. The co-expression pattern between patients with ischemic events (the events group) and patients without ischemic events (the no-events group) were compared. Then, we identified hub genes of each module. Finally, the DEG co-expression network was constructed and MCODE was used to identify crucial genes based on this co-expression network.
Results
In the study, 183 DEGs were screened and 8 and 6 modules were assessed in the events group and no-events group, respectively. Compared to the no-events group, genes associated with inflammation and immune response were clustered in the green-yellow module of the events group. The hub gene of the green-yellow module of the events group was KIR2DL5A. We obtained 1 DEG co-expression network, which has 16 nodes and 24 edges, and we detected 5 crucial genes: SIRT1, THRAP3, RBM43, PEX1, and KLHDC2. The upregulated genes (THRAP3 and RBM43) showed potential diagnostic and prognostic value for the occurrence of ischemic events.
Conclusions
We detected 8 modules for the events group and 6 modules for the no-events group. The hub genes for modules and crucial genes of the DEG co-expression network were also identified. These genes might serve as potential targets for medical therapies and biomarkers for diagnosis and prognosis. Further experimental and biological studies are needed to elucidate the role of these crucial genes in the progression of carotid atherosclerosis.
MeSH Keywords: Carotid Artery Diseases, Gene Expression Profiling, Gene Regulatory Networks, Microarray Analysis
Background
Atherosclerosis is an inflammatory disease that involves the accumulation of fibrous and/or fatty components in the intima of medium and large arteries such as the coronary artery, carotid artery, and peripheral artery, and the clinical manifestations vary with the arteries affected [1,2]. Ischemic strokes and transient ischemic attacks may occur if the carotid artery is involved, and carotid atherosclerotic disease accounts for approximately 18–25% of all ischemic strokes [3]. Prevention of stroke in patients with carotid atherosclerosis depends on the degree of carotid stenosis. These preventive methods mainly include carotid endarterectomy, carotid stenting, and medical management such as with statins and antiplatelet agents [4,5]. Although the medical management is effective and may even serve as an alternative to carotid endarterectomy in patients with asymptomatic carotid atherosclerosis, patients who are nonresponsive to medical therapies or not tolerant of the adverse effects may not benefit from present medical therapies [6–8]. Therefore, elucidating the molecular mechanism of the pathogenesis and progression of carotid atherosclerosis and identifying new potential molecular targets for medical therapies that can slow progression of carotid atherosclerosis and prevent ischemic events are quite important. The molecular mechanism mainly includes abnormal accumulation of lipids, immune response, and inflammation, and monocytes play an important role [1,9]. Induced by chemokines, circulating monocytes can bind to adhesion molecules expressed by endothelial cells, migrating into the arterial wall and differentiating into macrophages. Previous studies focused on the role of circulating monocytes in the pathogenesis and progression of carotid atherosclerosis; however, few researchers have used weighted (gene) correlation network analysis (WGCNA) to construct gene co-expression networks for carotid atherosclerosis based on high-throughput data of peripheral blood mononuclear cells (PBMCs) in patients.
Zhang and Horvath first developed the WGCNA algorithm in 2005, which can be used for gene co-expression network construction, gene module detection, and hub gene identification, based on gene expression data [10–12]. Furthermore, gene modules and hub genes can be correlated with clinical traits if these data are available. The WGCNA R package was developed on the official R website (https://cran.r-project.org/), making it more convenient for researchers to conduct WGCNA. Although WGCNA was first developed for analyzing gene expression data, it can also be used for miRNA, lncRNA, and even metabolome [13–15].
Previous studies screened differentially expressed genes (DEGs) using microarray data of carotid atherosclerotic plaques. For instance, Razuvaev et al. identified 11 downregulated genes and 19 upregulated genes by comparing the gene expression profile between symptomatic and asymptomatic patients [16]. However, DEG screening cannot reveal the interaction among genes or identify genes with crucial biological functions.
In the present study, we focused on the possible underlying molecular mechanism of the occurrence of ischemic events. The mRNA microarray data of the Biobank of Karolinska Endarterectomies (BiKE) were included. The expression data of peripheral blood mononuclear cells for patients with ischemic events (the events group) and patients without ischemic events (the no-events group) during follow-up [17] were used in our analysis. The genes in the gene modules were subjected to functional enrichment analysis. Then, we mapped DEGs into the co-expression network of events group and obtained 1 DEG co-expression network. Furthermore, we identified crucial genes based on the DEG co-expression network. The potential diagnostic and prognostics values of the upregulated crucial genes were identified.
Material and Methods
Datasets
The dataset GSE21545, from the Biobank of Karolinska Endarterectomy (BiKE), was selected from the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/). Series matrix file and platform data tables (GPL570) were downloaded.
DEG analysis
The series matrix file was annotated with GPL570 platform data tables, and the probe names in the matrix file were replaced by the gene symbols. Then, the 97 peripheral blood mononuclear cell (PBMC) samples were included in our analysis, in which 21 were samples of the events group and 76 were samples of the no-events group. Differentially expressed genes (DEGs) were screened using the “limma” R package. |log2(fold-change)|>2 and adjusted p<0.01 were set as the threshold of DEG screening.
Construction of co-expression network by WGCNA
Co-expression networks for both PBMC and plaque samples were constructed using the “WGCNA” R package. The algorithm filtered genes with the top 25% variance for further analysis, and WGCNA analysis was conducted for the events group (21 samples) and the no-events group (76 samples). The soft-power threshold β was chosen to ensure a scale-free topology. A topological overlap measure (TOM) matrix was created from the adjacency matrix to estimate the network’s connectivity property. A clustering dendrogram was constructed using average linkage hierarchical clustering based on the TOM matrix. The threshold for modules size was set as 50 for both groups to generate modules with proper size, and similar modules were merged.
GO and KEGG pathway enrichment of gene modules
Gene ontology (GO) and Kyoto Encyclopedia of Genes Genomes (KEGG) pathway analyses were conducted for genes in modules we detected using the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 (https://david.ncifcrf.gov/) to determine the biological function and signaling pathway involved in these modules. Count number >2 and p<0.05 were set as thresholds for the analysis. The differences between co-expression networks for the events group and no-events group were compared based on the results of functional enrichment analysis.
Identification of hub genes and crucial genes
Hub genes were considered to be the gene which had the largest intramodular connectivity in each module. Then, we mapped the DEGs into a co-expression network in the events group using Cytoscape v3.7.0, and we obtained 1 DEG co-expression network. Isolated nodes and isolated nodes pairs were removed from the network. The Molecular Complex Detection (MCODE), a plugin in Cytoscape to detect core subnetworks, was used to identify crucial gene clusters based on the DEG co-expression network. Receiver operating characteristic (ROC) analysis and survival analysis were also conducted using the combination of the upregulated genes in the crucial gene cluster by SPSS 25.0 to show the potential diagnostic and prognostic value of upregulated crucial genes.
Results
Flowchart
The flowchart of our study is shown in Figure 1. We constructed the co-expression networks for the events group and no-events group and detected gene modules. Then, DEG screening was conducted, and 183 DEGs were screened. The DEG co-expression network were constructed by mapping DEGs into the whole co-expression network of the events group. Based on the DEG co-expression network, crucial genes were identified, and their clinical significance was evaluated by ROC and survival analysis.
Figure 1.
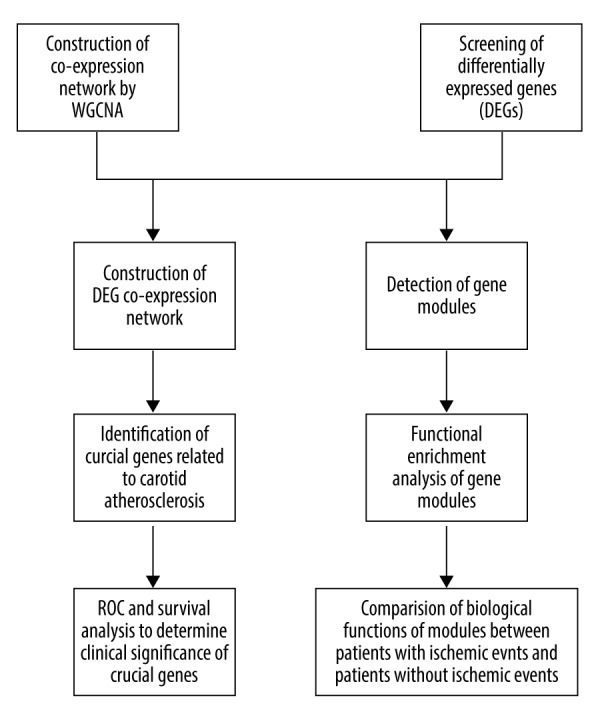
Flowchart for the study.
Screening of DEGs
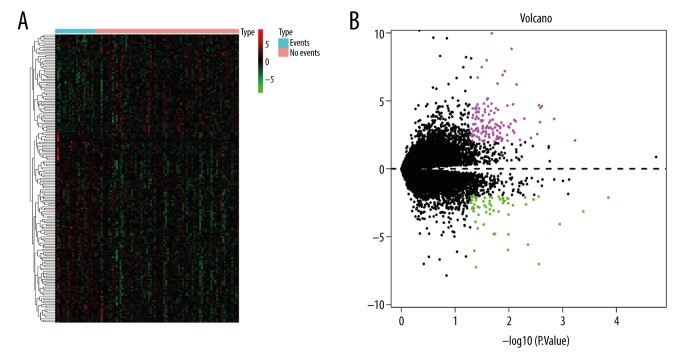
With the threshold of |log2(fold-change)|>2 and p<0.01, 183 DEGs were screened with 122 upregulated and 61 downregulated genes. The heatmap and the volcano plot showed the expression pattern of DEGs (Figure 2). Upregulated DEGs and downregulated DEGs with the top 10-fold-change are shown in Supplementary Table 1.
Figure 2.
DEG screening. (A) Heatmap for the DEGs we screened. (B) Volcano plots for the DEGs. The X-axis represents –log(P.val) and Y-axis represents logFC.
Construction of the co-expression network for the events group and no-events group
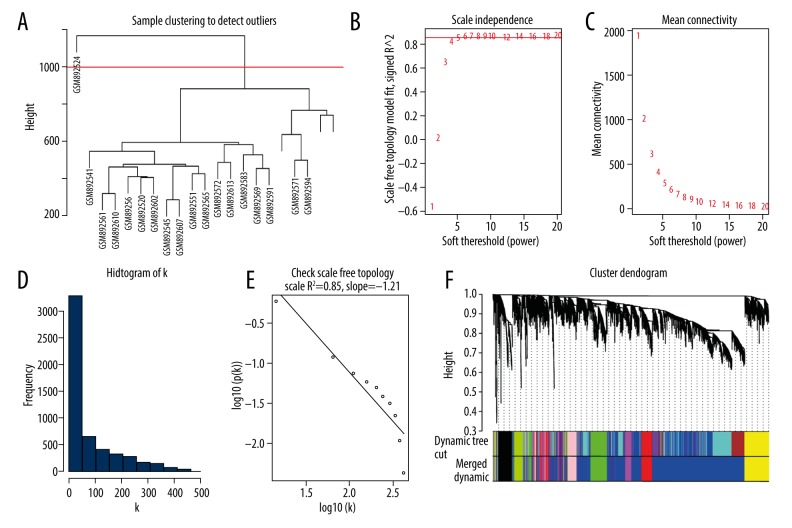
One outlier (GSM892524) in the events group was removed, while all samples in the no-events group were included for further analysis, as shown in the sample clustering dendrogram (Figure 3A and Supplementary Figure 1A). The power of β=10 and 16 were chosen as the soft-threshold for the network of the events group and no-events group, respectively (Figure 3B and Supplementary Figure 1B). And the both the co-expression networks we constructed met the requirements of scale-free topology (Figure 3C–3E and Supplementary Figure 1C–1E). We detected 8 gene modules for the events group and 6 gene modules for the no-events group (Figure 3F and Supplementary Figure 1F).
Figure 3.
WGCNA of event group. (A) One outlier (GSE89254) was delected by sample clustering. (B, C) Selection of soft-threshold β. (D, E) Fitness for scale free topology when β 10. (F) Cluster dendrogram. Each module was represented by WGCNA.
Comparison of co-expression patterns
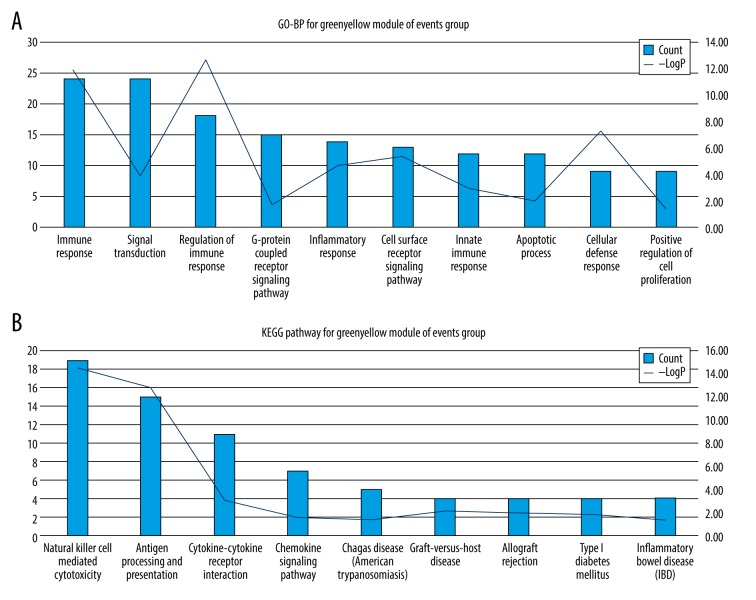
KEGG pathway and GO-BP analysis were used to assess the biological function of genes for modules. Results of GO-BP and KEGG analyses are shown in Supplementary Tables 2 and 3. The green-yellow module may be related to the occurrence of ischemic events. The green-yellow module is mainly associated with inflammation and immune response. Nonetheless, pathways associated with inflammation and immune response were scattered in modules of the no-events group. The KEGG pathway GO-BP terms with the top 10 count numbers for green-yellow modules of the events group are shown in Figure 4 and Table 1. These results indicate that PBMC might play a role in the occurrence of ischemic events through regulating inflammation and immune response.
Figure 4.
Enriched GO-BP terms and KEGG pathways with top10 count number for greenyellow module of events group. (A) GO-BP terms; (B) KEGG terms.
Table 1.
GO-BP KEGG pathways terms with top 10 count number of black module for events group.
| ID | Terms | Count | −LogP |
|---|---|---|---|
| GO-BP | |||
| GO: 0006955 | Immune response | 24 | 11.80 |
| GO: 0007165 | Signal transduction | 24 | 3.82 |
| GO: 0050776 | Regulation of immune response | 18 | 12.65 |
| GO: 0007186 | G-protein coupled receptor signaling pathway | 15 | 1.63 |
| GO: 0006954 | Inflammatory response | 14 | 4.60 |
| GO: 0007166 | Cell surface receptor signaling pathway | 13 | 5.34 |
| GO: 0045087 | Innate immune response | 12 | 2.89 |
| GO: 0006915 | Apoptotic process | 12 | 1.99 |
| GO: 0006968 | Cellular defense response | 9 | 7.24 |
| GO: 0008284 | Positive regulation of cell proliferation | 9 | 1.31 |
| KEGG | |||
| hsa04650 | Natural killer cell mediated cytotoxicity | 19 | 14.53 |
| hsa04612 | Antigen processing and presentation | 15 | 12.70 |
| hsa04060 | Cytokine-cytokine receptor interaction | 11 | 3.16 |
| hsa04062 | Chemokine signaling pathway | 7 | 1.59 |
| hsa05142 | Chagas disease (American trypanosomiasis) | 5 | 1.42 |
| hsa05332 | Graft-versus-host disease | 4 | 2.13 |
| hsa05330 | Allograft rejection | 4 | 1.99 |
| hsa04940 | Type I diabetes mellitus | 4 | 1.84 |
| hsa05321 | Inflammatory bowel disease (IBD) | 4 | 1.37 |
Hub genes in modules of the events group and no-events group
Hub genes for modules of the events group and no-events group are shown in Table 2. The hub genes of the green-yellow modules of the events group were killer cell immunoglobulin-like receptor, 2 Ig domains, and long cytoplasmic tail 5A (KIR2DL5A), which are killer cell immunoglobulin-like receptors (KIRs) and are mainly expressed by natural killer cells and subsets of T cells.
Table 2.
Hub genes of each module for events group and no-events group.
| Module | Gene symbol | Official full gene name |
|---|---|---|
| Events group | ||
| Black | ACRBP | Acrosin binding protein |
| Blue | AP2M1 | Adaptor related protein complex 2 subunit mu 1 |
| Green | DOCK10 | Dedicator of cytokinesis 10 |
| Greenyellow | KIR2DL5A | Killer cell immunoglobulin like receptor, two Ig domains and long cytoplasmic tail 5A |
| Magenta | GNS | Glucosamine (N-acetyl)-6-sulfatase |
| Pink | FHOD1 | Formin homology 2 domain containing 1 |
| Red | ITGA5 | Integrin subunit alpha 5 |
| Yellow | MPEG1 | Macrophage expressed 1 |
| No-events group | ||
| Black | CTTN | Cortactin |
| Green | PRKCSH | Protein kinase C substrate 80K-H |
| Magenta | MAPRE1 | Microtubule associated protein RP/EB family member 1 |
| Red | FAM103A1 | RNA Guanine-7 Methyltransferase Activating Subunit |
| Tan | ZHX1 | Zinc fingers and homeoboxes 1 |
| Yellow | ZBTB20 | Zinc finger and BTB domain containing 20 |
Identification of crucial genes mediating ischemic events
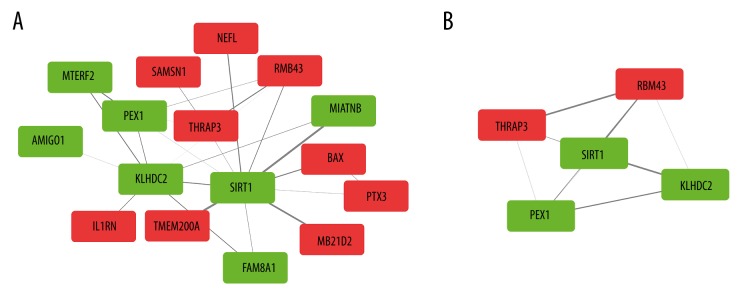
The DEG co-expression network was obtained by mapping DEGs into the whole co-expression network of the events group. The threshold for weighted edge was set as 0.1. After removing isolated nodes and isolated nodes pairs, a network with 16 nodes and 24 edges was generated (Figure 5A). MCODE detected 1 significant cluster consisting of 5 genes for the DEG co-expression network (Figure 5B, Table 3). Among these 5 genes, 2 genes were upregulated (THRAP3 and RBM43) and 3 genes were downregulated (SIRT1, PEX1, and KLHDC2). Sirtuin 1 (SIRT1), a member of the sirtuin family, had the highest connectivity among the 5 crucial genes.
Figure 5.
DEG co-expression network and crucial genes. (A) Red boxes represent up-regulated genes. Green boxes represent down-regulated genes. (B) Crucial genes generated by MCODE.
Table 3.
Crucial genes detected by MCODE.
| Entrez ID | Gene symbol | Official full gene name |
|---|---|---|
| 23411 | SIRT1 | Sirtuin 1 |
| 9967 | THRAP3 | Thyroid Hormone Receptor Associated Protein 3 |
| 375287 | RBM43 | RNA Binding Motif Protein 43 |
| 23588 | KLHDC2 | Kelch Domain Containing 2 |
| 5189 | PEX1 | Peroxisomal Biogenesis Factor 1 |
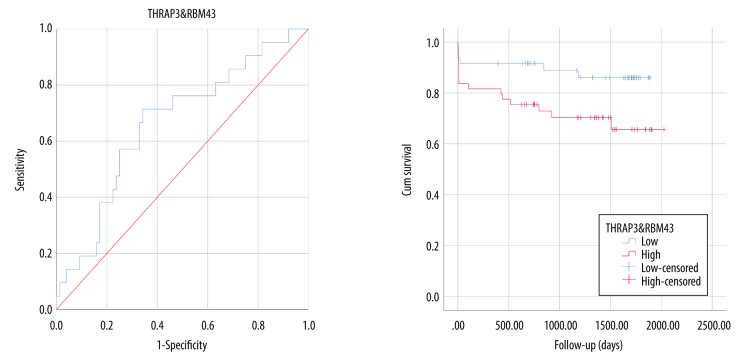
Combination of the 2 upregulated genes showed potential diagnostic and prognostic value (Figure 6).
Figure 6.
ROC and survival analysis of up-regulated crucial genes.
Discussion
We screened 183 DEGs, among which 122 were upregulated and 61 were downregulated (Figure 2 and Supplementary Table 1). Weighted co-expression networks were constructed using the WGCNA algorithm. We detected 8 modules for the events group and 6 modules for the no-events group.
We also conducted KEGG pathway and GO-BP analysis (Supplementary Tables 2 and 3) and found that pathways related to inflammation and immune response were mainly enriched in the green-yellow module of the events group. However, these pathways were dispersed in modules of the no-events group. Hub genes were considered to be genes which had the highest connectivity in each module (Table 2). Then, the DEG co-expression network was obtained by mapping DEGs into the whole co-expression network of the events group, and crucial genes were identified by MCODE based on the DEG co-expression network. These crucial genes were THRAP3, RBM43, SIRT1, PEX1, and KLHDC2. SIRT1 had the highest connectivity among the 5 crucial genes, and the combination of 2 upregulated genes (THRAP3 and RBM43) showed potential prognostic and diagnostic value.
Perisic et al. used the same dataset and analyzed the expression signature of PBMCs, and the DEGs they screened were different from the DEGs in our study. They grouped patients into a symptomatic group and an asymptomatic group. In the symptomatic group, patients already had plaque instability, which was defined as transient ischemic attack (TIA), minor stroke (MF), and amaurosis fugax (AF) [18]. However, unlike the previous study, we classified patients into an events group and a no-events group, depending on the occurrence of ischemic events during follow-up [17]. The difference in grouping patients may account for the difference in DEG screening results.
Several previous studies conducted WGCNA on expression data of atherosclerosis. Using aortic samples from Apobtm2SgyLdlrtm1Her knockout mice, Deshpande et al. discovered that inflammation and immune response might play a role in the pathogenesis and progression of atherosclerosis, and identified several related genes (TM9SF1, LEPR, WIF1, and SP1). In contrast to the sample Desphande et al. used, some researchers used human atherosclerotic samples from the GEO website and also found that inflammation and immune response might have important roles. Zhang et al. discovered crucial genes such as TNPO1 and ZDHHC17, while Wang et al. found that a lncRNA module was associated with inflammation and immune response. However, they did not elucidate the molecular mechanism based on the expression profiling of PBMC samples, and the grouping was also different [19–22].
The gene module detection and functional enrichment analysis indicated that the co-expression patterns in the events group and no-events group were different. We found that inflammation and immune response were clustered in the green-yellow module of the events group. A previous study showed that CD14+CD16– monocyte has a proinflammatory phenotype, and increased circulating proinflammatory monocytes were observed in the atherosclerotic models of ApoE–/– mice [23,24]. Belge et al. also discovered that proinflammatory cytokines such as TNF-α can be produced by activated CD14hiCD16+ monocytes, which might participate in atherosclerosis progression [25]. In addition, monocytes are involved in regulation of immune response in atherosclerosis. Some tissue macrophages and dendritic cells in the lesion originated from monocytes [26,27]. Evans et al. found that T cell response can be regulated by monocytes [28]. Furthermore, the TLR-4 expression in CD14hiCD16+ monocytes were correlated with occurrence of plaque progression and ischemic events in coronary artery disease [29]. In a recent experimental study, Bruen et al. showed that conjugated linoleic acid (CLA), which is an anti-inflammatory lipid, can induce regression of atherosclerosis in ApoE–/– mice. In mice fed CLA, more monocytes differentiated into anti-inflammatory M2 macrophages [30]. Sun et al. fed ApoE–/– model mice phenytoin, a non-selective voltage-gated sodium channels antagonist, and the mice subsequently exhibited increased levels of anti-inflammatory monocytes and decreased levels of proinflammatory monocytes [31]. Statins were also found to affect monocytes in atherosclerosis. Using samples from patients, Gasbarrino et al. discovered that intensive statins therapy can downregulate the expression of the anti-inflammatory adiponectin-AdipoR pathway in monocytes and macrophages, instead of positively regulating this pathway, which may explain part of the residual cardiovascular risk in patients using statins [32]. These studies, together with our findings, suggest that monocytes participate in the pathogenesis and progression of atherosclerosis via mediating inflammation and immune response, both directly and indirectly.
The hub gene of the green-yellow module was KIR2DL5A, belonging to the KIR family, and it is mainly expressed by natural killer cells and T cells. KIR2DL5A is an inhibitory receptor of immune response [33] and it is involved in immune response to viral infection and prognosis of certain malignant diseases. Shan et al. reported that patients with KIR2DL5A–/2DL5B+ genotype had increased HCV clearance [34]. In colorectal cancer, the presence of KIR2DL5A is related to increased complete response rate in patients treated with FOLFIRI chemotherapy [35], and KIR2DL5A is also a protective factor against breast cancer [36], while in pediatric leukemia patients after hematopoietic stem cell transplantation, the presence of KIR2DL5A is associated with higher relapse rate [37]. However, few studies had reported the role of KIR2DL5A in monocytes or its role in atherosclerosis, and it might be a promising target to elucidate the molecular mechanism for the progression of carotid atherosclerosis.
SIRT1 was the gene having the highest degree among the 5 crucial genes, and it was downregulated in the events group. SIRT1 is a type of NAD-dependent histone deacetylase [38] and participates in regulating inflammation, apoptosis, and cell senescence [39,40]. It also plays roles in stress response, aging, and longevity [41,42]. SIRT1 can also slow the progression of atherosclerosis by lipid modification, oxidative stress reduction, anti-inflammatory actions, foam cells, and autophagy regulation, and downregulation of SIRT1 was observed in a atherosclerotic mouse model [43], which is consistent with our findings. Recently, Lee et al. discovered that SIRT1 inhibits the adhesion of monocytes to vascular endothelia cells by suppressing MAC-1 expression in monocytes [44]. In addition, Nguyen et al. discovered that a dipeptidyl peptidase 4 inhibitor, evogliptin, can inhibit monocytes adhesion to vascular endothelial cells in an ApoE–/– mouse model, and this effect is associated with regulation of NF-κB by SIRT1 [45]. Therefore, SIRT1 might also slow the progression of atherosclerosis by preventing monocytes adhesion, which is the one of the initiation steps in the pathogenesis of atherosclerosis.
The upregulated genes, THRAP3 and RBM43, showed potential diagnostic and prognostic value. THRAP3, thyroid hormone receptor-associated protein 3, is an RNA-processing factors and can also participate in the DNA damage response (DDR) pathway and transcription regulation [46–49]. Mutations in THRAP3 may cause DNA damage repair defects, and Vohhodina reported that loss of THRAP3 made 293T and U2OS cells more susceptible to DNA-damaging factors [49]. Ino et al. used LNCaP and LNCaP-AI prostate cancer cell lines to demonstrate that THRAP3 phosphorylation can contribute to the acquisition of androgen independence in prostate cancer via transcriptional regulation [48]. Another study, using high-fat-fed mice, found that THRAP3 can act as a transcriptional regulator in diabetes and can control diabetic gene programming [47]. RBM43 is an RNA binding motif protein 43 and its detailed biological function is not known. At present, it is unclear whether THRAP3 and RBM43 participates the pathogenesis of atherosclerosis, although they were found to have potential clinical significance for the occurrence of ischemic events in carotid atherosclerosis patients.
In the present study, for the first time, we constructed a co-expression network, detected genes modules, and identified hub genes and crucial genes in carotid atherosclerosis using PBMC expression data. However, datasets in GEO lack clinical information; therefore, it is difficult to correlate traits with clinical importance with gene modules in WGCNA analysis.
The events group and no-events group had different co-expression patterns, and these differences suggest that monocytes are of vital importance in the pathogenesis and progression of carotid atherosclerosis via mediating inflammation and immune response. Then, we identified hub genes and crucial genes, which might have crucial biological functions in the pathogenesis of carotid atherosclerosis or potential diagnostic and prognostic value for ischemic events.
Conclusions
We detected 8 modules for the events group and 6 modules for the no-events group. The hub genes for each module and crucial genes of the DEG co-expression network were also identified. These genes might serve as potential targets for medical therapies and as biomarkers for diagnosis and prognosis. Further mechanism studies are needed to explore the biological function of these genes in the pathogenesis and progression of carotid atherosclerosis.
Supplementary Data
WGCNA of no-event. (A) No outlier was detected by sample clustering. (B, C) Selection of soft-threshold β. (D, E) Fitness of scale free topology when β-16. (F) Cluster dendrogram. Each module was represented by WGCNA.
Supplementary Table 1.
Top10 up-regulated and down-regulated DEGs.
| Gene symbol | Official full gene name | log2 (fold-change) (patients with events/patients without events) |
|---|---|---|
| Up-regulated | ||
| TNFAIP6 | TNF alpha induced protein 6 | 9.968020902 |
| PTX3 | Pentraxin 3 | 8.826641354 |
| RNASE2 | Ribonuclease A family member 2 | 7.978917622 |
| KCNJ2 | Potassium inwardly rectifying channel subfamily J member 2 | 7.481378497 |
| SERPINB2 | Serpin family B member 2 | 7.18837765 |
| PLA2G7 | Phospholipase A2 group VII | 6.901998631 |
| BCL2A1 | BCL2 related protein A1 | 6.719719825 |
| CLEC4D | C-type lectin domain family 4 member D | 6.272368641 |
| SAMSN1 | SAM domain, SH3 domain and nuclear localization signals 1 | 6.209244405 |
| GPR84 | G protein-coupled receptor 84 | 5.186451434 |
| Down-regulated | ||
| KLRC3 | Killer cell lectin like receptor C3 | −7.246559422 |
| BTN3A2 | Butyrophilin subfamily 3 member A2 | −7.003853494 |
| ANKRD20A11P | Ankyrin repeat domain 20 family member A11, pseudogene | −6.02418399 |
| ZNF600 | Zinc finger protein 600 | −5.985685983 |
| NLRC3 | NLR family CARD domain containing 3 | −5.579931019 |
| LCK | LCK proto-oncogene, Src family tyrosine kinase | −4.825468373 |
| GOLGA8N | Golgin A8 family member N | −4.799854266 |
| CEP78 | Centrosomal protein 78 | −4.795917474 |
| SLC9A3R1 | SLC9A3 regulator 1 | −4.381636674 |
| SEP1 | Septin 1 | −4.097540674 |
Supplemenatry Table 3.
KEGG pathways for modules of events group and no-events group.
| Events group | KEGG ID | KEGG pathway | Count | −logP | No-events group | KEGG ID | KEGG pathway | Count | −logP |
|---|---|---|---|---|---|---|---|---|---|
| Black | Black | ||||||||
| hsa05034 | Alcoholism | 14 | 4.58 | hsa04611 | Platelet activation | 11 | 5.10 | ||
| hsa05322 | Systemic lupus erythematosus | 13 | 5.13 | hsa05322 | Systemic lupus erythematosus | 11 | 4.98 | ||
| hsa04611 | Platelet activation | 11 | 3.80 | hsa05034 | Alcoholism | 11 | 3.94 | ||
| hsa05203 | Viral carcinogenesis | 10 | 1.82 | hsa04512 | ECM-receptor interaction | 8 | 3.83 | ||
| hsa05202 | Transcriptional misregulation in cancer | 9 | 1.87 | hsa05203 | Viral carcinogenesis | 8 | 1.71 | ||
| hsa04062 | Chemokine signaling pathway | 9 | 1.62 | hsa04510 | Focal adhesion | 8 | 1.70 | ||
| hsa04512 | ECM-receptor interaction | 6 | 1.62 | hsa04810 | Regulation of actin cytoskeleton | 8 | 1.66 | ||
| hsa04540 | Gap junction | 6 | 1.60 | hsa04540 | Gap junction | 6 | 2.20 | ||
| hsa05219 | Bladder cancer | 4 | 1.38 | hsa04670 | Leukocyte transendothelial migration | 6 | 1.73 | ||
| Blue | hsa05410 | Hypertrophic cardiomyopathy (HCM) | 5 | 1.70 | |||||
| hsa01100 | Metabolic pathways | 218 | 3.72 | hsa05414 | Dilated cardiomyopathy | 5 | 1.59 | ||
| hsa05166 | HTLV-I infection | 49 | 1.56 | hsa04640 | Hematopoietic cell lineage | 5 | 1.54 | ||
| hsa04144 | Endocytosis | 48 | 1.76 | hsa04530 | Tight junction | 5 | 1.54 | ||
| hsa04141 | Protein processing in endoplasmic reticulum | 45 | 4.34 | hsa05130 | Pathogenic Escherichia coli infection | 4 | 1.52 | ||
| hsa01130 | Biosynthesis of antibiotics | 45 | 2.15 | hsa00590 | Arachidonic acid metabolism | 4 | 1.32 | ||
| hsa05016 | Huntington’s disease | 38 | 1.42 | Green | |||||
| hsa04932 | Non-alcoholic fatty liver disease (NAFLD) | 36 | 2.60 | hsa04151 | PI3K-Akt signaling pathway | 16 | 1.30 | ||
| hsa05168 | Herpes simplex infection | 36 | 1.33 | hsa04144 | Endocytosis | 15 | 2.22 | ||
| hsa00190 | Oxidative phosphorylation | 31 | 2.13 | hsa04141 | Protein processing in endoplasmic reticulum | 13 | 2.66 | ||
| hsa04110 | Cell cycle | 30 | 2.30 | hsa05166 | HTLV-I infection | 13 | 1.35 | ||
| hsa04380 | Osteoclast differentiation | 30 | 1.96 | hsa04510 | Focal adhesion | 12 | 1.60 | ||
| hsa03040 | Spliceosome | 30 | 1.87 | hsa04380 | Osteoclast differentiation | 11 | 2.52 | ||
| hsa05012 | Parkinson’s disease | 30 | 1.51 | hsa04640 | Hematopoietic cell lineage | 9 | 2.61 | ||
| hsa05161 | Hepatitis B | 30 | 1.40 | hsa04722 | Neurotrophin signaling pathway | 9 | 1.78 | ||
| hsa04142 | Lysosome | 27 | 1.65 | hsa05220 | Chronic myeloid leukemia | 8 | 2.48 | ||
| hsa00240 | Pyrimidine metabolism | 25 | 2.09 | hsa05230 | Central carbon metabolism in cancer | 7 | 2.11 | ||
| hsa01200 | Carbon metabolism | 25 | 1.51 | hsa05212 | Pancreatic cancer | 7 | 2.08 | ||
| hsa04660 | T cell receptor signaling pathway | 23 | 1.58 | hsa05100 | Bacterial invasion of epithelial cells | 7 | 1.71 | ||
| hsa05132 | Salmonella infection | 22 | 2.21 | hsa05132 | Salmonella infection | 7 | 1.59 | ||
| hsa05323 | Rheumatoid arthritis | 20 | 1.36 | hsa04210 | Apoptosis | 6 | 1.57 | ||
| hsa03018 | RNA degradation | 19 | 1.62 | hsa04662 | B cell receptor signaling pathway | 6 | 1.40 | ||
| hsa04210 | Apoptosis | 18 | 2.26 | hsa04962 | Vasopressin-regulated water reabsorption | 5 | 1.50 | ||
| hsa05131 | Shigellosis | 18 | 2.11 | hsa00510 | N-Glycan biosynthesis | 5 | 1.36 | ||
| hsa00510 | N-Glycan biosynthesis | 16 | 2.54 | magenta | |||||
| hsa05221 | Acute myeloid leukemia | 15 | 1.60 | hsa04670 | Leukocyte transendothelial migration | 7 | 2.60 | ||
| hsa05134 | Legionellosis | 14 | 1.39 | hsa04142 | Lysosome | 7 | 2.48 | ||
| hsa00280 | Valine, leucine and isoleucine degradation | 13 | 1.50 | hsa04810 | Regulation of actin cytoskeleton | 7 | 1.39 | ||
| hsa00520 | Amino sugar and nucleotide sugar metabolism | 13 | 1.43 | hsa04015 | Rap1 signaling pathway | 7 | 1.39 | ||
| hsa05340 | Primary immunodeficiency | 12 | 2.19 | hsa03008 | Ribosome biogenesis in eukaryotes | 6 | 2.41 | ||
| hsa00071 | Fatty acid degradation | 12 | 1.49 | hsa05131 | Shigellosis | 5 | 2.14 | ||
| hsa00640 | Propanoate metabolism | 10 | 1.86 | hsa04520 | Adherens junction | 5 | 1.98 | ||
| hsa03060 | Protein export | 9 | 1.92 | hsa05100 | Bacterial invasion of epithelial cells | 5 | 1.84 | ||
| Green | hsa05132 | Salmonella infection | 5 | 1.75 | |||||
| hsa05166 | HTLV-I infection | 9 | 1.36 | hsa01200 | Carbon metabolism | 5 | 1.33 | ||
| hsa03040 | Spliceosome | 8 | 2.34 | hsa04710 | Circadian rhythm | 4 | 2.22 | ||
| hsa05010 | Alzheimer’s disease | 8 | 1.81 | hsa05130 | Pathogenic Escherichia coli infection | 4 | 1.63 | ||
| hsa04110 | Cell cycle | 6 | 1.36 | hsa04621 | NOD-like receptor signaling pathway | 4 | 1.53 | ||
| hsa00310 | Lysine degradation | 5 | 2.07 | Red | |||||
| hsa04115 | p53 signaling pathway | 5 | 1.70 | hsa01100 | Metabolic pathways | 36 | 3.34 | ||
| Greenyellow | hsa05010 | Alzheimer’s disease | 13 | 4.71 | |||||
| hsa04650 | Natural killer cell mediated cytotoxicity | 19 | 14.53 | hsa05016 | Huntington’s disease | 13 | 4.14 | ||
| hsa04612 | Antigen processing and presentation | 15 | 12.70 | hsa00190 | Oxidative phosphorylation | 12 | 4.96 | ||
| hsa04060 | Cytokine-cytokine receptor interaction | 11 | 3.16 | hsa05012 | Parkinson’s disease | 12 | 4.69 | ||
| hsa04062 | Chemokine signaling pathway | 7 | 1.59 | hsa04932 | Non-alcoholic fatty liver disease (NAFLD) | 9 | 2.46 | ||
| hsa05142 | Chagas disease (American trypanosomiasis) | 5 | 1.42 | hsa03010 | Ribosome | 8 | 2.14 | ||
| hsa05332 | Graft-versus-host disease | 4 | 2.13 | hsa03050 | Proteasome | 4 | 1.44 | ||
| hsa05330 | Allograft rejection | 4 | 1.99 | hsa00520 | Amino sugar and nucleotide sugar metabolism | 4 | 1.35 | ||
| hsa04940 | Type I diabetes mellitus | 4 | 1.84 | Tan | |||||
| hsa05321 | Inflammatory bowel disease (IBD) | 4 | 1.37 | hsa01100 | Metabolic pathways | 147 | 2.23 | ||
| Magenta | hsa04120 | Ubiquitin mediated proteolysis | 36 | 7.03 | |||||
| hsa04010 | MAPK signaling pathway | 7 | 1.45 | hsa03013 | RNA transport | 33 | 3.48 | ||
| hsa04664 | Fc epsilon RI signaling pathway | 4 | 1.55 | hsa04141 | Protein processing in endoplasmic reticulum | 30 | 2.64 | ||
| Pink | hsa03040 | Spliceosome | 25 | 2.57 | |||||
| hsa05168 | Herpes simplex infection | 8 | 1.91 | hsa04110 | Cell cycle | 22 | 1.99 | ||
| hsa04931 | Insulin resistance | 6 | 1,80 | hsa03018 | RNA degradation | 21 | 4,38 | ||
| hsa04145 | Phagosome | 7 | 1,78 | hsa05161 | Hepatitis B | 21 | 1,09 | ||
| hsa00190 | Oxidative phosphorylation | 6 | 1,46 | hsa04114 | Oocyte meiosis | 18 | 1,33 | ||
| Red | hsa03015 | mRNA surveillance pathway | 17 | 1,78 | |||||
| hsa01100 | Metabolic pathways | 37 | 2.07 | hsa04070 | Phosphatidylinositol signaling system | 17 | 1.50 | ||
| hsa04114 | Oocyte meiosis | 8 | 2.17 | hsa04668 | TNF signaling pathway | 17 | 1.20 | ||
| hsa04120 | Ubiquitin mediated proteolysis | 8 | 1.70 | hsa04066 | HIF-1 signaling pathway | 15 | 1.04 | ||
| hsa04668 | TNF signaling pathway | 7 | 1.69 | hsa04720 | Long-term potentiation | 14 | 1.93 | ||
| hsa01200 | Carbon metabolism | 7 | 1.59 | hsa04115 | p53 signaling pathway | 13 | 1.51 | ||
| hsa04722 | Neurotrophin signaling pathway | 7 | 1.48 | hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | 13 | 1.51 | ||
| hsa04666 | Fc gamma R-mediated phagocytosis | 6 | 1.58 | hsa04210 | Apoptosis | 12 | 1.40 | ||
| hsa05230 | Central carbon metabolism in cancer | 5 | 1.41 | hsa00562 | Inositol phosphate metabolism | 12 | 1.04 | ||
| hsa05211 | Renal cell carcinoma | 5 | 1.37 | hsa00520 | Amino sugar and nucleotide sugar metabolism | 11 | 1.75 | ||
| hsa00010 | Glycolysis / Gluconeogenesis | 5 | 1.35 | hsa05130 | Pathogenic Escherichia coli infection | 11 | 1.58 | ||
| hsa04662 | B cell receptor signaling pathway | 5 | 1.31 | hsa05110 | Vibrio cholerae infection | 11 | 1.52 | ||
| hsa00512 | Mucin type O-Glycan biosynthesis | 4 | 1.63 | hsa00510 | N-Glycan biosynthesis | 10 | 1.30 | ||
| hsa00620 | Pyruvate metabolism | 4 | 1.34 | hsa00280 | Valine, leucine and isoleucine degradation | 9 | 1.05 | ||
| Yellow | hsa03420 | Nucleotide excision repair | 9 | 1.05 | |||||
| hsa05152 | Tuberculosis | 21 | 4.97 | hsa03060 | Protein export | 8 | 2.26 | ||
| hsa04142 | Lysosome | 19 | 6.25 | hsa03430 | Mismatch repair | 6 | 1.15 | ||
| hsa04145 | Phagosome | 19 | 4.88 | Yellow | |||||
| hsa05164 | Influenza A | 17 | 3.05 | hsa05166 | HTLV-I infection | 16 | 2.96 | ||
| hsa05166 | HTLV-I infection | 17 | 1.50 | hsa05152 | Tuberculosis | 11 | 2.00 | ||
| hsa04380 | Osteoclast differentiation | 16 | 3.92 | hsa04010 | MAPK signaling pathway | 11 | 1.08 | ||
| hsa05162 | Measles | 15 | 3.31 | hsa04145 | Phagosome | 10 | 2.01 | ||
| hsa01130 | Biosynthesis of antibiotics | 15 | 1.52 | hsa05168 | Herpes simplex infection | 10 | 1.50 | ||
| hsa04640 | Hematopoietic cell lineage | 14 | 4.68 | hsa05203 | Viral carcinogenesis | 10 | 1.24 | ||
| hsa05140 | Leishmaniasis | 13 | 4.92 | hsa05161 | Hepatitis B | 9 | 1.64 | ||
| hsa05323 | Rheumatoid arthritis | 13 | 3.96 | hsa05164 | Influenza A | 9 | 1.24 | ||
| hsa05145 | Toxoplasmosis | 12 | 2.54 | hsa05140 | Leishmaniasis | 8 | 2.83 | ||
| hsa04064 | NF-kappa B signaling pathway | 11 | 2.80 | hsa04660 | T cell receptor signaling pathway | 8 | 2.00 | ||
| hsa05150 | Staphylococcus aureus infection | 10 | 3.77 | hsa05169 | Epstein-Barr virus infection | 8 | 1.57 | ||
| hsa04066 | HIF-1 signaling pathway | 10 | 1.99 | hsa05162 | Measles | 8 | 1.39 | ||
| hsa04620 | Toll-like receptor signaling pathway | 10 | 1.72 | hsa04612 | Antigen processing and presentation | 7 | 2.02 | ||
| hsa04612 | Antigen processing and presentation | 9 | 2.11 | hsa05145 | Toxoplasmosis | 7 | 1.32 | ||
| hsa04666 | Fc gamma R-mediated phagocytosis | 9 | 1.86 | hsa03040 | Spliceosome | 7 | 1.00 | ||
| hsa04660 | T cell receptor signaling pathway | 9 | 1.45 | hsa05332 | Graft-versus-host disease | 6 | 2.99 | ||
| hsa04672 | Intestinal immune network for IgA production | 8 | 2.75 | hsa05330 | Allograft rejection | 6 | 2.76 | ||
| hsa05134 | Legionellosis | 8 | 2.40 | hsa04940 | Type I diabetes mellitus | 6 | 2.51 | ||
| hsa05321 | Inflammatory bowel disease (IBD) | 8 | 1.99 | hsa03050 | Proteasome | 6 | 2.42 | ||
| hsa01230 | Biosynthesis of amino acids | 8 | 1.73 | hsa05320 | Autoimmune thyroid disease | 6 | 2.11 | ||
| hsa05133 | Pertussis | 8 | 1.64 | hsa05416 | Viral myocarditis | 6 | 1.94 | ||
| hsa05204 | Chemical carcinogenesis | 8 | 1.50 | hsa05323 | Rheumatoid arthritis | 6 | 1.22 | ||
| hsa00480 | Glutathione metabolism | 7 | 1.92 | hsa05310 | Asthma | 5 | 2.27 | ||
| hsa05416 | Viral myocarditis | 7 | 1.69 | hsa04672 | Intestinal immune network for IgA production | 5 | 1.59 | ||
| hsa05310 | Asthma | 6 | 2.31 | hsa05223 | Non-small cell lung cancer | 5 | 1.35 | ||
| hsa05332 | Graft-versus-host disease | 5 | 1.46 | hsa05321 | Inflammatory bowel disease (IBD) | 5 | 1.17 | ||
| hsa00920 | Sulfur metabolism | 4 | 2.42 | hsa04662 | B cell receptor signaling pathway | 5 | 1.08 | ||
| hsa00511 | Other glycan degradation | 4 | 1.54 | hsa01230 | Biosynthesis of amino acids | 5 | 1.03 | ||
| hsa03022 | Basal transcription factors | 4 | 1.03 |
Footnotes
Source of support: This work was supported by the Natural Science Foundation of China (81770481 and 51890894), the Natural Science Foundation of Beijing (7172171), and the CAMS Innovation Fund for Medical Sciences (CIFMS, 2017-I2M-1-008)
Supplemenatry Table 2: GO-BP terms for modules of events group and no events group.
Supplementary/raw data available from the corresponding author on request.
References
- 1.Libby P, Buring JE, Badimon L, et al. Atherosclerosis. Nat Rev Dis Primers. 2019;5:56. doi: 10.1038/s41572-019-0106-z. [DOI] [PubMed] [Google Scholar]
- 2.Hahn C, Schwartz MA. Mechanotransduction in vascular physiology and atherogenesis. Nat Rev Mol Cell Biol. 2009;10:53–62. doi: 10.1038/nrm2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saba L, Saam T, Jager HR, et al. Imaging biomarkers of vulnerable carotid plaques for stroke risk prediction and their potential clinical implications. Lancet Neurol. 2019;18:559–72. doi: 10.1016/S1474-4422(19)30035-3. [DOI] [PubMed] [Google Scholar]
- 4.Eckstein HH. European Society for Vascular Surgery guidelines on the management of atherosclerotic carotid and vertebral artery disease. Eur J Vasc Endovasc Surg. 2018;55:1–2. doi: 10.1016/j.ejvs.2017.06.026. [DOI] [PubMed] [Google Scholar]
- 5.Kernan WN, Ovbiagele B, Black HR, et al. Guidelines for the prevention of stroke in patients with stroke and transient ischemic attack: A guideline for healthcare professionals from the American Heart Association/American Stroke Association. Stroke. 2014;45:2160–236. doi: 10.1161/STR.0000000000000024. [DOI] [PubMed] [Google Scholar]
- 6.Spence JD. Asymptomatic carotid stenosis. Circulation. 2013;127:739–42. doi: 10.1161/CIRCULATIONAHA.112.153734. [DOI] [PubMed] [Google Scholar]
- 7.Stroes ES, Thompson PD, Corsini A, et al. Statin-associated muscle symptoms: Impact on statin therapy – European Atherosclerosis Society Consensus Panel Statement on Assessment, Aetiology and Management. Eur Heart J. 2015;36:1012–22. doi: 10.1093/eurheartj/ehv043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michelson AD, Cattaneo M, Eikelboom JW, et al. Aspirin resistance: Position paper of the Working Group on Aspirin Resistance. J Thromb Haemost. 2005;3:1309–11. doi: 10.1111/j.1538-7836.2005.01351.x. [DOI] [PubMed] [Google Scholar]
- 9.Weber C, Noels H. Atherosclerosis: Current pathogenesis and therapeutic options. Nat Med. 2011;17:1410–22. doi: 10.1038/nm.2538. [DOI] [PubMed] [Google Scholar]
- 10.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Dam S, Vosa U, van der Graaf A, et al. Gene co-expression analysis for functional classification and gene-disease predictions. Brief Bioinform. 2018;19:575–92. doi: 10.1093/bib/bbw139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 13.Hu Z, Yang D, Tang Y, et al. Five-long non-coding RNA risk score system for the effective prediction of gastric cancer patient survival. Oncol Lett. 2019;17:4474–86. doi: 10.3892/ol.2019.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly RS, Chawes BL, Blighe K, et al. An integrative transcriptomic and metabolomic study of lung function in children with asthma. Chest. 2018;154:335–48. doi: 10.1016/j.chest.2018.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma X, Tao R, Li L, et al. Identification of a 5microRNA signature and hub miRNAmRNA interactions associated with pancreatic cancer. Oncol Rep. 2019;41:292–300. doi: 10.3892/or.2018.6820. [DOI] [PubMed] [Google Scholar]
- 16.Razuvaev A, Ekstrand J, Folkersen L, et al. Correlations between clinical variables and gene-expression profiles in carotid plaque instability. Eur J Vasc Endovasc Surg. 2011;42:722–30. doi: 10.1016/j.ejvs.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 17.Folkersen L, Persson J, Ekstrand J, et al. Prediction of ischemic events on the basis of transcriptomic and genomic profiling in patients undergoing carotid endarterectomy. Mol Med (Cambridge, Mass) 2012;18:669–75. doi: 10.2119/molmed.2011.00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perisic L, Aldi S, Sun Y, et al. Gene expression signatures, pathways and networks in carotid atherosclerosis. J Intern Med. 2016;279:293–308. doi: 10.1111/joim.12448. [DOI] [PubMed] [Google Scholar]
- 19.Deshpande V, Sharma A, Mukhopadhyay R, et al. Understanding the progression of atherosclerosis through gene profiling and co-expression network analysis in Apob(tm2Sgy)Ldlr(tm1Her) double knockout mice. Genomics. 2016;107:239–47. doi: 10.1016/j.ygeno.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 20.Barr TL, VanGilder RL, Seiberg R, et al. Systemic transcriptional alterations of innate and adaptive immune signaling pathways in atherosclerosis, ischemia stroke, and myocardial infarction. J Bioanal Biomed. 2015;7:29–34. doi: 10.4172/1948-593X.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Sun R, Liu L. Potentially critical roles of TNPO1, RAP1B, ZDHHC17, and PPM1B in the progression of coronary atherosclerosis through microarray data analysis. J Cell Biochem. 2019;120:4301–11. doi: 10.1002/jcb.27715. [DOI] [PubMed] [Google Scholar]
- 22.Wang CH, Shi HH, Chen LH, et al. Identification of key lncRNAs associated with atherosclerosis progression based on public datasets. Front Genet. 2019;10:123. doi: 10.3389/fgene.2019.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogacev KS, Cremers B, Zawada AM, et al. CD14++CD16+ monocytes independently predict cardiovascular events: A cohort study of 951 patients referred for elective coronary angiography. J Am Coll Cardiol. 2012;60:1512–20. doi: 10.1016/j.jacc.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 24.Swirski FK, Libby P, Aikawa E, et al. Ly-6Chi monocytes dominate hypercholesterolemia-associated monocytosis and give rise to macrophages in atheromata. J Clin Invest. 2007;117:195–205. doi: 10.1172/JCI29950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belge KU, Dayyani F, Horelt A, et al. The proinflammatory CD14+CD16+DR++ monocytes are a major source of TNF. J Immunol (Baltimore, Md: 1950) 2002;168:3536–42. doi: 10.4049/jimmunol.168.7.3536. [DOI] [PubMed] [Google Scholar]
- 26.Boltjes A, van Wijk F. Human dendritic cell functional specialization in steady-state and inflammation. Front Immunol. 2014;5:131. doi: 10.3389/fimmu.2014.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu K, Victora GD, Schwickert TA, et al. In vivo analysis of dendritic cell development and homeostasis. Science (New York, NY) 2009;324:392–97. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans HG, Gullick NJ, Kelly S, et al. In vivo activated monocytes from the site of inflammation in humans specifically promote Th17 responses. Proc Natl Acad Sci USA. 2009;106:6232–37. doi: 10.1073/pnas.0808144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Justo-Junior AS, Villarejos LM, Lima XTV, et al. Monocytes of patients with unstable angina express high levels of chemokine and pattern-recognition receptors. Cytokine. 2019;113:61–67. doi: 10.1016/j.cyto.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 30.Bruen R, Curley S, Kajani S, et al. Different monocyte phenotypes result in proresolving macrophages in conjugated linoleic acid-induced attenuated progression and regression of atherosclerosis. FASEB J. 2019;33:11006–20. doi: 10.1096/fj.201900922R. [DOI] [PubMed] [Google Scholar]
- 31.Sun H, Jiang J, Gong L, et al. Voltage-gated sodium channel inhibitor reduces atherosclerosis by modulating monocyte/macrophage subsets and suppressing macrophage proliferation. Biomed Pharmacother. 2019;120:109352. doi: 10.1016/j.biopha.2019.109352. [DOI] [PubMed] [Google Scholar]
- 32.Gasbarrino K, Hafiane A, Zheng H, Daskalopoulou SS. Intensive statin therapy compromises the adiponectin–adipoR pathway in the human monocyte-macrophage lineage. Stroke. 2019;50:3609–17. doi: 10.1161/STROKEAHA.119.026280. [DOI] [PubMed] [Google Scholar]
- 33.Estefania E, Flores R, Gomez-Lozano N, et al. Human KIR2DL5 is an inhibitory receptor expressed on the surface of NK and T lymphocyte subsets. J Immunol (Baltimore, Md: 1950) 2007;178:4402–10. doi: 10.4049/jimmunol.178.7.4402. [DOI] [PubMed] [Google Scholar]
- 34.Shan Z, Huang J, Liao Q, et al. Association of killer cell immunoglobulin-like receptors with spontaneous clearance of hepatitis C virus in the Chinese population. Transfusion. 2018;58:1028–35. doi: 10.1111/trf.14527. [DOI] [PubMed] [Google Scholar]
- 35.De Re V, Caggiari L, De Zorzi M, et al. Genetic diversity of the KIR/HLA system and outcome of patients with metastatic colorectal cancer treated with chemotherapy. PLoS One. 2014;9:e84940. doi: 10.1371/journal.pone.0084940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alomar SY, Alkhuriji A, Trayhyrn P, et al. Association of the genetic diversity of killer cell immunoglobulin-like receptor genes and HLA-C ligand in Saudi women with breast cancer. Immunogenetics. 2017;69:69–76. doi: 10.1007/s00251-016-0950-x. [DOI] [PubMed] [Google Scholar]
- 37.Escudero A, Martinez-Romera I, Fernandez L, et al. Donor KIR genotype impacts on clinical outcome after T cell-depleted HLA matched related allogeneic transplantation for high-risk pediatric leukemia patients. Biol Blood Marrow Transplant. 2018;24:2493–500. doi: 10.1016/j.bbmt.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 38.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–38. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brunet A, Sweeney LB, Sturgill JF, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science (New York, NY) 2004;303:2011–15. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 40.Zeng HT, Fu YC, Yu W, et al. SIRT1 prevents atherosclerosis via liverXreceptor and NFkappaB signaling in a U937 cell model. Mol Med Rep. 2013;8:23–28. doi: 10.3892/mmr.2013.1460. [DOI] [PubMed] [Google Scholar]
- 41.Bonkowski MS, Sinclair DA. Slowing ageing by design: the rise of NAD(+) and sirtuin-activating compounds. Nat Rev Mol Cell Biol. 2016;17:679–690. doi: 10.1038/nrm.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imai S, Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014;24:464–71. doi: 10.1016/j.tcb.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sosnowska B, Mazidi M, Penson P, et al. The sirtuin family members SIRT1, SIRT3 and SIRT6: Their role in vascular biology and atherogenesis. Atherosclerosis. 2017;265:275–82. doi: 10.1016/j.atherosclerosis.2017.08.027. [DOI] [PubMed] [Google Scholar]
- 44.Lee SJ, Baek SE, Jang MA, Kim CD. SIRT1 inhibits monocyte adhesion to the vascular endothelium by suppressing Mac-1 expression on monocytes. Exp Mol Med. 2019;51:47. doi: 10.1038/s12276-019-0239-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen PA, Won JS, Rahman MK, et al. Modulation of Sirt1/NF-kappaB interaction of evogliptin is attributed to inhibition of vascular inflammatory response leading to attenuation of atherosclerotic plaque formation. Biochem Pharmacol. 2019;168:452–64. doi: 10.1016/j.bcp.2019.08.008. [DOI] [PubMed] [Google Scholar]
- 46.Beli P, Lukashchuk N, Wagner SA, et al. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol Cell. 2012;46:212–25. doi: 10.1016/j.molcel.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi JH, Choi SS, Kim ES, et al. Thrap3 docks on phosphoserine 273 of PPARgamma and controls diabetic gene programming. Genes Dev. 2014;28:2361–69. doi: 10.1101/gad.249367.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ino Y, Arakawa N, Ishiguro H, et al. Phosphoproteome analysis demonstrates the potential role of THRAP3 phosphorylation in androgen-independent prostate cancer cell growth. Proteomics. 2016;16:1069–78. doi: 10.1002/pmic.201500365. [DOI] [PubMed] [Google Scholar]
- 49.Vohhodina J, Barros EM, Savage AL, et al. The RNA processing factors THRAP3 and BCLAF1 promote the DNA damage response through selective mRNA splicing and nuclear export. Nucleic Acids Res. 2017;45:12816–33. doi: 10.1093/nar/gkx1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
WGCNA of no-event. (A) No outlier was detected by sample clustering. (B, C) Selection of soft-threshold β. (D, E) Fitness of scale free topology when β-16. (F) Cluster dendrogram. Each module was represented by WGCNA.
Supplementary Table 1.
Top10 up-regulated and down-regulated DEGs.
| Gene symbol | Official full gene name | log2 (fold-change) (patients with events/patients without events) |
|---|---|---|
| Up-regulated | ||
| TNFAIP6 | TNF alpha induced protein 6 | 9.968020902 |
| PTX3 | Pentraxin 3 | 8.826641354 |
| RNASE2 | Ribonuclease A family member 2 | 7.978917622 |
| KCNJ2 | Potassium inwardly rectifying channel subfamily J member 2 | 7.481378497 |
| SERPINB2 | Serpin family B member 2 | 7.18837765 |
| PLA2G7 | Phospholipase A2 group VII | 6.901998631 |
| BCL2A1 | BCL2 related protein A1 | 6.719719825 |
| CLEC4D | C-type lectin domain family 4 member D | 6.272368641 |
| SAMSN1 | SAM domain, SH3 domain and nuclear localization signals 1 | 6.209244405 |
| GPR84 | G protein-coupled receptor 84 | 5.186451434 |
| Down-regulated | ||
| KLRC3 | Killer cell lectin like receptor C3 | −7.246559422 |
| BTN3A2 | Butyrophilin subfamily 3 member A2 | −7.003853494 |
| ANKRD20A11P | Ankyrin repeat domain 20 family member A11, pseudogene | −6.02418399 |
| ZNF600 | Zinc finger protein 600 | −5.985685983 |
| NLRC3 | NLR family CARD domain containing 3 | −5.579931019 |
| LCK | LCK proto-oncogene, Src family tyrosine kinase | −4.825468373 |
| GOLGA8N | Golgin A8 family member N | −4.799854266 |
| CEP78 | Centrosomal protein 78 | −4.795917474 |
| SLC9A3R1 | SLC9A3 regulator 1 | −4.381636674 |
| SEP1 | Septin 1 | −4.097540674 |
Supplemenatry Table 3.
KEGG pathways for modules of events group and no-events group.
| Events group | KEGG ID | KEGG pathway | Count | −logP | No-events group | KEGG ID | KEGG pathway | Count | −logP |
|---|---|---|---|---|---|---|---|---|---|
| Black | Black | ||||||||
| hsa05034 | Alcoholism | 14 | 4.58 | hsa04611 | Platelet activation | 11 | 5.10 | ||
| hsa05322 | Systemic lupus erythematosus | 13 | 5.13 | hsa05322 | Systemic lupus erythematosus | 11 | 4.98 | ||
| hsa04611 | Platelet activation | 11 | 3.80 | hsa05034 | Alcoholism | 11 | 3.94 | ||
| hsa05203 | Viral carcinogenesis | 10 | 1.82 | hsa04512 | ECM-receptor interaction | 8 | 3.83 | ||
| hsa05202 | Transcriptional misregulation in cancer | 9 | 1.87 | hsa05203 | Viral carcinogenesis | 8 | 1.71 | ||
| hsa04062 | Chemokine signaling pathway | 9 | 1.62 | hsa04510 | Focal adhesion | 8 | 1.70 | ||
| hsa04512 | ECM-receptor interaction | 6 | 1.62 | hsa04810 | Regulation of actin cytoskeleton | 8 | 1.66 | ||
| hsa04540 | Gap junction | 6 | 1.60 | hsa04540 | Gap junction | 6 | 2.20 | ||
| hsa05219 | Bladder cancer | 4 | 1.38 | hsa04670 | Leukocyte transendothelial migration | 6 | 1.73 | ||
| Blue | hsa05410 | Hypertrophic cardiomyopathy (HCM) | 5 | 1.70 | |||||
| hsa01100 | Metabolic pathways | 218 | 3.72 | hsa05414 | Dilated cardiomyopathy | 5 | 1.59 | ||
| hsa05166 | HTLV-I infection | 49 | 1.56 | hsa04640 | Hematopoietic cell lineage | 5 | 1.54 | ||
| hsa04144 | Endocytosis | 48 | 1.76 | hsa04530 | Tight junction | 5 | 1.54 | ||
| hsa04141 | Protein processing in endoplasmic reticulum | 45 | 4.34 | hsa05130 | Pathogenic Escherichia coli infection | 4 | 1.52 | ||
| hsa01130 | Biosynthesis of antibiotics | 45 | 2.15 | hsa00590 | Arachidonic acid metabolism | 4 | 1.32 | ||
| hsa05016 | Huntington’s disease | 38 | 1.42 | Green | |||||
| hsa04932 | Non-alcoholic fatty liver disease (NAFLD) | 36 | 2.60 | hsa04151 | PI3K-Akt signaling pathway | 16 | 1.30 | ||
| hsa05168 | Herpes simplex infection | 36 | 1.33 | hsa04144 | Endocytosis | 15 | 2.22 | ||
| hsa00190 | Oxidative phosphorylation | 31 | 2.13 | hsa04141 | Protein processing in endoplasmic reticulum | 13 | 2.66 | ||
| hsa04110 | Cell cycle | 30 | 2.30 | hsa05166 | HTLV-I infection | 13 | 1.35 | ||
| hsa04380 | Osteoclast differentiation | 30 | 1.96 | hsa04510 | Focal adhesion | 12 | 1.60 | ||
| hsa03040 | Spliceosome | 30 | 1.87 | hsa04380 | Osteoclast differentiation | 11 | 2.52 | ||
| hsa05012 | Parkinson’s disease | 30 | 1.51 | hsa04640 | Hematopoietic cell lineage | 9 | 2.61 | ||
| hsa05161 | Hepatitis B | 30 | 1.40 | hsa04722 | Neurotrophin signaling pathway | 9 | 1.78 | ||
| hsa04142 | Lysosome | 27 | 1.65 | hsa05220 | Chronic myeloid leukemia | 8 | 2.48 | ||
| hsa00240 | Pyrimidine metabolism | 25 | 2.09 | hsa05230 | Central carbon metabolism in cancer | 7 | 2.11 | ||
| hsa01200 | Carbon metabolism | 25 | 1.51 | hsa05212 | Pancreatic cancer | 7 | 2.08 | ||
| hsa04660 | T cell receptor signaling pathway | 23 | 1.58 | hsa05100 | Bacterial invasion of epithelial cells | 7 | 1.71 | ||
| hsa05132 | Salmonella infection | 22 | 2.21 | hsa05132 | Salmonella infection | 7 | 1.59 | ||
| hsa05323 | Rheumatoid arthritis | 20 | 1.36 | hsa04210 | Apoptosis | 6 | 1.57 | ||
| hsa03018 | RNA degradation | 19 | 1.62 | hsa04662 | B cell receptor signaling pathway | 6 | 1.40 | ||
| hsa04210 | Apoptosis | 18 | 2.26 | hsa04962 | Vasopressin-regulated water reabsorption | 5 | 1.50 | ||
| hsa05131 | Shigellosis | 18 | 2.11 | hsa00510 | N-Glycan biosynthesis | 5 | 1.36 | ||
| hsa00510 | N-Glycan biosynthesis | 16 | 2.54 | magenta | |||||
| hsa05221 | Acute myeloid leukemia | 15 | 1.60 | hsa04670 | Leukocyte transendothelial migration | 7 | 2.60 | ||
| hsa05134 | Legionellosis | 14 | 1.39 | hsa04142 | Lysosome | 7 | 2.48 | ||
| hsa00280 | Valine, leucine and isoleucine degradation | 13 | 1.50 | hsa04810 | Regulation of actin cytoskeleton | 7 | 1.39 | ||
| hsa00520 | Amino sugar and nucleotide sugar metabolism | 13 | 1.43 | hsa04015 | Rap1 signaling pathway | 7 | 1.39 | ||
| hsa05340 | Primary immunodeficiency | 12 | 2.19 | hsa03008 | Ribosome biogenesis in eukaryotes | 6 | 2.41 | ||
| hsa00071 | Fatty acid degradation | 12 | 1.49 | hsa05131 | Shigellosis | 5 | 2.14 | ||
| hsa00640 | Propanoate metabolism | 10 | 1.86 | hsa04520 | Adherens junction | 5 | 1.98 | ||
| hsa03060 | Protein export | 9 | 1.92 | hsa05100 | Bacterial invasion of epithelial cells | 5 | 1.84 | ||
| Green | hsa05132 | Salmonella infection | 5 | 1.75 | |||||
| hsa05166 | HTLV-I infection | 9 | 1.36 | hsa01200 | Carbon metabolism | 5 | 1.33 | ||
| hsa03040 | Spliceosome | 8 | 2.34 | hsa04710 | Circadian rhythm | 4 | 2.22 | ||
| hsa05010 | Alzheimer’s disease | 8 | 1.81 | hsa05130 | Pathogenic Escherichia coli infection | 4 | 1.63 | ||
| hsa04110 | Cell cycle | 6 | 1.36 | hsa04621 | NOD-like receptor signaling pathway | 4 | 1.53 | ||
| hsa00310 | Lysine degradation | 5 | 2.07 | Red | |||||
| hsa04115 | p53 signaling pathway | 5 | 1.70 | hsa01100 | Metabolic pathways | 36 | 3.34 | ||
| Greenyellow | hsa05010 | Alzheimer’s disease | 13 | 4.71 | |||||
| hsa04650 | Natural killer cell mediated cytotoxicity | 19 | 14.53 | hsa05016 | Huntington’s disease | 13 | 4.14 | ||
| hsa04612 | Antigen processing and presentation | 15 | 12.70 | hsa00190 | Oxidative phosphorylation | 12 | 4.96 | ||
| hsa04060 | Cytokine-cytokine receptor interaction | 11 | 3.16 | hsa05012 | Parkinson’s disease | 12 | 4.69 | ||
| hsa04062 | Chemokine signaling pathway | 7 | 1.59 | hsa04932 | Non-alcoholic fatty liver disease (NAFLD) | 9 | 2.46 | ||
| hsa05142 | Chagas disease (American trypanosomiasis) | 5 | 1.42 | hsa03010 | Ribosome | 8 | 2.14 | ||
| hsa05332 | Graft-versus-host disease | 4 | 2.13 | hsa03050 | Proteasome | 4 | 1.44 | ||
| hsa05330 | Allograft rejection | 4 | 1.99 | hsa00520 | Amino sugar and nucleotide sugar metabolism | 4 | 1.35 | ||
| hsa04940 | Type I diabetes mellitus | 4 | 1.84 | Tan | |||||
| hsa05321 | Inflammatory bowel disease (IBD) | 4 | 1.37 | hsa01100 | Metabolic pathways | 147 | 2.23 | ||
| Magenta | hsa04120 | Ubiquitin mediated proteolysis | 36 | 7.03 | |||||
| hsa04010 | MAPK signaling pathway | 7 | 1.45 | hsa03013 | RNA transport | 33 | 3.48 | ||
| hsa04664 | Fc epsilon RI signaling pathway | 4 | 1.55 | hsa04141 | Protein processing in endoplasmic reticulum | 30 | 2.64 | ||
| Pink | hsa03040 | Spliceosome | 25 | 2.57 | |||||
| hsa05168 | Herpes simplex infection | 8 | 1.91 | hsa04110 | Cell cycle | 22 | 1.99 | ||
| hsa04931 | Insulin resistance | 6 | 1,80 | hsa03018 | RNA degradation | 21 | 4,38 | ||
| hsa04145 | Phagosome | 7 | 1,78 | hsa05161 | Hepatitis B | 21 | 1,09 | ||
| hsa00190 | Oxidative phosphorylation | 6 | 1,46 | hsa04114 | Oocyte meiosis | 18 | 1,33 | ||
| Red | hsa03015 | mRNA surveillance pathway | 17 | 1,78 | |||||
| hsa01100 | Metabolic pathways | 37 | 2.07 | hsa04070 | Phosphatidylinositol signaling system | 17 | 1.50 | ||
| hsa04114 | Oocyte meiosis | 8 | 2.17 | hsa04668 | TNF signaling pathway | 17 | 1.20 | ||
| hsa04120 | Ubiquitin mediated proteolysis | 8 | 1.70 | hsa04066 | HIF-1 signaling pathway | 15 | 1.04 | ||
| hsa04668 | TNF signaling pathway | 7 | 1.69 | hsa04720 | Long-term potentiation | 14 | 1.93 | ||
| hsa01200 | Carbon metabolism | 7 | 1.59 | hsa04115 | p53 signaling pathway | 13 | 1.51 | ||
| hsa04722 | Neurotrophin signaling pathway | 7 | 1.48 | hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | 13 | 1.51 | ||
| hsa04666 | Fc gamma R-mediated phagocytosis | 6 | 1.58 | hsa04210 | Apoptosis | 12 | 1.40 | ||
| hsa05230 | Central carbon metabolism in cancer | 5 | 1.41 | hsa00562 | Inositol phosphate metabolism | 12 | 1.04 | ||
| hsa05211 | Renal cell carcinoma | 5 | 1.37 | hsa00520 | Amino sugar and nucleotide sugar metabolism | 11 | 1.75 | ||
| hsa00010 | Glycolysis / Gluconeogenesis | 5 | 1.35 | hsa05130 | Pathogenic Escherichia coli infection | 11 | 1.58 | ||
| hsa04662 | B cell receptor signaling pathway | 5 | 1.31 | hsa05110 | Vibrio cholerae infection | 11 | 1.52 | ||
| hsa00512 | Mucin type O-Glycan biosynthesis | 4 | 1.63 | hsa00510 | N-Glycan biosynthesis | 10 | 1.30 | ||
| hsa00620 | Pyruvate metabolism | 4 | 1.34 | hsa00280 | Valine, leucine and isoleucine degradation | 9 | 1.05 | ||
| Yellow | hsa03420 | Nucleotide excision repair | 9 | 1.05 | |||||
| hsa05152 | Tuberculosis | 21 | 4.97 | hsa03060 | Protein export | 8 | 2.26 | ||
| hsa04142 | Lysosome | 19 | 6.25 | hsa03430 | Mismatch repair | 6 | 1.15 | ||
| hsa04145 | Phagosome | 19 | 4.88 | Yellow | |||||
| hsa05164 | Influenza A | 17 | 3.05 | hsa05166 | HTLV-I infection | 16 | 2.96 | ||
| hsa05166 | HTLV-I infection | 17 | 1.50 | hsa05152 | Tuberculosis | 11 | 2.00 | ||
| hsa04380 | Osteoclast differentiation | 16 | 3.92 | hsa04010 | MAPK signaling pathway | 11 | 1.08 | ||
| hsa05162 | Measles | 15 | 3.31 | hsa04145 | Phagosome | 10 | 2.01 | ||
| hsa01130 | Biosynthesis of antibiotics | 15 | 1.52 | hsa05168 | Herpes simplex infection | 10 | 1.50 | ||
| hsa04640 | Hematopoietic cell lineage | 14 | 4.68 | hsa05203 | Viral carcinogenesis | 10 | 1.24 | ||
| hsa05140 | Leishmaniasis | 13 | 4.92 | hsa05161 | Hepatitis B | 9 | 1.64 | ||
| hsa05323 | Rheumatoid arthritis | 13 | 3.96 | hsa05164 | Influenza A | 9 | 1.24 | ||
| hsa05145 | Toxoplasmosis | 12 | 2.54 | hsa05140 | Leishmaniasis | 8 | 2.83 | ||
| hsa04064 | NF-kappa B signaling pathway | 11 | 2.80 | hsa04660 | T cell receptor signaling pathway | 8 | 2.00 | ||
| hsa05150 | Staphylococcus aureus infection | 10 | 3.77 | hsa05169 | Epstein-Barr virus infection | 8 | 1.57 | ||
| hsa04066 | HIF-1 signaling pathway | 10 | 1.99 | hsa05162 | Measles | 8 | 1.39 | ||
| hsa04620 | Toll-like receptor signaling pathway | 10 | 1.72 | hsa04612 | Antigen processing and presentation | 7 | 2.02 | ||
| hsa04612 | Antigen processing and presentation | 9 | 2.11 | hsa05145 | Toxoplasmosis | 7 | 1.32 | ||
| hsa04666 | Fc gamma R-mediated phagocytosis | 9 | 1.86 | hsa03040 | Spliceosome | 7 | 1.00 | ||
| hsa04660 | T cell receptor signaling pathway | 9 | 1.45 | hsa05332 | Graft-versus-host disease | 6 | 2.99 | ||
| hsa04672 | Intestinal immune network for IgA production | 8 | 2.75 | hsa05330 | Allograft rejection | 6 | 2.76 | ||
| hsa05134 | Legionellosis | 8 | 2.40 | hsa04940 | Type I diabetes mellitus | 6 | 2.51 | ||
| hsa05321 | Inflammatory bowel disease (IBD) | 8 | 1.99 | hsa03050 | Proteasome | 6 | 2.42 | ||
| hsa01230 | Biosynthesis of amino acids | 8 | 1.73 | hsa05320 | Autoimmune thyroid disease | 6 | 2.11 | ||
| hsa05133 | Pertussis | 8 | 1.64 | hsa05416 | Viral myocarditis | 6 | 1.94 | ||
| hsa05204 | Chemical carcinogenesis | 8 | 1.50 | hsa05323 | Rheumatoid arthritis | 6 | 1.22 | ||
| hsa00480 | Glutathione metabolism | 7 | 1.92 | hsa05310 | Asthma | 5 | 2.27 | ||
| hsa05416 | Viral myocarditis | 7 | 1.69 | hsa04672 | Intestinal immune network for IgA production | 5 | 1.59 | ||
| hsa05310 | Asthma | 6 | 2.31 | hsa05223 | Non-small cell lung cancer | 5 | 1.35 | ||
| hsa05332 | Graft-versus-host disease | 5 | 1.46 | hsa05321 | Inflammatory bowel disease (IBD) | 5 | 1.17 | ||
| hsa00920 | Sulfur metabolism | 4 | 2.42 | hsa04662 | B cell receptor signaling pathway | 5 | 1.08 | ||
| hsa00511 | Other glycan degradation | 4 | 1.54 | hsa01230 | Biosynthesis of amino acids | 5 | 1.03 | ||
| hsa03022 | Basal transcription factors | 4 | 1.03 |