Abstract
Phosphofurin acidic cluster sorting protein-1 (PACS-1) is a multifunctional membrane traffic regulator that plays important roles in organ homeostasis and disease. In this study, we elucidate a novel nuclear function for PACS-1 in maintaining chromosomal integrity. PACS-1 progressively accumulates in the nucleus during cell cycle progression, where it interacts with class I histone deacetylases 2 and 3 (HDAC2 and HDAC3) to regulate chromatin dynamics by maintaining the acetylation status of histones. PACS-1 knockdown results in the proteasome-mediated degradation of HDAC2 and HDAC3, compromised chromatin maturation, as indicated by elevated levels of histones H3K9 and H4K16 acetylation, and, consequently, increased replication stress-induced DNA damage and genomic instability.
Keywords: PACS-1, DNA replication, replication stress, DNA damage response, histones, histone deacetylases, HDAC2, HDAC3
INTRODUCTION
The human genome is packaged with histones to create tightly wrapped and highly organized structures collectively referred to as chromatin 1. Chromatin undergoes dynamic structural changes during DNA replication, gene transcription and DNA repair, each of which requires the coordinated opening and closing of chromatin structures. Post-translational modifications of histones such as acetylation and methylation govern the conformational states of the chromatin, there by determining the openness and thus accessibility of the DNA to chromatin modifiers 2. Central to this process are histone acetyltransferases (HATs) and histone deacetylases (HDACs), which acetylate or deacetylate lysine residues in the N-terminal tails of histones in a highly coordinated manner. These histone modifications regulate access to genomic DNA by proteins involved in DNA replication, transcription and repair. Notably, proteins involved in DNA replication form a multi-protein complex known as the replisome, which requires an open chromatin structure to initiate the replicative process 3
HDACs play essential roles in both DNA replication and maintenance of genome integrity. Histone content must be doubled to accommodate duplicated chromosomal DNA at each cell division. Newly synthesized histones are in an acetylated form before their incorporation on to the nascent DNA 4. The most commonly found acetylated residues associated with newly synthesized DNA are histone H4K5 and H4K12, which assist histone chaperones to correctly assemble nucleosomes 5. HDACs then deacetylate the histones during chromatin compaction 6. These processes are evolutionarily conserved from yeast to humans. The class I HDACs, comprised of HDAC1, HDAC2 and HDAC3, are present in replisomes 7,8. HDAC3 is essential for chromatin organization during replication and its deficiency impairs S-phase progression, induces replication-associated DNA double strand breaks and causes cell death 9–11. HDAC1 and HDAC2 also function during replication 12. However, their roles in replication are redundant. Thus, only cells deficient for both HDAC1 and HDAC2 display increased histone H4K5 and H4K12 acetylation and S-phase arrest.
Deregulated DNA replication and cell division are hallmarks of cancer. Chromatin remodeling enzymes, including HDACs, are critical for cell survival because they maintain chromatin integrity during uncontrolled cell division. Data from tumor studies convincingly demonstrate upregulation of class I HDACs in tumor tissue compared to adjacent normal tissue 13. Increased HDAC activity has been associated with closed chromatin assembly and inhibition of gene expression, a characteristic feature of malignantly transformed cells. Owing to their importance in cancer, several HDAC inhibitors have been approved for cancer treatment 14. Thus, understanding the regulation of these HDACs during cell cycle progression and in tumorigenesis is critical for optimization of cancer therapies that target this class of enzymes.
Phosphofurin acidic cluster sorting protein-1 (PACS-1) is a multifunctional membrane traffic regulator that plays an important role in organ homeostasis 15,16. PACS-1 regulates the function of several acidic cluster-containing proteins by shuttling or transporting them between endosomes and trans-golgi network (TGN). Some well-studied clients of PACS-1 include the proprotein convertase furin, the cation-independent mannose 6-phosphate receptor (CI-MPR), and the HIV-1 accessory protein Nef, and thus PACS-1 has been implicated in diverse pathological conditions such as neurological and metabolic disorders as well as viral pathogenesis 17–23.
Although furin is upregulated in cancers and is associated with aggressive disease and poor prognosis, a direct role for PACS-1 in cancer has yet to be established 16. Nevertheless, genomic studies on cervical cancer cell lines and primary tumors recognized rearrangements at chromosome 11q13 showing a 5.5kb homozygous deletion that also localizes to the 1st intron of PACS-1 gene 24–27. Here we show that PACS-1 is distributed in both the cytosolic and nuclear compartments and localizes to the nucleus during cell cycle progression. In response to DNA damage, nuclear PACS-1 promotes stabilization of HDAC2 and HDAC3, which is necessary for DNA damage repair and genomic stability.
RESULTS
PACS-1 regulates cell cycle progression and promotes genomic stability
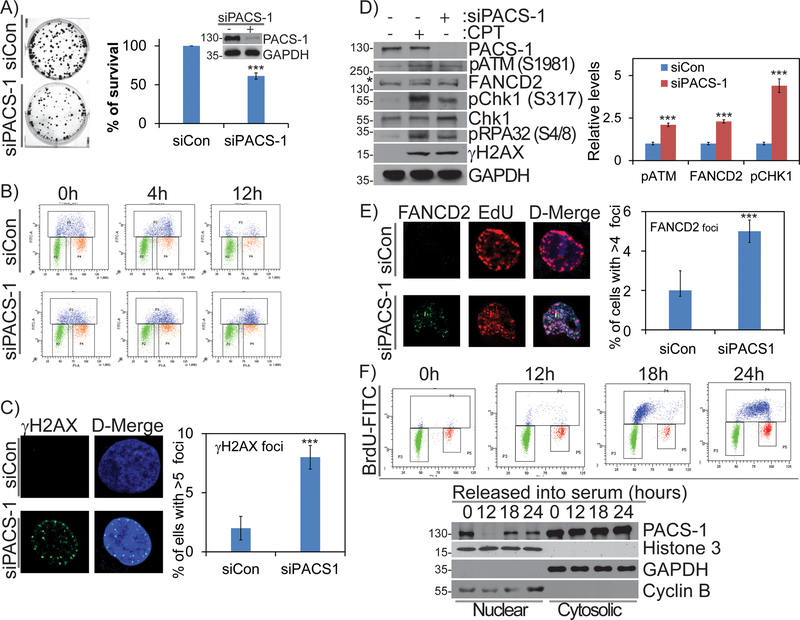
To date, studies on PACS-1 have focused on its evolutionally conserved roles in localization of cargo proteins between secretory pathway compartments. To examine potential roles for PACS-1 in the nucleus, we first assessed the effect of PACS-1 siRNA knockdown on HeLa cell viability and cell cycle progression. We found that PACS-1 knockdown attenuated clonogenic survival (Figure 1A). This decreased cell viability correlated with altered cellular cycling of PACS-1 knockdown cells, which accumulated in S-phase as determined by flow cytometry analysis (Figure S1A). To further examine the role of PACS-1 in cell cycle progression, we pulse-labeled PACS-1 knockdown or control cells with 5-Bromodeoxyuridine (BrdU) and monitored BrdU+ cells using flow cytometry. By 12 hours post-labeling, cell cycle progression in the PACS-1 knockdown cells was markedly altered, with a large accumulation of cells in late S-phase (Figure 1B). This delayed exit from S-phase in the PACS-1 knockdown cells correlated with increased level of γH2AX, suggesting an increase in double strand breaks (DSBs) (Figure 1C). We therefore compared the effect of PACS-1 knockdown to treatment of the cells with the DNA topoisomerase 1 inhibitor camptothecin (CPT) (500 nM), which induces replication stress and DSB formation 28. CPT and PACS-1 knockdown each caused DNA DSBs (as indicated by increased γH2AX) and induced the DDR as determined by activation of ATM (pSer1981-ATM), Chk1 (pSer317-Chk1), RPA32 (pSer4/8-RPA32) and mono-ubiquitinated FANCD2 (Figure 1D). A subpopulation of mono-ubiquitinated FANCD2 co-localized with EdU+ foci, which represent sites of active DNA synthesis (Figure 1E). These cells also exhibited increased phosphorylation of replication protein A (RPA) foci, which results from the recruitment of this protective single strand DNA (ssDNA) binding protein at single-stranded discontinuities during replication stress (Figure S1B). Collectively, these observations suggest PACS-1 deficiency induces genomic instability and replication stress.
Figure 1: PACS-1 nuclear accumulation increases during cell cycle progression and its deficiency induces replication stress and cell death.
A) Clonogenic survival in HeLa cells treated with control or PACS-1 siRNA shows PACS-1 is necessary for cancer cell survival. Insert represents WB data to confirm PACS-1 knockdown in colony assay. B) FACS analysis of BrdU labeled HeLa cells show S-phase specific cell cycle arrest in PACS-1 siRNA treated cells. C) Confocal image shows increased DNA damage (γH2AX foci formation) in the siPACS-1 HeLa cells, compared to control (siCon) cells. Foci number up to 5 is considered as the basal level. More than 100 cells from three independent experiments were scored. D) Western blot analysis shows activation of DDR proteins, such as monoubiquitination of FANCD2, phosphorylation of ATM, Chk1, RPA32 and H2AX. E) Confocal image indicates that a subset of FANCD2 foci co-localizes with EdU in siPACS-1 HeLa cells. F) FACS analysis of serum starved HDF cells and their corresponding western blots in nuclear and cytoplasmic fraction show that PACS-1 which is mostly in the cytoplasm, accumulates into nucleus during S-phase. All the experiments were repeated three times and their S.E. is denoted in the bar graph. *** p < 0.001.
PACS-1 accumulates in the nucleus in a cell cycle-dependent manner
The increased replicative stress in PACS-1 knockdown cells led us to ask whether PACS-1 accumulates at sites of DNA replication. Normal human dermal fibroblasts (HDFs) were synchronized in G0 by serum starvation and then stimulated with serum to re-enter the cell cycle. At increasing times following serum addition, the cells were harvested and separated into nuclear and cytosolic fractions and the presence of PACS-1 in each fraction was determined by western blot (Figure 1F). PACS-1 was detected in the nuclear fraction from G0-arrested cells but not from cells stimulated with serum for 12 hours. By 18 or 24 hours, PACS-1 reappeared in the nucleus, which coincided with re-entry of the cells into S-phase as determined by BrdU labeling (Figure 1F). The presence of PACS-1 in the nucleus in non-cycling G0 cells may be due to stress caused by serum starvation. Additionally, nuclear accumulation of PACS-1 in cells that were actively synthesizing DNA (S-phase) was also confirmed in normal human fibroblast (BJ) cells (Figure S1C). Collectively, these findings suggest a role for PACS-1 during DNA replication.
PACS-1 protects DNA replication fork stability and kinetics
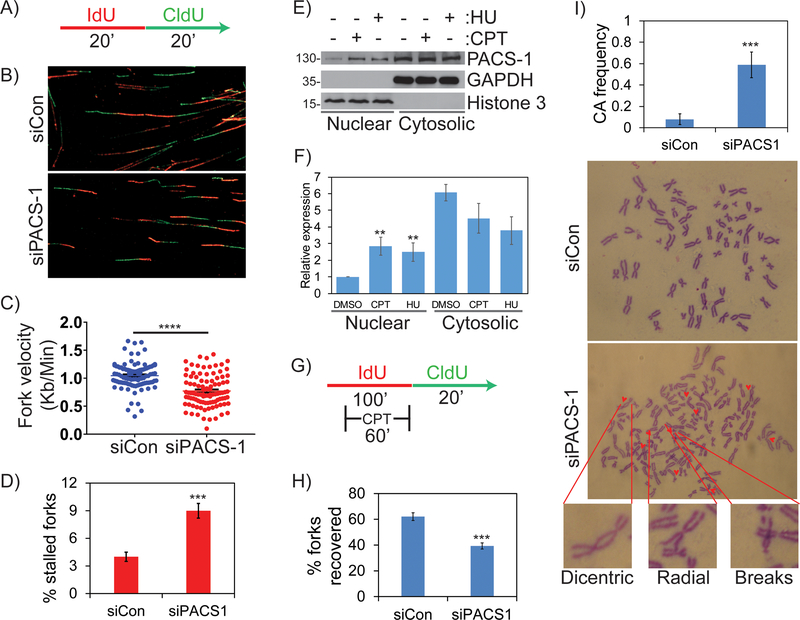
The appearance of PACS-1 in the nucleus of EdU+ cells led us to ask if it is involved in DNA replication or replication fork progression. To test these possibilities, we conducted single cell DNA fiber assays. PACS-1 knockdown or control cells were pulse-labeled with the nucleoside analogue IdU (5-iodo-2’-deoxyuridine) for 20 minutes followed by pulse-labeling with the nucleoside analog CldU (5-chloro-2’-deoxyuridine) for an additional 20 minutes (Figure 2A). To visualize the newly replicated DNA fibers and measure fork velocities, the DNA was stained with fluorescent-labeled antibodies specific to IdU (red) and CldU (green), respectively (Figure 2B and S1D). The average fork velocity in control cells was 1.04 kb/minute whereas in PACS-1 knockdown cells the fork velocity was reduced to 0.77 kb/minute (Figure 2C). PACS-1 knockdown also resulted in stalled or terminated replication forks, which were IdU+ and CldU− (Figure 2D). These results suggest an important role for PACS-1 in DNA replication as well as the integrity and progression of replication forks. In support of this possibility, subcellular fractionation studies showed nuclear PACS-1 levels rapidly increased in response to replication stress induced by 10 μM hydroxyurea (HU) for overnight or 500 nM CPT for 2 hours (Figure 2E and 2F). This suggests that PACS-1 enters nucleus during DNA damage responses and might be required to protect stalled forks to maintain genomic integrity. We therefore asked whether PACS-1 might function in recovery of stalled or collapsed forks. Control or PACS-1 knockdown cells were pulse-labeled with IdU alone for 40 minutes and then together with 1 μM CPT for an additional 60 minutes to stall replication forks. IdU and CPT were washed out and the cells were labeled with CldU for 20 minutes (Figure 2G). Analysis of the DNA fiber tracks showed PACS-1 knockdown markedly reduced replication fork recovery (Figure 2H and S2A), suggesting that PACS-1 is recruited to sites of DNA damage where it may modulate the repair of stalled or collapsed forks. In support of this observation, PACS-1 knockdown led to multiple chromosomal abnormalities, including chromatid breaks, gaps, dicentrics and radials (Figure 2I). These findings suggest an important role for PACS-1 in recovery of perturbed replication forks, which protects against increased chromosomal aberrations and decreased cell proliferation.
Figure 2: PACS-1 regulates replication progression during normal and perturbed conditions and its deficiency leads to genomic instability.
A-D) Molecular combing studies reveal decreased fork velocity and increased stalled forks in PACS-1 deficient HeLa cells. More than 100 replication structures from three independent experiments were scored. E, F) Western blot showing increased subcellular distribution of PACS-1 into the nucleus after CPT or HU treatment. G, H) PACS-1 is important for efficient fork recovery from CPT-induced replication stress. More than 300 replication structures from three independent experiments were scored. I) Metaphase spread from HeLa cells show chromosomal abnormalities in PACS-1 downregulated cells in comparison to control siRNA treated cells. Chromosomal abnormalities include gaps, breaks, deletions and dicentric chromosomes. A total of 15 to 25 metaphase spreads from three independent experiments were scored. All the experiments were repeated three times and their S.E. is denoted in the bar graph. ** p < 0.01; *** p < 0.001.
PACS-1 associates with chromatin and promotes HDAC2- and HDAC3-mediated chromatin maturation
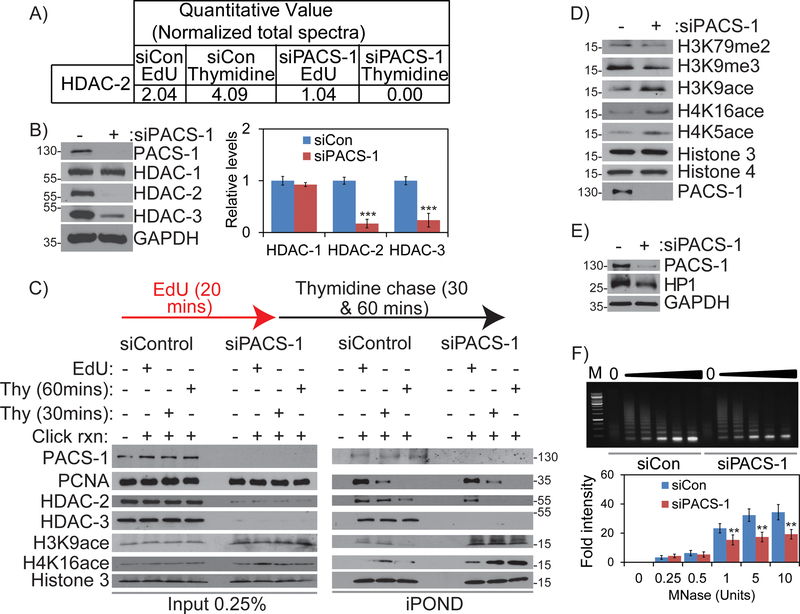
Our results led us to ask if PACS-1 associates with replisomes or chromatin to modulate chromatin-associated proteins during replication progression. To study this, we purified proteins associated with replisomes by iPOND (isolation of proteins on nascent DNA) assay. PACS-1 knockdown and control HeLa cells were pulse-labeled with EdU for 20 minutes and then chased with thymidine (Thy) for 30 minutes to label proteins associated with either replisomes (EdU) or with newly formed chromatin not associated with the replication machinery (Thy) (Figure S2B). Proteomic analysis of iPOND proteins identified several replisome-associated proteins reported in previous studies 7 (see also Table S1). Notably, this analysis detected reduced levels of HDAC2 in both the replisomes and in nascent chromatin fractions from PACS-1 knockdown cells (Figure 3A). We therefore asked if PACS-1 knockdown affected the expression of class I HDACs. Western blot analysis of HeLa cell lysates showed PACS-1 knockdown resulted in reduced levels of HDAC2 and HDAC3 but not HDAC1 (Figure 3B). Similar findings were observed in H1299 (lung cancer), HaCaT (normal keratinocyte) and BJ (human normal fibroblast) cells, and several other cancer cells (HCT116 and HT-29 colon cancer cells and MCF-7 breast cancer cells), suggesting this effect was not cell line dependent (Figure S2C and S4B). To rule out the possibility that the reduced HDAC expression was due to an off-target effect of the siRNA, we tested multiple PACS-1 siRNAs and found that each similarly reduced levels of HDAC2 and HDAC3 (Figure S2D). Next, we interrogated the iPOND samples for HDAC2 or HDAC3 by western blot. Reproducibly, PACS-1 knockdown resulted in decreased HDAC2 levels in the nascent chromatin fraction and reduced HDAC3 in both the replisome and chromatin fractions (Figure 3C). Comparatively, PACS-1 levels were similar in the EdU and Thy chase pulldown, suggesting PACS-1 is enriched in chromatin compared to replisomes. We therefore asked if PACS-1 knockdown altered the acetylation state of histones. The acetylation of the HDAC2/ HDAC3 substrates acetyl H3K9, acetyl H4K5 and acetyl H4K16 were selectively increased in PACS-1 knockdown cells (Figure 3D). This increased H3K9 acetylation correlated with reduced levels of tri-methylated H3K9me3 and di-methylated H3K79me2. The increase in acetylated H3K9 and H4K16 that accompanied the reduced levels of replisome-associated HDAC2 and HDAC3 in PACS-1 knockdown cells was also found using iPOND (Figure 3C).
Figure 3: PACS-1 localizes to chromatin and regulates the stability of HDAC2 and HDAC3 proteins.
A) iPOND followed by analysis with mass spectrometry reveals decreased HDAC2 in siPACS-1 cells. B) Level of class I HDACs in control and PACS-1 downregulated cells. C) iPOND assay reveals PACS-1 localization to the sites of DNA replication and it is important for HDAC2 and HDAC3 stability in the replication forks and to maintain chromatin regulation. D) Acetylation status of the HDAC2 and HDAC3 specific histone modification in control and PACS-1 down regulated cells. E) Depletion of PACS-1 leads to decreased HP1, marker for heterochromatin. F) MNase assay results of nuclei isolated from control vs PACS-1 knockdown cells. **p < 0.01.
Similar to PACS-1 siRNA knockdown, treatment of cells with the HDAC3 inhibitor RGFP996 for 2 hours increased acetylation of H3K9, H4K16 and H4K5, as well as phosphorylation of Chk1 (pSer317) and H2AX (pSer139) (Figure S2E). Moreover, siRNA knockdown of HDAC3 and, to a lesser extent, HDAC2, led to increased acetyl H3K9 (Figure S3A). These studies suggest the effects of PACS-1 knockdown on replication stress resulted, at least in part, from attenuated HDAC3 and, to a lesser extent, attenuated HDAC2.
During DNA synthesis, di-acetylated H4K16 is recruited to newly synthesized DNA and then deacetylated by class I HDACs during chromatin compaction 29. We therefore asked if PACS-1 knockdown altered the status of H4K16 in the iPOND assay. We found that H4K16 acetylation persisted in the nascent chromatin fraction (Figure 3C). Similar results were obtained for tri-acetylated H3K9. Consistent with these histone marks for open chromatin, we observed decreased HP1, a closed chromatin marker (Figure 3E), and also the chromatin from the PACS-1-deficient cells was more sensitive to the exo-endonuclease micrococcal nuclease (MNase) compared to control cells (Figure 3F). Moreover, the level of DNA associated with mono-nucleosomes did not increase even with the increased concentrations of MNase in PACS-1-deficient cells compared to control cells. The reduced levels of HDAC2 and HDAC3 in the PACS-1 knockdown cells detected by western blot correlated with their reduced enzyme activities (Figure S3B and C).
PACS-1 protects HDAC2 and HDAC3 proteins from proteasomal degradation
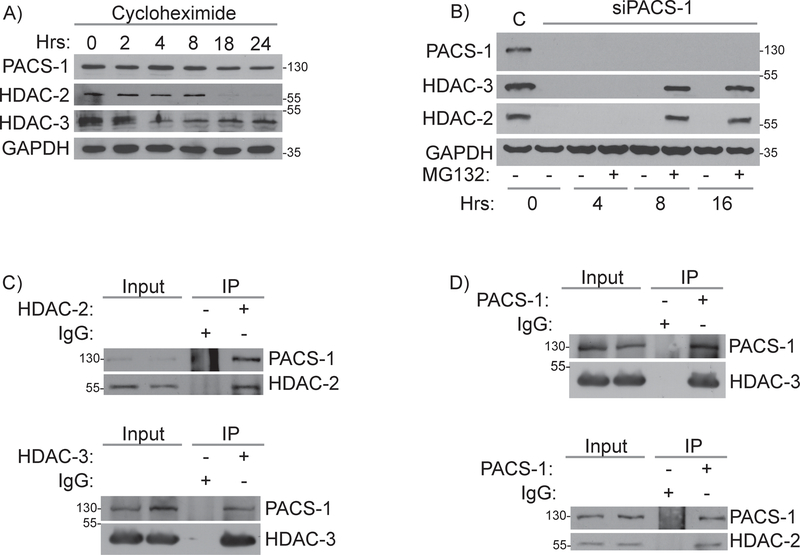
In order to examine the mechanisms by which PACS-1 regulates HDAC2 and HDAC3 protein levels, we first evaluated the effect of PACS-1 knockdown on HDAC2 and HDAC3 gene expression. As expected, PACS-1 siRNAs downregulated the PACS-1 transcript ~90% compared to control cells but had no effect on the levels of the HDAC2 or HDAC3 transcripts (Figure S3 D–F). To assess the stability of PACS-1, HDAC2 and HDAC3, HeLa cells were exposed to cycloheximide and levels of each protein was assessed over 24 hours by western blot. Inhibition of protein synthesis gradually decreased PACS-1 levels, with only about 20% of the protein remaining at 24 hours. The half-lives of HDAC2 and HDAC3 were shorter relative to PACS-1, consistent with a role for PACS-1 in their stabilization (Figure 4A and S4A). Treatment of the PACS-1 knockdown cells with MG132 for 8 hours restored HDAC2 and HDAC3 proteins levels, suggesting PACS-1 protects the deacetylases from proteasomal degradation (Figure 4B). Co-immunoprecipitation (co-IP) assays were performed to test whether PACS-1 interacts with HDAC 2 or 3. Markedly, PACS-1 was captured in HDAC2 and HDAC3 immunoprecipitates (Figure 4C). Similarly, HDAC2 and HDAC3 were captured in PACS-1 immunoprecipitates (Figure 4D).
Figure 4: PACS-1 binds to HDAC2 and HDAC3 proteins and regulates their stabilities.
A) Cycloheximide treatment and the level of PACS-1, HDAC2 and HDAC3 in various time points. B) MG-132 treatment and the level of PACS-1, HDAC2 and HDAC3 in various time points. C) Co-IP studies of HeLa cell lysates using PACS-1 antibody showed PACS-1 binds to HDAC2 and HDAC3 and similarly (D) HDAC2 and HDAC3 antibodies pull down PACS-1.
The PACS-1 FBR interacts with HDAC3
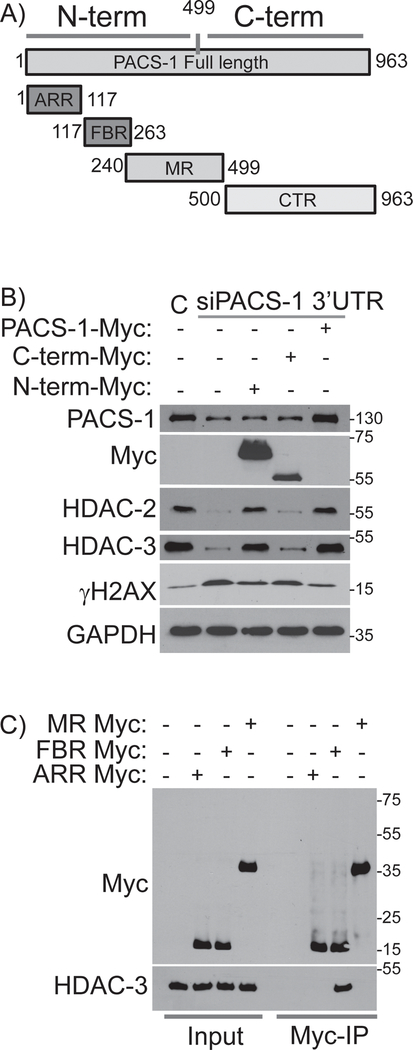
In order to identify the regions of PACS-1 responsible for its interaction with HDACs and their stability, we examined four previously mapped structural domains of PACS-1 (Figure 5A). A proline/glutamine and serine/alanine rich N-terminal atropin-1 related region (ARR); followed by the 140-residue furin (cargo) binding region (FBR), which binds client proteins and cellular trafficking machinery, the middle region (MR), which contains an autoregulatory domain, and the C-terminal region (CTR). We generated Myc-tagged PACS-1 N- and C-terminal domain truncation mutants in pcDNA3.1-Myc vectors. PACS-11−499 contains the ARR, FBR and MR regions whereas PACS-1500−963 contains only the CTR (Figure 5A). We pre-treated HeLa cells with siRNAs targeting the PACS-1 3’UTR region for 48 hours, which, as with PACS-1 coding region siRNAs, reduced HDAC2 and HDAC3 levels and triggered the DDR (increased γH2AX, Figure 5B). The knockdown cells were then transfected with plasmids expressing siRNA-resistant full-length PACS-1, PACS-11−499 or PACS-1500–963. Western blot analysis showed that only cells expressing full-length PACS-1 or PACS-11−499 prevented loss of HDAC2 and HDAC3 and repressed induction of γH2AX (Figure 5B). These results suggest that the PACS-1 N-terminal segment containing the ARR, FBR and MR is sufficient to stabilize HDAC2 and HDAC3 and, in turn, suppress the DDR (γH2AX).
Figure 5: FBR region of PACS-1 is essential for its interaction with HDACs.
A) Full length (963a.a.) PACS-1 protein is divided in to N-term (1–499a.a.) and C-term (500–963a.a.). N-term is further divided into ARR (1–117a.a.), FBR (117–263a.a.) and MR (240–499a.a.). B) PACS-1 downregulated HeLa cells were complemented with Myc-tagged N-terminal, C-terminal and full length PACS-1. Interestingly, full length PACS-1-Myc and N-terminal PACS-1-Myc brought back the HDAC2 and HDAC3 levels and reduced the γH2AX formation in PACS-1 downregulated cells. C) IP results show that FBR region of PACS-1-Myc binds to the HDAC3.
To determine which of the three N-terminal domains of PACS-1 interacts with HDAC3, we expressed ARR, FBR and MR domains in pcDNA3.1-Myc vectors (Figure 5C). HeLa cells were transfected with Myc-tagged ARR, FBR and MR vectors and whole cell lysates were incubated with anti-Myc antibody or an IgG control and pull down assays were performed. These results demonstrated that HDAC3 interacts with the PACS-1 FBR but not the ARR or MR (Figure 5C).
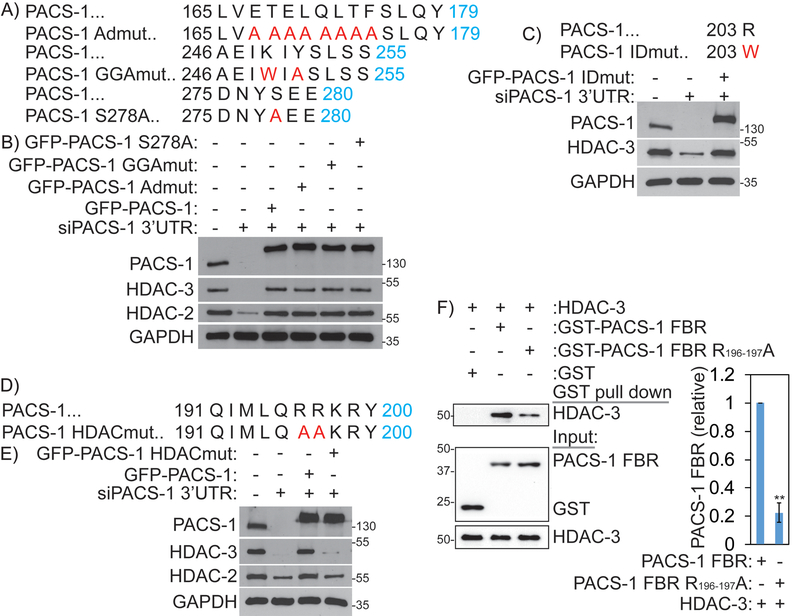
Intriguingly, the FBR is important for the cytoplasmic sorting functions of PACS-1. Previous studies identified two motifs in this region that interact with the monomeric adaptor GGA3 (Golgi Associated, Gamma Adaptin Ear Containing, ARF Binding Protein 3), and the heterotetrameric sorting adapter (AP-1), which are important for mediating trafficking of cargo proteins. Mutation of amino acid residues K249 and Y252 to W and A in PACS-1 (PACS-1GGAmut), precludes interaction with GGA3 and sorting of CI-MPR 30. Similarly, PACS-1Admut (E168TELQLTF175→A168AAAAAAA175) fails to bind to AP-1 and sorting of CI-MPR, furin and HIV-1 Nef 31. Phosphorylation of serine 278 (S278) in the PACS-1 MR regulates cargo binding to the FBR, such that a PACS-1 S278A mutant blocks the trafficking of PACS-1 cargo23. To determine whether cytoplasmic sorting functions of PACS-1 are important for nuclear HDAC2 and HDAC3 protein stability, we generated these three PACS-1 mutants, PACS-1Admut, PACS-1-GGAmut and PACS-1 S278A, in pEGFP-C3 vectors (Figure 6A). HeLa cells were transfected with siPACS-1 3’-UTR to downregulate endogenous PACS-1 and, consequently, reduce expression of HDAC2 and HDAC3 (Figure 6B). The cells were then complemented with either empty vector, GFP-PACS-1 or GFP-PACS-1 mutants (GFP-PACS-1GGAmut, GFP-PACS-1Admut and GFP-PACS-1 S278A), and assessed their ability to complement HDAC2 and HDAC3 stability. In addition, to determine whether the recently identified R203W neurodevelopmental mutation in PACS-1 32, may be involved in stability of HDAC2 or HDAC3, we generated a GFP-tagged PACS-1-R203W plasmid (hereafter called PACS-1IDmut, Figure 6C and 6D). Complementation with each of the above mutants defective in PACS-1 sorting functions efficiently complemented stability of HDAC2 and HDAC3 proteins to near normal levels (Figure 6B and 6C). These results indicate the existence of additional motifs within the PACS-1FBR that are important for its interaction with HDAC2 and HDAC3. Towards the goal of finding the motif that is responsible for PACS-1 interaction with HDAC3, we looked into highly conserved residues within the FBR region 191QIMLQRRKRY200, which is involved in binding protein kinase CK2 30. We generated a mutant GFP-PACS-1HDACmut (RR196,197 →AA) and assessed this mutant in complementation studies as described above (Figure 6D). As expected, expression of wild type GFP-PACS-1 stabilized HDAC3 in PACS-1 downregulated cells. By contrast, GFP-PACS-1HDACmut failed to complement HDAC2 and HDAC3 stabilization (Figure 6E). Together, these results indicate that a conserved motif in the PACS-1 FBR is important for PACS-1 interactions with HDAC3 and enzyme stability. To confirm that PACS-1 and HDAC3 interact directly, purified HDAC3 was mixed with GST-tagged PACS-1 FBR, PACS-1 RR196,197AA-FBR or GST alone as a negative control. GST proteins were captured and bound HDAC3 was detected by western blot. We found that HDAC3 directly bound PACS-1 FBR and that the RR196,197AA FBR mutation greatly reduced binding (Figure 6F).
Figure 6: PACS-1 interactions with HDACs are distinct from its cytosolic TGN functions.
A and B) ADmut, GGAmut and S278A of GFP-PACS-1 are able to complement HDAC2 and HDAC3 expression. C) IDmut of GFP-PACS-1 is able to complement HDAC2 and HDAC3 expression. D and E) HDACmut of GFP-PACS-1 is unable to complement HDAC2 and HDAC3 expression. F) Affinity purified HDAC was incubated with GST or GST-PACS-1 FBR or GST-PACS-1 FBR HDACmut. GST proteins were captured and bound HDAC3 was detected by western blot. **p < 0.01.
PACS-1 HDACmut induces genome instability
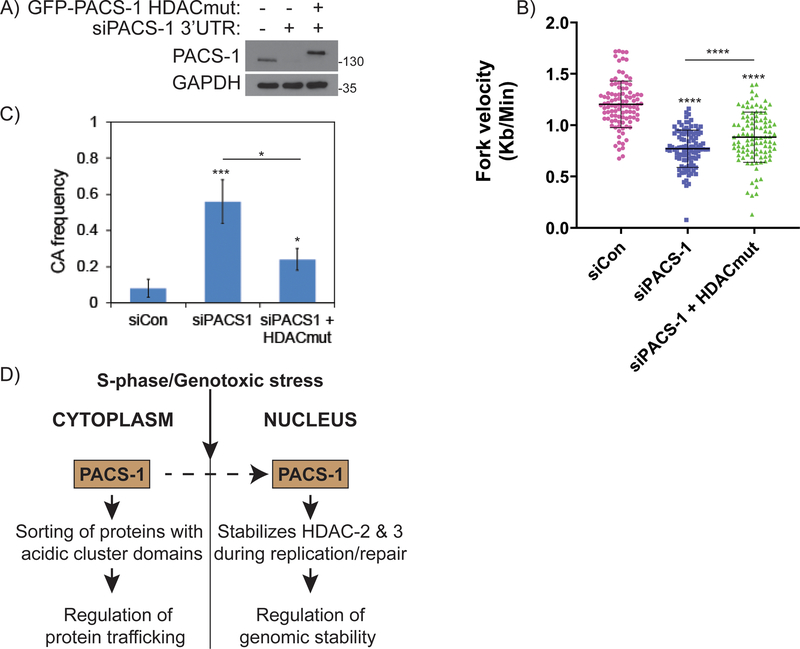
To test whether PACS-1 HDACmut results in replication stress and genomic instability, we performed DNA fiber and chromosomal aberration assays. We used siPACS-1 3’UTR to downregulate endogenous PACS-1 in HeLa cells, and then complemented these cells with GFP-PACS-1HDACmut (Figure 7A). Inhibition of PACS-1 by 3’ UTR siRNAs showed reduced fork velocity (Figure 7B), similar to the siRNAs targeting the PACS-1 coding region (Figure 2C). However, complementation of PACS-1 deficient cells with GFP-PACS-1HDACmut did not rescue the slow progression of replication forks. The fork velocities relatively increased in GFP-PACS-1HDACmut expressing cells compared to PACS-1 deficient cells. Similarly, downregulation of PACS-1 using siPACS-1 3’ UTR siRNAs led to increased chromosomal aberration frequency (Figure 7C), as observed with siRNAs targeting to PACS-1 coding region (Figure 2I). However, complementation of PACS-1 deficient cells with GFP-PACS-1HDACmut failed to rescue the chromosomal aberration frequency. Moreover, the partial complementation of replication fork velocities and chromosomal aberration frequency by GFP-PACS-1HDACmut expression in PACS-1 deficient cells may be attributed to its weaker binding to HDACs (Figure 6F), or possibly to additional PACS-1 clients that may be affected by the HDACmut mutation. Nevertheless, these data collectively demonstrate a novel and important role for PACS-1 in timely progression of cell cycle and in maintaining genomic integrity, at least by partly regulating stabilities of HDAC2 and HDAC3.
Figure 7: PACS-1 promotes malignant behavior of cancer cells and a poor prognostic marker.
A) PACS-1 downregulated HeLa cells were complemented with GFP-PACS-1 HDACmut. B) Fiber Assay and C) Chromosomal Aberrations in PACS-1 downregulated HeLa cells complemented with GFP-PACS-1 HDACmut. D) A hypothetical model showing PACS-1 protein, which is mostly present in the cytoplasm, accumulates into the nucleus during S-phase of the cell cycle and during genotoxic stress. Nuclear localized PACS-1 then interacts and stabilizes HDAC2 and HDAC3, which otherwise induces replication stress and genomic instability. *p < 0.05; ***p < 0.001; **** p < 0.0001.
DISCUSSION
DNA replication is a complex and exquisitely regulated process. Any errors can lead to mutations, genome instability or epigenetic changes that contribute to pathological conditions ranging from developmental disabilities to cancer 33. Many factors, including disorganized chromatin and deregulated oncogenes impair DNA replication progression leading to replication stress, DNA damage and genomic instability 34. Class I HDACs associate with replisomes and are important for maintaining chromatin structure and genomic stability during replication (Summers et al., 2013). Our observations suggest that PACS-1 plays a critical role in chromatin maintenance and genome integrity by mediating the stability of HDAC2 and HDAC3 (Figure 7D). Together these data suggest multifunctional PACS-1 is a previously unknown epigenetic regulator that has important roles in proper progression during DNA replication.
Following DNA replication, the nascent DNA strands are naked and must be properly prepared by histones to package into chromatin. Acetylated histones are deposited on newly replicated DNA strands and then deacetylated by class I HDACs during chromatin maturation 35. Acetylated H4K16 and H3K9 are substrates for HDAC2 and HDAC3 and their deacetylation is necessary for histone compaction 36–43. Likewise, increased H3K9 acetylation impairs S-phase progression and induces replication stress 9–11,44. PACS-1 knockdown was coupled with reduced levels of replisome-associated HDAC2 and HDAC3 and, in turn, increased acetylation of H4K5, H4K16 and H3K9. This increased acetylation sensitized the chromatin to nuclease attack, suggesting an important role for PACS-1 in regulation of chromatin organization (Figure 3). Intriguingly, PACS-1 deficiency led to reduced levels of HDAC2 and HDAC3 (Figure 3B). Since HDAC3 deficiency leads to S-phase arrest and genome instability 36, the defects in replication progression and chromatin maturation in PACS-1-deficient cells observed here may be attributed to a deficiency in HDAC3 (Figures 2 and 3).
The ubiquitin-proteasome pathway regulates the turnover of many proteins involved in cell division and DNA repair 45. Mechanistic studies revealed that PACS-1 binds HDACs and protects them from proteasome-mediated degradation (Figures 4 and 5). While most studies of PACS-1 have focused on its many roles in cytoplasmic protein traffic, the role of PACS-1 in the nucleus has not been addressed 15,46. Consistent with this finding, HDAC3 binds the PACS-1 FBR in vitro, which was reduced by an R196R197→AA mutation within the FBR. Accordingly, PACS-1HDACmut failed to interact with HDAC3 and rescue HDAC3 stability in PACS-1 knockdown cells (Figures 6D–F). These observations suggest that nuclear PACS-1 acts as a scaffold protein to bind and stabilize the class I HDACs HDAC2 and HDAC3. While the PACS-1 RR196,197AA mutation blocked the interaction of PACS-1 with HDACs, mutations in PACS-1 that block its cytoplasmic protein trafficking functions had little effect. These findings suggest distinct motifs on PACS-1 are critical for its cytoplasmic versus nuclear roles. Further studies are needed to identify the ubiquitin ligases involved and how PACS-1 influences their interactions with HDAC2 and HDAC3.
PACS-1 levels increased in the nucleus during S-phase replication and in response to replication stress (Figures 1 and 2). Similarly, PACS-1 deficiency induced replication stress and gross chromosomal aberrations, suggesting that upregulated PACS-1 may facilitate oncogenic replication by stabilizing HDAC2 and HDAC3, thereby promoting tumor cell survival and progression (Figure 7). Interestingly, HDAC3 is upregulated in actively proliferating cells during S-phase and in many cancer cells where it promotes multiple oncogenic pathways 47. Our findings that PACS-1 is enriched within the nucleus during S-phase (Figures 1 and S1), and binds and stabilizes HDAC3 (Figures 4 and 5), suggest an important role for PACS-1 during replication and in maintaining genomic stability. PACS-1 depletion leads to proteasome-mediated degradation of HDAC2 and HDAC3, which results in compromised chromatin maturation, replication stress and genomic instability.
Materials and methods
Cell lines and reagents
HeLa (cervical cancer), H1299 (lung cancer), HaCaT (normal keratinocyte), BJ (human normal fibroblast), HDF (primary human dermal fibroblast), HCT116 (colorectal cancer), HT-29 (colorectal cancer), MCF-7 (breast cancer) were procured from ATCC. All the cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) and supplemented with 10% FBS (Omega Scientific, Tarzana, CA) and 100 μg/ml streptomycin sulfate (Corning, Manassas, VA) and 100 U/ml penicillin (Corning, Manassas, VA) and cells less than 15 passages were used. All the cells were regularly monitored for mycoplasma contamination. Following antibodies were used in this study. BrdU (347580) and BrdU-FITC conjugate (347583) were procured BD biosciences (San Jose, CA). Rat-BrdU (6326), Histone 3 (1791), HDAC1 (7028), HDAC2 (7029), HDAC3 (32629), Dimethyl H3K79 (2621), Acetyl H4K5 (519997), Histone 4 (177840) were procured from Abcam (Cambridge, MA). H2AX-S139 (05636), Acetyl H3K9 (6942), Acetyl H3K16 (7329) and Tri-methyl H3K9 (7442) were procured from Millipore (Billerica, MA). PACS-1 (HPA038914), Anti-FLAG (F7425) were procured from Sigma (St Louis, MO). ATM-S1981 (4526), CHK1-S317 (2344) and Cyclin B (4138) were procured from Cell Signaling (Danvers, MA). RPA32-S4/8 (A300–245A-2) was procured from Bethyl (Montgomery, TX), Anti-GST (05–311) was procured from Upstate Biotechnology (Lake Placid, NY). Anti-Myc (SC-40), HP1 (28735), PCNA (SC-56), FANCD2 (SC-20022), CHK1 (SC-8408) and GAPDH (SC-32233) were procured from Santa Cruz Biotechnology (Santa Cruz, CA). Cycloheximide (C7698, Sigma, St Louis, MO), G132 (C2211, Sigma, St Louis, MO), Camptothecin (C9911, Sigma, St Louis, MO), 3X FLAG peptide (F4799, Sigma, St Louis, MO), anti-FLAG M2 agarose affinity gel (A2220, Sigma, St Louis, MO) and Glutathione Sepharose 4B (20182003–3, Bioworld) were used in this study.
siRNA and plasmid transfection
siRNAs were purchased from Dharmacon, Lafayette, CO. Following are the sequence or catalog number used in this study. sicontrol (5′-UAGCGACUAAACACAUCAAUU-3′), siPACS-1: (5’-ACTCAGTGGTCATCGCTGTGAA-3’) (Crump et al., 2003), siPACS-1 (2) (D-006697–01-0005, Dharmacon, Lafayette, CO), siPACS-1 (3) (D-006697–02-0005, Dharmacon, Lafayette, CO), siPACS-1 (smart pool) (M-006697–01, Dharmacon, Lafayette, CO), siPACS-1 (3’UTR): (5’-CUAGGAUGCAGCUGCCAGAUU-3’), siHDAC2: (5′-UCCGUAAUGUUGCUCGAUGTT-3′) (Senese et al., 2007), siHDAC3: (5-AAUAUCCCUCUACUCGUGCUGA-3) 48.
Plasmids pEGFP-C3 GFP-PACS-1 ADmut, pEGFP-C3 GFP-PACS-1 GGAmut, pEGFP-C3 GFP-PACS-1 HDACmut, pEGFP-C3 GFP-PACS-1 S278A and pEGFP-C3 GFP-PACS-1 IDmut were created from pEGFP-C3 GFP-PACS-146 using Q5® Site-Directed Mutagenesis Kit (E0554S, NEB, Carlsbad, CA). Full length PACS-1 Myc, PACS-1 N-term-Myc, PACS-1 C-term-Myc, PACS-1 MR-Myc, PACS-1 FBR-Myc, PACS-1 AR-Myc were cloned into pcDNA3.1 vector. pGEX-3X -PACS-1- FBR and pGEX-3X- PACS-1 FBR R196–197A were made from pGEX-4T1 (GST) (GE Healthcare).
For the experiments involving co-transfection with siRNAs and plasmids, we first transfected the cells transiently with siRNA for 48 hours, and the last 24 hours were co-transfected with plasmid DNA before evaluating the cells for endpoints.
Cell Cycle synchronization
For cell cycle synchronization, BJ and HDF cells were grown in 0.05% serum for 48 hours to synchronize in G0 phase, and then washed and release into medium with 10% FBS to synchronously release into cell cycle. The time points were selected based on the cell lines progression through each phase G1 and S-phase of the cell cycle (as given in the figures) the cells were fixed and analyzed using immunofluorescence and flow cytometry.
Analysis of Cell Cycle Progression by PI and/or BrdU Incorporation
After 48 hours of siRNA transfection, HeLa cells were trypsinized, fixed in 70% methanol and analyzed by flow cytometry after propidium iodide (PI) staining (P3566, Invitrogen, Eugene, OR). BrdU incorporation was measured in exponentially growing cells 48 hours after siPACS-1 transfection. HeLa cells were pulsed with 10 μM BrdU (B5002, Sigma, St Louis, MO) for 30 minutes. Cells were harvested and fixed in 70% ethanol, up to 12 hours after the BrdU pulse at an interval of 0, 4, 8, 12 hours. BrdU content was analyzed after DNA denaturation with 2N HCl plus 0.1% Triton X-100 (45 minutes) at room temperature followed by neutralization with 0.1 M Na2B4O7 buffer, pH 8.5. The cells were labeled using specific anti-BrdU-FITC monoclonal antibody (347583, BD, San Jose, CA) and suspended in 600 μl of PBS containing 2 μg of PI per ml and 200 μg of RNase 1 (EN0531, Invitrogen, Vilnius, LT) for 30 minutes. Bivariate histograms of BrdU/DNA content were obtained using the BD flow cytometer, collecting FITC green fluorescence (log scale) and red PI fluorescence (linear scale) for 10,000 events in each sample 49
RT-PCR
Total RNA from cells was isolated by Trizol following manufacturer’s protocol (15596026, Invitrogen, Eugene, OR). Tumor sample RNA was prepared as outlined above. cDNA was prepared from total RNA using cDNA preparation kit (4387406, Applied Biosystems, Foster City, CA). Prime PCR for PACS-1 (10025636), HDAC2 (10025636), HDAC3 (10025636) were procured from Bio-Rad, USA. Gene-specific primers (Prime PCR; Bio-Rad) were used for semi-quantitative RT-PCR analysis as per manufacturer’s instructions.
Immunoprecipitation assay
For immunoprecipitation assays (IP), cells were lysed with cold IP buffer (87787, Thermo, Rockford, IL) supplemented immediately before use with phosphatase inhibitors (04906837001, Roche, Germany) and protease inhibitors (05892970001, Roche, Germany). The precleared samples were immunoprecipitated with primary antibody followed by protein A/G sepharose beads (sc-2003, Santa Cruz, CA) and the bound proteins were analyzed by western blot technique as described earlier 49. To avoid the influence of light and heavy chain bands, IPs involved with HDAC3 and Myc were analyzed using Clean-Blot™ IP Detection Kit (HRP) (21232, Thermo, Rockford, IL).
Nuclear and cytosolic fractions
Approximately 80–90% confluent HeLa cells were treated with DMSO or 500 nM CPT for 2 hours. Cells were washed with ice clod PBS thrice and proteins were extracted using cytoskeletal (CSK) buffer (10 mM PIPES (pH 6.8), 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, 0.1 mM ATP, 1 mM Na3VO4, 10 mM NaF and 0.1% Triton X-100, freshly supplemented with protease and phosphatase inhibitors (Roche). Detergent-insoluble nuclei and soluble fractions were separated by centrifugation at 3,000 g for 5 minutes. The soluble fractions were removed, and the remaining insoluble nuclear pellets were washed twice with 0.5 ml of CSK buffer. The resulting washed nuclei were suspended in an approximately equal volume of CSK 50. Protein concentrations of soluble and nuclear fractions were determined immediately and the samples were analyzed by western blot technique as described below.
Clonogenic survival assay
Cells were transfected with control or PACS-1 siRNAs and 400 cells/well were plated in 6- well plates in triplicate and allowed to attach and form colonies in complete growth medium. After 10 to 14 days, colonies were fixed in methanol, stained with crystal violet (C581–25, Fisher, Fair Lawn, NJ) and the colonies with >25 cells were counted using Gene Tools, Syngene 51
Immunofluorescence
Cells transfected with siRNAs were seeded into glass bottom 35 mm dishes. The resulting cells were treated with CPT (or DMSO for controls) at the concentrations specified in the figures or figure legends. Cells were fixed in 3% formaldehyde for 10 minutes and then in 100% methanol (−20°C) for 10 minutes at room temperature. Fixed cells were blocked in 10% FBS for 30 minutes. After three washes with PBS, cells were incubated overnight at 4°C with primary antibodies in PBS containing 5% bovine serum albumin (BSA), Sigma, St Louis, MO and 0.1% Triton X-100 (PBS-T). The slides were washed three times with PBS-T containing 1% BSA then incubated with anti-mouse IgG-Cy3 or anti-mouse IgG-FITC antibodies (Molecular Probes, USA) for 2 hours at room temperature and mounted with DAPI (H-1500, Vectasheild, USA) 52
Western blot
For Western blotting, cell lysates were prepared after washing extensively with PBS. Cells were lysed in ice-cold CSK buffer freshly supplemented with protease and phosphatase inhibitors (Roche). After normalizing the protein concentrations, samples were prepared in 2x SDS-PAGE sample buffer and heated to 100°C for 15 minutes. Denatured samples were resolved by SDS-PAGE and transferred to nitrocellulose membranes 49
DNA Fiber Assay
DNA fiber labeling analysis was used to assess DNA replication fork progression. 48 hours after siRNA transfection, HeLa cells were labeled for 20 minutes with 25 μM IdU (I7125, Sigma, St Louis, MO) followed by 20 minutes labeling with 250 μM CldU (C6891, Sigma, St Louis, MO). Cells were harvested and suspended in ice cold PBS. Next 2 μl of the cell suspension was deposited over the slide and 10 μl of lysis buffer (0.5% SDS, 200 mM Tris-HCl pH 7.4, 50 mM EDTA) was added. The slides were tilted to 15° to stretch the DNA fibers. Slides were then air dried, fixed in 3:1 methanol: acetic acid, denatured in 2.5M HCl and blocked with 5% BSA in PBS. Then slides were incubated with rat anti-CldU and mouse anti-IdU for one hour followed by goat anti-mouse 568 and goat anti-rat Alexa Fluor 488. Fork velocity and stalled replication fork (only red) were measured and statistical analysis was performed using Prism 6 (GraphPad Software).
For fork recovery, HeLa cells were labeled for 100 minutes with 25 μM IdU and the last 60 minutes was added with 1 μM CPT. Both the IdU and CPT were washed, then followed by 20 minutes labeling with 250 μM CldU. Cells were then processed for fiber combing as explained before. Recovered forks (red followed by green) and unrecovered forks (only red) were counted for 300 replication structures and their percentage was calculated 52
iPOND assay (Mass spectrometry and Western blot)
Analysis of proteins associated with DNA replication forks was performed using the iPOND method. HeLa cells were pulse labelled with 10 μM EdU (A10044, Molecular probes, USA) for 15 minutes. For the pulse-chase experiment, the EdU was removed, washed and replaced with media containing 10 μM thymidine (T9250, Sigma, St Louis, MO) for 30 and 60 minutes. The cells were then fixed with 1% formaldehyde for 20 minutes and quenched with 0.125 mM glycine (pH 7) for 5 minutes. Cell pellets were suspended in 0.25% Triton X-100 in PBS, washed and incubated in click reaction buffer (2 mM CuSO4, 10 mM biotin azide (B10184, Invitrogen, Eugene, OR) and 10 mM sodium ascorbate) for 2 hours. A no click reaction sample that did not include biotin azide was used as a negative control. Nascent DNA was conjugated to biotin using click chemistry, and Streptavidin beads (69203, Novagen, USA) were used to capture the biotin conjugated DNA-protein complexes. The eluted proteins were either sent to MS Bioworks LLC, Ann Arbor, MI for proteome analysis or suspended in Laemelli buffer (161-0737, Biorad, USA) and analyzed by western blot.
HDAC activity assays
HDAC activity was measured with the fluorometric HDAC3 Activity Assay kit (EP1004, Sigma, St Louis, MO) and kinetic HDAC2 assay kit (53002, BPS biosciences, USA) according to the manufacturer’s instructions.
RGFP966 treatment
HeLa cells were treated with 10 mM RGFP966 (S7229, Sellechem, USA), a HDAC3 specific inhibitor for 0, 2, 4, 6, 8 and 24 hours.
Micrococcal Nuclease (MNase) assay
Nucleosome DNA assay was done with EZ nucleosomal kit (D5220) supplied form Zymo Research according to the manufacturer’s instructions. In brief, nuclei were isolated from control or siPACS-1 transfected HeLa cells, and treated with 0, 0.25, 0.5, 1.0, 5.0 and 10 U (unit) micrococcal nuclease for the 20 minutes at 42 °C. DNA was subsequently resolved in a 2% agarose gel.
Analysis of chromosomal aberrations
HeLa cells were transfected with control siRNA or siPACS-1 for 48 hours. After 48 hours, cells were washed and treated with demecolcine solution (D1925, Sigma, St Louis, MO) for 2 hours. Cells were then harvested and suspended in hypotonic solution (0.075 M KCl) and kept at room temperature for 20 minutes. Cells were fixed in cold Carnoy’s fixative (methanol: acetic acid, 3:1) and deposited on slides and stained with Giemsa (G146–10, Fisher, USA) and analyzed as described previously 52
Statistical analysis
Statistical analysis was done using GraphPad Prism version 6.0 (GraphPad Software Inc). All statistical analysis was carried out using Student’s t test, unless stated. *p < 0.05; **p < 0.01; *** p < 0.001; **** p < 0.0001; as stated within the figure.
Supplementary Material
Acknowledgements
We thank Steve McClellan, Mitchell Cancer Institute for flow cytometry. This work is supported by grants from Abraham Mitchell Cancer Research Scholar Endowment grant, and partly by NIH grants R01GM098956 and R01CA219187 to K.P., the NIH grants DK114855 and DK112844 to G.T., the NIH grant R01GM108648 and DOD BRCP Breakthrough Award BC151728 to A.V.
Footnotes
Competing interests
Authors declare that there are no competing interests.
References
- 1.Gilbert N, Gilchrist S, Bickmore WA. Chromatin organization in the mammalian nucleus. Int Rev Cytol 2005; 242: 283–336. [DOI] [PubMed] [Google Scholar]
- 2.Peterson CL, Laniel M-A. Histones and histone modifications. Curr Biol CB 2004; 14: R546–551. [DOI] [PubMed] [Google Scholar]
- 3.Leman AR, Noguchi E. The replication fork: understanding the eukaryotic replication machinery and the challenges to genome duplication. Genes 2013; 4: 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lande-Diner L, Zhang J, Cedar H. Shifts in replication timing actively affect histone acetylation during nucleosome reassembly. Mol Cell 2009; 34: 767–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci U S A 1995; 92: 1237–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol 2014; 6: a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev 2011; 25: 1320–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stengel KR, Hiebert SW. Class I HDACs Affect DNA Replication, Repair, and Chromatin Structure: Implications for Cancer Therapy. Antioxid Redox Signal 2015; 23: 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhaskara S, Chyla BJ, Amann JM, Knutson SK, Cortez D, Sun Z-W et al. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell 2008; 30: 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Summers AR, Fischer MA, Stengel KR, Zhao Y, Kaiser JF, Wells CE et al. HDAC3 is essential for DNA replication in hematopoietic progenitor cells. J Clin Invest 2013; 123: 3112–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wells CE, Bhaskara S, Stengel KR, Zhao Y, Sirbu B, Chagot B et al. Inhibition of histone deacetylase 3 causes replication stress in cutaneous T cell lymphoma. PloS One 2013; 8: e68915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhaskara S, Jacques V, Rusche JR, Olson EN, Cairns BR, Chandrasekharan MB. Histone deacetylases 1 and 2 maintain S-phase chromatin and DNA replication fork progression. Epigenetics Chromatin 2013; 6: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene 2007; 26: 5420–5432. [DOI] [PubMed] [Google Scholar]
- 14.De Souza C, Chatterji BP. HDAC Inhibitors as Novel Anti-Cancer Therapeutics. Recent Patents Anticancer Drug Discov 2015; 10: 145–162. [DOI] [PubMed] [Google Scholar]
- 15.Thomas G, E., Aslan J, Thomas L, Shinde P, Shinde U, Simmen T Caught in the act: Protein adaptation and the expanding roles of the PACS proteins in tissue homeostasis and disease. J Cell Science 2017; : In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan L, Molloy SS, Thomas L, Liu G, Xiang Y, Rybak SL et al. PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell 1998; 94: 205–216. [DOI] [PubMed] [Google Scholar]
- 17.Blagoveshchenskaya AD, Thomas L, Feliciangeli SF, Hung CH, Thomas G. HIV-1 Nef downregulates MHC-I by a PACS-1- and PI3K-regulated ARF6 endocytic pathway. Cell 2002; 111: 853–866. [DOI] [PubMed] [Google Scholar]
- 18.Crump CM, Hung C-H, Thomas L, Wan L, Thomas G. Role of PACS-1 in trafficking of human cytomegalovirus glycoprotein B and virus production. J Virol 2003; 77: 11105–11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinners I, Wendler F, Fei H, Thomas L, Thomas G, Tooze SA. AP-1 recruitment to VAMP4 is modulated by phosphorylation-dependent binding of PACS-1. EMBO Rep 2003; 4: 1182–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenkins PM, Zhang L, Thomas G, Martens JR. PACS-1 mediates phosphorylation-dependent ciliary trafficking of the cyclic-nucleotide-gated channel in olfactory sensory neurons. J Neurosci Off J Soc Neurosci 2009; 29: 10541–10551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piguet V, Wan L, Borel C, Mangasarian A, Demaurex N, Thomas G et al. HIV-1 Nef protein binds to the cellular protein PACS-1 to downregulate class I major histocompatibility complexes. Nat Cell Biol 2000; 2: 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt V, Sporbert A, Rohe M, Reimer T, Rehm A, Andersen OM et al. SorLA/LR11 regulates processing of amyloid precursor protein via interaction with adaptors GGA and PACS-1. J Biol Chem 2007; 282: 32956–32964. [DOI] [PubMed] [Google Scholar]
- 23.Scott GK, Gu F, Crump CM, Thomas L, Wan L, Xiang Y et al. The phosphorylation state of an autoregulatory domain controls PACS-1-directed protein traffic. EMBO J 2003; 22: 6234–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendonca MS, Farrington DL, Mayhugh BM, Qin Y, Temples T, Comerford K et al. Homozygous deletions within the 11q13 cervical cancer tumor-suppressor locus in radiation-induced, neoplastically transformed human hybrid cells. Genes Chromosomes Cancer 2004; 39: 277–287. [DOI] [PubMed] [Google Scholar]
- 25.Srivatsan ES, Bengtsson U, Manickam P, Benyamini P, Chandrasekharappa SC, Sun C et al. Interstitial deletion of 11q13 sequences in HeLa cells. Genes Chromosomes Cancer 2000; 29: 157–165. [DOI] [PubMed] [Google Scholar]
- 26.Srivatsan ES, Chakrabarti R, Zainabadi K, Pack SD, Benyamini P, Mendonca MS et al. Localization of deletion to a 300 Kb interval of chromosome 11q13 in cervical cancer. Oncogene 2002; 21: 5631–5642. [DOI] [PubMed] [Google Scholar]
- 27.Zainabadi K, Jain AV, Donovan FX, Elashoff D, Rao NP, Murty VV et al. One in four individuals of African-American ancestry harbors a 5.5kb deletion at chromosome 11q13.1. Genomics 2014; 103: 276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pommier Y Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer 2006; 6: 789–802. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 2009; 138: 1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott GK, Fei H, Thomas L, Medigeshi GR, Thomas G. A PACS-1, GGA3 and CK2 complex regulates CI-MPR trafficking. EMBO J 2006; 25: 4423–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crump CM, Xiang Y, Thomas L, Gu F, Austin C, Tooze SA et al. PACS-1 binding to adaptors is required for acidic cluster motif-mediated protein traffic. EMBO J 2001; 20: 2191–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stern D, Cho MT, Chikarmane R, Willaert R, Retterer K, Kendall F et al. Association of the missense variant p.Arg203Trp in PACS1 as a cause of intellectual disability and seizures. Clin Genet 2017; 92: 221–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaillard H, García-Muse T, Aguilera A. Replication stress and cancer. Nat Rev Cancer 2015; 15: 276–289. [DOI] [PubMed] [Google Scholar]
- 34.Khurana S, Oberdoerffer P. Replication Stress: A Lifetime of Epigenetic Change. Genes 2015; 6: 858–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayakawa T, Nakayama J. Physiological Roles of Class I HDAC Complex and Histone Demethylase. J Biomed Biotechnol 2011; 2011. doi: 10.1155/2011/129383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhaskara S, Knutson SK, Jiang G, Chandrasekharan MB, Wilson AJ, Zheng S et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell 2010; 18: 436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 2011; 331: 1315–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hagelkruys A, Lagger S, Krahmer J, Leopoldi A, Artaker M, Pusch O et al. A single allele of Hdac2 but not Hdac1 is sufficient for normal mouse brain development in the absence of its paralog. Dev Camb Engl 2014; 141: 604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson CA, White DA, Lavender JS, O’Neill LP, Turner BM. Human class I histone deacetylase complexes show enhanced catalytic activity in the presence of ATP and co-immunoprecipitate with the ATP-dependent chaperone protein Hsp70. J Biol Chem 2002; 277: 9590–9597. [DOI] [PubMed] [Google Scholar]
- 40.Ma P, Schultz RM. Histone Deacetylase 2 (HDAC2) Regulates Chromosome Segregation and Kinetochore Function via H4K16 Deacetylation during Oocyte Maturation in Mouse. PLoS Genet 2013; 9. doi: 10.1371/journal.pgen.1003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mullican SE, Gaddis CA, Alenghat T, Nair MG, Giacomin PR, Everett LJ et al. Histone deacetylase 3 is an epigenomic brake in macrophage alternative activation. Genes Dev 2011; 25: 2480–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner FF, Zhang Y-L, Fass DM, Joseph N, Gale JP, Weïwer M et al. Kinetically Selective Inhibitors of Histone Deacetylase 2 (HDAC2) as Cognition Enhancers. Chem Sci 2015; 6: 804–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Wharton W, Yuan Z, Tsai S-C, Olashaw N, Seto E. Activation of the Growth-Differentiation Factor 11 Gene by the Histone Deacetylase (HDAC) Inhibitor Trichostatin A and Repression by HDAC3. Mol Cell Biol 2004; 24: 5106–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conti C, Leo E, Eichler GS, Sordet O, Martin MM, Fan A et al. Inhibition of histone deacetylase in cancer cells slows down replication forks, activates dormant origins, and induces DNA damage. Cancer Res 2010; 70: 4470–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ciechanover A The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J 1998; 17: 7151–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atkins KM, Thomas LL, Barroso-González J, Thomas L, Auclair S, Yin J et al. The multifunctional sorting protein PACS-2 regulates SIRT1-mediated deacetylation of p53 to modulate p21-dependent cell-cycle arrest. Cell Rep 2014; 8: 1545–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Godman CA, Joshi R, Tierney BR, Greenspan E, Rasmussen TP, Wang H-W et al. HDAC3 impacts multiple oncogenic pathways in colon cancer cells with effects on Wnt and vitamin D signaling. Cancer Biol Ther 2008; 7: 1570–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoon H-G, Chan DW, Huang Z-Q, Li J, Fondell JD, Qin J et al. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J 2003; 22: 1336–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tripathi K, Mani C, Barnett R, Nalluri S, Bachaboina L, Rocconi RP et al. Gli1 protein regulates the S-phase checkpoint in tumor cells via Bid protein, and its inhibition sensitizes to DNA topoisomerase 1 inhibitors. J Biol Chem 2014; 289: 31513–31525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clark DW, Tripathi K, Dorsman JC, Palle K. FANCJ protein is important for the stability of FANCD2/FANCI proteins and protects them from proteasome and caspase-3 dependent degradation. Oncotarget 2015; 6: 28816–28832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tripathi K, Hussein UK, Anupalli R, Barnett R, Bachaboina L, Scalici J et al. Allyl isothiocyanate induces replication-associated DNA damage response in NSCLC cells and sensitizes to ionizing radiation. Oncotarget 2015; 6: 5237–5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tripathi K, Mani C, Clark DW, Palle K. Rad18 is required for functional interactions between FANCD2, BRCA2, and Rad51 to repair DNA topoisomerase 1-poisons induced lesions and promote fork recovery. Oncotarget 2016; 7: 12537–12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.