Abstract
Machine-learning techniques have led to remarkable advances in data extraction and analysis of medical imaging. Applications of machine learning to breast MRI continue to expand rapidly as increasingly accurate 3D breast and lesion segmentation allows the combination of radiologist-level interpretation (eg, BI-RADS lexicon), data from advanced multiparametric imaging techniques, and patient-level data such as genetic risk markers. Advances in breast MRI feature extraction have led to rapid dataset analysis, which offers promise in large pooled multiinstitutional data analysis. The object of this review is to provide an overview of machine-learning and deep-learning techniques for breast MRI, including supervised and unsupervised methods, anatomic breast segmentation, and lesion segmentation. Finally, it explores the role of machine learning, current limitations, and future applications to texture analysis, radiomics, and radiogenomics.
DYNAMIC CONTRAST-ENHANCED BREAST magnetic resonance imaging (MRI) plays an integral role in the detection and characterization of breast cancer, along with mammography and ultrasound. The main indications for a breast MRI examination are screening to detect occult breast cancer in women at increased risk, preoperative assessment of the extent of disease in women with a known breast cancer, and assessment of treatment response to neoadjuvant chemotherapy.1,2
Breast MRI is the most sensitive imaging modality to detect breast cancer, including ductal carcinoma in situ (DCIS). However, lesion identification can be limited by background enhancement, which may mask or mimic lesions. Additionally, errors in perception, interpretation, and management contribute to false-negative examinations, as retrospective reviews of screen-detected cancers demonstrate that 34–47% of cancers were present on prior studies.3 In addition, specificity of MRI is only moderate, with positive predictive values of 35–64% for screening MR in high-risk women.4 Note that these results are from high-volume centers, and may not be generalizable to different levels of expertise. The high cost and long exam time are also barriers to wider clinical implementation of breast MRI. Machine learning (ML) is poised to address some or all of these limitations. Similar to artificial intelligence (AI) tools that increase the diagnostic performance of screening mammography, ML tools will likely increase the diagnostic performance of breast MRI by shifting away from the subjective and qualitative assessment of images. Until now, radiologist interpretation involved visual interpretation of a static picture. Emerging ML techniques, in contrast, allow for higher-order statistical analysis of patterns within the image, converting images into data and allowing for subsequent, high-volume analysis of the pooled data extracted from hundreds of thousands of images. As ML techniques bring together data from large numbers of studies, they may first be of use in standardizing MR interpretation across radiologist levels of experience and geographic practice patterns. ML tools then may be used as computer vision to see beyond what is apparent to the radiologist; highlighting lesions from background enhancement to improve sensitivity, and separating benign from malignant lesions to improve specificity. ML tools may sufficiently improve accuracy from interpretation of one or a few MR sequences, such that the number of necessary sequences is reduced, overall improving scan time, and cost.
Given the growing number of publications on ML in breast MRI, this article aims to review the current literature with respect to techniques and clinical applications. Specifically, we describe supervised and unsupervised methods for anatomic breast segmentation and lesion segmentation, and techniques for texture analysis. Additionally, we discuss possible clinical applications with their current limitations and areas for future study.
Machine-Learning Methods
The emergence of AI tools that may learn and continuously improve their diagnostic performance has generated enormous interest in the medical imaging community. Therefore, it is important to understand how these machine-learning tools work and how they can be adapted to perform a variety of functions.
ML, which falls under the umbrella of AI, is a branch of data science that enables computers to learn from existing “training” data without explicit programming. ML applications for medical imaging are divided into two broad paradigms: unsupervised learning and supervised learning. Unsupervised learning aims to discover the structure in the data that has no labels or categories assigned to training examples. The most common unsupervised learning task is clustering, which consists of grouping similar examples together according to some predefined similarity metric. The goal is to discover novel patterns in the data, which would otherwise be difficult to notice.
In contrast, supervised ML methods arrive at a classification decision without learning any intermediate representation of the data. Therefore, these methods can work well only if the input features are very predictive to begin with; for example, being trained on “ground truth labels.” These methods are not based on neural networks and are routinely used in the medical and nonmedical community. Examples of supervised ML classifiers include logistic regression, decision trees, and support vector machines (SVM).
Neural Networks and Deep Learning
Neural networks are ML models that consist of many layers and are more structurally complex than the supervised ML models described above. The study of neural networks is often referred to as deep learning. Three recent technological advances have allowed for the rapid growth of deep-learning applications in medical imaging. First, graphical processing units (GPUs) exponentially increased the computing power and allowed neural networks to be trained quickly. Second, the introduction of backpropagation allowed for an efficient way of computing the gradient of the training loss with respect to the parameters of the model.5 Third, neural networks are flexible models and allow for complex relationships between inputs and outputs. This flexibility allows an ML model to learn how to make difficult predictions. However, this flexibility also means that, with a small amount of data, there is a risk of overfitting where a neural network will simply memorize the training sample cases, rather than learn patterns relevant to the prediction problem. The easiest way to alleviate overfitting is to collect more data and to have properly “held out” validation samples. However, most available datasets are too small for neural networks to be useful. A big catalyst to the AI field was the introduction of the large ImageNet dataset.6 GPUs and improved mathematical optimization methods enabled upscaling the architectures of these neural network models with enough intermediate layers to achieve superhuman performance, as demonstrated by the ImageNet challenge in 2012.7
Recently, deep-learning-based methods have surpassed more traditional machine-learning segmentation methods for image-based learning problems. Convolutional neural networks (CNNs) are a type of neural network that has a special connectivity structure in its hidden layers, which aids in image recognition tasks. The special property of CNNs is the introduction of the convolutional layer and the pooling layer. Within the convolutional layer, neurons in two consecutive layers are only connected if they are spatially close to each other. The parameters in the convolutional layer are also shared between spatial locations, which accelerates learning. The pooling layer does not have any parameters. It is only averaging values of hidden neurons in spatially adjacent locations. More complex versions of CNNs, deep convolutional neural networks (DCNN),7 improved their diagnostic performance even further. DCNNs vary in their total number of layers, the architecture of convolutional and pooling layers, the number of parameters (depending on the shape of the filter), and their hyperparameters (such as dropout and learning rate). Examples of these DCNNs, in order of increasing number of layers, are AlexNet, VGG, GoogleNet, and ResNet. These DCNNs have shown increasing accuracy in image classification, as each has successively won the ImageNet competition.
For example, AlexNet, which won the ImageNet challenge in 2012, has five convolutional layers alternating with three pooling layers, doing the work of feature extraction. The final three layers are fully connected, which are the layers where the classification task occurs. A unique feature of AlexNet is that it was the first DCNN to use the Rectified Linear Unit activation function. The activation function simulates the firing rate of a neuron and can be modeled using different functions. Historically, sigmoid functions were used, but AlexNet uses a linear function that therefore has the advantage of being nonsaturating. The AlexNet architecture has 60 million parameters. To minimize overfitting, heavy data augmentation was used, increasing the training set by a factor of 2048. Dropout of 0.5 was also used to minimize overfitting; in this technique half of the hidden neurons have an output of zero and this forces adjacent neurons to learn more robust features than those simply related to the presence of particular other neurons.7
VGGNet improved on AlexNet by using more layers (16 instead of 8) and smaller filters, for more than double the number of parameters (138 million).8
GoogleNet surprisingly improved classification with more layers22 but much fewer parameters (5 million total). This architecture made use of an Inception module, which is a network within a network that can be stacked, increasing the depth and width of the network.9
ResNet staggeringly increased depth of the network to 152 layers. Historically, deeper networks had higher training errors. ResNet addressed this problem by using a deep residual learning framework, in which layers were given residual identity maps from earlier layers.10
DenseNet addresses the problem of very deep networks, as information passes through many layers it washes out by the time it reaches the end of the network. The DenseNet architecture therefore connects all layers directly with each other. This decreases the number of overall parameters and makes the network easier to train.11
The DCNNs listed above are rich feature extractors, and can be used for any computer vision task. However, V-Net and U-Net are particularly suited to the task of image segmentation. These have a contracting path and a symmetric expanding path connected by a bottleneck, giving them a U or V shape.12,13 Both the contracting and expanding paths are composed of multiple blocks of layers. The contracting path captures context but loses its location information, so the features extracted from this path are forwarded to the expanding path via horizontal connections. The expanding path enables precise localization along with the contextual information fed from the contracting path. These methods use heavy data augmentation by applying elastic deformations to the training images, and therefore can train from only a few (20–35) annotated images. A difference between V-Net and U-Net is that U-Net is a nonresidual learning network, while V-Net has a residual function learned at each stage. In addition, V-Net uses volumetric convolutions (as opposed to processing input volumes slicewise) to process 3D data. U-Net applications to segmentation are further discussed below.
Emerging DCNN models are profoundly different from traditional computer-aided detection (CAD). Since the multi-layered DCCN is capable of extracting salient features directly from the data, manual feature design and its associated challenges are now obviated. Through incorporation of digital data beyond imaging, such as patient-level information, and tumor-level information, DCCN models are capable of identifying not only known correlations but also novel imaging biomarkers that have enormous potential to enhance clinical performance. This has become possible because of breakthroughs in computer processing, data storage, and algorithm design in recent years, reenergizing the field of computerized image analysis, as reflected by an exponential rise in the number of publications on CAD, machine learning, and deep learning in PubMed since the year 2000.14
Radiomics
Radiomics is the field in which large numbers of quantitative features are extracted from medical images and pooled in large-scale analysis to create decision support models.15 The image data may be combined with patient-level data; known as radiogenomics when the imaging data is combined with genetic data. Radiomics studies overall follow a methodology of 1) image acquisition, 2) image segmentation, 3) feature extraction, 4) feature selection, and 5) predictive modeling. An example of this workflow is shown in Fig. 1. Supervised and unsupervised ML methods may be used at any step of this process. In this review we focus on some of the steps where there is sufficient literature. We will also highlight some of the more common clinical scenarios where radiomics and ML tools have been used, such as predicting the likelihood of malignancy and developing imaging biomarkers associated with tumor aggressiveness. It is important to emphasize that all the studies show an association and not causation.
FIGURE 1:
Example of radiomics study workflow. In all, 163 breast cancer patients with DCE-MRI scans were included in this study. The ROIs were identified on (a) the first postcontrast image; (b) the corresponding intratumoral and peritumoral ROIs: the yellow region is the original intratumoral ROI covering the enhancing tumor drawn by the radiologist, while the red region indicates the peritumoral ROI after dilation; radiomic features were extracted from (c) washin map, (d) washout map, and (e) SER map. The dataset was then randomly separated into a training set (~67%) and a validation set (~33%). The prediction model was built in the training set after combining clinical and histopathological information and was further tested in the completely independent validation set. (Reprinted and adapted with permission from Liu et al. J Magn Reson Imaging 2019;49:131–140.)
Breast Anatomic Segmentation
Large-scale imaging analysis of breast MRI requires a number of steps, often varying across medical centers and hardware. MR images must be acquired and reconstructed to the requirements of the clinical protocol, processed for viewing with assistance from technologists and/or vendor software, and then have anatomic boundaries delineated by computer-assisted contouring, ie, segmentation. Anatomic segmentation allows for analysis of what is of interest to the breast imager (the breast tissue, immediate chest wall, and axilla) while discarding that which is not clinically useful (surrounding air, thoracic cavity, and abdomen). After identifying and discarding irrelevant information, the segmented breast image is smaller in size, decreasing postprocessing time and computing power. It can be further dissected and refined as required for analysis.
Breast MRI segmentation can be divided into three consecutive tasks: the delineation of breast–chest wall and breast–air, the separation of breast fibroglandular tissue (FGT) from fat, and distinguishing abnormal enhancement from normal background parenchymal enhancement (BPE) (Fig. 2). Unlike breast boundary and breast FGT segmentation, breast lesion evaluation requires T1-weighted pre- and postcontrast sequences, which offer specific technical challenges, including B0 and B1 inhomogeneity across the parenchyma at the breast–air boundary and across the coil gradient.16 This must be corrected as part of image postprocessing. Finally, routine fat-suppression in many clinical breast MRI protocols introduces additional artifact that must also be corrected.
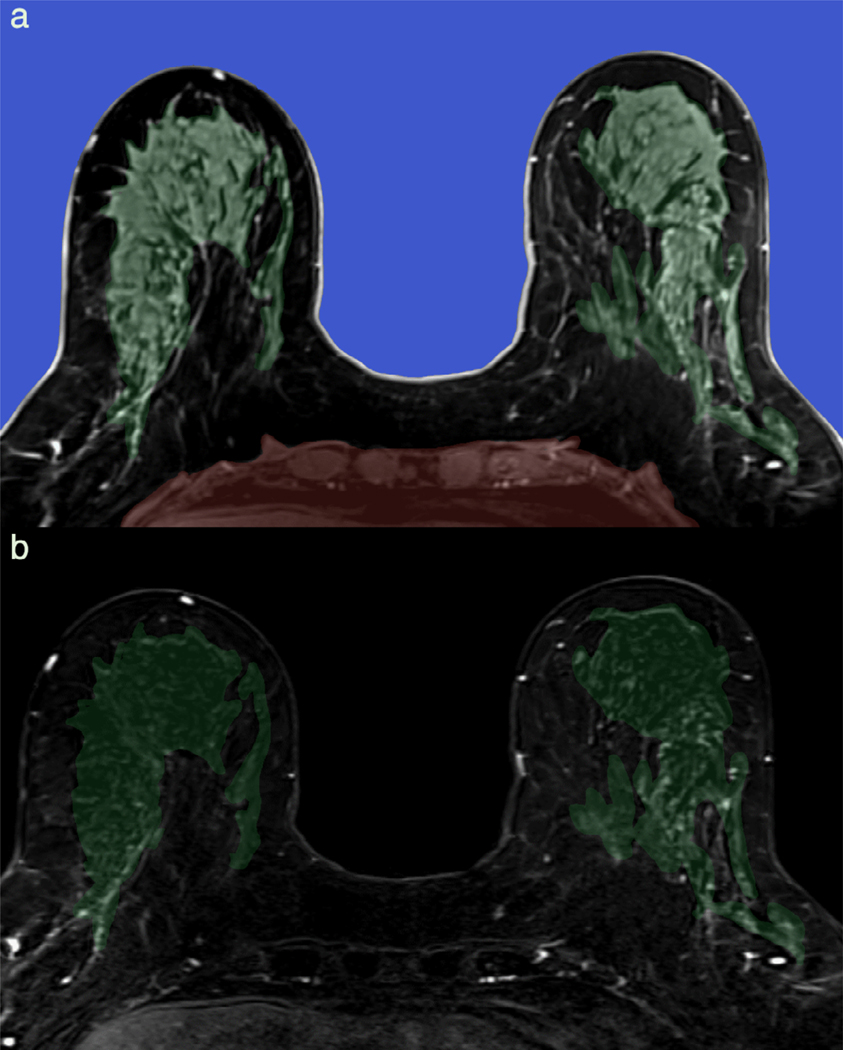
FIGURE 2:
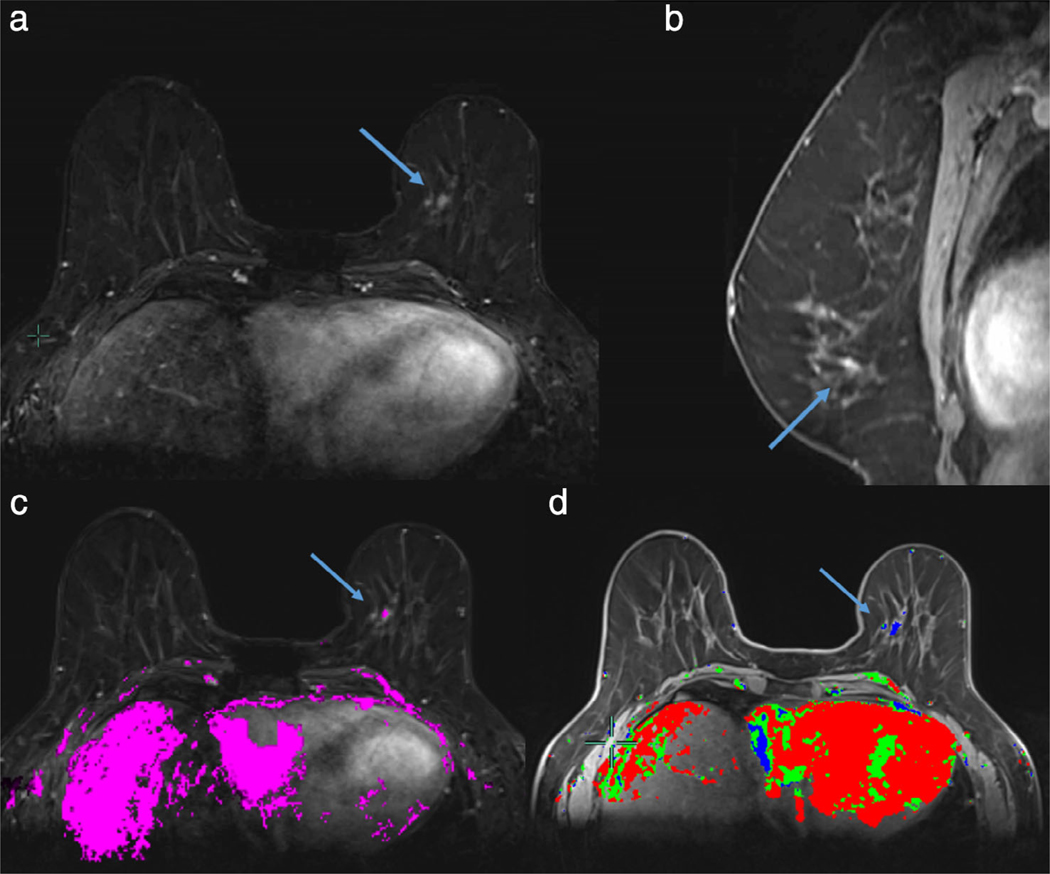
Automatic breast segmentation pipeline incorporating machine learning. Axial T1-weighted precontrast images are automatically segmented at breast-air (blue) and breast-chest wall (red) boundary (a). Breast is further subdivided into fibroglandular breast tissue (FGT, green) and fat. Background parenchymal enhancement (BPE) is calculated as FGT enhancement over baseline and is 12% (minimal) in this 66-year-old screening patient, as demonstrated on the first postcontrast subtraction images (b).
Delineation of the breast FGT in pre- and postcontrast imaging allows quantification of breast BPE. Mammographic breast density, which correlates with FGT, is a known risk factor for breast cancer risk and decreased mammographic sensitivity for breast cancer.17 FGT is a 3D volumetric measurement of breast density, allowing more accurate quantification than mammographic density. BPE, which measures the physiologic postcontrast degree of enhancement within FGT, has been shown to be sensitive to physiological changes in estrogen and to estrogen suppression.18,19 As a biomarker for systemic estrogen levels within the breast, BPE has been shown to be an independent risk factor for breast cancer.18,19 Since radiologist assessments of BPE are subject to interreader variability,20 precise quantification similar to mammographic density evaluation is of interest for large-scale analysis.
Previous approaches to breast MRI segmentation of FGT and BPE often used a hybrid approach of atlas-based and statistical methods, which result in highly accurate correlation when compared with manual segmentation.21 Fat-water separation approaches have also demonstrated high accuracy for FGT separation22 but are not routinely used in many clinical breast MRI protocols, limiting availability.
Anatomic breast FGT/BPE segmentation using deep-learning techniques has resulted in high Dice similarity coefficients and relatively fast processing times. Early work using a hierarchical SVM in a small cohort demonstrated statistically significant improvement in overlap ratios compared to FCM segmentation.23 More recent breast segmentation work has relied primarily on U-net approaches, which can allow for whole image processing without the need for use of smaller “patches,” where the image is divided into much smaller sections for analysis. Dalmis et al compared a two consecutive (2C) U-net approach to a single three class (3C) U-net approach and found that the consecutive U-net approach outperformed 3C and conventional segmentation methods for FGT segmentation, but 3C U-net segmentation results correlated better with breast density on mammography.24 Of interest, no inhomogeneity artifact correction was applied, suggesting that the use of deep learning may eliminate the need for initial bias correction. However, the routine use of bias correction prior to segmentation may speed processing times.25 Similar high similarity metrics have been seen in various U-net approaches for FGT and BPE segmentation25,26 with processing times of 0.42–8.3 seconds per case and no significant difference between scanner types.27 Although many U-nets use a combination of fat-suppressed and nonfat-suppressed images, studies have demonstrated comparable results for breast FGT segmentation with or without fat suppression.28
Current approaches have primarily compared segmentation results to ground truth as defined by manual segmentation (Table 1). Future directions in BPE segmentation include the application of large-scale, proven anatomic segmentation techniques to evaluate quantified BPE as an imaging biomarker for breast cancer risk (Fig. 3).29
TABLE 1.
Review of Machine-Learning Breast Anatomic Segmentation Techniques
| Study | N= | Sequences | Segmentation | Deep-learning method | Similarity metrics |
|---|---|---|---|---|---|
| Wang 2013 | 4 | T1, T2, PD, Dixon | FGT | Hierarchical SVM | 1. Overlap ratios93.25–94.08% |
| Dalm1ș 2017 | 66 | Pre T1W WOFS | Anatomic, FGT | 1. 2C U-Net x 2 2. 3C U-Net |
1. FGT DSC = 0.811 2. FGT DSC = 0.850 |
| Xu 2018 | 50 | Pre, post T1W | Anatomic | 2D U-Net | DSC 0.9744 |
| Fashandi 2019 | 85 | Pre T1W WOFS, FS T1W | Anatomic | 1. 2CU-Net with WOFS, FS, mixed, multi-channel input 2. 3C U-Net, same inputs |
1. 2C U-net NonFS DSC = 0.96 2. 3D multi-channel DSC = 0.96 |
| Ha 2019 | 137 | Pre, post, sub | FGT, BPE | Modified 3D CNN/2D U-Net | 1. FGT DSC 0.813 2. BPE DSC 0.829 |
| Ivanovska 2019 | 40 | Pre T1W WOFS | Anatomic, FGT | 2C 2D U-Net x 2 | 3. Anat DSC 0.98 4. FGT DSC 0.932 |
| Zhang 2019 | 114 | Pre T1W WOFS | Anatomic, FGT | U-Net x 2 | 1. Anat DSC 0.86 2. FGT DSC 0.83 |
FS = fat saturated; WOFS = without fat saturation; +FGT = fibroglandular tissue; BPE = background parenchymal enhancement; Anat = anatomic segmentation (breast/chest wall/air).
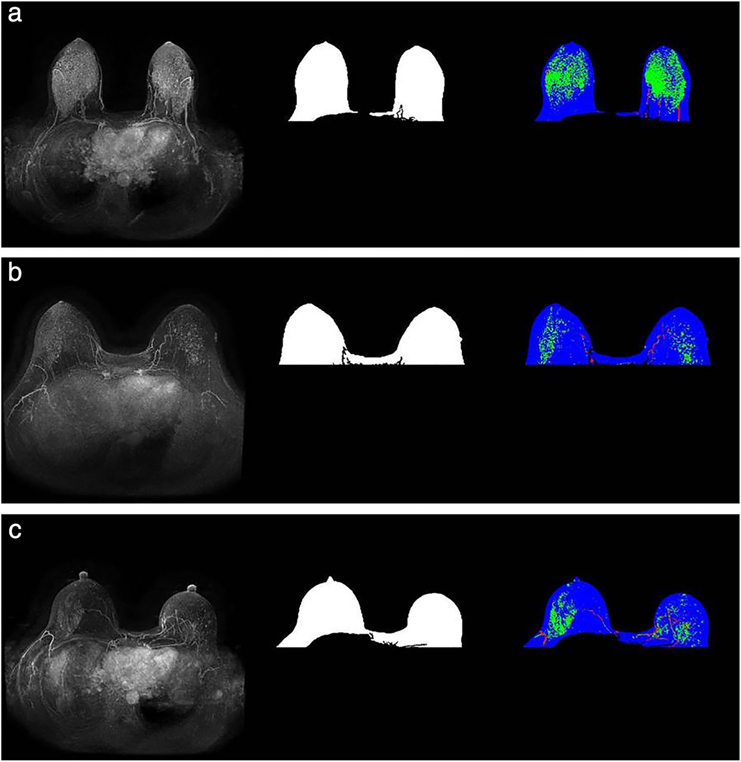
FIGURE 3:
Images from a patient who subsequently developed cancer (a) and the two matched controls (b,c, respectively) The MIPs are presented in the first column and the breast masks are shown in the second column. The third column represents enhanced FGT on the MIP in green, vessels in red, and remaining breast mask in blue. The FGT for these images was extracted from the corresponding T1 nonfat-saturated sequence. (Reprinted and adapted with permission from Saha et al. J Magn Reson Imaging 2019.)
Lesion Segmentation
Breast lesion segmentation is an emerging technique in machine learning. Early approaches to lesion segmentation often used region-growing algorithms, where a seed region of interest (ROI) was selected by an experienced radiologist and adjacent pixels matching seed ROI intensity were automatically included by the algorithm. Later refinements included statistical-based techniques, with improved similarity metrics. Fuzzy c-means (FCM) approaches in particular have been widely popularized due to ease of implementation and accuracy of results,30 although different approaches, including level set techniques, may outperform FCM in overall accuracy.31 In general, these segmentation methods work best with lesions that display a sharp contrast between lesion borders and surrounding BPE, and are less accurate with lesions with low enhancement, lesions with indistinct or vague borders (ie, diffuse nonmass enhancement), or lesions in the setting of moderate to marked BPE. Many studies using deep learning in breast analysis have therefore used statistical-based algorithms or manual annotation by radiologists to select the lesion of interest before feature extraction and model training, in what can be termed a CADx approach to lesion analysis (Fig. 4).
FIGURE 4:
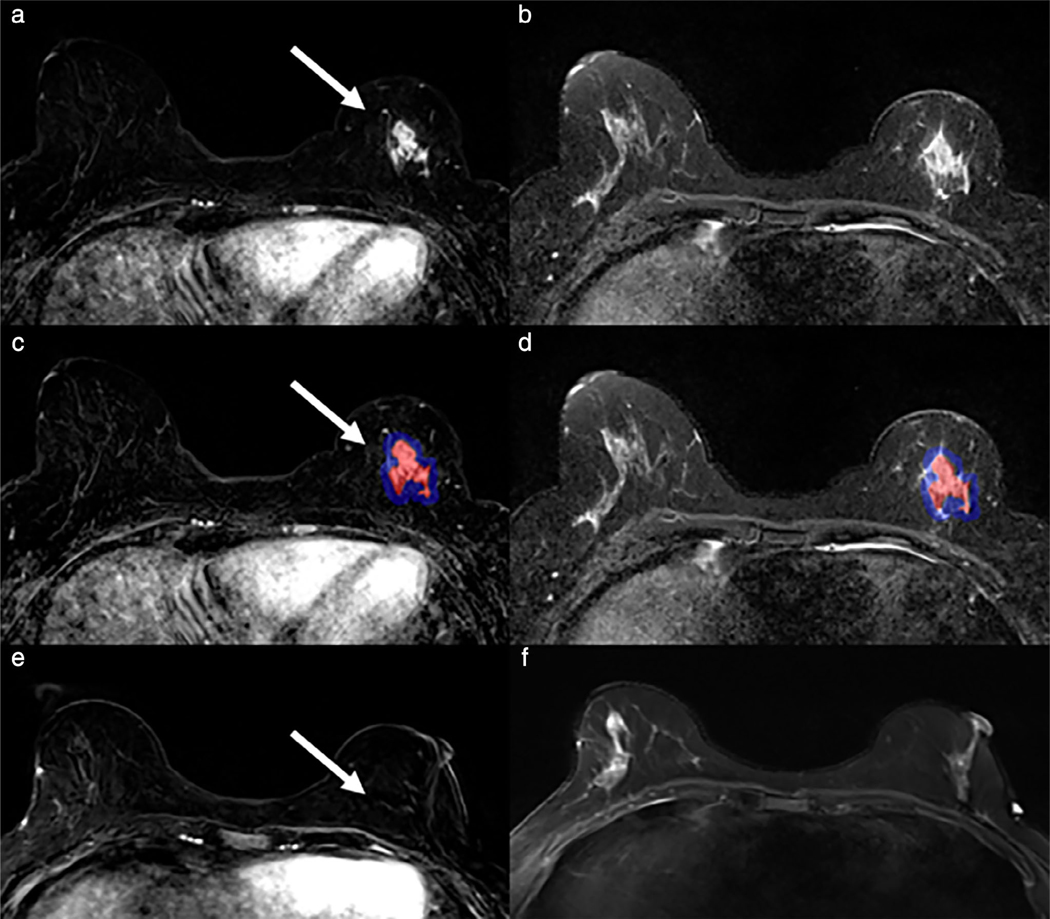
SVM analysis of known cancers offers promise in evaluating extent of disease. Maximum washin-slope and peak enhancement were associated with malignancy in SVM analysis of predictors of malignancy in ipsilateral and contralateral breast lesions. A 63-year-old woman had a new diagnosis of 8 mm right retroareolar papillary carcinoma with planned breast conservation. Breast MRI demonstrated two irregular masses and two additional foci (blue arrows) of abnormal enhancement (a, first postcontrast images). Manual volumes of interest annotating these foci on high temporal resolution MRI (b, high temporal resolution T1-weighted subtraction images at 45 seconds postcontrast) demonstrated early peak enhancement (c). Subsequent MR-directed ultrasound and MRI biopsies yielded additional papillary carcinoma and papillary lesions, leading to surgical decision for mastectomy instead of breast conservation.
While FCM and similar lesion segmentation techniques offer high accuracy30 and allow for relatively quick annotation of large datasets, many rely on the initial placement of an ROI around the lesion borders or a small enhancing portion of the lesion, usually on a single slice of interest. Such radiologist-driven ROI approaches raise the concern for the introduction of interreader bias. An evaluation of interreader reliability of radiomic features generated by such techniques found that the placement of initial ROI by expert radiologists resulted in a moderate change in the resulting extracted radiomic features.32 This suggests that initial seed ROI selection by the radiologist may be subject to bias and therefore have decreased interreader reliability. In contrast, deep-learning lesion segmentation techniques offer greater reliability and the possibility of high reproducibility across different machines33 and institutions, allowing for larger dataset analysis (Table 2).
TABLE 2.
Review of Machine-Learning Studies Evaluating Lesion Segmentation
| Study | N= | Lesion type | Initial segmentation training | Machine-learning method for segmentation | Segmentation similarity metrics |
|---|---|---|---|---|---|
| Dalm1ș 2018 | 385 | Mass, NME | Manual annotations | 3-D CNN | CPM = 0.6429 |
| Gallego-Ortiz 2018 | 792 | NME | 2D bounding box | ANN | NS |
| Herent 2019 | 50* | NS | 2D bounding box | CNN (ResNet-50) | NS |
| Zhou 2019 | 1537 | Mass | Manual annotations; image-level classification method | CNN (classification activation map + DenseNet | DSC 0.501 |
Evaluated only a single slice from each breast MRI; other studies listed evaluated the lesion using multiple slices.
NS = not specified; NME = nonmass enhancement; CNN = convolutional neural network; ANN = artificial neural network; CPM = computation performance metric; DSC = Dice similarity coefficient.
A less subjective approach to lesion segmentation includes boundary box approaches, which allow for faster dataset labeling34,35 and are less prone to bias than ROIs placed within the lesion itself. In this CADx-style approach, a radiologist denotes a general ROI by selecting a box that includes both the lesion and nonlesion surrounding tissue. A deep-learning method is then used to identify and segment the lesion within the bounding box. While U-net whole image analysis still predominates for this type of segmentation, patch-based approaches are more feasible given the smaller area to be analyzed. A limitation of this approach is that boundary boxes are often placed on single slices and may therefore still be prone to error when the choice of slice differs between radiologists.
One limitation of breast lesion segmentation is the analysis of nonmass enhancement. Machine-learning approaches, including patch-based CNNs and U-net approaches, are traditionally trained with annotations of mass-type lesions. Many studies specifically exclude NME and asymmetric BPE lesions from these training sets,34 leading to a paucity of studies evaluating the more difficult to segment NME. These may require much larger training datasets or novel approaches. One novel graph-modeling approach to NME using CNN segmentation requires CADx-style placement of an initial ROI, but offers improved accuracy compared to prior attempts at NME identification and segmentation.35
As techniques continue to evolve, it is likely that lesion-level analysis will give way to image-level analysis. In image-level analysis, the training sets are designated as “benign” or “malignant” but the precise lesion in question is not delineated for training purposes. Given sufficient processing power and layers, CNNs can be trained to identify the area of interest and then analyze features to determine percentage of malignancy. Image level annotations have been rarely tested to date, but a recent large study of 1537 patients using a 3D DenseNet CNN and image level annotations identified breast cancer with 83.7% accuracy (Fig. 5).36 The chief advantage of image-level annotations as a training set is that the time-consuming step of expert lesion annotation can be bypassed; the drawback of such an approach is that patch-based approaches that incorporate lesion annotations generally produce higher accuracy. Additional limitations of this study included its exclusion of breast MRIs with multiple findings (a more clinically common scenario) and asymmetric BPE.
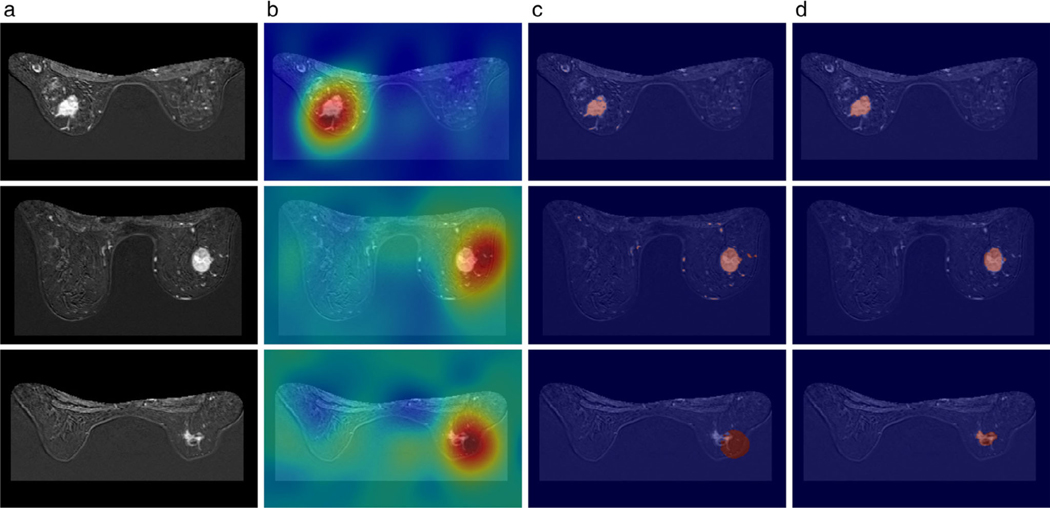
FIGURE 5:
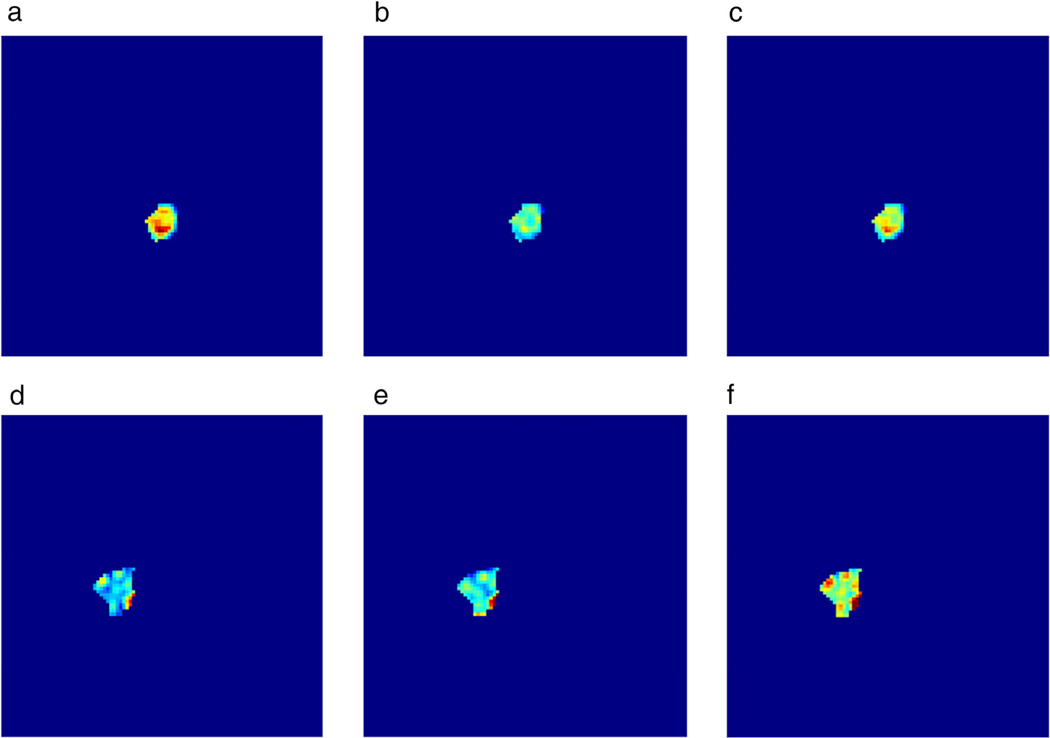
Visualization of (a) the MRI slices from three different samples, (b) the corresponding heatmap obtained from the GMP model, (c) the corresponding refined weak label using DenseCRF, and (d) the manual annotation. Fired color indicates higher values for the activations in (b). Red color indicates the annotation by model and human in (c,d). The Dice coefficients of each sample were: 0.823, 0.683, and 0.091, respectively. (Reprinted and adapted with permission from Zhou et al. J Magn Reson Imaging 2019.)
Texture Analysis
Texture analysis in radiomics refers to mathematically extracted quantitative statistical features of an image, which span a large group of related features. First-order statistical texture features, often referred to as histogram features, evaluate the grayscale intensity of pixels. Second-order and higher-order texture features evaluate the relationship between these pixels in the x and y direction (Laws energy), edge detection after filter application (Gabor), co-occurrence matrix (Haralick), or dominant intensity gradient orientations (CoLIAGe), all essentially measuring various pixel relationships in terms of heterogeneity, correlation, and entropy.37 Texture features can further be subdivided into static texture features and textural kinetics; the former usually measured from peak signal intensity of a lesion or on first postcontrast images, and the latter evaluating the change in texture parameters over time.38 First- and higher-order texture features have been shown in prior works to correlate with tumor molecular subtypes39 and pathologic complete response after neoadjuvant chemotherapy.40 These studies have demonstrated that measurements of margin irregularity and entropy correlate with malignancy. However, texture analysis is not standardized across different machine parameters and field strengths.
Breast lesions can potentially have hundreds of texture features extracted after segmentation. As a result, overfitting is a common concern and sufficient sample size for testing and validating a model is necessary. Deep learning is particularly helpful in analysis of the large volume of data generated from computer-extracted imaging features, and often incorporated with other features in radiomic analysis. In most texture studies to date, lesions have been manually annotated by radiologists or segmented by semiautomated or automated statistical algorithms, particularly FCM-based techniques.
Texture analysis is most often applied to T1-weighted imaging, including precontrast, postcontrast, and/or generated subtraction images.40–42 While texture features appear to show promise in distinguishing benign from malignant, it is also clear that they are complementary to, but do not replace, current BI-RADS descriptors of internal enhancement patterns and lesion margin.42,43 This may reflect the previous emphasis on static texture, as textural kinetics over time may offer more important dynamic information. For example, in one study, triple-negative breast cancers demonstrated increased heterogeneity at peak contrast enhancement (a static texture feature), but also increased homogeneity over time (a textural kinetic feature), when compared with other lesion types.38 While texture as evaluated on T1-weighted postcontrast imaging has shown to have high discrimination between benign and malignant lesions, the additional information from multiparametric texture approaches incorporating T2-weighted and other imaging sequences improves accuracy.44
The chief advantage of breast MRI over other breast imaging modalities is the functional information offered by the washin of contrast. Most deep-learning texture analysis therefore evaluates postcontrast T1-weighted imaging. However, recent studies of noncontrast breast imaging, particularly T2-weighted and diffusion-weighted imaging, have shown the utility of texture analysis using noncontrast imaging alone (Fig. 6).45 It is likely that these imaging sequences are best utilized when combined with postcontrast imaging for further analysis.
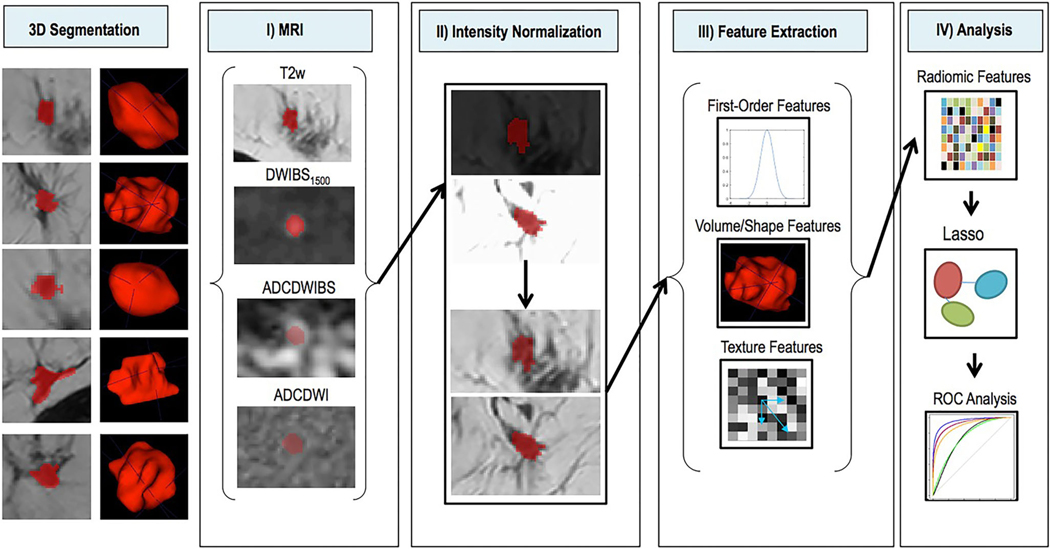
FIGURE 6:
Schematic depiction of image processing. Left: 3D segmentations of lesions shown on single T2w slices (left) and as surface shaded 3D renderings (right). I: Segmentations (red) are shown overlaid on the four imaging sequences (T2w, DWIBS1500, ADCDWIBS, and ADCDWI). II: Intensity normalization transforms variable MR signal intensities on T2w and DWI images (top) into comparable image intensities (bottom). III: Radiomic feature extraction uses first-order statistics, volumetric and texture features as defined in Data Supplement S1 to generate a multidimensional imaging signature. IV: The radiomic feature matrix and corresponding outcome data (histopathological results) are combined and used for supervised training of the Lasso regularized logistic regression model. Performance of the constructed model is compared to the performance of known standard parameters using ROC analysis. T2w = T2-weighted; DWI = diffusion-weighted imaging; DWIBS = diffusion-weighted imaging with background suppression (DWIBS); ADC = apparent diffusion coefficient: ROC = receiver operating characteristics. (Reprinted and adapted with permission from Bickelhaupt et al. J Magn Reson Imaging 2017;46:604–616.)
Traditional limitations of texture analysis have included the exclusion of lesions less than 1 cm3 in size. However, in screening breast MRI of high-risk populations, many suspicious lesions are smaller than this. Recent studies specifically evaluating lesions smaller46 than 1 cm3 (Fig. 7) or with a mean lesion size of less than 1.2 cm45 have nonetheless found texture features can discriminate between benign and malignant lesions, suggesting size may not be a limitation in future analysis.
FIGURE 7:
Initial enhancement (a,d), overall enhancement (b,e), and area under the enhancement curve (c,f) maps for a benign papilloma (a–c) and a malignant invasive ductal carcinoma (d–f). It is evident that spatially heterogeneous enhancement is present, which can thus be quantified using texture analysis. (Reprinted and adapted with permission from Gibbs et al. J Magn Reson Imaging 2019.)
As whole breast segmentation and lesion segmentation deep-learning approaches continue to evolve, evaluation of breast FGT and BPE may offer contextual clues to the breast tumor microenvironment, helping predict pathologic complete response (pCR) and risk of recurrence (Fig. 8). Braman et al cross-correlated peritumoral texture features to pathology analysis from core biopsy specimens, finding an association between high peritumoral heterogeneity in higher-order texture features and densely packed stromal tumor-infiltrating lymphocytes, with these tumors more likely to achieve pCR after neoadjuvant chemotherapy.40 Similar studies of peritumoral FGT/BPE on T1-weighted images have demonstrated a correlation between peritumoral texture features and pCR41 and tumor molecular subtype.47
FIGURE 8:
A 52-year-old woman with triple positive (ER, PR, and HER2+) high-grade invasive ductal carcinoma evaluated with breast MRI before and after four cycles neoadjuvant chemotherapy (a: first postcontrast subtraction, b: T2-weighted). 3D whole lesion volume of interest (VOI) was annotated using a seed-growing semiautomated segmentation technique on subtraction images (c, red lesion) with peritumoral region (c, blue lesion) automatically generated based on VOI. Tumor and peritumoral VOIs were then propagated to coregistered T2 images (c) and first-order texture features were analyzed. Lesions that demonstrated high T2 whole lesion entropy, T1 core lesion entropy, and T2 peritumoral skewness and kurtosis were more likely to exhibit pathologic complete response (pCR; accuracy = 74%). The patient demonstrated complete imaging response on post-neoadjuvant therapy imaging (e,f) and had pCR at final surgical pathology. (Reprinted and adapted with permission from Heacock et al. RSNA 2017.)
The role of textural kinetics remains underexplored. Preliminary data have suggested that washin textural kinetics may be more important than washout textural kinetics,46 particularly in small lesions. Initial studies of ultrafast temporal kinetics show similar high accuracy of washin kinetics (Fig. 9) and texture features.48 This remains to be confirmed in larger-scale studies of larger lesions, but is of interest as abbreviated/FAST breast MRI becomes more widespread.
FIGURE 9:
A 52-year-old woman with CHEK2 mutation undergoing high-risk screening MRI. A new 4 mm focus of enhancement (blue arrow) at left 7:00 (a,b, first postcontrast axial subtraction and sagittal images) was manually segmented in a 3D volume of interest. The segmented lesion demonstrated early washin on high temporal resolution sequences acquired in the first 60 seconds (c) but persistent temporal kinetics on washout curve analysis (d). MRI-guided biopsy yielded high-grade invasive ductal carcinoma. Early maximum slope on high temporal resolution images is associated with malignancy in SVM analysis.
Future directions in deep-learning texture analysis involve the incorporation of both multiparametric imaging sequences and whole-breast analysis (lesion, peritumoral region, and BPE). Standardization of texture parameters and large-scale, multiinstitutional trials will allow for generalizability of future results as texture analysis increasingly becomes incorporated into radiomics analysis.
Lesion Classification
Dynamic contrast-enhanced MRI is highly sensitive (89–100%) but variably specific (35–64%) in the detection of breast cancer.4 There is strong interest in developing imaging models to differentiate benign from malignant lesions to decrease the number of benign biopsies.
Radiomics studies classifying lesions into benign or malignant have most frequently used dynamic contrast-enhanced (DCE)-MRI features.31,42,49–51 These include morphological features including 2D and 3D features such as compactness and sphericity,31,42,49–51 and texture features such as GLCM,31,42,49,50 and Laws’ features.49 Pre- and multiple postcontrast time points can also be treated as a 4D dataset from which kinetic features can be extracted.50,51
Antropova et al52 used only the maximum intensity projection (MIP) for classification. By using the MIP image, the investigators sought to incorporate volumetric information in a single image, since most pretrained CNN models demand a 2D image as an input. They found that training the CNN with the MIP images improved classification as compared with training on single slices from the postcontrast sequence.
Other than DCE-MRI, MR sequences such as nonenhanced T1-weighted images,53 diffusion-weighted images,45,54 and T2-weighted images45,55 have been used to improve lesion characterization. Investigators have also developed multiparametric models combining diffusion-weighted imaging with DCE-MR44,56–58 for lesion discrimination, with accuracies up to 0.93. Specific features that were significantly different between benign and malignant lesions included entropy44 and signal enhancement ratio.58 Jiang et al56 found that adding additional parameters to a multiparametric model improved accuracy of discrimination between benign and malignant lesions. For example, adding kinetic features to a model using shape and texture features from DCE-MR improved accuracy, and adding an ADC threshold to this model improved accuracy again.
While most studies investigate tumor features only, Kim et al59 incorporated features from tumor and background parenchyma and found that the Ktrans in the fibroglandular (nontumor) tissue was significantly different between the malignant and benign groups, and was as predictive of malignancy as lesion Ktrans.
The accuracy and generalizability of the classifier will depend on the types of benign and malignant lesions that are included in the study. Whitney et al51 narrowed the classification task to benign lesions vs. luminal A breast cancers, which are a subtype of cancer that typically manifests as a spiculated and irregular mass. This is potentially an easier task than classifying benign from malignant lesions that include different histologies such as invasive ductal, invasive lobular, mixed ductal lobular, and DCIS.52,58 For the classification task to be clinically meaningful, the dataset should include malignancies that can mimic benign lesions, and benign lesions that mimic malignancy. For example, a study evaluating breast cancers vs. fibroadenomas specifically evaluated the accuracy of classifying fibroadenomas from triple-negative cancers, which are known to sometimes manifest as round or oval masses with circumscribed margins.60 It is therefore important to be explicit about what lesions were analyzed. Only Truhn et al61 explicitly included a significant number of nonmass enhancement cases in addition to masses. Their results suggest that, with a sufficiently large number of training cases, a deep-learning algorithm could learn to correctly classify a wide range of lesions.
The classifiers used in these studies include linear discriminant analysis,51 Bayesian,50 difference-weighted local hyperplane,56 and SVM.31,44,52–54 Cai et al58 evaluated four machine-learning models and found that SVM was the best classifier among the machine-learning models (less accurate algorithms were Naïve Bayes, k-nearest neighbors, and logistic regression).
Studies comparing radiomics-style handcrafted feature extraction against deep-learning-based feature extraction shows that the deep-learning techniques outperform the radiomics features in classifying benign vs. malignant tumors.61,62 Truhn et al61 noted that the performance of their deep-learning classifier improved when the number of training cases increased, although the accuracy of the radiomics algorithm did not. This suggests that the radiomics approach may reach a ceiling of attainable accuracy, but that a deep-leaning model, with its more complex and expandable structures, may continue to improve with larger datasets.
Predicting Occult Invasive Cancer in DCIS
DCIS is a noninvasive lesion bound by the mammary duct basement membrane. As it is most commonly identified by core-needle biopsy, there is a risk that associated invasive disease will be discovered at the time of surgical excision. A review of the literature showed that the upgrade rate ranges widely, reported as 3.5–56%.63 Multiple studies have attempted to predict upgrade to invasion using descriptive MR features.63,64 Radiomic and machine-learning models have the potential to improve our ability to predict invasion from MR images.
This is a relatively underexplored area. Harowicz et al,65 using radiomics features of morphology, texture, and enhancement, found that a textural feature had the highest predictive value of upstaging. A more homogeneous texture predicted invasion, which reflects the “sponge-like” appearance of DCIS as opposed to the more mass-like appearance of invasion. This same group66 used deep-learning techniques to predict upstaging, with only moderate success, with an area under the curve (AUC) of 0.68. Both of these studies used postcontrast MR images only, and required image annotation by a radiologist.
Rather than predict invasion, radiomics analysis has been used to evaluate high-risk features of DCIS.67 For example, nuclear grade is an important clinical factor, as high nuclear grade DCIS has a different prognosis than low and intermediate nuclear grade DCIS, and is more likely to progress to invasive cancer. Chou et al showed that there was no difference in MRI parameters or BI-RADS descriptors between high-risk DCIS and low-risk DCIS.67 However, a single heterogeneity metric of surface-to-volume ratio was significantly smaller in high-risk DCIS, suggesting that it is a more compact and consolidated disease. This study demonstrates that unaided visual perception is unable to decode the underpinning biological variability of disease, for which deep learning shows promise.
Lymph Node Status and Markers of Aggressiveness
There are multiple clinical predictors of tumor aggressiveness and patient outcome. These include stage, lymph node involvement, Ki-67 expression, and the presence of tumor-infiltrating lymphocytes. These are clinically important, as they are prognostic and may help determine the patient’s therapy.
Lymph node involvement is among the most important prognostic markers, as axillary lymph nodes are usually the first site of metastasis from breast cancer. Investigators have sought to predict axillary lymph node metastasis based on the radiomics of the primary tumor.50,68–70 DCE-MRI features that were predictive of lymph node involvement included morphologic irregularity,68 texture homogeneity,68 and peritumoral texture.70 In addition to DCE-MRI, noncontrast texture features obtained from T2-weighted imaging and DWI can be predictive.69 Incorporation of clinicopathologic features as well as radiomics feature may result in the most accurate model.70
While the previous studies extracted imaging features from the primary tumors, several studies have evaluated imaging features of the lymph nodes themselves.71–73 All of these studies evaluated postcontrast T1-weighted images on which radiologists identified index lymph nodes. Morphologic features were found to be more predictive than kinetic and texture features.71,72 Only one study followed a deep-learning approach73 in which handcrafted radiomics features were not obtained. A strength of this study is its independent test set that was not used for training of the neural network, which is a more robust evaluation than the leave-one-out cross validation technique.
Ki-67 is a protein associated with cell proliferation and has been found to be a predictive and prognostic marker in breast cancer. Radiomics features from DCE-MRI have been used to demonstrate that tumors with high Ki-67 expression are larger and more homogeneous than those with low expression,74 and to demonstrate that imaging features of intratumoral subregions are more predictive than whole-tumor features.75 A study showing that a classifier based on T2-MR features was better able to predict Ki-67 status, compared than one with DCE-MR features,76 suggesting that additional work needs to be done regarding which MR sequences would result in best feature selection.
Tumor-infiltrating lymphocytes (TILs) are an immunological biomarker that are associated with improved response and prognosis after neoadjuvant chemotherapy.77 They are also associated with improved prognosis after adjuvant chemotherapy in certain tumor subtypes. Wu et al78 sought to investigate the appearance of DCE-MR and TILs. In addition to imaging features, they included the mean expression of two genes related to immune infiltration and DC8+ T-cell activation. The addition of the genetic information improved the accuracy of the model. They tested the prognostic significant of their model in a group of patients with triple-negative disease, and found that lower TILs predicted by the model was associated with worse recurrence-free survival, demonstrating the predictive value of this work.
Predicting Prognosis and Likelihood of Recurrence
Breast cancer is a heterogeneous disease, and long-term survival is not well predicted for any individual. The recurrence rate is ~20% at 10 years, and the majority of patients with recurrence present with metastatic disease.79 Recurrence is itself an important prognostic indicator, as 10-year overall survival is 82% for patients without recurrence, 61% for women with local recurrence, 41% for regional recurrence, and 20% for distant recurrence (metastatic disease). There are numerous patient and tumor features associated with recurrence, including tumor molecular subtype. An imaging feature that has been associated with recurrence is an increase in relative tumor enhancement rate (relative to background).80
Quantitative radiomics approaches to predicting survival and recurrence-free survival have evaluated texture features from T2-weighted and DCE-MR.81,82 Park et al82 demonstrated that adding clinicopathological variables such as lymph node involvement and molecular subtype improved accuracy of the model. In these studies, the time span for follow-up of recurrence-free survival should be correlated with the molecular subtype, as estrogen receptor (ER)-positive tumors can recur up to 20 years after diagnosis, while human epidermal growth factor receptor (HER2)-enriched and triple-negative tumors recur much earlier.83
In patients undergoing neoadjuvant chemotherapy, the most enhancing tumor volume on either the pretreatment or early posttreatment MR was found to predict recurrence-free survival.84 Interestingly, the change in volume from baseline to early posttreatment MR did not have any predictive ability for recurrence-free survival. This is in contrast to the change in tumor size that has been used as an indicator of response to therapy, which is a short-term measure.85 The more longterm measure of treatment success in terms of recurrence-free survival can be measured from a single timepoint.
Dashevsky et al86 correlated quantitative MR features with surgical outcome. They specifically focused on resectability of HER2-positive breast tumors undergoing breast conservation therapy. They found that shape-based features were associated with reexcision; as can be expected, a more irregularly shaped tumor was more likely to require reexcision. The investigators suggest that MR features may assist surgical planning and encourage wide margins in patients who are at risk for reexcision.
Radiogenomics
Molecular Subtypes of Breast Cancer
Using gene expression patterns, Perou et al87 classified breast cancer into four intrinsic molecular subtypes: luminal A, luminal B, HER2-enriched tumors, and triple-negative/basallike tumors. Immunohistochemical surrogates, consisting of estrogen and progesterone positivity, HER2 positivity, and Ki-67 positivity are commonly used to correlate with the molecular subtypes. Luminal A patients have the most favorable prognosis, followed by Luminal B patients, who have an intermediate prognosis, while the triple-negative subtype is associated with an unfavorable prognosis.88 HER2-enriched subtypes are associated with response to targeted therapy. All subtypes have unique responses to therapy, disease-free survival, and overall survival. As a result, conventional systemic therapies are implemented based on the molecular subtype.
As imaging features are related to gene signatures, the goal is to classify genetic breast cancer subtypes from MRI. Radiomics-style approaches using large numbers of quantitative imaging features were shown to be superior to the qualitative approach.89 Naturally, the methodology has evolved to using ML models to improve classification.
These studies commonly evaluate DCE-MR images,extracting morphologic and texture features.39,60,90 Morphologic features are most useful for differentiating subtypes that are commonly circumscribed (such as triple negative) from subtypes that are commonly spiculated (such as HER2-enriched).60 Texture is most useful in separating subtypes with similar morphology but different internal heterogeneity, such as luminal A from luminal B.90 Adding commonly-obtained clinical information such as nuclear and histologic grade can improve the accuracy of the model.39 In addition to postcontrast imaging, precontrast T1 images90 and DW-MR images54 have been evaluated, both of which have shown promise in representing intratumor heterogeneity.
In addition to tumor features, several investigators have included background parenchymal features in their evaluation.47,91–93 Wang et al47 found that background parenchymal enhancement (BPE) texture features were more accurate than clinical and tumor features. They found a significant association between BPE heterogeneity and triple-negative breast cancers. Fan et al92 found that fusing intratumoral and peritumoral characteristics increased prediction accuracy.
These radiomics-style studies depended on feature engineering using semiautomated feature extraction. In contrast, deep-learning approaches have been attempted in which features are automatically extracted.94,95 Both of these studies used postcontrast MR images that were annotated by a radiologist. Ha et al94 used a customized neural network as feature extractor and classifier. Zhu et al95 used VGGNet, which is a neural network that is freely available and can be pretrained with an online image database, as a feature extractor and then an SVM as a classifier.
While the prior studies have used imaging to predict underlying tumor biology, the integration of radiomic and genomic features may allow more sophisticated and accurate modeling of tumor biology. A study96 of radiomics features obtained from DCE-MRI as well as genomic features obtained from 70 breast cancer genes attempted to model the radiogenomic features against predictive clinical outcomes. The radiomic features predicted pathologic stage, while the genomic features predicted ER and PR status. This suggests that radiomic and genomic features are complementary in describing tumor biology.
Radiogenomics also shows promise in the discovery of new genetic signatures.97–100 In these studies, correlations between radiomic and genomic profiles are explored in an unsupervised way. These can lead to new associations; for example, that a certain MR feature (enhancing rim fraction score) is associated with expression of a long noncoding RNA that is known to be associated breast cancer progression and metastasis.98,101 This reveals a possible link between a molecular pathway and its manifestation at a macroscopic imaging level.
Genomic Predictors of Recurrence
Advances in adjuvant chemotherapy, hormonal agents, and radiotherapy have contributed to improved breast cancer mortality over the past 30 years. However, on an individual level, there is concern for overtreatment of patients who may have excellent long-term survival without chemotherapy. The personalization of breast cancer treatment is therefore an essential topic as researchers and clinicians seek to predict each patient’s risk of relapse and individually tailored therapy.
Commercially available gene expression assays can now predict prognosis and/or effectiveness of treatment in certain breast cancers. Commonly used assays are MammPrint, Oncotype DX, and PAM50. These are in current clinical use for identifying patients with low-risk tumors who would not benefit from chemotherapy. The downsides of these tests are their cost and the time needed to wait for results. There has therefore been interest in using MRI to similarly predict patient outcome and tumor genomics.
Quantitative radiomics approaches have compared imaging features with recurrence scores from commercially available assays.102–105 Features found to be associated with higher risk of recurrence include large size and more heterogeneous enhancement,105 and increased kurtosis on postcontrast series.103
Saha et al104 compared computer-extracted features from contrast-enhanced MR to Oncotype DX recurrence scores. They trained ML models to predict high and low scores, yielding an AUC of 0.77. The strength of this study was its larger number of patients (261 total) and the use of separate training and test sets.
Response to Neoadjuvant Chemotherapy
Neoadjuvant or preoperative chemotherapy use can have several potential advantages, including shrinking tumor size to permit breast-conserving surgery in patients who would have needed mastectomy, as well as a prognostic indicator, since patients with pathologic complete response (pCR) after therapy have improved survival compared with those with residual disease.106
Response to neoadjuvant therapy can be mixed, with large studies showing an average pCR rate of 19%.107 The pCR rate depends on cancer subtype, ranging from 0.3–38.7%. Given these low pCR rates, and that pCR can only be evaluated once the patient has had surgery, there has been great interest in using imaging as a surrogate marker for response to therapy. DCE-MRI is considered more accurate and sensitive for the detection of residual disease than other imaging modalities, with sensitivity 83–87% and specificity of 54–83%.108 More important, MRI shows promise in predicting response to therapy. The landmark I-SPY trial compared the tumor volume on pretreatment MR to volume after some cycles of chemotherapy and showed a change in tumor volume early in therapy could predict pCR.109
Supervised ML studies comparing pretreatment MR to MR performed after one or two cycles of chemotherapy have found that changes in lesion size in three dimensions,110 percentage change in DCE-MRI parametric maps,111 and change in heterogeneity of the most-enhancing tumor subregion112 were good predictors of early response to therapy. Other studies evaluating pCR found that models incorporating data from pretreatment MR and early posttreatment MR outperformed models using only pretreatment data.113,114
Despite this, the ability to predict the response to therapy on pretherapy imaging alone is the ultimate goal. This would allow clinicians to better plan the timing of surgery and chemotherapy, and to provide more individualized prognosis for the patient. Multiple supervised ML studies have sought to predict pCR using pretreatment MR images alone (Table 3). Most of these have focused on DCE-MR imaging, evaluating kinetic, textural, and morphologic tumor features.41,115,116 In an example of the large number of features that can be extracted, Braman et al40 used Gavor wavelet, co-occurrence measures and energy measures to generate 1980 features from DCE-MR images.
TABLE 3.
Review of Machine-Learning Studies Predicting Response to Neoadjuvant Chemotherapy
| Study | Patients (N, subgroups) | MR sequences | Features | Machine-learning classifier | Results |
|---|---|---|---|---|---|
| Braman (2017) | 117 HR+, TN, HER2+ | DCE, pretherapy | Intratumoral and peritumoral texture | LDA, DLDA,* quadratic discriminant analysis, naive Bayes,* SVM | AUC 0.78 all patients; AUC 0.93 for TN/HER2+ |
| Cain (2019) | 288 HR+, TN/HER2+ | 1st postcontrast subtraction, pretherapy | Fibroglandular tissue (nontumor) and tumor volume, | enhancement, texture | Logistic regression,* SVM |
| AUC 0.707 in TN/HER2+ Tahmassebi (2019) | 38 | T2, DCE, DWI, pretherapy | BIRADS descriptors, | ||
| pharmacokinetic, ADC values | SVM, LDA, logistic regression, random forests, stochastic gradient descent, decision tree, adaptive boosting, XGBoost* | AUC 0.86 for RCB class | |||
| Machireddy (2019) | 55 | DCE, pretherapy and after 1st cycle | Texture, | multiresolution fractal analysis | SVM |
| AUC 0.91 Banerjee (2018) | 53, TN | DCE, pre and posttherapy | Intensity, texture, shape, Riesz wavelets | Lasso, SVM | AUC 0.83 |
| Johansen (2007) | 24 | DCE, pre and after 1st cycle of therapy | Pre and | posttreatment change in signal intensity | Probabilistic neural network and Kohonen neural network |
| Significant difference between pCR and non-pCR groups | |||||
| Aghaei (2015) | 68 | DCE, pretherapy | Kinetics of necrotic and enhancing tumor, background parenchyma | ANN | AUC 0.96 |
| Giannini (2017) | 44 | 1st postcontrast subtraction, pretherapy | Texture | Bayesian | 70% accuracy |
| Wu (2016) | 35 | DCE, before and after first cycle of chemo | Texture within tumor subregions | LASSO and logistic regression | AUC 0.79 |
| Liu (2019) | 414, HR+, TN, HER2+ | T2, DWI, postcontrast, pretherapy | Morphology, texture, wavelet | SVM | AUC 0.79 |
| Braman (2019) | 209, HR+, TN, HER2+ | DCE, pretherapy | Intratumoral and peritumoral texture | DLDA | AUC 0.89 |
| Aghaei (2016) | 151 | DCE, pretherapy | Global kinetic (both breasts) | ANN | AUC 0.83 |
| Fan (2017) | 57 | DCE, pretherapy | Morphology, dynamic, texture | Wrapper Subset Evaluator | AUC 0.874 |
| Ha (2018) | 141, HR+, triple positive, TN, HER2+ | First postcontrast T1, pretherapy | (unsupervised learning) | CNN | 88% accuracy |
| Ravichandran (2018) | 168, HER2 status | Pre and postcontrast, pretherapy | (unsupervised learning) | CNN | AUC 0.85 |
HR = hormone receptor; LDA = linear discriminant analysis; DLDA = diagonal linear discriminant analysis; SVM = support vector machine; CNN = convolutional neural network; TN = triple negative.
Better-performing classifier.
In using kinetic features of the tumor itself to predict pCR, Aghaei et al115 found that including and excluding the necrotic regions of the tumor resulted in different performance, and that the features computed from the active enhancing tumor are most salient. Similarly, Wu et al112 partitioned the tumor into subregions with similar enhancement patterns and found that a subregion associated with rapid washout of contrast agent played a dominant role in predicting response to therapy.
Adding multiparametric data to DCE-MRI data increases the type and diversity of features available for modeling and shows promise in increasing accuracy.117 A study including T2-weighted and DWI imaging as well as DCE demonstrated that T2 features (peritumoral edema) and DWI features (minimum ADC) were among the most relevant features for predicting residual cancer burden.110 There is also evidence that precontrast imaging may be more predictive than postcontrast imaging.118
Evaluating peritumoral features or background parenchymal features in addition to tumor-specific features has been found to improve performance. Braman et al40,119 found that combining texture features from the intratumoral and peritumoral spaces yielded the best performance in predicting pCR. In a study of peritumoral features of HER2-positive tumors, the authors posit that the peritumoral microenvironment may be among the most important factors in breast cancer and development, particularly since tumor-infiltrating lymphocytes in the stroma surrounding these tumors have been strongly associated with improved therapeutic outcomes.119 A study of 10 global kinetic image features, which included both breasts and were not specific to the tumor, showed that several features from the global breast MR images including average contrast enhancement and standard deviation of the contrast enhancement improved accuracy when added to a parameter that reflected the tumor itself.120 Similarly, a study of morphologic, kinetic, and texture features of the tumor and background parenchyma found that the most accurate model included six tumoral and six background features, of which five were from the contralateral breast.121
In evaluating response to therapy, patient selection is important, as the different tumor subtypes have different probabilities of response to therapy and different underlying biology of response to therapy. A study of textural features obtained from DCE-MRI showed that separating hormone receptor-positive tumors and triple-negative/HER2-positive tumors into two groups improved accuracy.40 Interestingly, the radiomic features most predictive of response vary across different receptor subtypes.40,41 In a study of pCR in hormone receptor-positive, HER2 positive, and triple-negative subgroups, the model was most accurate in the triple-negative subgroup.117
Attention to the outcome classifier is important, as some studies have used imaging response or RECIST criteria to classify patients.115,120,121 However, the gold standard should be comparison with surgical pathology, as MRI is only 83–87% sensitive in detecting residual disease.108 It may also be useful to separate the category of partial response from nonresponder/progressive disease, as these categories are clinically relevant.122
A limitation of multiple supervised ML studies is their small sample size that precludes using separate training and test sets. The larger studies of 117 patients,40 288 patients,41 and 414 patients117 did use independent test sets, meaning that those tumors were not used in the training of the ML classifier and could therefore be more reliable tests of model performance and generalizability. Use of an independent validation cohort from a different institution can be used to demonstrate generalizability of the ML model performance.117,119 Even a relatively small study can create an independent test set, as a study of 55 patients used 15 as its validation set.111
Beyond the supervised ML studies described, a few investigators have used unsupervised ML techniques to predict response to therapy.123,124 Using deep learning in this way allows investigators to skip the selection of radiomics features, their extraction, and subsequent analysis to select the most predictive features. Using computing power and MR images as training data, a deep neural network can be trained to identify predictive imaging features in an unsupervised fashion. In these studies, a radiologist either segmented the tumor or chose the DCE-MR slice with the largest tumor area, and these images were then fed to the CNN for training. A separate validation set is required to evaluate the model. Clinical information can also be added to the model, and HER2 status has been found to improve accuracy.124 This is particularly important, as these studies evaluated a mix of breast cancer subtypes, and either separating the subtypes or providing clinical information to reveal the underlying subtype will be important to further refine the models.
A last area of study is whether imaging can predict response to therapy in the axillary lymph nodes. This is an emerging field of study, as sentinel lymph node surgery is becoming accepted after neoadjuvant therapy in node-positive patients.125 Ha et al126 trained a CNN on the pretreatment MRI of the primary tumor in patients who had a biopsy-proven lymph node metastasis prior to neoadjuvant chemotherapy. 38.6% of patients achieved a pCR in the axilla, and the CNN achieved an accuracy of 83% with AUC of 0.93 in predicting axillary response.
Discussion
The field of ML in breast MRI is rapidly evolving, with advances in lesion detection, lesion classification, radiogenomics, and prediction of response to neoadjuvant chemotherapy. Both supervised and unsupervised ML techniques require continued study, as they have not yet achieved clinical applicability. A major hurdle is the current lack of standardization; as we have demonstrated in this review of the current literature, there is no standard method of segmentation, feature extraction, feature selection, or classification. In addition, the clinical relevance of these techniques has not yet been demonstrated; this will require large datasets, with subgroup analysis by patient group and/or tumor type, and subsequent independent testing. The problem of small sample sizes is notable, as ML techniques require large datasets for training, particularly when the image class to be identified (ie, malignancy) is rare compared with the other classes (ie, benign lesions). An inherent limitation of ML is that the decision-making process of the model is so complex that why or how a model gave a certain answer is difficult to understand, resulting in these models sometimes being called a “black box.” Future directions include: 1) decreasing scan time by using ML in MR acquisition and reconstruction techniques, 2) unsupervised ML from raw (unprocessed) MR images, 3) expanding the uses of noncontrast MRI, possibly to replace DCE-MRI, 4) improving personalized risk assessment with BPE texture analysis, 5) incorporating more clinical, genetic, and pathologic information including genetic and pathologic data extracted by ML,127 and 6) prediction of patient’s response to therapy and outcome, for individualized treatment planning.
Acknowledgments
Contract grant sponsor: NIH; Contract grant number: R21CA225175-01
References
- 1.Sardanelli F, Boetes C, Borisch B, et al. Magnetic resonance imaging of the breast: Recommendations from the EUSOMA working group. Eur J Cancer 2010;46:1296–1316. [DOI] [PubMed] [Google Scholar]
- 2.ACR practice parameter for the performance of contrast-enhanced magnetic resonance imaging of the breast Resolution 34 American College of Radiology Website; 2018. [Google Scholar]
- 3.Mann RM, Kuhl CK, Moy L. Contrast-enhanced MRI for breast cancer screening. J Magn Reson Imaging 2019. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuhl C. The current status of breast MR imaging. Part I. Choice of technique, image interpretation, diagnostic accuracy, and transfer to clinical practice. Radiology 2007;244:356–378. [DOI] [PubMed] [Google Scholar]
- 5.Rumelhart DE, McClelland JL. Learning internal representations by error propagation In: Parallel distributed processing, volume 1: Explorations in the microstructure of cognition; MITP; 1987. [Google Scholar]
- 6.Deng J DW, Socher R, Li L, Li K, Fei-Fei L. ImageNet: A large-scale hierarchical image database. IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR) Miami, FL; 2009. p 248–255. [Google Scholar]
- 7.Krizhevsky A, Sutskeyer I, Hinton GE. ImageNet classification with deep convolutional neural networks. NIPS’12 Proceedings of the 25th International Conference on Neural Information Processing Systems. Volume 1 Lake Tahoe, NV; 2012. p 1097–1105. [Google Scholar]
- 8.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. 2014. arXiv 1409.1556. [Google Scholar]
- 9.Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. 2014. arXiv 1409.4842. [Google Scholar]
- 10.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016. p 770–778. [Google Scholar]
- 11.Huang G LZ, van der Maatn L, Weinberg KQ. Densely connected convolutional networks. IEEE Conference on Computer Vision and Pattern Recognition; 2017. p 2261–2269. [Google Scholar]
- 12.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional networks for biomedical image segmentation In: Navab NHJ, Wells W, Frangi A, editor. Medical image computing and computer-assisted intervetion — MICCAI Lecture Notes in Computer Science, vol 9351: Berlin: Springer; 2015. [Google Scholar]
- 13.Milletari F, Navab N, Ahmadi S. V-Net: Fully convolutional neural networks for volumetric medical image segmentation. Fourth International Conference on 3D Vision Stanford, CA; 2016. p 565–571. [Google Scholar]
- 14.Giger ML. Machine learning in medical imaging. J Am Coll Radiol 2018;15(3 Pt B):512–520. [DOI] [PubMed] [Google Scholar]
- 15.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images are more than pictures, they are data. Radiology 2016;278:563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhl CK, Kooijman H, Gieseke J, Schild HH. Effect of B1 inhomogeneity on breast MR imaging at 3.0 T. Radiology 2007;244:929–930. [DOI] [PubMed] [Google Scholar]
- 17.Boyd NF, Guo H, Martin LJ, et al. Mammographic density and the risk and detection of breast cancer. N Engl J Med 2007;356:227–236. [DOI] [PubMed] [Google Scholar]
- 18.Dontchos BN, Rahbar H, Partridge SC, et al. Are qualitative assessments of background parenchymal enhancement, amount of fibroglandular tissue on MR images, and mammographic density associated with breast cancer risk? Radiology 2015;276:371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King V, Brooks JD, Bernstein JL, Reiner AS, Pike MC, Morris EA. Background parenchymal enhancement at breast MR imaging and breast cancer risk. Radiology 2011;260:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bignotti B, Calabrese M, Signori A, et al. Background parenchymal enhancement assessment: Inter- and intra-rater reliability across breast MRI sequences. Eur J Radiol 2019;114:57–61. [DOI] [PubMed] [Google Scholar]
- 21.Wu S, Weinstein SP, Conant EF, Kontos D. Automated fibroglandular tissue segmentation and volumetric density estimation in breast MRI using an atlas-aided fuzzy C-means method. Med Phys 2013;40: 122302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wengert GJ, Helbich TH, Vogl WD, et al. Introduction of an automated user-independent quantitative volumetric magnetic resonance imaging breast density measurement system using the Dixon sequence: Comparison with mammographic breast density assessment. Invest Radiol 2015;50:73–80. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Morrell G, Heibrun ME, Payne A, Parker DL. 3D multiparametric breast MRI segmentation using hierarchical support vector machine with coil sensitivity correction. Acad Radiol 2013;20: 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalmis MU, Litjens G, Holland K, et al. Using deep learning to segment breast and fibroglandular tissue in MRI volumes. Med Phys 2017;44:533–546. [DOI] [PubMed] [Google Scholar]
- 25.Ivanovska T, Jentschke TG, Daboul A, Hegenscheid K, Volzke H, Worgotter F. A deep learning framework for efficient analysis of breast volume and fibroglandular tissue using MR data with strong artifacts. Int J Comput Assist Radiol Surg 2019. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 26.Ha R, Chang P, Mema E, et al. Fully automated convolutional neural network method for quantification of breast MRI fibroglandular tissue and background parenchymal enhancement. J Digit Imaging 2019;32: 141–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Chen JH, Chang KT, et al. Automatic breast and fibroglandular tissue segmentation in breast MRI using deep learning by a fully-convolutional residual neural network U-Net. Acad Radiol 2019. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fashandi H, Kuling G, Lu Y, Wu H, Martel AL. An investigation of the effect of fat suppression and dimensionality on the accuracy of breast MRI segmentation using U-nets. Med Phys 2019;46:1230–1244. [DOI] [PubMed] [Google Scholar]
- 29.Saha A, Grimm LJ, Ghate SV, et al. Machine learning-based prediction of future breast cancer using algorithmically measured background parenchymal enhancement on high-risk screening MRI. J Magn Reson Imaging 2019. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen W, Giger ML, Bick U. A fuzzy c-means (FCM)-based approach for computerized segmentation of breast lesions in dynamic contrast-enhanced MR images. Acad Radiol 2006;13:63–72. [DOI] [PubMed] [Google Scholar]
- 31.Pang Z, Zhu D, Chen D, Li L, Shao Y. A computer-aided diagnosis system for dynamic contrast-enhanced MR images based on level set segmentation and relief feature selection. Comput Math Methods Med 2015;2015:450531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saha A, Harowicz MR, Mazurowski MA. Breast cancer MRI radiomics: An overview of algorithmic features and impact of inter-reader variability in annotating tumors. Med Phys 2018;45:3076–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dalmis MU, Vreemann S, Kooi T, Mann RM, Karssemeijer N, Gubern-Merida A. Fully automated detection of breast cancer in screening MRI using convolutional neural networks. J Med Imaging (Bellingham) 2018;5:014502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herent P, Schmauch B, Jehanno P, et al. Detection and characterization of MRI breast lesions using deep learning. Diagn Interv Imaging 2019;100:219–225. [DOI] [PubMed] [Google Scholar]
- 35.Gallego-Ortiz C, Martel AL. A graph-based lesion characterization and deep embedding approach for improved computer-aided diagnosis of nonmass breast MRI lesions. Med Image Anal 2019;51:116–124. [DOI] [PubMed] [Google Scholar]
- 36.Zhou J, Luo LY, Dou Q, et al. Weakly supervised 3D deep learning for breast cancer classification and localization of the lesions in MR images. J Magn Reson Imaging 2019. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 37.Chitalia RD, Kontos D. Role of texture analysis in breast MRI as a cancer biomarker: A review. J Magn Reson Imaging 2019;49:927–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agner SC, Soman S, Libfeld E, et al. Textural kinetics: A novel dynamic contrast-enhanced (DCE)-MRI feature for breast lesion classification. J Digit Imaging 2011;24:446–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sutton EJ, Dashevsky BZ, Oh JH, et al. Breast cancer molecular subtype classifier that incorporates MRI features. J Magn Reson Imaging 2016;44:122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braman NM, Etesami M, Prasanna P, et al. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res 2017;19:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cain EH, Saha A, Harowicz MR, Marks JR, Marcom PK, Mazurowski MA. Multivariate machine learning models for prediction of pathologic response to neoadjuvant therapy in breast cancer using MRI features: A study using an independent validation set. Breast Cancer Res Treat 2019;173:455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nie K, Chen JH, Yu HJ, Chu Y, Nalcioglu O, Su MY. Quantitative analysis of lesion morphology and texture features for diagnostic prediction in breast MRI. Acad Radiol 2008;15:1513–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sutton EJ, Huang EP, Drukker K, et al. Breast MRI radiomics: Comparison of computer- and human-extracted imaging phenotypes. Eur Radiol Exp 2017;1:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parekh VS, Jacobs MA. Integrated radiomic framework for breast cancer and tumor biology using advanced machine learning and multiparametric MRI. NPJ Breast Cancer 2017;3:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bickelhaupt S, Paech D, Kickingereder P, et al. Prediction of malignancy by a radiomic signature from contrast agent-free diffusion MRI in suspicious breast lesions found on screening mammography. J Magn Reson Imaging 2017;46:604–616. [DOI] [PubMed] [Google Scholar]
- 46.Gibbs P, Onishi N, Sadinski M, et al. Characterization of sub-1 cm breast lesions using radiomics analysis. J Magn Reson Imaging 2019. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Kato F, Oyama-Manabe N, et al. Identifying triple-negative breast cancer using background parenchymal enhancement heterogeneity on dynamic contrast-enhanced MRI: A pilot radiomics study. PLoS One 2015;10:e0143308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Milenkovic J, Dalmis MU, Zgajnar J, Platel B. Textural analysis of early-phase spatiotemporal changes in contrast enhancement of breast lesions imaged with an ultrafast DCE-MRI protocol. Med Phys 2017;44:4652–4664. [DOI] [PubMed] [Google Scholar]
- 49.McLaren CE, Chen WP, Nie K, Su MY. Prediction of malignant breast lesions from MRI features: A comparison of artificial neural network and logistic regression techniques. Acad Radiol 2009;16:842–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhooshan N, Giger ML, Jansen SA, Li H, Lan L, Newstead GM. Cancerous breast lesions on dynamic contrast-enhanced MR images: Computerized characterization for image-based prognostic markers. Radiology 2010;254:680–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whitney HM, Taylor NS, Drukker K, et al. Additive benefit of radiomics over size alone in the distinction between benign lesions and luminal A cancers on a large clinical breast MRI dataset. Acad Radiol 2019;26:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Antropova N, Abe H, Giger ML. Use of clinical MRI maximum intensity projections for improved breast lesion classification with deep convolutional neural networks. J Med Imaging (Bellingham) 2018;5:014503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Juntu J, Sijbers J, De Backer S, Rajan J, Van Dyck D. Machine learning study of several classifiers trained with texture analysis features to differentiate benign from malignant soft-tissue tumors in T1-MRI images. J Magn Reson Imaging 2010;31:680–689. [DOI] [PubMed] [Google Scholar]
- 54.Vidic I, Egnell L, Jerome NP, et al. Support vector machine for breast cancer classification using diffusion-weighted MRI histogram features: Preliminary study. J Magn Reson Imaging 2018;47:1205–1216. [DOI] [PubMed] [Google Scholar]
- 55.Gallego-Ortiz C, Martel AL. Using quantitative features extracted from T2-weighted MRI to improve breast MRI computer-aided diagnosis (CAD). PLoS One 2017;12:e0187501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang X, Xie F, Liu L, Peng Y, Cai H, Li L. Discrimination of malignant and benign breast masses using automatic segmentation and features extracted from dynamic contrast-enhanced and diffusion-weighted MRI. Oncol Lett 2018;16:1521–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu B, Xu K, Zhang Z, Chai R, Li S, Zhang L. A radiomic nomogram based on an apparent diffusion coefficient map for differential diagnosis of suspicious breast findings. Chin J Cancer Res 2018;30:432–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai H, Peng Y, Ou C, Chen M, Li L. Diagnosis of breast masses from dynamic contrast-enhanced and diffusion-weighted MR: A machine learning approach. PLoS One 2014;9:e87387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim SG, Freed M, Leite APK, Zhang J, Seuss C, Moy L. Separation of benign and malignant breast lesions using dynamic contrast enhanced MRI in a biopsy cohort. J Magn Reson Imaging 2017;45:1385–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agner SC, Rosen MA, Englander S, et al. Computerized image analysis for identifying triple-negative breast cancers and differentiating them from other molecular subtypes of breast cancer on dynamic contrast-enhanced MR images: A feasibility study. Radiology 2014; 272:91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Truhn D, Schrading S, Haarburger C, Schneider H, Merhof D, Kuhl C. Radiomic versus convolutional neural networks analysis for classification of contrast-enhancing lesions at multiparametric breast MRI. Radiology 2019;290:290–297. [DOI] [PubMed] [Google Scholar]
- 62.Antropova N, Huynh BQ, Giger ML. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets. Med Phys 2017;44:5162–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee CW, Wu HK, Lai HW, et al. Preoperative clinicopathologic factors and breast magnetic resonance imaging features can predict ductal carcinoma in situ with invasive components. Eur J Radiol 2016;85: 780–789. [DOI] [PubMed] [Google Scholar]
- 64.Wisner DJ, Hwang ES, Chang CB, et al. Features of occult invasion in biopsy-proven DCIS at breast MRI. Breast J 2013;19:650–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harowicz MR, Saha A, Grimm LJ, et al. Can algorithmically assessed MRI features predict which patients with a preoperative diagnosis of ductal carcinoma in situ are upstaged to invasive breast cancer? J Magn Reson Imaging 2017;46:1332–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu Z HM, Zhang J, Ashirbani S, Grimm LJ, Hwang S, Mazurowski MA. Deep learning-based features of breast MRI for prediction of occult invasive disease following a diagnosis of ductal carcinoma in situ: Preliminary data. SPIE Medical Imaging. Volume 10575. Houston, TX; 2018. [Google Scholar]
- 67.Chou SS, Gombos EC, Chikarmane SA, Giess CS, Jayender J. Computer-aided heterogeneity analysis in breast MR imaging assessment of ductal carcinoma in situ: Correlating histologic grade and receptor status. J Magn Reson Imaging 2017;46:1748–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burnside ES, Drukker K, Li H, et al. Using computer-extracted image phenotypes from tumors on breast magnetic resonance imaging to predict breast cancer pathologic stage. Cancer 2016;122:748–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dong Y, Feng Q, Yang W, et al. Preoperative prediction of sentinel lymph node metastasis in breast cancer based on radiomics of T2-weighted fat-suppression and diffusion-weighted MRI. Eur Radiol 2018;28:582–591. [DOI] [PubMed] [Google Scholar]
- 70.Liu C, Ding J, Spuhler K, et al. Preoperative prediction of sentinel lymph node metastasis in breast cancer by radiomic signatures from dynamic contrast-enhanced MRI. J Magn Reson Imaging 2019;49: 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schacht DV, Drukker K, Pak I, Abe H, Giger ML. Using quantitative image analysis to classify axillary lymph nodes on breast MRI: A new application for the Z 0011 Era. Eur J Radiol 2015;84:392–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fusco R, Sansone M, Granata V, et al. Use of quantitative morphological and functional features for assessment of axillary lymph node in breast dynamic contrast-enhanced magnetic resonance imaging. Biomed Res Int 2018;2018:2610801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ha R, Chang P, Karcich J, et al. Axillary lymph node evaluation utilizing convolutional neural networks using MRI dataset. J Digit Imaging 2018;31:851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Juan MW, Yu J, Peng GX, Jun LJ, Feng SP, Fang LP. Correlation between DCE-MRI radiomics features and Ki-67 expression in invasive breast cancer. Oncol Lett 2018;16:5084–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fan M, Cheng H, Zhang P, et al. DCE-MRI texture analysis with tumor subregion partitioning for predicting Ki-67 status of estrogen receptor-positive breast cancers. J Magn Reson Imaging 2018;48: 237–247. [DOI] [PubMed] [Google Scholar]
- 76.Liang C, Cheng Z, Huang Y, et al. An MRI-based radiomics classifier for preoperative prediction of Ki-67 status in breast cancer. Acad Radiol 2018;25:1111–1117. [DOI] [PubMed] [Google Scholar]
- 77.Dieci MV, Radosevic-Robin N, Fineberg S, et al. Update on tumor-infiltrating lymphocytes (TILs) in breast cancer, including recommendations to assess TILs in residual disease after neoadjuvant therapy and in carcinoma in situ: A report of the International Immuno-Oncology Biomarker Working Group on Breast Cancer. Semin Cancer Biol 2018;52(Pt 2):16–25. [DOI] [PubMed] [Google Scholar]
- 78.Wu J, Li X, Teng X, et al. Magnetic resonance imaging and molecular features associated with tumor-infiltrating lymphocytes in breast cancer. Breast Cancer Res 2018;20:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Geurts YM, Witteveen A, Bretveld R, et al. Patterns and predictors of first and subsequent recurrence in women with early breast cancer. Breast Cancer Res Treat 2017;165:709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mazurowski MA, Grimm LJ, Zhang J, et al. Recurrence-free survival in breast cancer is associated with MRI tumor enhancement dynamics quantified using computer algorithms. Eur J Radiol 2015;84:2117–2122. [DOI] [PubMed] [Google Scholar]
- 81.Huang SY, Franc BL, Harnish RJ, et al. Exploration of PET and MRI radiomic features for decoding breast cancer phenotypes and prognosis. NPJ Breast Cancer 2018;4:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Park H, Lim Y, Ko ES, et al. Radiomics signature on magnetic resonance imaging: Association with disease-free survival in patients with invasive breast cancer. Clin Cancer Res 2018;24:4705–4714. [DOI] [PubMed] [Google Scholar]
- 83.Rueda OM, Sammut SJ, Seoane JA, et al. Dynamics of breast-cancer relapse reveal late-recurring ER-positive genomic subgroups. Nature 2019;567:399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Drukker K, Li H, Antropova N, Edwards A, Papaioannou J, Giger ML. Most-enhancing tumor volume by MRI radiomics predicts recurrence-free survival “early on” in neoadjuvant treatment of breast cancer. Cancer Imaging 2018;18:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marinovich ML, Sardanelli F, Ciatto S, et al. Early prediction of pathologic response to neoadjuvant therapy in breast cancer: Systematic review of the accuracy of MRI. Breast 2012;21:669–677. [DOI] [PubMed] [Google Scholar]
- 86.Dashevsky BZ, Oh JH, Apte AP, et al. MRI features predictive of negative surgical margins in patients with HER2 overexpressing breast cancer undergoing breast conservation. Sci Rep 2018;8:315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature 2000;406:747–752. [DOI] [PubMed] [Google Scholar]
- 88.Iborra S, Stickeler E. HER2-orientated therapy in early and metastatic breast cancer. Breast Care (Basel) 2016;11:392–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu J, Sun X, Wang J, et al. Identifying relations between imaging phenotypes and molecular subtypes of breast cancer: Model discovery and external validation. J Magn Reson Imaging 2017;46:1017–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Holli-Helenius K, Salminen A, Rinta-Kiikka I, et al. MRI texture analysis in differentiating luminal A and luminal B breast cancer molecular subtypes — A feasibility study. BMC Med Imaging 2017;17:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fan M, Li H, Wang S, Zheng B, Zhang J, Li L. Radiomic analysis reveals DCE-MRI features for prediction of molecular subtypes of breast cancer. PLoS One 2017;12:e0171683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fan M, Zhang P, Wang Y, et al. Radiomic analysis of imaging heterogeneity in tumours and the surrounding parenchyma based on unsupervised decomposition of DCE-MRI for predicting molecular subtypes of breast cancer. Eur Radiol 2019. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 93.Saha A, Harowicz MR, Grimm LJ, et al. A machine learning approach to radiogenomics of breast cancer: A study of 922 subjects and 529 DCE-MRI features. Br J Cancer 2018;119:508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ha R, Mutasa S, Karcich J, et al. Predicting breast cancer molecular subtype with MRI dataset utilizing convolutional neural network algorithm. J Digit Imaging 2019;32:276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhu Z AE, Saha A, Zhang J, Harowicz M, Mazurowski MA. Breast cancer molecular subtype classification using deep features: Preliminary results. SPIE Medical Imaging. Volume 10575. Houston, TX; 2018. [Google Scholar]
- 96.Guo W, Li H, Zhu Y, et al. Prediction of clinical phenotypes in invasive breast carcinomas from the integration of radiomics and genomics data. J Med Imaging (Bellingham) 2015;2:041007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yamamoto S, Maki DD, Korn RL, Kuo MD. Radiogenomic analysis of breast cancer using MRI: A preliminary study to define the landscape. AJR Am J Roentgenol 2012;199:654–663. [DOI] [PubMed] [Google Scholar]
- 98.Lehrer M, Bhadra A, Aithala S, et al. High-dimensional regression analysis links magnetic resonance imaging features and protein expression and signaling pathway alterations in breast invasive carcinoma. Oncoscience 2018;5:39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu J, Li B, Sun X, et al. Heterogeneous enhancement patterns of tumor-adjacent parenchyma at MR imaging are associated with dysregulated signaling pathways and poor survival in breast cancer. Radiology 2017;285:401–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu J, Cui Y, Sun X, et al. Unsupervised clustering of quantitative image phenotypes reveals breast cancer subtypes with distinct prognoses and molecular pathways. Clin Cancer Res 2017;23:3334–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yamamoto S, Han W, Kim Y, et al. Breast cancer: Radiogenomic biomarker reveals associations among dynamic contrast-enhanced MR imaging, long noncoding RNA, and metastasis. Radiology 2015;275:384–392. [DOI] [PubMed] [Google Scholar]
- 102.Ashraf AB, Daye D, Gavenonis S, et al. Identification of intrinsic imaging phenotypes for breast cancer tumors: Preliminary associations with gene expression profiles. Radiology 2014;272:374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sutton EJ, Oh JH, Dashevsky BZ, et al. Breast cancer subtype intertumor heterogeneity: MRI-based features predict results of a genomic assay. J Magn Reson Imaging 2015;42:1398–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Saha A, Harowicz MR, Wang W, Mazurowski MA. A study of association of Oncotype DX recurrence score with DCE-MRI characteristics using multivariate machine learning models. J Cancer Res Clin Oncol 2018;144:799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li H, Zhu Y, Burnside ES, et al. MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of MammaPrint, Oncotype DX, and PAM50 gene assays. Radiology 2016;281:382–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Teshome M, Hunt KK. Neoadjuvant therapy in the treatment of breast cancer. Surg Oncol Clin N Am 2014;23:505–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Haque W, Verma V, Hatch S, Suzanne Klimberg V, Brian Butler E, Teh BS. Response rates and pathologic complete response by breast cancer molecular subtype following neoadjuvant chemotherapy. Breast Cancer Res Treat 2018;170:559–567. [DOI] [PubMed] [Google Scholar]
- 108.Marinovich ML, Houssami N, Macaskill P, et al. Meta-analysis of magnetic resonance imaging in detecting residual breast cancer after neoadjuvant therapy. J Natl Cancer Inst 2013;105:321–333. [DOI] [PubMed] [Google Scholar]
- 109.Hylton NM, Blume JD, Bernreuter WK, et al. Locally advanced breast cancer: MR imaging for prediction of response to neoadjuvant chemotherapy—Results from ACRIN 6657/I-SPY TRIAL. Radiology 2012;263:663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tahmassebi A, Wengert GJ, Helbich TH, et al. Impact of machine learning with multiparametric magnetic resonance imaging of the breast for early prediction of response to neoadjuvant chemotherapy and survival outcomes in breast cancer patients. Invest Radiol 2019;54:110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Machireddy A, Thibault G, Tudorica A, et al. Early prediction of breast cancer therapy response using multiresolution fractal analysis of DCE-MRI parametric maps. Tomography 2019;5:90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wu J, Gong G, Cui Y, Li R. Intratumor partitioning and texture analysis of dynamic contrast-enhanced (DCE)-MRI identifies relevant tumor subregions to predict pathological response of breast cancer to neoadjuvant chemotherapy. J Magn Reson Imaging 2016;44:1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Banerjee I, Malladi S, Lee D, et al. Assessing treatment response in triple-negative breast cancer from quantitative image analysis in perfusion magnetic resonance imaging. J Med Imaging (Bellingham) 2018;5:011008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Johansen R, Jensen LR, Rydland J, et al. Predicting survival and early clinical response to primary chemotherapy for patients with locally advanced breast cancer using DCE-MRI. J Magn Reson Imaging 2009;29:1300–1307. [DOI] [PubMed] [Google Scholar]
- 115.Aghaei F, Tan M, Hollingsworth AB, Qian W, Liu H, Zheng B. Computer-aided breast MR image feature analysis for prediction of tumor response to chemotherapy. Med Phys 2015;42:6520–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Giannini V, Mazzetti S, Marmo A, Montemurro F, Regge D, Martincich L. A computer-aided diagnosis (CAD) scheme for pretreatment prediction of pathological response to neoadjuvant therapy using dynamic contrast-enhanced MRI texture features. Br J Radiol 2017;90:20170269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu Z, Li Z, Qu J, et al. Radiomics of multi-parametric MRI for pretreatment prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer: A multicenter study. Clin Cancer Res 2019. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 118.Huynh BQ AN, Giger ML. Comparison of breast DCE-MRI contrast time points for predicting response to neoadjuvant chemotherapy using deep convolutional neural network features with transfer learning. SPIE Medical Imaging. Volume 10134, Medical Imaging 2017: Computer-Aided Diagnosis; 101340U Orlando, FL; 2017. [Google Scholar]
- 119.Braman N, Prasanna P, Whitney J, et al. Association of peritumoral radiomics with tumor biology and pathologic response to preoperative targeted therapy for HER2 (ERBB2)-positive breast cancer. JAMA Netw Open 2019;2:e192561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Aghaei F, Tan M, Hollingsworth AB, Zheng B. Applying a new quantitative global breast MRI feature analysis scheme to assess tumor response to chemotherapy. J Magn Reson Imaging 2016;44: 1099–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fan M, Wu G, Cheng H, Zhang J, Shao G, Li L. Radiomic analysis of DCE-MRI for prediction of response to neoadjuvant chemotherapy in breast cancer patients. Eur J Radiol 2017;94:140–147. [DOI] [PubMed] [Google Scholar]
- 122.Symmans WF, Peintinger F, Hatzis C, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol 2007;25:4414–4422. [DOI] [PubMed] [Google Scholar]
- 123.Ha R, Chin C, Karcich J, et al. Prior to initiation of chemotherapy, can we predict breast tumor response? Deep learning convolutional neural networks approach using a breast MRI tumor dataset. J Digit Imaging 2018. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ravichandran K, Braman N, Janowczyk A, Madabhushi A. A deep learning classifier for prediction of pathological complete response to neoadjuvant chemotherapy from baseline breast DCE-MRI. SPIE Medical Imaging. Volume 10575, Medical Imaging 2018: Computer-Aided Diagnosis; 105750C Houston, TX; 2018. [Google Scholar]
- 125.Boughey JC, Suman VJ, Mittendorf EA, et al. Sentinel lymph node surgery after neoadjuvant chemotherapy in patients with node-positive breast cancer: The ACOSOG Z1071 (Alliance) clinical trial. JAMA 2013;310:1455–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ha R, Chang P, Karcich J, et al. Predicting post neoadjuvant axillary response using a novel convolutional neural network algorithm. Ann Surg Oncol 2018;25:3037–3043. [DOI] [PubMed] [Google Scholar]
- 127.Klauschen F, Muller KR, Binder A, et al. Scoring of tumor-infiltrating lymphocytes: From visual estimation to machine learning. Semin Cancer Biol 2018;52(Pt 2):151–157. [DOI] [PubMed] [Google Scholar]
- 128.Heacock L MA, Wadera A, Moy L, Kim SG, Rusinek H. DCE-MRI texture features of locally advanced breast cancer: Correlation with tumor biomarkers, post-treatment imaging and pathologic complete response. RSNA; Chicago, IL; 2017. [Google Scholar]