Abstract
Transcriptomics has become an important tool for identification of biological pathways dysregulated in Alzheimer’s disease (AD). We performed a network-based gene expression analysis of blood-based microarray gene expression profiles using two independent cohorts, Alzheimer’s Disease Neuroimaging Initiative (ADNI; N=661) and AddNeuroMed (N=674). Weighted gene co-expression network analysis identified 17 modules from ADNI and 13 from AddNeuroMed. Four of the modules derived in ADNI were significantly related to AD; 5 modules in AddNeuroMed were significant. Gene-set enrichment analysis of the AD-related modules identified and replicated three biological pathways including the Fc gamma receptor-mediated phagocytosis pathway. Module-based association analysis showed the AD-related module, which has the three pathways, to be associated with cognitive function and neuroimaging biomarkers. Gene-based association analysis identified PRKCD in the Fc gamma receptor-mediated phagocytosis pathway as being significantly associated with cognitive function and CSF biomarkers. The identification of the Fc gamma receptor-mediated phagocytosis pathway implicates the peripheral innate immune system in the pathophysiology of AD. PRKCD is known to be related to neurodegeneration induced by amyloid-β.
Keywords: Alzheimer’s disease, Transcriptome, PRKCD, Network, Co-expression, Phagocytosis
1. Introduction
Alzheimer’s disease (AD) is the most common form of dementia (undefined author, 2018). Although AD is classically viewed primarily as a neurodegenerative CNS disease, many systemic manifestations suggest that AD is a multifactorial disease that affects both brain and periphery (Morris et al., 2014). The systemic manifestations generally parallel the progressive functional decline associated with neurodegeneration (Morris et al., 2014). However, some systemic manifestations are also observable prior to the presence of clinical symptoms in AD (Vidoni et al., 2011).
Because blood interacts with every organ in the body, including the brain, blood-based profiles may provide an accessible and effective tool for assessing the complex interplay between the brain and the periphery in the pathogenesis of AD (Mohr and Liew, 2007). Among blood-based biomarkers, the transcriptome uniquely reflects both fixed genetic effects and dynamic environmental effects (Gladkevich et al., 2004; Tylee et al., 2013). There have been several studies investigating blood transcriptomic profiles in AD, which reported biological pathways including stress and immune responses (Bai et al., 2014; Booij et al., 2011; Chen et al., 2011; Fehlbaum-Beurdeley et al., 2012; Han et al., 2013; Kálmán et al., 2005; Lunnon et al., 2012, 2013; Maes et al., 2007; Naughton et al., 2015; Roed et al., 2013; Rye et al., 2011). However, many of the studies analyzed only a small number of samples and their findings were not replicated in independent cohorts. Furthermore, most of the aforementioned studies evaluated differential expressions only at the level of individual genes. Because genes with similar function tend to have correlated expression (Eisen et al., 1998), network-based approaches can better elucidate the molecular mechanisms underlying complex brain disorders (Dragomir et al., 2018).
In this study, using two independent cohorts, we performed blood-based gene co-expression network analysis to identify AD-related modules. We then performed pathway-based enrichment analysis to determine biological functions characteristic of these AD-related modules and association analysis of the AD-related modules and genes belonging to the biological pathways with fluid and neuroimaging biomarkers for AD. For fluid biomarkers for AD, we used the concentration of amyloid-β42 (Aβ42), phosphorylated tau181p (p-tau) and total tau (t-tau) in cerebrospinal fluid (CSF) (Kang et al., 2015). For neuroimaging biomarkers for AD, we used a global cortical measure of amyloid burden measured from [18F] florbetapir PET scans (Ramanan et al., 2014) and hippocampal volume measured from MRI scans (Potkin et al., 2009). Genetic data were also used for gene-based association analysis and expression quantitative trait locus (eQTL) analysis (Rockman and Kruglyak, 2006).
2. Material and methods
2.1. Participants
Data used in the study were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) and AddNeuroMed cohorts as discovery and replication samples, respectively. The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to accurately capture the progression of mild cognitive impairment (MCI) and early AD. The AddNeuroMed is a cross European, public/private consortium developed for AD biomarker discovery (Lovestone et al., 2009). The diagnosis of AD was made clinically using NINCDS/ADRDA criteria for probable AD (McKhann et al., 1984). The diagnosis of MCI was made according to the presence of objective memory impairment but without meeting the criteria for dementia. Written informed consent was obtained at the time of enrollment and included permission for analysis and data sharing. The protocol and informed consent forms were approved by each participating sites’ Institutional Review Board.
2.2. Genotyping and imputation
Genotyping for ADNI and AddNeuroMed was performed using blood genomic DNA samples and Illumina GWAS array platforms (Illumina Human610-Quad BeadChip, Illumina HumanOmni Express BeadChip, and Illumina HumanOmni 2.5M BeadChip) (Furney et al., 2011; Saykin et al., 2015). APOE genotyping was separately conducted using standard methods as described previously to yield the APOE ε4 allele defining single nucleotide polymorphisms (SNPs) (rs429358, rs7412) (Furney et al., 2011; Saykin et al., 2015). Using PLINK 1.9 (www.cog-genomics.org/plink2/) (Purcell et al., 2007), we performed standard quality control (QC) procedures for samples and SNPs as described previously (Lee et al., 2018): (1) for SNP, SNP call rate < 95%, Hardy-Weinberg p-value < 1×10−6, and minor allele frequency (MAF) < 1%; (2) for sample, sex inconsistencies, and sample call rate < 95%. Furthermore, in order to prevent spurious association due to population stratification, we selected only non-Hispanic participants of European ancestry that clustered with HapMap CEU (Utah residents with Northern and Western European ancestry from the CEPH collection) or TSI (Toscani in Italia) populations using multidimensional scaling analysis (Price et al., 2006; Thorisson et al., 2005). After QC procedures, as ADNI and AddNeuroMed used different genotyping platforms, we imputed ungenotyped SNPs separately in each platform using MaCH with the Haplotype Reference Consortium data as a reference panel (Li et al., 2010; McCarthy et al., 2016). After the imputation, we imposed an r2 value equal to 0.30 as the threshold to accept the imputed genotypes.
2.3. Imaging and cerebrospinal fluid biomarkers in ADNI
To measure hippocampal and intracranial volumes from T1-weighted brain MRI scans, we used FreeSurfer version 5.1 (surfer.nmr.mgh.harvard.edu) (Jack et al., 2010). For assessment of cortical amyloid accumulation, we used pre-processed (co-registered, averaged, standardized image and voxel size, uniform resolution) [18F] florbetapir PET scans (Jagust et al., 2015) and calculated mean standardized uptake values using a whole cerebellum reference region as previously described (Risacher et al., 2015). The concentration of CSF Aβ42, p-tau and t-tau were measured by the validated and highly automated Roche Elecsys electrochemiluminescence immunoassays (Roche Diagnostics, Mannheim, Germany) (Bittner et al., 2016).
2.4. Blood-based RNA expression microarray profiling
For the ADNI and AddNeuroMed samples, the PAXgene Blood RNA Kit (Qiagen Inc., Valencia, CA, USA) was used to purify total RNA from whole blood collected in a PAXgene Blood RNA Tube (Lunnon et al., 2012; Saykin et al., 2015). The Affymetrix Human Genome U219 Array (Affymetrix, Santa Clara, CA, USA) and the Illumina Human HT-12 v3 Expression BeadChips (Illumina Inc., San Diego, CA, USA) were used for expression profiling in ADNI and AddNeuroMed, respectively (Lunnon et al., 2012; Saykin et al., 2015). Raw expression values were pre-processed using the robust multi-chip average normalization method in ADNI (Choe et al., 2005) and the robust spline normalization method in AddNeuroMed (Du et al., 2008). We checked discrepancies between the reported sex and sex determined from sex-specific gene expression data including XIST and USP9Y. We also evaluated whether SNP genotypes were matched with genotypes predicted from gene expression data (Schadt et al., 2012). After QC, the RNA expression profiles contained 21,150 and 5,141 probes, in ADNI and AddNeuroMed, respectively. The RNA expression profiles were pre-adjusted with RNA integrity number (RIN) values and batch effects using linear regression.
2.5. Gene co-expression network analysis & identification of AD-related modules
We constructed clusters (modules) of highly co-expressed genes from RNA expression profiles of all participants (cognitively normal older adults (CN) and patients with MCI and AD) using the weighted correlation network analysis (WGCNA) software (Langfelder and Horvath, 2008), which calculates the network adjacency matrix based on co-expression similarity and identifies gene modules using unsupervised hierarchical clustering. Modules were represented by a weighted average expression profile, the module eigengene (ME), which is defined as the first principal component of the expression matrix in each module. We then performed a correlation analysis between ME and AD diagnosis (CN vs. AD) and presented this as a color-coded correlation map. We also performed a linear regression analysis with AD diagnosis, age and sex as independent variables and ME as an outcome to identify modules that were dysregulated in AD using R version 3.6.0 (www.R-project.org). Multiple testing correction was performed using the false discovery rate (FDR) with the Benjamini-Hochberg procedure (Benjamini and Hochberg, 1995).
2.6. Pathway-based enrichment analysis of AD-related modules
We performed enrichment analysis to identify the biological pathways of genes assigned to each of the AD-related modules. The DAVID bioinformatic resource was used to evaluate whether genes in a particular biological pathway were significantly more enriched in a given module than would be expected by random chance (FDR-corrected p-value < 0.05) (Huang et al., 2009). KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways were used as a reference for functional annotation (Kanehisa et al., 2016). We then checked whether the biological pathways identified in ADNI were replicated in AddNeuroMed. We defined pathways as replicated only when the ME of the corresponding module in AddNeuroMed showed also a significant diagnosis group difference in the same direction (positively or negatively) as the ME of the matching ADNI module.
2.7. Module-based association analysis of modules with replicated biological pathways with AD biomarkers
We performed a linear regression analysis to evaluate whether the ME of AD-related modules that had the replicated biological pathways were also associated with the following AD biomarkers in all participants (CN, MCI and AD) from the ADNI cohort (FDR-corrected p-value < 0.05): composite scores for memory and executive function (Crane et al., 2012; Gibbons et al., 2012), hippocampal volume on MRI, CSF Aβ42, CSF p-tau, CSF t-tau, CSF p-tau/Aβ42, CSF ttau/Aβ42 and averaged cortical uptake of [18F] florbetapir PET. Covariates included age and sex. Intracranial volume and MRI field strength were also used as covariates for hippocampal volume.
2.8. Gene-based association analysis of target genes in replicated biological pathways
We selected common genes that belonged to each replicated biological pathway in both ADNI and AddNeuroMed. For each pathway, we identified genes that showed differential expression between AD and CN in ADNI and examined whether the findings were replicated in AddNeuroMed (FDR-corrected p-value < 0.05). For those differentially expressed genes, gene-based association analysis with AD biomarkers was performed using a set-based test in PLINK 1.9. SNPs in the coding region, 5’ untranslated region, 3’ untranslated region, regulatory region and intronic region (±20 kb of upstream and downstream regions) with MAF greater than 0.05 were used for analysis. The same AD biomarkers in the module-based association analysis were used for this analysis as well. An empirical p-value (20,000 permutation) was calculated for each gene.
2.9. eQTL analysis of differentially expressed genes
We performed eQTL analysis on differentially expressed genes in each replicated biological pathway using PLINK 1.9 (FDR-corrected p-value < 0.05) (Rockman and Kruglyak, 2006). The GWAS and RNA expression data from all participants (CN, MCI and AD) with the same set of SNPs for each gene in the gene-based association analysis were used for eQTL analysis. We then checked whether the significantly associated SNPs were replicated in AddNeuroMed. In addition, we performed a meta-analysis using METAL (Willer et al., 2010), which weighted the effect size estimates by their estimated standard errors. Results of the eQTL analysis were plotted using LocusZoom (Pruim et al., 2010).
2.10. Hub genes in AD-related modules with the replicated biological pathways
We identified hub genes (top 10% of genes with the highest intramodular connectivity) and the overlapping hub genes for the AD-related modules with replicated biological pathways in ADNI and AddNeuroMed using WGCNA.
3. Results
A total of 661 participants from ADNI and 674 participants from AddNeuroMed were included in the present study (Table 1). Gene co-expression network analysis using WGCNA yielded 17 and 13 modules of highly co-expressed genes in ADNI and AddNeuroMed, respectively (Figure 1). Following a linear regression analysis, 4 modules (lightgreen, red, brown and darkturquoise) in ADNI and 5 modules (turquoise, yellow, black, tan and green) in AddNeuroMed were significantly dysregulated in AD compared to control samples (FDR-corrected p-value < 0.05) (Table A.1).
Table 1.
Demographics of study population
| Cohort | Diagnosis | No. of participants | Female (%) | Age, mean (SD) | RIN, mean (SD) |
|---|---|---|---|---|---|
| ADNI (N=661) | CN | 213 | 107 (50%) | 76.4 (6.4) | 6.91 (0.51) |
| MCI | 345 | 144 (42%) | 73.2 (7.9) | 6.98 (0.55) | |
| AD | 103 | 38 (37%) | 77.6 (7.8) | 6.98 (0.64) | |
| AddNeuroMed (N=674) | CN | 243 | 147 (60%) | 74.2 (6.6) | 8.96 (0.73) |
| MCI | 208 | 120 (58%) | 75.5 (6.5) | 8.50 (0.59) | |
| AD | 223 | 146 (65%) | 76.8 (6.8) | 8.43 (0.64) |
Key: AD: Alzheimer’s disease; CN: cognitively normal older adults; MCI: mild cognitive impairment; RIN: RNA integrity number; SD: standard deviation
Figure 1. Clustering dendrogram with correlation between modules and AD diagnosis (CN vs. AD).
Weighted correlation network analysis constructed 17 and 13 modules in ADNI (A; discovery data) and AddNeuroMed (B; replication data) cohorts, respectively. The modules are represented in the rows with the clustering dendrogram. Colors were assigned to the modules arbitrarily according to their size by the WGCNA software. Each cell contains the correlation between the corresponding module and AD diagnosis (CN vs. AD) with its FDR-corrected p-value. It is color-coded by the correlation according to the color legend. The p-value was derived from a linear regression analysis with AD diagnosis, age and sex as independent variables and module eigengene as an outcome.
Abbreviation: AD: Alzheimer’s disease; CN: cognitively normal older adults; FDR: false discovery rate; WGCNA: weighted correlation network analysis
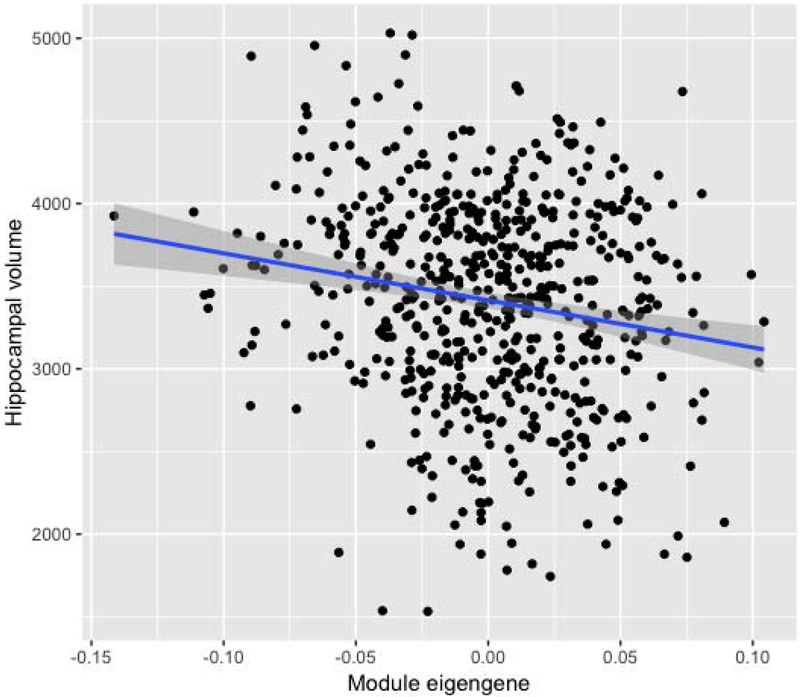
Pathway-based enrichment analysis of the four AD-related modules in ADNI identified a total of 10 enriched biological pathways (Table 2). Among them, 3 pathways linked to the brown module (containing 1580 genes) in ADNI were replicated in the yellow module (containing 466 genes) of AddNeuroMed (FDR-corrected p-value < 0.05): Fc gamma receptor (FcγR)-mediated phagocytosis, osteoclast differentiation, and tuberculosis. The ribosome pathway was also enriched in both the red module of ADNI and the turquoise module of AddNeuroMed. However, it was not considered to be replicated because the direction of the relationship between the ME and AD diagnosis was opposite in ADNI and AddNeuroMed. Therefore, only the brown module in ADNI and the yellow module in AddNeuroMed had the replicated biological pathways in the enrichment analysis. Module-based association analysis revealed that the brown module in ADNI was positively associated with mean cortical amyloid-β (Aβ) accumulation measured using [18F] florbetapir PET scans, and negatively associated with composite scores for memory and executive function as well as hippocampal volume (Table 3 and Figure 2).
Table 2.
Biological pathways identified in enrichment analysis
| Module (Direction of association between ME and diagnosis) | KEGG pathway | FDR- corrected p-value |
| Lightgreen (↓ ME in AD) | B cell receptor signaling | 7.44×10−5 |
| Red (↓ ME in AD) | Ribosome | 3.35×10−21 |
| Brown (↑ ME in AD) | Fc gamma receptor-mediated phagocytosis | 8.49×10−4 |
| Osteoclast differentiation | 1.58×10−5 | |
| Tuberculosis | 2.42×10−6 | |
| Endocytosis | 2.51×10−2 | |
| B cell receptor signaling | 2.25×10−2 | |
| Hepatitis B | 1.89×10−2 | |
| Lysosome | 2.22×10−3 | |
| Estrogen signaling | 1.98×10−2 | |
| Yellow (↑ ME in AD) | Fc gamma receptor-mediated phagocytosis | 4.58×10−5 |
| Osteoclast differentiation | 2.31×10−7 | |
| Tuberculosis | 4.26×10−5 | |
| Turquoise (↑ ME in AD) | Ribosome | 3.25×10−17 |
Values were derived from the DAVID bioinformatic resources (Huang et al., 2009).
No KEGG pathways were enriched at darkturquoise module in ADNI and black, green and tan modules in AddNeuroMed (FDR-corrected p-value < 0.05).
In AddNeuroMed, we only demonstrated KEGG pathways which were identified in ADNI and were significantly enriched (FDR-corrected p-value < 0.05).
Key: FDR: false discovery rate; KEGG: Kyoto Encyclopedia of Genes and Genomes
Table 3.
Association between the brown module of ADNI and various AD biomarkers
| AD biomarker | t-value | FDR-corrected p-value |
|---|---|---|
| ADNI-EFa | −2.779 | 1.71×10−2 |
| ADNI-MEM | −2.677 | 1.71×10−2 |
| Hippocampal volume in MRIb | −3.148 | 1.55×10−2 |
| Averaged cortical uptake of [18F] florbetapir PETc | 2.684 | 1.71×10−2 |
t-values and p-values were derived from a linear regression analysis between module eigengene of the brown module and AD biomarkers.
Data for 2 participants were unavailable.
Data for 44 participants were unavailable.
Data for 99 participants were unavailable.
Key: AD: Alzheimer’s disease; ADNI-EF: Composite score of executive function in ADNI; ADNI-MEM: Composite score of memory in ADNI; FDR: false discovery rate
Figure 2. Relationship between hippocampal volume and the brown module in ADNI.
The relationship between hippocampal volume and module eigengene (ME) of the brown module in ADNI was represented in a scatter plot. The blue line was obtained from a linear regression analysis (FDR-corrected p-value = 1.55×10−2), and the gray zone around the blue line indicates 95% confidence interval.
Abbreviation: FDR: false discovery rate
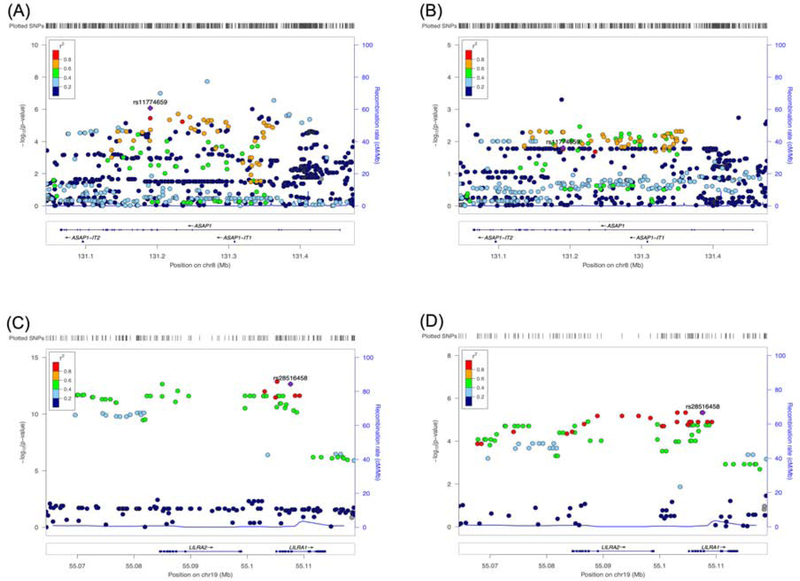
In the FcγR-mediated phagocytosis pathway, we identified 11 genes (ASAP1, GAB2, LYN, MAPK1, MAPK3, MARCKS, PAK1, PLCG2, PRKCD, SYK, and VASP) that were common in both the brown module of ADNI and the yellow module of AddNeuroMed. In ADNI, ASAP1, PRKCD and VASP were significantly overexpressed in AD compared to CN. Among the 3 genes, ASAP1 and PRKCD were replicated in AddNeuroMed. Gene-based association analysis of SNPs within ASAP1 and PRKCD with AD biomarkers identified PRKCD as being significantly associated with cognitive functions and CSF p-tau/Aβ42 (Table 4). eQTL analysis identified several SNPs in ASAP1 as being associated with expression levels of ASAP1, which were replicated in AddNeuroMed. rs11774659 had the lowest p-value of 4.81×10−9 in the meta-analysis. We plot each SNP in the region on the chromosome and indicate their association with ASAP1 expression levels (Figure 3). No SNPs were identified and replicated in PRKCD.
Table 4.
Gene-based association analysis with AD biomarkers
| AD biomarker | Gene set | No. of SNP | No. of significant SNP | Empirical p-value | Independent significant SNPs |
|---|---|---|---|---|---|
| ADNI-MEM | PRKCD | 132 | 30 | 2.55×10−3 | rs6764111, rs11130350 |
| ADNI-EFa | PRKCD | 132 | 50 | 2.61×10−2 | rs6764111, rs62254274, rs3821689, rs2358617, rs12495976 |
| CSF p-tau/Aβ42b | PRKCD | 132 | 43 | 4.18×10−2 | rs17052826, rs1872037, rs6778939, rs11130347, rs55685362 |
| ADNI-MEM | LILRB3 | 216 | 9 | 6.25×10−3 | rs11878556, rs17841905, rs117107587 |
| ADNI-EFa | LILRB3 | 216 | 22 | 1.50×10−2 | rs6509855, rs620207, rs11084325, rs4442928, rs16960152 |
| CSF p-tau/Aβ42b | LILRB3 | 216 | 10 | 3.88×10−2 | rs6509855, rs34810796, rs606851, rs202131060 |
Data for 2 participants were unavailable.
Data for 206 participants were unavailable.
Key: AD: Alzheimer’s disease; Aβ42: amyloid-β42; ADNI-EF: Composite score of executive function in ADNI; ADNI-MEM: Composite score of memory in ADNI; p-tau: phosphorylated tau181p
Figure 3. LocusZoom plot for eQTL analysis of ASAP1 and LILRA2.
SNPs’ positions on chromosome and their color-coded association with expression levels of ASAP1 are plotted in ADNI (A) and AddNeuroMed (B). The SNP with the lowest p-value in a meta-analysis (rs11774659) is indicated. The eQTL results of LILRA2 are plotted for ADNI (C) and AddNeuroMed (D) with an indication of the SNP with the lowest p-value (rs28516458) in a meta-analysis.
Abbreviation: eQTL: expression quantitative trait locus
In the osteoclast differentiation pathway, we identified 15 genes (GAB2, GRB2, IFNAR1, LILRA2, LILRB2, LILRB3, MAPK1, MAPK3, OSCAR, PLCG2, SIRPA, SPI1, SYK, TNFRSF1A, and TYROBP) that were common in both the brown module of ADNI and the yellow module of AddNeuroMed. In ADNI, LILRA2, LILRB3, SPI1, and TYROBP were significantly overexpressed in AD compared to CN. Among them, 3 genes (LILRA2, LILRB3, and TYROBP) were replicated in AddNeuroMed. Gene-based association analysis of SNPs with AD biomarkers identified only LILRB3 as being significantly associated with cognitive functions and CSF p-tau/Aβ42 (Table 4). eQTL analysis identified SNPs in LILRA2 as significantly associated with expression levels of LILRA2, which was replicated in AddNeuroMed. rs28516458 had the smallest p-value of 5.42×10−18 in the meta-analysis (Figure 3). No SNPs were identified and replicated in LILRB3 and TYROBP.
In the tuberculosis pathway, we identified 14 common genes (APAF1, ATP6V0B, ATP6V0D1, CAMK2G, CREBBP, CTSD, FADD, LSP1, MAPK1, MAPK3, RAB5C, RAB7A, SYK, and TNFRSF1A) in both the brown module of ADNI and the yellow module of AddNeuroMed. APAF1 were significantly overexpressed in AD compared to CN in ADNI, which was replicated in AddNeuroMed. Gene-based association analysis of SNPs within APAF1 did not identify any associations between SNPs and AD biomarkers. No SNPs in APAF1 were identified and replicated in eQTL analysis.
In the intramodular connectivity analysis, we identified 160 and 58 hub genes from the brown module of ADNI and the yellow module of AddNeuroMed, respectively. A total of 30 genes, including PRKCD and LILRB3, overlapped between the two modules (Table A.2).
4. Discussion
In this study, we constructed modules from gene co-expression network analysis in peripheral blood and determined that 3 biological pathways (FcγR-mediated phagocytosis, osteoclast differentiation, and tuberculosis) were dysregulated in AD patients. The brown module in ADNI in which these three pathways were found, was associated with AD biomarkers including cognitive functions, hippocampal volume, and cortical amyloid-β accumulation. This is the first blood-based transcriptomic study in AD that used two independent cohorts (discovery and replication samples) to demonstrate consistent findings.
FcγRs, which are cellular receptors for IgG, play an important role in various types of immune responses. One function is to couple the specificity of the antibody response to innate effector pathways, such as phagocytosis, antibody-dependent cellular cytotoxicity, and the recruitment and activation of inflammatory cells (Nimmerjahn and Ravetch, 2006). It is known that FcγRs are necessary for the intracerebral immune complex to induce inflammation in the brain (Teeling et al., 2012). The increased expression of FcRs on microglia has been observed in both animal models and patients with AD (Fuller et al., 2014). Indeed, it is through this mechanism that antibodies against Aβ peptide may trigger microglia to clear Aβ plaques and thus cause the serious side effects called ARIA (amyloid-related imaging abnormalities) when using such immunotherapy strategies (Gu et al., 2016; Lai et al., 2017; Le Page et al., 2018). It is possible to prevent these outcomes by neutralizing the Fc arm of the antibody without loss of efficacy as Aβ plaques can be neutralized by Aβ antibodies without the action of Fc receptors (Das et al., 2003; Golde et al., 2009).
Interestingly, in-vitro phagocytic activity by peripheral blood monocytes was significantly increased in participants with amyloid-positive PET scans compared to participants with amyloid-negative PET scans (Gu et al., 2016). Peripheral inflammatory markers were found to be elevated in patients with AD as well (Lai et al., 2017). Inflammation initiated in the brain may lead to the release of soluble inflammatory mediators that migrate to the periphery and activate peripheral immune cells (Le Page et al., 2018). On the other hand, these peripheral immune cells may also migrate to the brain and perpetuate brain inflammation through the disrupted blood-brain barrier in AD, sustaining a vicious cycle between the brain and periphery (Le Page et al., 2018; Sweeney et al., 2018).
We also found that ASAP1 and PRKCD were significantly overexpressed in AD patients compared to CN among genes in the FcγR-mediated phagocytosis pathway. In linkage analysis of autopsy-confirmed familial AD, it was reported that some SNPs near ASAP1 were associated with AD (Sillén et al., 2011). In addition, PRKCD encodes protein kinase C-δ which is activated by Aβ and phosphorylates myristoylated alanine-rich C kinase substrate (MARCKS) (Nakai et al., 2001). The phosphorylation of MARCKS is known to induce neurite degeneration via instability of the actin network in human and mouse brains (Fujita et al., 2016).
Osteoclasts are bone-resorbing cells that regulate bone turnover (Cappariello et al., 2014). It was known that low bone mineral density was associated with the risk of AD (Tan et al., 2005). Previously the association between AD and osteoporosis was thought to be related to reduced daily activities, low Vitamin D or low estrogen exposure (Ensrud and Crandall, 2017; Mokry et al., 2016; Tan et al., 2005). However, it was also reported that differentiation of osteoclasts is increased in an AD mouse model, which favors loss of bone mineral (Cui et al., 2011). Furthermore, Aβ peptides were elevated in human osteoporotic bone tissues and enhanced osteoclast function (Li et al., 2014). The altered osteoclast differentiation pathway that we identified in peripheral blood of AD patients might be associated with increased risk of osteoporosis in AD patients.
Differentially expressed genes between AD and CN in the osteoclast differentiation pathway were related to the pathogenesis of AD patients. TYRO protein tyrosine kinase-binding protein, encoded by TYROBP, is a signaling adaptor protein that plays important roles in signal transduction in microglia, osteoclasts, macrophages and dendritic cells, and enhances phagocytic activity of microglia (Ma et al., 2015). It was reported that TYROBP acted as a key regulator in an immune- and microglia-specific module that was constructed from the brain transcriptomic network of AD patients (Zhang et al., 2013). Leukocyte Ig-like receptors are expressed on innate and adaptive immune cells and maintain immune homeostasis (Takeda and Nakamura, 2017). LILRA2 was known to act as one of the hub genes in the Fc receptor system in the brain transcriptomic network of AD patients (Zhang et al., 2013).
The tuberculosis pathway was also identified in the enrichment analysis. Some of the sub-pathways of the tuberculosis pathway, such as the toll-like receptor signaling pathway and the mitogen-activated protein kinase signaling pathway, are known to be associated with the pathogenesis of AD and may be of some influence (Huang et al., 2017; Kim and Choi, 2010).
For the past few decades, the amyloid cascade hypothesis has been accepted as the main pathophysiological mechanism of AD (Selkoe and Hardy, 2016). It states that the deposition of Aβ protein is the causative agent of Alzheimer’s pathology and that the neurofibrillary tangles, cell loss, and cognitive decline follow as a direct result of this deposition (Hardy and Higgins, 1992). However, clinical trials that reduce brain amyloid in AD patients have repeatedly failed, suggesting that another key mechanism, in addition to the Aβ deposition, is necessary to explain the occurrence and progression of AD (Mullane and Williams, 2018). Previously, neuroinflammation was considered to be merely a late stage phenomenon in AD (Wyss-Coray, 2006); however, preclinical, genetic, and bioinformatic studies have shown that it actively contributes to AD pathogenesis and is now recognized as a core feature of AD (Heppner et al., 2015). Brain transcriptomic studies as well as several small blood transcriptomic studies have implicated the innate immune pathway in the pathogenesis of AD (International Genomics of Alzheimer’s Disease Consortium, 2015; Zhang et al., 2013), which is summarized in a review article (Song et al., 2016).
This present study has some limitations. First, the regression coefficients observed in blood are relatively small in magnitude, compared to what is typically observed in post-mortem brain studies. However, our findings were replicated in two independent data sets. Furthermore, while access to brain tissue from living patients is extremely limited, blood is easily accessible, non-invasive, inexpensive, and can be obtained longitudinally, providing a useful potential biomarker for AD. Second, blood-based transcriptomic profiles could be influenced by confounding factors such as medication and blood collection, processing and storage procedures (Hampel et al., 2018; Mohr and Liew, 2007). However, we checked RNA quality using RIN values and also filtered out genes with low expression values. Importantly, we replicated our results using an independent dataset. Third, transcriptome profiling was performed on different microarray platforms in ADNI and AddNeuroMed. Therefore, in this study, we did not perform a mega-analysis but constructed network-based modules in ADNI and AddNeuroMed separately. Although the present replicated results are robust, we may have not detected other potentially important peripheral transcriptomic changes due to methodological differences between cohorts. Fourth, we employed expression profile technology. Newer RNA-sequencing methods in the future may better quantitate expression and clarify the role of other factors such as alternative splicing sites. Finally, we analyzed cross-sectionally collected gene expression data. Thus, our findings represent association not causality. Longitudinal studies are needed to understand the role of altered pathways in the onset of AD as well as cause and effect relationships.
In summary, network-based blood gene expression analysis showed that dysregulated pathways, including FcγR-mediated phagocytosis, were observed in patients with AD. Transcriptomics can provide a comprehensive understanding of biological processes and hold great promise for personalized and precision medicine (Li et al., 2016). With more sophisticated blood transcriptome profiling methods and integrative analysis approaches with validation using other multi-omics data, blood-based transcriptomics profiles may become suitable for use in clinical practice for predicting, diagnosing, and personalizing treatment for AD.
Supplementary Material
Verification.
All authors did not have any actual or potential conflicts of interest including any financial, personal or other relationships with other people or organizations within three years that could inappropriately influence this work.
This work was supported by several grants from various Institutes including NIA. However, the funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript, and decision to submit the manuscript for publication.
The data contained in the manuscript being submitted have not been previously published, have not been submitted elsewhere and will not be submitted elsewhere while under consideration at Neurobiology of Aging.
Written informed consent was obtained at the time of enrollment and/or genetic sample collection and protocols were approved by each participating study and sites’ Institutional Review Board.
All authors have reviewed the contents of the manuscript being submitted, approve of its contents and validate the accuracy of the data.
Acknowledgements
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. The collection and analysis of AddNeuroMed samples was supported by InnoMed (Innovative Medicines in Europe) an Integrated Project funded by the European Union of the Sixth Framework program priority FP6-2004-LIFESCIHEALTH-5, the Alzheimer’s Research Trust, the John and Lucille van Geest Foundation and the NIHR Biomedical Research Centre for Mental Health at the South London and Maudsley NHS Foundation Trust and [Institute of Psychiatry] Kings College London.
Additional support for data analysis was provided by NLM R01 LM012535, NIA R03 AG054936, NIA R01 AG19771, NIA P30 AG10133, NLM R01 LM011360, DOD W81XWH-14-2-0151, NIGMS P50GM115318, NCATS UL1 TR001108, NIA K01 AG049050, the Alzheimer’s Association, the Indiana Clinical and Translational Science Institute, and the IU Health-IU School of Medicine Strategic Neuroscience Research Initiative.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none
Reference
- Bai Z, Stamova B, Xu H, Ander BP, Wang J, Jickling GC, Zhan X, Liu D, Han G, Jin LW, Decarli C, Lei H, Sharp FR, 2014. Distinctive RNA expression profiles in blood associated with Alzheimer disease after accounting for white matter hyperintensities. Alzheimer Dis Assoc Disord 28, 226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc 57, 289–300. [Google Scholar]
- Bittner T, Zetterberg H, Teunissen CE, Ostlund RE, Militello M, Andreasson U, Hubeek I, Gibson D, Chu DC, Eichenlaub U, Heiss P, Kobold U, Leinenbach A, Madin K, Manuilova E, Rabe C, Blennow K, 2016. Technical performance of a novel, fully automated electrochemiluminescence immunoassay for the quantitation of β-amyloid (1–42) in human cerebrospinal fluid. Alzheimers Dement 12, 517–526. [DOI] [PubMed] [Google Scholar]
- Booij BB, Lindahl T, Wetterberg P, Skaane NV, Sæbø S, Feten G, Rye PD, Kristiansen LI, Hagen N, Jensen M, Bårdsen K, Winblad B, Sharma P, Lönneborg A, 2011. A gene expression pattern in blood for the early detection of Alzheimer’s disease. J Alzheimers Dis 23, 109–119. [DOI] [PubMed] [Google Scholar]
- Cappariello A, Maurizi A, Veeriah V, Teti A, 2014. The Great Beauty of the osteoclast. Arch Biochem Biophys 558, 70–78. [DOI] [PubMed] [Google Scholar]
- Chen KD, Chang PT, Ping YH, Lee HC, Yeh CW, Wang PN, 2011. Gene expression profiling of peripheral blood leukocytes identifies and validates ABCB1 as a novel biomarker for Alzheimer’s disease. Neurobiol Dis 43, 698–705. [DOI] [PubMed] [Google Scholar]
- Crane PK, Carle A, Gibbons LE, Insel P, Mackin RS, Gross A, Jones RN, Mukherjee S, Curtis SM, Harvey D, Weiner M, Mungas D, Alzheimer’s Disease Neuroimaging Initiative, 2012. Development and assessment of a composite score for memory in the Alzheimer’s Disease Neuroimaging Initiative (ADNI). Brain Imaging Behav 6, 502–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Xiong F, Hong Y, Jung JU, Li XS, Liu JZ, Yan R, Mei L, Feng X, Xiong WC, 2011. APPswe/Aβ regulation of osteoclast activation and RAGE expression in an age-dependent manner. J Bone Miner Res 26, 1084–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P, Howard V, Loosbrock N, Dickson D, Murphy MP, Golde TE, 2003. Amyloid-beta immunization effectively reduces amyloid deposition in FcR gamma(−/−) knock-out mice. J Neurosci 23, 8532–8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragomir A, Vrahatis A, Bezerianos A, 2019. A Network-Based Perspective in Alzheimer’s Disease: Current State and an Integrative Framework. IEEE J Biomed Health Inform 13, 14–25. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D, 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensrud KE, Crandall CJ, 2017. Osteoporosis. Ann. Intern. Med 167, ITC17–ITC32. [DOI] [PubMed] [Google Scholar]
- Fehlbaum-Beurdeley P, Sol O, Désiré L, Touchon J, Dantoine T, Vercelletto M, Gabelle A, Jarrige AC, Haddad R, Lemarié JC, Zhou W, Hampel H, Einstein R, Vellas B, EHTAD/002 study group, 2012. Validation of AclarusDx™, a blood-based transcriptomic signature for the diagnosis of Alzheimer’s disease. J Alzheimers Dis 32, 169–181. [DOI] [PubMed] [Google Scholar]
- Fujita K, Motoki K, Tagawa K, Chen X, Hama H, Nakajima K, Homma H, Tamura T, Watanabe H, Katsuno M, Matsumi C, Kajikawa M, Saito T, Saido T, Sobue G, Miyawaki A, Okazawa H, 2016. HMGB1, a pathogenic molecule that induces neurite degeneration via TLR4-MARCKS, is a potential therapeutic target for Alzheimer’s disease. Sci Rep 6, 31895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller JP, Stavenhagen JB, Teeling JL, 2014. New roles for Fc receptors in neurodegeneration-the impact on Immunotherapy for Alzheimer’s Disease. Front Neurosci 8, 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furney SJ, Simmons A, Breen G, Pedroso I, Lunnon K, Proitsi P, Hodges A, Powell J, Wahlund LO, Kloszewska I, Mecocci P, Soininen H, Tsolaki M, Vellas B, Spenger C, Lathrop M, Shen L, Kim S, Saykin AJ, Weiner MW, Lovestone S, 2011. Genome-wide association with MRI atrophy measures as a quantitative trait locus for Alzheimer’s disease. Mol Psychiatry 16, 1130–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons LE, Carle AC, Mackin RS, Harvey D, Mukherjee S, Insel P, Curtis SM, Mungas D, Crane PK, Alzheimer’s Disease Neuroimaging Initiative, 2012. A composite score for executive functioning, validated in Alzheimer’s Disease Neuroimaging Initiative (ADNI) participants with baseline mild cognitive impairment. Brain Imaging Behav 6, 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladkevich A, Kauffman HF, Korf J, 2004. Lymphocytes as a neural probe: potential for studying psychiatric disorders. Prog Neuropsychopharmacol Biol Psychiatry 28, 559–576. [DOI] [PubMed] [Google Scholar]
- Golde TE, Das P, Levites Y, 2009. Quantitative and mechanistic studies of Abeta immunotherapy. CNS Neurol Disord Drug Targets 8, 31–49. [DOI] [PubMed] [Google Scholar]
- Gu BJ, Huang X, Ou A, Rembach A, Fowler C, Avula PK, Horton A, Doecke JD, Villemagne VL, Macaulay SL, Maruff P, Fletcher EL, Guymer R, Wiley JS, Masters CL, 2016. Innate phagocytosis by peripheral blood monocytes is altered in Alzheimer’s disease. Acta Neuropathol 132, 377–389. [DOI] [PubMed] [Google Scholar]
- Hampel H, O’Bryant SE, Molinuevo JL, Zetterberg H, Masters CL, Lista S, Kiddle SJ, Batrla R, Blennow K, 2018. Blood-based biomarkers for Alzheimer disease: mapping the road to the clinic. Nat Rev Neurol 14, 639–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G, Wang J, Zeng F, Feng X, Yu J, Cao HY, Yi X, Zhou H, Jin LW, Duan Y, Wang YJ, Lei H, 2013. Characteristic transformation of blood transcriptome in Alzheimer’s disease. J Alzheimers Dis 35, 373–386. doi: 10.3233/JAD-121963 [DOI] [PubMed] [Google Scholar]
- Hardy JA, Higgins GA, 1992. Alzheimers disease: the amyloid cascade hypothesis. Science 256, 184–185. [DOI] [PubMed] [Google Scholar]
- Heppner FL, Ransohoff RM, Becher B, 2015. Immune attack: the role of inflammation in Alzheimer disease. Nat Rev Neurosci 16, 358–372. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA, 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Huang NQ, Jin H, Zhou SY, Shi JS, Jin F, 2017. TLR4 is a link between diabetes and Alzheimer’s disease. Behav Brain Res 316, 234–244. [DOI] [PubMed] [Google Scholar]
- International Genomics of Alzheimer’s Disease Consortium, 2015. Convergent genetic and expression data implicate immunity in Alzheimer’s disease. Alzheimers Dement 11, 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Bernstein MA, Borowski BJ, Gunter JL, Fox NC, Thompson PM, Schuff N, Krueger G, Killiany RJ, DeCarli CS, Dale AM, Carmichael OW, Tosun D, Weiner MW, Alzheimer’s Disease Neuroimaging Initiative, 2010. Update on the magnetic resonance imaging core of the Alzheimer’s disease neuroimaging initiative. Alzheimers Dement 6, 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagust WJ, Landau SM, Koeppe RA, Reiman EM, Chen K, Mathis CA, Price JC, Foster NL, Wang AY, 2015. The Alzheimer’s Disease Neuroimaging Initiative 2 PET Core: 2015. Alzheimers Dement 11, 757–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M, 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kálmán J, Kitajka K, Pákáski M, Zvara A, Juhász A, Vincze G, Janka Z, Puskás LG, 2005. Gene expression profile analysis of lymphocytes from Alzheimer’s patients. Psychiatr Genet 15, 1–6. [DOI] [PubMed] [Google Scholar]
- Kang JH, Korecka M, Figurski MJ, Toledo JB, Blennow K, Zetterberg H, Waligorska T, Brylska M, Fields L, Shah N, Soares H, Dean RA, Vanderstichele H, Petersen RC, Aisen PS, Saykin AJ, Weiner MW, Trojanowski JQ, Shaw LM, 2015. The Alzheimer’s Disease Neuroimaging Initiative 2 Biomarker Core: A review of progress and plans. Alzheimers Dement 11, 772–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EK, Choi EJ, 2010. Pathological roles of MAPK signaling pathways in human diseases. Biochim Biophys Acta 1802, 396–405. [DOI] [PubMed] [Google Scholar]
- Lai KSP, Liu CS, Rau A, Lanctôt KL, Köhler CA, Pakosh M, Carvalho AF, Herrmann N, 2017. Peripheral inflammatory markers in Alzheimer’s disease: a systematic review and meta-analysis of 175 studies. J Neurol Neurosurg Psychiatry 88, 876–882. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S, 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Page A, Dupuis G, Frost EH, Larbi A, Pawelec G, Witkowski JM, Fulop T, 2018. Role of the peripheral innate immune system in the development of Alzheimer’s disease. Exp Gerontol 107, 59–66. [DOI] [PubMed] [Google Scholar]
- Lee Y, Han S, Kim D, Kim D, Horgousluoglu E, Risacher SL, Saykin AJ, Nho K, 2018. Genetic variation affecting exon skipping contributes to brain structural atrophy in Alzheimer’s disease. AMIA Jt Summits Transl Sci Proc 2017, 124–131. [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu B, Zhang L, Rong L, 2014. Amyloid beta peptide is elevated in osteoporotic bone tissues and enhances osteoclast function. Bone 61, 164–175. [DOI] [PubMed] [Google Scholar]
- Li S, Todor A, Luo R, 2016. Blood transcriptomics and metabolomics for personalized medicine. Comput Struct Biotechnol J 14, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR, 2010. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 34, 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovestone S, Francis P, Kloszewska I, Mecocci P, Simmons A, Soininen H, Spenger C, Tsolaki M, Vellas B, Wahlund LO, Ward M, 2009. AddNeuroMed--the European collaboration for the discovery of novel biomarkers for Alzheimer’s disease. Ann N Y Acad Sci 1180, 36–46. [DOI] [PubMed] [Google Scholar]
- Lunnon K, Ibrahim Z, Proitsi P, Lourdusamy A, Newhouse S, Sattlecker M, Furney S, Saleem M, Soininen H, Kłoszewska I, Mecocci P, Tsolaki M, Vellas B, Coppola G, Geschwind D, Simmons A, Lovestone S, Dobson R, Hodges A, 2012. Mitochondrial dysfunction and immune activation are detectable in early Alzheimer’s disease blood. J Alzheimers Dis 30, 685–710. [DOI] [PubMed] [Google Scholar]
- Lunnon K, Sattlecker M, Furney SJ, Coppola G, Simmons A, Proitsi P, Lupton MK, Lourdusamy A, Johnston C, Soininen H, Kłoszewska I, Mecocci P, Tsolaki M, Vellas B, Geschwind D, Lovestone S, Dobson R, Hodges A, 2013. A blood gene expression marker of early Alzheimer’s disease. J Alzheimers Dis 33, 737–753. [DOI] [PubMed] [Google Scholar]
- Ma J, Jiang T, Tan L, Yu JT, 2015. TYROBP in Alzheimer’s disease. Mol Neurobiol 51, 820–826. [DOI] [PubMed] [Google Scholar]
- Maes OC, Xu S, Yu B, Chertkow HM, Wang E, Schipper HM, 2007. Transcriptional profiling of Alzheimer blood mononuclear cells by microarray. Neurobiol Aging 28, 1795–1809. [DOI] [PubMed] [Google Scholar]
- McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, Luo Y, Sidore C, Kwong A, Timpson N, Koskinen S, Vrieze S, Scott LJ, Zhang H, Mahajan A, Veldink J, Peters U, Pato C, van Duijn CM, Gillies CE, Gandin I, Mezzavilla M, Gilly A, Cocca M, Traglia M, Angius A, Barrett JC, Boomsma D, Branham K, Breen G, Brummett CM, Busonero F, Campbell H, Chan A, Chen S, Chew E, Collins FS, Corbin LJ, Smith GD, Dedoussis G, Dorr M, Farmaki A-E, Ferrucci L, Forer L, Fraser RM, Gabriel S, Levy S, Groop L, Harrison T, Hattersley A, Holmen OL, Hveem K, Kretzler M, Lee JC, McGue M, Meitinger T, Melzer D, Min JL, Mohlke KL, Vincent JB, Nauck M, Nickerson D, Palotie A, Pato M, Pirastu N, McInnis M, Richards JB, Sala C, Salomaa V, Schlessinger D, Schoenherr S, Slagboom PE, Small K, Spector T, Stambolian D, Tuke M, Tuomilehto J, van den Berg LH, Van Rheenen W, Volker U, Wijmenga C, Toniolo D, Zeggini E, Gasparini P, Sampson MG, Wilson JF, Frayling T, de Bakker PIW, Swertz MA, McCarroll S, Kooperberg C, Dekker A, Altshuler D, Willer C, Iacono W, Ripatti S, Soranzo N, Walter K, Swaroop A, Cucca F, Anderson CA, Myers RM, Boehnke M, McCarthy MI, Durbin R, Haplotype Reference Consortium, 2016. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 48, 1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM, 1984. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 34, 939–944. [DOI] [PubMed] [Google Scholar]
- Mohr S, Liew CC, 2007. The peripheral-blood transcriptome: new insights into disease and risk assessment. Trends Mol Med 13, 422–432. [DOI] [PubMed] [Google Scholar]
- Mokry LE, Ross S, Morris JA, Manousaki D, Forgetta V, Richards JB, 2016. Genetically decreased vitamin D and risk of Alzheimer disease. Neurology 87, 2567–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JK, Honea RA, Vidoni ED, Swerdlow RH, Burns JM, 2014. Is Alzheimer’s disease a systemic disease? Biochim Biophys Acta 1842, 1340–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullane K, Williams M, 2018. Alzheimer’s disease (AD) therapeutics - 1: Repeated clinical failures continue to question the amyloid hypothesis of AD and the current understanding of AD causality. Biochem Pharmacol 158, 359–375. [DOI] [PubMed] [Google Scholar]
- Nakai M, Tanimukai S, Yagi K, Saito N, Taniguchi T, Terashima A, Kawamata T, Yamamoto H, Fukunaga K, Miyamoto E, Tanaka C, 2001. Amyloid beta protein activates PKC-delta and induces translocation of myristoylated alanine-rich C kinase substrate (MARCKS) in microglia. Neurochem Int 38, 593–600. [DOI] [PubMed] [Google Scholar]
- Naughton BJ, Duncan FJ, Murrey DA, Meadows AS, Newsom DE, Stoicea N, White P, Scharre DW, Mccarty DM, Fu H, 2015. Blood genome-wide transcriptional profiles reflect broad molecular impairments and strong blood-brain links in Alzheimer’s disease. J Alzheimers Dis 43, 93–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn F, Ravetch JV, 2006. Fcgamma receptors: old friends and new family members. Immunity 24, 19–28. [DOI] [PubMed] [Google Scholar]
- Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, Saykin AJ, Orro A, Lupoli S, Salvi E, Weiner M, Macciardi F, 2009. Hippocampal Atrophy as a Quantitative Trait in a Genome-Wide Association Study Identifying Novel Susceptibility Genes for Alzheimer’s Disease. PLoS ONE 4, e6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D, 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38, 904–909. [DOI] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ, 2010. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC, 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan VK, Risacher SL, Nho K, Kim S, Swaminathan S, Shen L, Foroud TM, Hakonarson H, Huentelman MJ, Aisen PS, Petersen RC, Green RC, Jack CR, Koeppe RA, Jagust WJ, Weiner MW, Saykin AJ, 2014. APOE and BCHE as modulators of cerebral amyloid deposition: a florbetapir PET genome-wide association study. Mol Psychiatry 19, 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risacher SL, Kim S, Nho K, Foroud T, Shen L, Petersen RC, Jack CR, Beckett LA, Aisen PS, Koeppe RA, Jagust WJ, Shaw LM, Trojanowski JQ, Weiner MW, Saykin AJ, Alzheimer’s Disease Neuroimaging Initiative (ADNI), 2015. APOE effect on Alzheimer’s disease biomarkers in older adults with significant memory concern. Alzheimers Dement 11, 1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L, 2006. Genetics of global gene expression. Nat Rev Genet 7, 862–872. [DOI] [PubMed] [Google Scholar]
- Roed L, Grave G, Lindahl T, Rian E, Horndalsveen PO, Lannfelt L, Nilsson C, Swenson F, Lönneborg A, Sharma P, Sjögren M, 2013. Prediction of mild cognitive impairment that evolves into Alzheimer’s disease dementia within two years using a gene expression signature in blood: a pilot study. J Alzheimers Dis 35, 611–621. [DOI] [PubMed] [Google Scholar]
- Rye PD, Booij BB, Grave G, Lindahl T, Kristiansen L, Andersen HM, Horndalsveen PO, Nygaard HA, Naik M, Hoprekstad D, Wetterberg P, Nilsson C, Aarsland D, Sharma P, Lönneborg A, 2011. A novel blood test for the early detection of Alzheimer’s disease. J Alzheimers Dis 23, 121–129. [DOI] [PubMed] [Google Scholar]
- Saykin AJ, Shen L, Yao X, Kim S, Nho K, Risacher SL, Ramanan VK, Foroud TM, Faber KM, Sarwar N, Munsie LM, Hu X, Soares HD, Potkin SG, Thompson PM, Kauwe JSK, Kaddurah-Daouk R, Green RC, Toga AW, Weiner MW, 2015. Genetic studies of quantitative MCI and AD phenotypes in ADNI: Progress, opportunities, and plans. Alzheimers Dement 11, 792–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ, Hardy J, 2016. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med 8, 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillén A, Brohede J, Forsell C, Lilius L, Andrade J, Odeberg J, Kimura T, Winblad B, Graff C, 2011. Linkage analysis of autopsy-confirmed familial Alzheimer disease supports an Alzheimer disease locus in 8q24. Dement Geriatr Cogn Disord 31, 109–118. [DOI] [PubMed] [Google Scholar]
- Sweeney MD, Sagare AP, Zlokovic BV, 2018. Blood-brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat Rev Neruol 14, 133–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Nakamura A, 2017. Regulation of immune and neural function via leukocyte Ig-like receptors. J Biochem 162, 73–80. [DOI] [PubMed] [Google Scholar]
- Tan ZS, Seshadri S, Beiser A, Zhang Y, Felson D, Hannan MT, Au R, Wolf PA, Kiel DP, 2005. Bone mineral density and the risk of Alzheimer disease. Arch Neurol 62, 107–111. [DOI] [PubMed] [Google Scholar]
- Teeling JL, Carare RO, Glennie MJ, Perry VH, 2012. Intracerebral immune complex formation induces inflammation in the brain that depends on Fc receptor interaction. Acta Neuropathol 124, 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorisson GA, Smith AV, Krishnan L, Stein LD, 2005. The International HapMap Project Web site. Genome Res 15, 1592–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tylee DS, Kawaguchi DM, Glatt SJ, 2013. On the outside, looking in: a review and evaluation of the comparability of blood and brain “-omes”. Am J Med Genet B Neuropsychiatr Genet 162B, 595–603. [DOI] [PubMed] [Google Scholar]
- Vidoni ED, Townley RA, Honea RA, Burns JM, 2011. Alzheimer disease biomarkers are associated with body mass index. Neurology 77, 1913–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR, 2010. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss-Coray T, 2006. Inflammation in Alzheimer disease: driving force, bystander or beneficial response? Nat Med 12, 1005–1015. [DOI] [PubMed] [Google Scholar]
- Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, Zhang C, Xie T, Tran L, Dobrin R, Fluder E, Clurman B, Melquist S, Narayanan M, Suver C, Shah H, Mahajan M, Gillis T, Mysore J, MacDonald ME, Lamb JR, Bennett DA, Molony C, Stone DJ, Gudnason V, Myers AJ, Schadt EE, Neumann H, Zhu J, Emilsson V, 2013. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153, 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.