Abstract
Most replicated genetic determinants for type 1 diabetes are common (minor allele frequency [MAF] >5%). We aimed to identify novel rare or low-frequency (MAF <5%) single nucleotide polymorphisms with large effects on risk of type 1 diabetes. We undertook deep imputation of genotyped data followed by genome-wide association testing and meta-analysis of 9,358 type 1 diabetes case and 15,705 control subjects from 12 European cohorts. Candidate variants were replicated in a separate cohort of 4,329 case and 9,543 control subjects. Our meta-analysis identified 27 independent variants outside the MHC, among which 3 were novel and had MAF <5%. Three of these variants replicated with Preplication < 0.05 and Pcombined < Pdiscovery. In silico analysis prioritized a rare variant at 2q24.3 (rs60587303 [C], MAF 0.5%) within the first intron of STK39, with an effect size comparable with those of common variants in the INS and PTPN22 loci (combined [from the discovery and replication cohorts] estimate of odds ratio [ORcombined] 1.97, 95% CI 1.58–2.47, Pcombined = 2.9 × 10−9). Pharmacological inhibition of Stk39 activity in primary murine T cells augmented effector responses through enhancement of interleukin 2 signaling. These findings provide insight into the genetic architecture of type 1 diabetes and have identified rare variants having a large effect on disease risk.
Introduction
Type 1 diabetes accounts for 5–10% (1) of diabetes cases, affecting ∼20 million individuals worldwide, and represents the majority of diabetes diagnoses in individuals <20 years of age (2,3). Its prevalence is consistently rising worldwide at an annual rate of ∼2–5% (3,4) and it is estimated that by 2050, 5 million people, among whom more than half a million youths, will be living with type 1 diabetes in the U.S. (5). Type 1 diabetes is a chronic autoimmune disorder caused by destruction of the insulin-producing pancreatic β-cell, which generally occurs in genetically susceptible individuals (6) and may be precipitated by environmental factors (7). Thus, understanding the genetic determinants of type 1 diabetes and their effects can provide a better understanding of the pathophysiology of the disease and enable new treatment approaches, with potentially important public health benefits.
Our knowledge on the genetic architecture of type 1 diabetes has expanded substantially with the emergence of genome-wide association studies (GWAS). In the pre-GWAS era, the few genetic loci established to contribute to type 1 diabetes risk were identified primarily through linkage studies, such as the human leukocyte antigen (HLA) class II genes in the MHC, or through candidate gene studies, such as the genes encoding insulin (INS), cytotoxic T-cell–associated protein 4 (CTLA4) (8), protein tyrosine phosphatase, nonreceptor type 22 (PTPN22) (9), and interleukin 2 receptor α (IL2RA) (10). In comparison, recent GWAS comprising thousands of type 1 diabetes cases have identified 57 loci associated with risk of type 1 diabetes (11–13). However, with a few exceptions (14), the identified variants by GWAS are common (defined as variants with a minor allele frequency [MAF] >5%) and have modest effects on type 1 diabetes risk (odds ratios [ORs] <1.5) (13) compared with those of the previously identified HLA, INS, and PTPN22 variants (ORs >6 for the HLA and >2 for the INS and PTPN22 loci, respectively). Evaluation of rare and low-frequency variants for association with type 1 diabetes is an important area, and a recent study (15) showed the utility of rare variants for identifying potential causal genes in type 2 diabetes. In addition, understanding the biology of most GWAS loci remains challenging, as fine mapping and functional validation are typically lacking (13). Furthermore, the small effect sizes attributed to most common variants decreases the ability to dissect their function through approaches such as genome editing via CRISPR-Cas9, since the anticipated effect sizes on relevant phenotypes would be smaller than variants of larger effect. Therefore, larger effect size variants may help us to better understand the genetic pathways that increase risk of type 1 diabetes.
It has been shown that known loci explain ∼80% of the heritability of type 1 diabetes (16), with the HLA variants accounting for the largest portion of this variance (17). Therefore, about 20% of the genetic risk of type 1 diabetes remains unexplained (13). The rest of this unidentified genetic component has been attributed to undetected susceptibility loci (17), possibly due to the inability of past studies to interrogate loci containing low-frequency or rare genetic variants (defined as variants with an MAF ≤5% and >1% or ≤1%, respectively), despite the strong effects that some of them may have. Interestingly, among the 57 known type 1 diabetes loci, only 3 loci include low-frequency or rare variants, specifically in the tyrosine kinase 2 (TYK2), interferon induced with helicase C domain 1 (IFIH1), and RNA binding motif protein 17 (RBM17) (in the same locus as IL2RA) genes, which were identified mainly through fine mapping of previously known autoimmune loci (14,18). The recent availability of large human whole-genome sequencing data sets has enabled the interrogation of rare genetic variation by imputation from directly genotyped data, allowing identification of rare single nucleotide polymorphisms (SNPs) with large effects on complex traits (19,20). Here, we sought to apply these recent advances in genomics to type 1 diabetes, with the aim of identifying additional low-frequency and rare loci of large effect. To do so, we undertook a GWAS of ∼9 million SNPs in 9,358 type 1 diabetes case and 15,705 control subjects from 12 cohorts of predominantly European descent and replicated our findings using de novo genotyping in a separate cohort of 4,329 type 1 diabetes case subjects from the Type 1 Diabetes Genetics Consortium (T1DGC) and 9,543 control subjects from the UK Biobank. In addition, potential biological effects of the top low-frequency locus were explored using in vitro functional experiments.
Research Design and Methods
Cohorts and Overview of GWAS
For our ascertained case-control association study, similar to a previous type 1 diabetes GWAS (11), we used data from multiple type 1 diabetes case cohorts and control cohorts. In the discovery stage, the cohorts who were genotyped on Affymetrix or Illumina platforms were pooled separately and genotypes were imputed using the Haplotype Reference Consortium reference panel (21), which allows improved imputation of rare variants. Next, association studies were performed separately for each one of the two platform types. The summary statistics of the two association studies were then meta-analyzed to generate the final discovery GWAS estimates (Supplementary Fig. 1). Individual-level genotype data for the discovery meta-analysis were drawn from 12 cohorts of predominantly European descent totaling, before quality control, 9,684 type 1 diabetes case and 17,153 control subjects. Specifically, among 3,173 type 1 diabetes case subjects genotyped on the Affymetrix 500K array, 1,173 subjects were from the Genetics of Kidneys in Diabetes (GoKinD) study and 2,000 were from the Wellcome Trust Case Control Consortium (WTCCC). These were then compared with 5,998 similarly genotyped control subjects from WTCCC, specifically, 1,999 subjects from the type 2 diabetes (T2D) study, 2,001 subjects from the hypertension (HT) study, and 1,998 individuals from the bipolar disorder (BD) study. Among the 6,511 type 1 diabetes case subjects genotyped on Illumina arrays (HumanHap550 or Human1-2M-DuoCustom_v1_A), 514 individuals were from McGill University, 483 from Children's Hospital of Philadelphia (CHOPTDT), 1,385 from the Diabetes Control and Complications Trial–Epidemiology of Diabetes Interventions and Complications (DCCT-EDIC) cohort, and 4,129 from T1DGC. These were compared with 9,745 similarly genotyped control subjects recruited from TwinsUK (n = 2,249), and two WTCCC studies (2,836 individuals from the 1958 British Cohort [58BC] and 2,633 individuals from the U.K. National Blood Service sample [NBS], respectively). In the replication stage, we used 4,329 type 1 diabetes case subjects from T1DGC and 9,543 randomly selected control subjects of white-British ancestry from the UK Biobank (22).
All participating studies in the GWAS meta-analysis were approved by the institutional review boards and the ethics committees of the respective institutions. Written informed consent was obtained from each participant in accordance with institutional requirements and the Declaration of Helsinki principles.
For a detailed description of the discovery study cohorts, see Supplementary Data and Supplementary Table 1.
Quality Control and Assessment of Population Stratification
Genotype quality assessment was performed according to published guidelines (23). This quality control removed individuals with problematic sex assignment, increased genotype missingness, increased proportion of identity by descent, and extreme heterozygosity rate. It also removed low-quality genetic variants, by retaining only array SNPs with an MAF >1%, missingness <0.05, and Hardy-Weinberg equilibrium P > 1 × 10−5. In the CHOPTDT case-parent trios cohort, we retained the single type 1 diabetes case per trio for association analysis, as the model used does not adjust for familial relatedness. A detailed description of the quality-control pipeline appears in the Supplementary Data. All individuals from cohorts genotyped on the same platform (Affymetrix or Illumina) were merged into a single data set, with only SNPs retained that were in common between all cohorts being merged (337,727 and 456,168 markers for Affymetrix- and Illumina-based arrays, respectively). Population stratification was assessed using EIGENSTRAT, version 6.0.1 (24). This assessment was performed for the merged cohorts genotyped by the same platform that passed quality control (Supplementary Table 1 [N(total) = 25,063]). For the Affymetrix- and Illumina-based data sets, 52,598 and 99,742 SNPs, respectively, were used as input into the analysis for population stratification, which were those that passed the aforementioned genotype quality control, followed by further filtering steps described in the Supplementary Data. Low-frequency and rare variant associations can be prone to bias from population stratification; therefore, additional steps were taken to decrease such potential effects. Specifically, analysis for population stratification removed 268 and 545 individuals identified as ancestral outliers from the Affymetrix- and Illumina-based arrays, respectively. The top 10 principal components for the remaining individuals were used as covariates in the GWAS to adjust for potential residual population stratification. An additional principal component analysis was conducted by projecting study samples to 2,504 samples from 1000 Genomes phase 3. This additional analysis revealed that the EIGENSTRAT-based population stratification analysis successfully removed individuals of non-European ancestry and that remaining individuals across case/control status or across individual cohorts are of European ancestry (Supplementary Fig. 2). The quality-control and population stratification analysis resulted in a total of 24,250 individuals used for genotype imputation. This included 2,903/5,678 case/control subjects and 6,093/9,576 case/control subjects for the Affymetrix- and Illumina-based data sets, respectively (Supplementary Table 1).
Genotype Imputation, GWAS, Meta-analysis, and Conditional Analysis
The Sanger Institute online service (25) was used to impute the available genotypes to the Haplotype Reference Consortium, version R1.1, on ∼39 million SNPs, separately in the Illumina and Affymetrix data sets. Association testing was undertaken using logistic regression as implemented in SNPTEST, version 2.5.2 (26), on samples from the same genotyping platform and including the top 10 principal components from EIGENSTRAT (24,27) to adjust for population stratification. We then combined the association results from the Affymetrix and Illumina cohorts in a fixed effects inverse variance meta-analysis using METAL (28) to generate the discovery cohort totaling 25,063 type 1 diabetes case and control subjects. We retained SNPs with an MAF in control subjects >0.5% and those with an imputation quality score (INFO) >0.3, resulting in 9,061,522 SNPs. The MAF cutoff was decided based on a power calculation using the genetic analysis calculator (https://zzz.bwh.harvard.edu/gpc/) and the following assumptions: perfect LD between the causative variant and the markers that were genotyped, an additive genetic model, a disease prevalence of 0.0033, and an α of 1 × 10−5 (similar to previous GWAS [11,12]). We declared genome-wide statistical significance at P ≤ 1.2 × 10−8 to account for the increased number of statistical tests compared with a common variant–based GWAS (29). Conditional analysis was performed using the GCTA-COJO package (30). Among the SNPs that remained genome-wide significant after conditional analysis, we considered as candidates for replication only variants that were not within 1 Mb of MHC and that had at least three SNPs with discovery P value <0.0001 within 40 kb, henceforth referred to as satellite SNPs (Supplementary Data).
Replication of Discovery Association Signals
Replication de novo genotyping was performed in a cohort consisting of 4,329 type 1 diabetes case subjects from the sibling-pair set of T1DGC and 9,543 control subjects from UK Biobank. A detailed description of the replication cohort and of its assessment for population stratification is provided in the Supplementary Data. De novo genotyping of the lead SNPs for the replication in type 1 diabetes case subjects from the T1DGC was performed by LGC Genomics applying KASP genotyping. Association testing for the replication cohort was performed using GMMAT (31), using a relatedness matrix built using GEMMA (32). SNPs achieving a P value ≤0.05 in the replication cohort and same direction of effect were combined with the discovery cohort in a fixed effects inverse variance meta-analysis (33) to create combined estimates of OR (ORcombined) and P values (Pcombined) and heterogeneity statistics from the discovery and replication cohorts.
Admixture Analysis
Since low-frequency and rare variants can be strongly influenced by subtle differences in frequency across ancestral populations, we undertook an admixture analysis using PCAdmix (34).
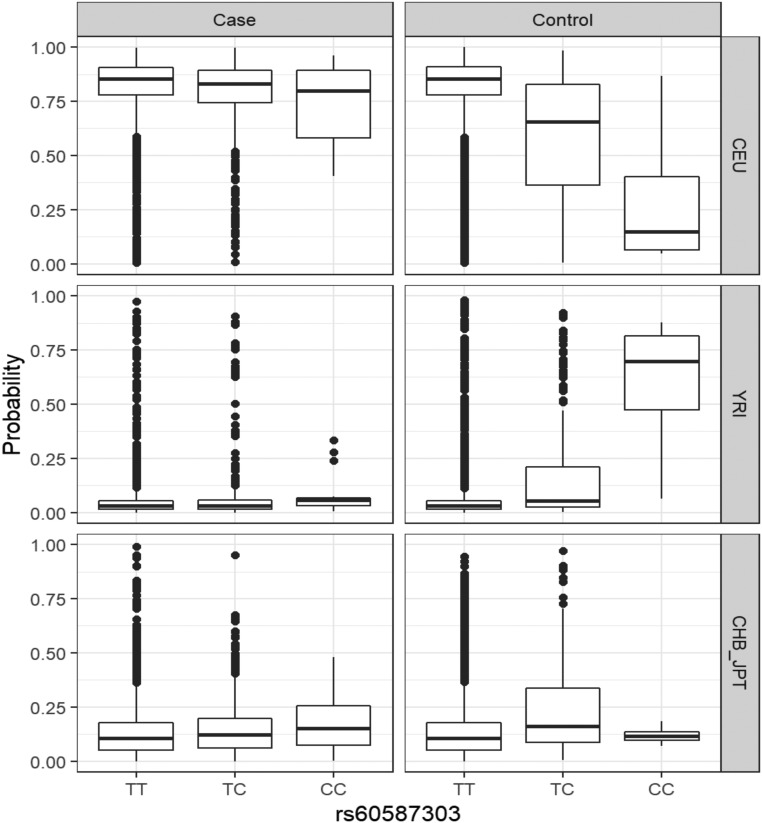
Specifically, the local admixture analysis was performed on 16,124 Illumina-based imputed genotypes on a 2 Mb region surrounding rs60587303 (STK39) (21,380 SNPs). Admixture of each individual was determined in comparison with genotype data from the European (CEU) (N = 99), African (YRI) (N = 108), and Asian (CHB/JPT) (N = 207) populations in 1000 Genomes phase 3. The distribution of probabilities of CEU, YRI, and CHB/JPT ancestries was stratified by dosage of the effect allele (C) at rs60587303 and type 1 diabetes case/control status.
Fine Mapping and In Silico Functional Exploration
We undertook fine mapping using FINEMAP (35) to prioritize variants at all GCTA-COJO conditionally independent loci. Putative functional targets were identified in silico as genes mapping in the same topological domain as the fine mapped SNPs or genes with fine mapped SNPs overlapping transcription factor binding sites and DNase I hypersensitivity clusters (Supplementary Data).
Exploration of In Vitro Function of STK39
Murine T cells
Given the commercial availability of an antiparasitic agent, closantel, which has been shown to specifically inhibit stk39 activity in cell lines and in mice (36), we tested the effects of this agent on cytokine responses in murine CD4+ T cells. Cytokine secretion assays for interferon γ (IFNγ) and interleukin-2 (IL-2) were performed on activated CD4+ T cells in the presence or absence of closantel at titrated doses (see Supplementary Data for further details).
Data and Resource Availability
The GWAS summary-level results will become available through GRASP (Genome-Wide Repository of Associations Between SNPs and Phenotypes) (https://grasp.nhlbi.nih.gov/).
Results
GWAS
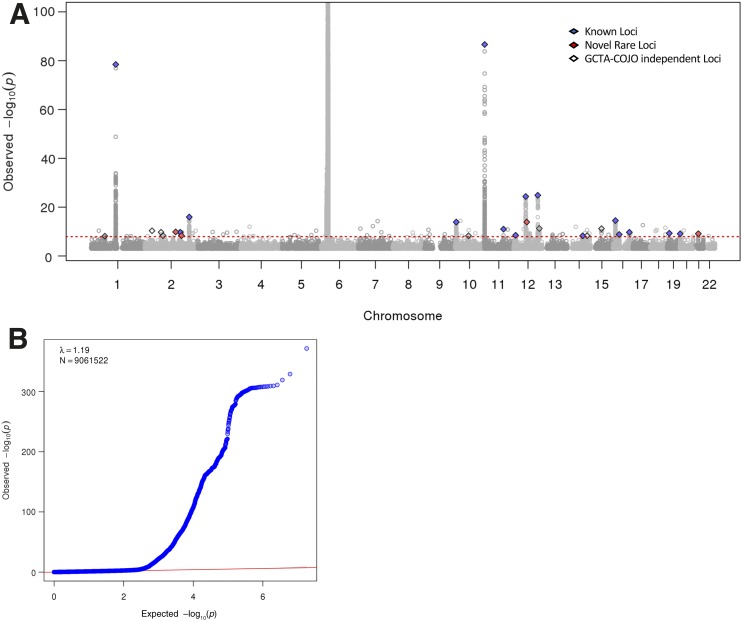
After quality control, imputation to 9,061,522 SNPs, and platform-specific case-control GWAS performed separately for the Affymetrix and Illumina genotyped samples, our discovery meta-analysis included a total of 9,358 type 1 diabetes case and 15,705 control subjects. The power of our meta-analysis to find variants with an MAF of 0.5% and a relative risk of 2.0 was 98% at an α of 1 × 10−5. After controlling for population stratification, the λ for this meta-analysis was 1.19 (Fig. 1), which is similar to that reported in previous type 1 diabetes GWAS (11). Based on our criteria for prioritization of our independent lead variants (MAF in control subjects >0.5%, INFO >0.3, P value <1.2 × 10−8 after conditional analysis, not being in the MHC region, and presence of at least three satellite SNPs), our list of lead variants included 27 independent variants in 27 loci (Supplementary Table 3). Among these, 23 were common, mostly at known autoimmune loci (n = 15) reported in ImmunoBase (available from https://genetics.opentargets.org/immunobase). Five were low-frequency or rare variants, four of which were located in loci not previously described to be associated with an autoimmune trait. One low-frequency variant (rs34536443) was located in the TYK2 locus, which has previously been associated with type 1 diabetes risk (11).
Figure 1.
Discovery meta-analysis. A: Manhattan plot depicting genome-wide association of 9.06 million genetic variants with type 1 diabetes risk [y-axis is truncated at –log10(P value) of 100]. Blue diamonds represent lead independent genetic variants at known type 1 diabetes loci, red diamonds represent the novel low-frequency/rare loci, and white diamonds represent the remaining GCTA-COJO independent loci. B: Quantile-quantile plot depicting the observed versus expected −log10 P values for the genome-wide association of 9.06 million genetic variants with type 1 diabetes risk.
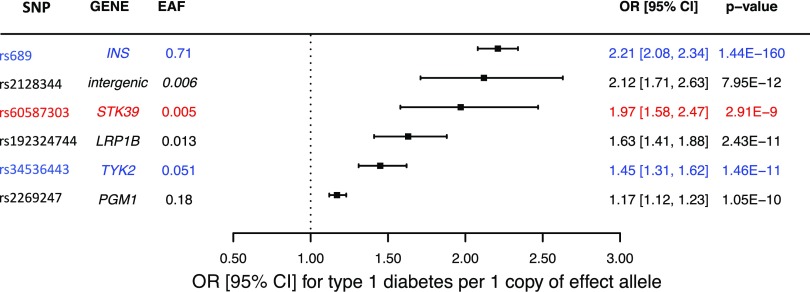
The four novel low-frequency/rare variants were brought forward to the replication stage (Supplementary Table 4). In addition, two novel common variants with >40 satellite SNPs were tested for replication by de novo genotyping in 4,329 case and 9,543 control subjects. We used two variants, in TYK2 (rs34536443) and INS (rs689), as positive controls in the replication stage. One of the two novel common SNPs (rs61944716) failed replication genotyping. Four of the five novel SNPs replicated at Preplication < 0.05. For these four variants, we combined the “discovery” and “replication” P values to create a Pcombined. We considered as “replicated” the SNPs that presented a Preplication < 0.05, Pcombined < Pdiscovery, and Pcombined ≤ 1.2 × 10−8 with consistent direction of association. Based on these criteria, three out of the four novel low-frequency/rare SNPs and the single novel common variant replicated (Fig. 2, Table 1, and Supplementary Table 5).
Figure 2.
Forest plot with the effects on type 1 diabetes of the variants from the discovery and replication meta-analysis. The variants used as positive controls appear in blue. For facilitation of comparison, the novel STK39 variant appears in red. EAF, effect allele frequency; OR, OR of type 1 diabetes after combining discovery and replication results.
Table 1.
Main findings from the combined discovery and replication meta-analysis
| SNP | Candidate gene(s) | EA | EAFcontrol subjects | Pdiscovery | Preplication | Pcombined | ORcombined (95% CI) |
|---|---|---|---|---|---|---|---|
| rs689* | INS | T | 0.71 | 2.3 × 10−87 | 2.3 × 10−96 | 1.4 × 10−160 | 2.21 (2.08–2.34) |
| rs2128344 | Intergenic variant | A | 0.0055 | 7.4 × 10−10 | 2.5 × 10−3 | 8.0 × 10−12 | 2.12 (1.71–2.63) |
| rs60587303 | STK39 | C | 0.0051 | 4.4 × 10−9 | 2.4 × 10−2 | 2.9 × 10−9 | 1.97 (1.58–2.47) |
| rs192324744 | LRP1B | G | 0.0131 | 1.4 × 10−10 | 5.6 × 10−3 | 2.4 × 10−11 | 1.63 (1.41–1.88) |
| rs34536443* | TYK2 | G | 0.0515 | 4.8 × 10−10 | 7.0 × 10−4 | 1.5 × 10−11 | 1.45 (1.31–1.62) |
| rs2269247 | PGM1 | T | 0.18 | 7.3 × 10−9 | 1.7 × 10−3 | 1.1 × 10−10 | 1.17 (1.12–1.23) |
Only variants with Preplication < 0.05 and a Pcombined < Pdiscovery appear in the table. EA, effect allele; EAF, effect allele frequency.
*Variants used as positive controls.
In Silico Functional Exploration and Fine Mapping
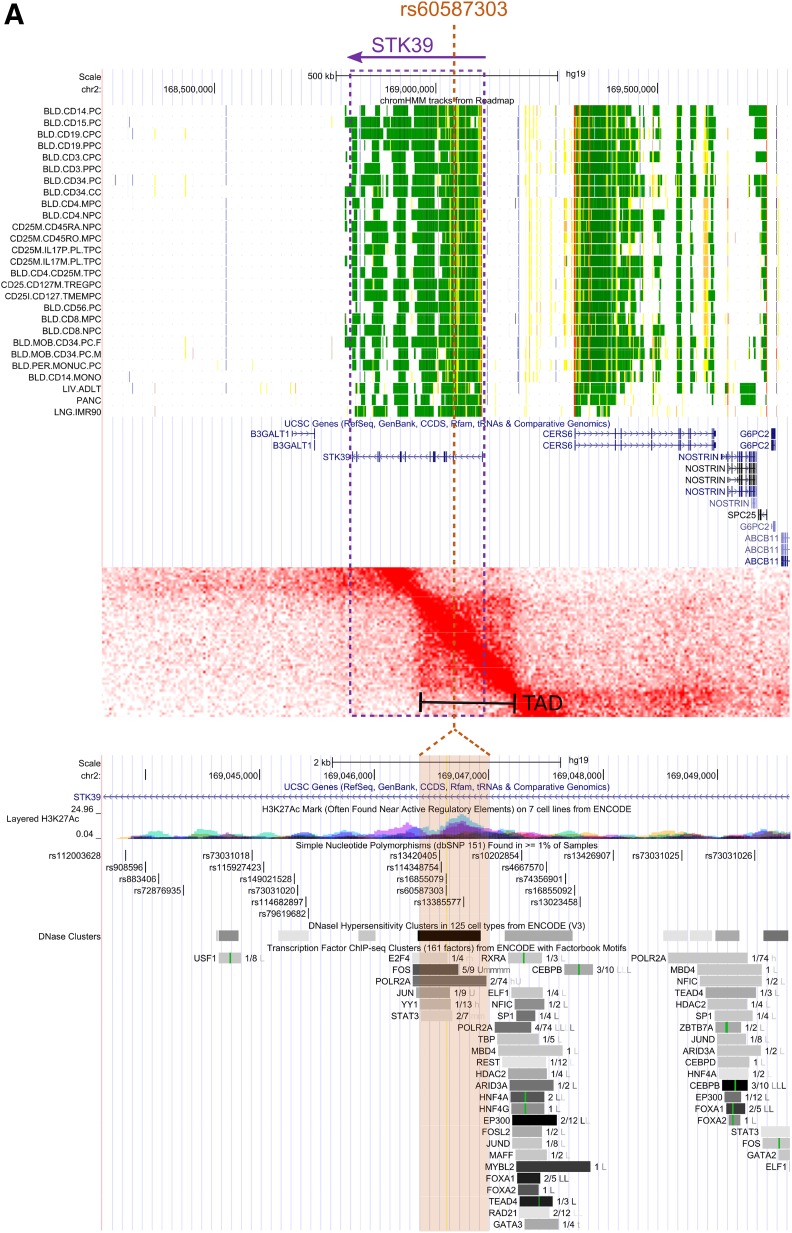
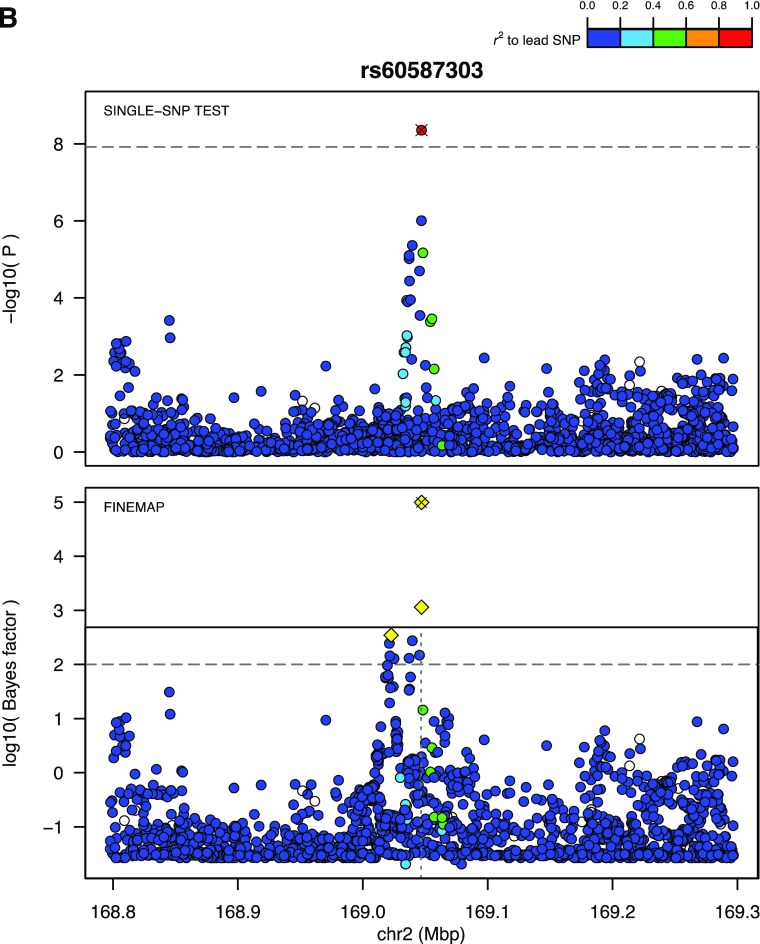
To prioritize variants for in vitro and in vivo functional exploration among the four novel variants that successfully replicated, we performed an in silico analysis of the four SNPs by considering their spatial proximity to putative functional elements and target genes (Fig. 3). Specifically, the top candidate genes were determined as genes within a region of elevated chromatin interactions with the lead SNP or, if lacking, nearby genes. Further, expression of these genes was assessed in type 1 diabetes–relevant cell types. These approaches converged upon STK39, since it resides in a region of elevated chromatin interaction with the lead SNP. The local chromatin interaction map for the STK39 locus and its proximity to regulatory elements are shown in Fig. 3.
Figure 3.
The STK39 locus. A, top: A regional view of the 2q24.3 locus in the UCSC Genome Browser with tracks showing chromatin state in various immune cell types (green means active). From this, we can see that STK39 is active in immune cells but also nearby genes such as CERS6. A, middle: Topological associated domains for the STK39 region. Deeper red color indicates that there is increased pairwise interaction within the genomic interval. A topologically associated domain, a region of increased interaction, is clearly visible that encompasses both the associated variant, rs60587303, and the promoter of STK39 but excludes CERS6. A, bottom: Zoomed-in view of region around rs60587303 shows clear overlap with active regulatory elements containing numerous transcription factor binding sites and open chromatin sites in 68 cell types. B: Regional Manhattan and fine mapping plots centered on rs60587303. Top panel depicts the results from the discovery meta-analysis of a region centered on rs60587303. This genetic variant is the lead signal and is supported by multiple genome-wide suggestive genetic variants. In the bottom panel, statistical fine mapping of this further locus supports rs60587303 as the lead putatively causal genetic variant (yellow diamond with ×), accompanied by two highly correlated genetic variants of lower log10 Bayes factor (open yellow diamonds). chr, chromosome.
The lead variant with the most compelling in silico functional evidence was rs60587303 (C), a rare variant (MAFcontrol subjects 0.5%) that resides in the first intron of the STK39 gene. With an ORcombined of 1.97 (95% CI 1.58–2.47, Pcombined = 2.9 × 10−9), the magnitude of its effect is comparable with that of the lead INS common variant in our analysis (rs689 [A], MAF 14%, OR 2.21, 95% CI 2.26–2.67, Pcombined = 1.44 × 10−160). rs60587303 overlaps a DNase I hypersensitivity cluster of 68 cell types, as well as a cluster of transcription factor binding sites (Fig. 3). Statistical fine mapping experiments also prioritized rs60587303 as one of the five top plausibly causal SNPs at the 2q24.3 locus (Fig. 3). Another low-frequency variant was located in the second intron of the LDL receptor–related protein 1B gene (LRP1B). With an MAFcontrol subjects of 1.3%, rs192324744 (G) had an ORcombined of 1.63 (95% CI 1.41–1.87, Pcombined = 2.4 × 10−11). Fine mapping also prioritized this variant in the 2q22.2 locus (Supplementary Fig. 4), but there was less compelling in silico functional evidence from the TAD (topologically associating domain) analysis. The third low-frequency variant that successfully replicated was intergenic (rs2128344 [A]) at the 21q22.1 locus, with an MAFcontrol subjects of 0.55% and an ORcombined of 2.12 (95% CI 1.71–2.63, Pcombined = 8.0 × 10−12). In silico functional evidence in support of this variant was lacking. Finally, the single common variant that successfully replicated was an SNP in the 7th intron of the phosphoglucomutase 1 (PGM1) gene. rs2269247 (T) had an MAFcontrol subjects of 18% and an ORcombined of 1.19 (95% CI 1.12–1.26, Pcombined = 7.3 × 10−9). Fine mapping prioritized this variant at the 1.p31.3 locus, and this SNP appeared to be in the same TAD as PGM1 (Supplementary Fig. 4).
Assessment for Population Stratification and Admixture Analysis
Since rare and low-frequency variants are more prone to confounding by population stratification than common variants, we undertook a principal component analyses separately in the Affymetrix and Illumina discovery cohorts and in the replication cohort. While the population structure between case and control subjects is unlikely to be similar, given that case and control subjects come from different cohorts, our analysis found no evidence for ancestral differences between case and control subjects in the discovery Affymetrix data (Supplementary Fig. 2A), in the Illumina data (Supplementary Fig. 2B), or in the replication cohort (Supplementary Fig. 3). Moreover, using seeds from the 1000 Genomes phase 3 data set, our principal component analysis demonstrated that the outliers removed by EIGENSTRAT lie outside the 1000 Genomes European superpopulation, whereas the inliers largely overlap or are in close proximity to the European superpopulation in the Affymetrix, Illumina, and replication data (Supplementary Figs. 2 and 3).
We also undertook an admixture analysis, where we queried the MAF of the rs60587303 (C) in different ethnicities in the 1000 Genome phase 3 release V3+. We found that its MAF varied by ancestry (East Asian MAF 0.250, American MAF 0.030, African MAF 0.248, European MAF 0.007, and South Asian MAF 0.053). Since this variant is common in East Asians and Africans, but almost absent in Europeans, we undertook an admixture analysis in our GWAS population to interrogate whether there was an enrichment of Asian or African descent among case subjects, which could have influenced the association signal from the STK39 variant. The distributions of probabilities of CEU, YRI, and CHB/JPT ancestries were stratified by dosage of the effect allele (C) at rs60587303 and case/control status (Fig. 4). This analysis showed that the type 1 diabetes case subjects of our GWAS had less African or Asian admixture compared with control subjects. This strongly decreases the probability that this finding has arisen due to population stratification or admixture because these effects would bias results at rs60587303 near STK39 toward the null. Interestingly, when we looked at the MAF of rs60587303 in European subpopulations available from the gnomAD database (version 2) (https://gnomad.broadinstitute.org/variant/2-169046632-T-C?dataset=gnomad_r2_1) we obtained the following results: Ashkenazi Jewish, 0.03793; European (Finnish), 0.01900; and European (non-Finnish), 0.009853. In the newest version, version 3, of gnomAD (https://gnomad.broadinstitute.org/variant/2-168190122-T-C?dataset=gnomad_r3), which is not overlapping with the previous version, we obtained the following results: Ashkenazi Jewish, 0.03101; European (Finnish), 0.01518; European (non-Finnish), 0.006926; and Amish, 0.000. These results suggest differences in MAF in different European subpopulations, and in particular, the MAF is double in the Finnish population compared with the non-Finnish European population. This is an interesting observation, considering Finland has the highest rate of type 1 diabetes in the world.
Figure 4.
Admixture analysis for rs60587303, showing the probability of European (CEU), African (YRI), and Asian (CHB/JPT) ancestry of the individual from the Illumina platform from our discovery meta-analysis, further stratified by type 1 diabetes case/control status and alleles at rs60587303. Admixture analysis was conducted for a 2 Mbp region centered on rs60587303. We observed limited non-CEU ancestry for case subjects, whereas there is an elevated number of individuals with likely YRI ancestry among control subjects who carry one or more minor alleles at rs60587303.
In Vitro Functional Exploration of Differential Stk39 Activity in T Cells
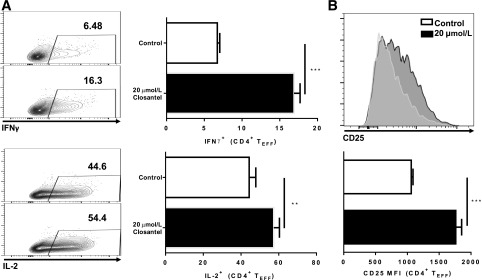
We next sought to explore the functional effects of Stk39 using a commercially available specific STK39 inhibitor (closantel) (36) to determine whether inhibition of Stk39 activity would affect murine T cell activation and functions in vitro. In T-cell receptor–activated, primary freshly isolated murine CD4+ T cells, treated with titrated concentrations of closantel, STK39 inhibition had no significant impact on T-cell activation or proliferation at doses that did not affect cell viability. However, we observed that closantel treatment enhanced the secretion of IFNγ and IL-2 by effector T (TEFF) cells. Closantel also increased the level of expression of the IL-2 receptor α chain (CD25), the high-affinity binding component of the functional IL-2 receptor, indicating that closantel might augment the sensitivity of activated T cells to IL-2 (Fig. 5).
Figure 5.
STK39 inhibition regulates TEFF functions. A: Secretion of IFNγ and IL-2 by TEFF cells 72 h following anti-CD3 activation in the presence of indicated concentrations of closantel. Gated on viable CD4+ Foxp3− cells. B: Expression of cell surface CD25 on TEFF cells 72 h following activation in the presence of indicated concentration of closantel. Gated on viable CD4+ Foxp3− cells. Plots shown from one representative experiment of three, done in triplicate. Error bars represent SD for that experiment. Unpaired parametric t test done for all comparisons. *P ≤ 0.05; **P ≤ 0.005; ***P < 0.0005.
Discussion
In an effort to discover new loci harboring not previously identified low-frequency and rare variants, we have carried out a large GWAS for type 1 diabetes, including the largest number of SNPs to date. Our analysis revealed the presence of three novel low-frequency/rare variants, the intergenic variant rs2128344, the rs60587303 in STK39, and the rs192324744 in LRP1B—all validated by replication using de novo genotyping and all with ORs ≥1.5, effects comparable with those of the lead non-MHC common variants at INS and PTPN22 loci. Functional exploration of the STK39 locus provided evidence supporting a role for STK39 in type 1 diabetes.
Our in silico exploration approach prioritized rs60587303 in the STK39 gene for our in vitro experiments. Specifically, carrying one risk allele of this variant appears to approximately double the risk of type 1 diabetes in individuals of European ancestry. This gene encodes a serine/threonine kinase that is thought to function in the cellular stress response pathway (37). Although not previously associated with autoimmunity, STK39 is linked in protein-protein interaction networks to protein kinase C θ (PRKCQ), which then interacts with CD28 (38). The IL2RA and PRKCQ loci on chromosome 10 are well-replicated type 1 diabetes GWAS gene loci. Our in vitro experiments using a cellular murine model of immunity provided evidence that STK39 could be involved in T-cell activation and effector functions. Pharmacological inhibition of STK39 in vitro appeared to enhance inflammatory responses from TEFF, providing a mechanistic hypothesis for a possible role of STK39 in type 1 diabetes. We emphasize that further functional validation of STK39’s role in immunity is required.
Additional association signals were identified from variants near LRP1B and PGM1—genes with known roles in metabolism: specifically, LRP1B encodes the LDL receptor protein 1B, which has been previously involved in tumor suppression pathways (39). PGM1 encodes the phosphoglucomutase-1 enzyme, which catalyzes the transfer of phosphate between the 1 and 6 positions of glucose (40). The novel PGM1 variant (rs2269247) identified in our study is in linkage disequilibrium with a variant (rs2269241) in the same locus, previously shown to present a genome-wide suggestive P value for type 1 diabetes (12). These findings provide rationale for further functional studies investigating the role of both LRP1B and PGM1 genes in type 1 diabetes pathophysiology.
Our study has limitations. Similar to previous type 1 diabetes genetic association studies (11,18), the case-control design of our GWAS meta-analysis did not allow for matching of case subjects to control subjects within the same European population because of the lack of availability of control samples in each participating case cohort. Instead, we matched case subjects to control subjects from different cohorts of similar ancestry. Also, case and control samples were not matched for demographics, such as age and sex. This approach could induce bias from population stratification; however, control for population stratification was applied. Also, most studies included adult individuals, and most cases of type 1 diabetes are manifested in childhood and early adulthood, making the likelihood of misclassification of control subjects very low. Moreover, the likelihood of similar population stratification issues being present in discovery and replication is low. Thus, given that both discovery and replication cohorts showed similar effects, the probability that both were biased by population stratification in the same direction is unlikely. Further, in the case of STK39, the increased allele frequency in non-Europeans should have biased our results toward the null, since control subjects were demonstrated to have a higher component of admixture from non-Europeans. On the other hand, differences in allele frequency of the STK39 variant within European populations (for instance, in the Finnish population compared with the general European population) could present residual population stratification, but the likelihood that this phenomenon has biased both the discovery and replication results is very low. Although we applied a liberal imputation INFO cutoff of 0.3 to select variants to include in our GWAS, all results replicated in separate cohorts using direct genotyping, which does not depend on imputation. Finally, ideally, the design of this study should have included, along with imputed GWAS data, exome chip or whole-exome sequencing data, which provide more insight on rare variants. Unfortunately, to date, large-scale exome studies, providing adequate statistical power to study less common diseases, such as type 1 diabetes, are not available.
In summary, our findings demonstrate the utility of deep imputation to enable the characterization of low-frequency and rare genetic variation, offering new insights into the pathophysiology of type 1 diabetes and enabling the identification of new potential drug targets.
Supplementary Material
Article Information
Acknowledgments. The authors acknowledge the T1DGC. This research has been conducted using the UK Biobank Resource under Application Number 27449. A complete list of participants in the DCCT/EDIC Research Group is presented in the Supplementary Material published online for the article in N Engl J Med 2017,376:1507–1516. The authors also thank the RI-MUHC Immunophenotyping Platform for excellent services.
Funding. The DCCT/EDIC has been supported by cooperative agreement grants (1982-1993, 2012-2017, 2017-2022) and contracts (1982-2012) with the Division of Diabetes, Endocrinology, and Metabolic Diseases of the National Institute of Diabetes and Digestive and Kidney Diseases (current grant numbers U01 DK094176 and U01 DK094157) and has been supported by the National Eye Institute, the National Institute of Neurological Disorders and Stroke, the General Clinical Research Centers Program (1993–2007), and the Clinical Translational Science Center Program (2006–present), Bethesda, MD. The DCCT/EDIC is registered at ClinicalTrials.gov, clinical trial reg. nos. NCT00360815 and NCT00360893. The Richards laboratory is supported by the Canadian Institutes of Health Research (CIHR), the Canadian Foundation for Innovation, and Fonds de Recherche Santé Québec (FRQS). J.B.R. is supported by a FRQS Clinical Research Scholarship. S.F.A.G. is supported by the National Institutes of Health (R01 DK085212) and the Daniel B. Burke Endowed Chair for Diabetes Research. H.H. is supported by The Children’s Hospital of Philadelphia Endowed Chair in Genomic Research. C.Pi. received financial support from the Canadian Institutes of Health Research (CIHR) operating grants (PJT-148821) and the Anna Maria Solinas Laroche Career Award in Immunology. D.M. is supported by JDRF (award no. 3-PDF-2017-370-A-N). Industry contributors have provided free or discounted supplies or equipment to support participants’ adherence to DCCT/EDIC: Abbott Diabetes Care (Alameda, CA), Animas (Westchester, PA), Bayer Diabetes Care (North America Headquarters, Tarrytown, NY), Becton Dickinson (Franklin Lakes, NJ), Eli Lilly (Indianapolis, IN), Extend Nutrition (St. Louis, MO), Insulet Corporation (Bedford, MA), LifeScan (Milpitas, CA), Medtronic Diabetes (Minneapolis, MN), Nipro Home Diagnostics (Ft. Lauderdale, FL), Nova Diabetes Care (Billerica, MA), Omron (Shelton, CT), Perrigo Diabetes Care (Allegan, MI), Roche Diabetes Care (Indianapolis, IN), and Sanofi (Bridgewater, NJ).
These industry contributors have had no role in DCCT/EDIC.
Duality of Interest. S.F.A.G. has received support from GlaxoSmithKline for research that is not related to the study presented in this article. H.H. has received support from Sanofi for type 1 diabetes research that is unrelated to this study. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. V.F. collected data and undertook all the bioinformatics analyses. D.M. wrote the first draft of the manuscript. S.R. collected data. L.M. and M.-C. T. were involved in the replication experiment. R.I. undertook the in vitro follow-up studies. H.-Q.Q., J.P.B., S.F., A.G., H.H., and C.Po. contributed study samples. J.B.R., C.Po., and C.Pi. conceived the experiments. All co-authors reviewed and edited the manuscript. J.B.R. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
A complete list of participants in the DCCT/EDIC Research Group is presented in the Supplementary Material published online for the article in N Engl J Med 2017;376:1507–1516.
This article contains Supplementary Data online at https://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-0831/-/DC1.
References
- 1.American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care 2009;32(Suppl. 1):S62–S67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Menke A, Orchard TJ, Imperatore G, Bullard KM, Mayer-Davis E, Cowie CC. The prevalence of type 1 diabetes in the United States. Epidemiology 2013;24:773–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.International Diabetes Federation IDF Diabetes Atlas. Brussels, Belgium, International Diabetes Federation, 2013 [Google Scholar]
- 4.Tuomilehto J. The emerging global epidemic of type 1 diabetes. Curr Diab Rep 2013;13:795–804 [DOI] [PubMed] [Google Scholar]
- 5.Imperatore G, Boyle JP, Thompson TJ, et al.; SEARCH for Diabetes in Youth Study Group . Projections of type 1 and type 2 diabetes burden in the U.S. population aged <20 years through 2050: dynamic modeling of incidence, mortality, and population growth. Diabetes Care 2012;35:2515–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hyttinen V, Kaprio J, Kinnunen L, Koskenvuo M, Tuomilehto J. Genetic liability of type 1 diabetes and the onset age among 22,650 young Finnish twin pairs: a nationwide follow-up study. Diabetes 2003;52:1052–1055 [DOI] [PubMed] [Google Scholar]
- 7.van Belle TL, Coppieters KT, von Herrath MG. Type 1 diabetes: etiology, immunology, and therapeutic strategies. Physiol Rev 2011;91:79–118 [DOI] [PubMed] [Google Scholar]
- 8.Nisticò L, Buzzetti R, Pritchard LE, et al. The CTLA-4 gene region of chromosome 2q33 is linked to, and associated with, type 1 diabetes. Belgian Diabetes Registry. Hum Mol Genet 1996;5:1075–1080 [DOI] [PubMed] [Google Scholar]
- 9.Bottini N, Musumeci L, Alonso A, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet 2004;36:337–338 [DOI] [PubMed] [Google Scholar]
- 10.Vella A, Cooper JD, Lowe CE, et al. Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. Am J Hum Genet 2005;76:773–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradfield JP, Qu HQ, Wang K, et al. A genome-wide meta-analysis of six type 1 diabetes cohorts identifies multiple associated loci. PLoS Genet 2011;7:e1002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett JC, Clayton DG, Concannon P, et al.; Type 1 Diabetes Genetics Consortium . Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009;41:703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pociot F. Type 1 diabetes genome-wide association studies: not to be lost in translation. Clin Transl Immunology 2017;6:e162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science 2009;324:387–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flannick J, Mercader JM, Fuchsberger C, et al.; Broad Genomics Platform; DiscovEHR Collaboration; CHARGE; LuCamp; ProDiGY; GoT2D; ESP; SIGMA-T2D; T2D-GENES; AMP-T2D-GENES . Exome sequencing of 20,791 cases of type 2 diabetes and 24,440 controls. Nature 2019;570:71–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wray NR, Yang J, Goddard ME, Visscher PM. The genetic interpretation of area under the ROC curve in genomic profiling. PLoS Genet 2010;6:e1000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Todd JA, Walker NM, Cooper JD, et al.; Genetics of Type 1 Diabetes in Finland; Wellcome Trust Case Control Consortium . Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet 2007;39:857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Onengut-Gumuscu S, Chen WM, Burren O, et al.; Type 1 Diabetes Genetics Consortium . Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet 2015;47:381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manousaki D, Dudding T, Haworth S, et al. Low-frequency synonymous coding variation in CYP2R1 has large effects on vitamin D levels and risk of multiple sclerosis. Am J Hum Genet 2017;101:227–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng HF, Forgetta V, Hsu YH, et al.; AOGC Consortium; UK10K Consortium . Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature 2015;526:112–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loh PR, Danecek P, Palamara PF, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet 2016;48:1443–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sudlow C, Gallacher J, Allen N, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc 2010;5:1564–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet 2006;2:e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarthy S, Das S, Kretzschmar W, et al.; Haplotype Reference Consortium . A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 2016;48:1279–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007;39:906–913 [DOI] [PubMed] [Google Scholar]
- 27.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006;38:904–909 [DOI] [PubMed] [Google Scholar]
- 28.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu C, Tachmazidou I, Walter K, Ciampi A, Zeggini E, Greenwood CM; UK10K Consortium . Estimating genome-wide significance for whole-genome sequencing studies. Genet Epidemiol 2014;38:281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011;88:76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen H, Wang C, Conomos MP, et al. Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am J Hum Genet 2016;98:653–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet 2012;44:821–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mägi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics 2010;11:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brisbin A. Linkage Analysis for Categorical Traits and Ancestry Assignment in Admixed Individuals. Ithaca, Cornell University, 2010 [Google Scholar]
- 35.Benner C, Spencer CC, Havulinna AS, Salomaa V, Ripatti S, Pirinen M. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 2016;32:1493–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.AlAmri MA, Kadri H, Alderwick LJ, Simpkins NS, Mehellou Y. Rafoxanide and closantel inhibit SPAK and OSR1 kinases by binding to a highly conserved allosteric site on their C-terminal domains. ChemMedChem 2017;12:639–645 [DOI] [PubMed] [Google Scholar]
- 37.Li Z, Zhu W, Xiong L, Yu X, Chen X, Lin Q. Role of high expression levels of STK39 in the growth, migration and invasion of non-small cell type lung cancer cells. Oncotarget 2016;7:61366–61377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Hu J, Vita R, Sun B, Tabata H, Altman A. SPAK kinase is a substrate and target of PKCtheta in T-cell receptor-induced AP-1 activation pathway. EMBO J 2004;23:1112–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu CX, Li Y, Obermoeller-McCormick LM, Schwartz AL, Bu G. The putative tumor suppressor LRP1B, a novel member of the low density lipoprotein (LDL) receptor family, exhibits both overlapping and distinct properties with the LDL receptor-related protein. J Biol Chem 2001;276:28889–28896 [DOI] [PubMed] [Google Scholar]
- 40.Gururaj A, Barnes CJ, Vadlamudi RK, Kumar R. Regulation of phosphoglucomutase 1 phosphorylation and activity by a signaling kinase. Oncogene 2004;23:8118–8127 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.