Abstract
Estrogen receptor (ER) positive breast cancers often contain subpopulations of cells that express the intermediate filament protein cytokeratin 5 (CK5). CK5+ cells are enriched in cancer stem cell (CSC) properties, can be induced by progestins, and predict poor prognosis in ER+ breast cancer. We established through CK5 knockout and overexpression in ER+ breast cancer cell lines that CK5 is important for tumorsphere formation, prompting us to speculate that CK5 has regulatory activity in CSCs. To interrogate CK5 interacting proteins that may be functionally cooperative, we performed immunoprecipitation-mass spectrometry for CK5 in ER+ breast cancer cells. Focusing on proteins with signaling activity, we identified β-catenin, a key transcription factor of the Wnt signaling pathway and cell adhesion molecule, as a CK5 interactor, which we confirmed by co-immunoprecipitation in several breast cancer models. We interrogated the dual functions of β-catenin in relation to CK5. Knockout or knockdown of CK5 ablated β-catenin transcriptional activity in response to progestins and Wnt stimuli. Conversely, CK5 induced by progestins or overexpression was sufficient to promote loss of β-catenin at the cell membrane and total E-cadherin loss. A breast cancer patient-derived xenograft showed similar loss of membrane β-catenin and E-cadherin in CK5+ but not intratumoral CK5− cells and single cell RNA sequencing found the top enriched pathways in the CK5+ cell cluster were cell junction remodeling and signaling. This report highlights that CK5 actively remodels cell morphology and that blockade of CK5-β-catenin interaction may reverse the detrimental properties of CK5+ breast cancer cells.
INTRODUCTION
Over three quarters of newly diagnosed breast cancers are estrogen receptor ⍺ (ER) positive based on immuno-detection in 1–99% cells (1, 2). Such heterogeneity in ER expression is poorly understood and may be a contributing factor in the õne third of patients that acquire resistance to standard endocrine therapies (3). In fact, intratumoral heterogeneity in ER expression was recently linked to worse prognosis (4), and little is known about the co-existent ER− cell populations. Roughly half of ER+ tumors contain a predominantly ER− subpopulation that expresses intermediate filament protein cytokeratin 5 (CK5) (5). Our group and others have shown that CK5+ cells exhibit all the hallmarks of breast cancer stem cells (CSCs) including enhanced tumor initiation, tumorsphere formation, and drug resistance compared to intratumoral CK5− cells (6–10). CK5 expression can be preexisting or acquired in breast cancer through hormone regulation. Either long-term estrogen withdrawal or progestins increase the CK5+ population in ER+ breast cancer cell lines (6, 9, 10). In clinical samples treated with neoadjuvant endocrine therapy, the number of CK5+ cells increased in post- compared to pre-treatment samples (9). Progestin-activated progesterone receptors (PR) bind to the proximal promoter of the CK5 gene (KRT5) and upregulate CK5 transcripts and protein (6, 7). Progestins and PR promote tumorsphere formation in ER+ breast cancer cell lines (7, 11) and shRNA knockdown of CK5 ablates this effect (7). This suggests that CK5 may have a functional role in promoting CSC properties.
Cytokeratins (CK) are often used as lineage and differentiation markers in the normal breast and in histological characterization of breast cancer. In the normal breast, CKs 8, 18, and 19 are typically found in the luminal epithelial layer while CKs 5, 14, and 17 are found in the basal epithelial layer (12, 13). However, sporadic CK5+ cells are present in luminal cells in the terminal ductal lobular unit (TDLU) and have been described as bi-potent stem/progenitor cells (14–16). CK5+ cells are also implicated as the potential origin of BRCA1 mutant basal-like breast cancer (BLBC) (17, 18), and CK5 is a defining marker of this aggressive subset of triple negative breast cancer (19). However, CK5 is not restricted to BLBC and its expression is associated with poor prognosis across all breast cancer subtypes (19–23). The defining features that make CK5+ cells aggressive are unknown.
Cytokeratins (CKs) are the major structural proteins of epithelial cells that protect the cell from mechanical and non-mechanical stress (24, 25). They have a conserved structure consisting of a central rod domain that facilitates hetero-dimerization between Type I (9–23) and Type II (1–8) CKs and a head and tail domain that controls CK dynamics, post-translational modifications, and protein-protein interactions (26, 27). Several reports implicate that CK5 and its common dimeric partner CK17 regulate cell signaling. In keratinocytes, CK17 interacts with the scaffolding protein 14–3-3σ and is necessary for mTOR activation and cell proliferation (28, 29). Likewise, CK17 regulates cell size in oral squamous carcinoma cells through a 14–3-3σ complex (30). In breast, cervix, and pancreatic cancer cells CK17 binds to p27KIP1 to induce its nuclear export and subsequent G1-S-phase transition, implicating CK17 as an oncoprotein (31). A CK5/CK17/14–3-3σ complex interacts with the actin cytoskeleton to increase invasiveness of BLBC cells (32). These studies highlight the emerging dual structural/signaling activities of CKs in cancer cells through key protein-protein interactions.
In this study we investigated the contribution of CK5 to CSC properties in breast cancer cells. We found that manipulation of CK5 through knockout or overexpression in ER+ breast cancer cell lines was sufficient reduce or enhance tumorsphere formation, respectively. We therefore interrogated potential CK5 interacting proteins by immunoprecipitation-mass spectrometry (IP-MS) and identified β-catenin, a Wnt signaling pathway downstream protein and adhesion molecule, as an interacting protein. Herein we demonstrate that CK5 alters both the transcription factor and adhesion functions of β-catenin. Strikingly, CK5 expression is sufficient to translocate β-catenin to the cytosol, where it is primed for transcriptional activity, and reduce E-cadherin localization to the cell membrane in cell line and tumor models. Disruption of CK5-mediated β-catenin/E-cadherin remodeling may be an effective way to therapeutically target this population of poor prognostic cells.
RESULTS
CK5 is required for progestin-induced and sufficient for de novo tumorsphere formation
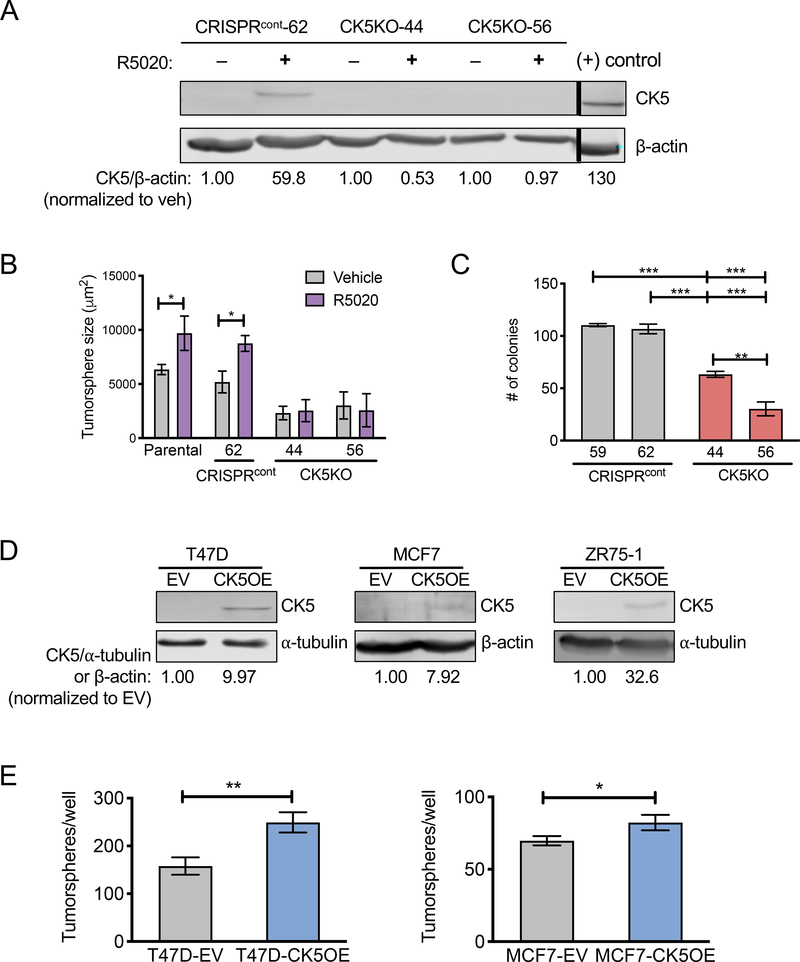
We previously demonstrated that shRNA knockdown of CK5 blocked progesterone- induced tumorsphere formation in ER+ breast cancer cells (7), and therefore speculated that CK5 may have a functional role in regulating the CSC property of self-renewal. To further test this hypothesis, we generated CRISPR-Cas9-mediated CK5 knockout (CK5KO) T47D breast cancer cells. CK5 expression was induced by the synthetic progestin R5020 in control cell lines (that underwent unsuccessful gene editing of CK5, CRISPRcont), but not in CK5KO cell lines (Figure 1A). Progestin treatment increased tumorsphere size in CRISPRcont but not CK5KO cell lines (Figure 1B, Supplemental Figure 1A), confirming our previous studies with CK5 shRNA (7). Proliferation in 2D under standard media conditions found slowed growth of CK5KO-56 but not CK5KO-44 compared to CRISPRcont cells (Supplemental Figure 1B). Both CK5KO cell lines had significantly impaired colony formation compared to CRISPRcont cell lines (Figure 1C, Supplemental Figure 1C).
Figure 1. CK5 is required for progestin induced and sufficient for de novo tumorsphere formation in ER+ breast cancer cells.
A. T47D CRISPRcont (#62) and CK5KO (#44, 56) cells were treated with vehicle (ethanol) or 10 nM R5020 for 48 h. Cell lysates were collected and analyzed by immunoblot for CK5 expression with β-actin used as a loading control. Normalized CK5/β-actin levels are indicated as fold change of R5020 over vehicle within each cell line. Positive (+) control is lysate from the cell line MDA-MB-468. Positive control is normalized to vehicle treated CRISPRcont-62. Black bar indicates omitted lanes on the same gel. B. T47D CRISPRcont and CK5KO cells stably expressing ZsGreen were seeded at 100 cells/well in Mammocult media with 1% methylcellulose, treated with vehicle or 10 nM R5020, and cultured for two weeks. Data were quantified using IncuCyte Zoom. Average tumorsphere size (um2) is indicated ± s.e.m. Vehicle vs. R5020 groups were compared by Student’s T-test. *P<0.05. C. T47D CRISPRcont or CK5KO cells were seeded at 400 cells per well into 6 well plates in complete media. Colonies were grown for two weeks, stained with Crystal violet, and counted. Average colonies/well ± s.e.m are shown. All groups were compared via ANOVA/Tukey. **P<0.01, ***P<0.001. D. T47D, MCF7, and ZR75–1 cells were transduced with lentivirus containing CK5 cDNA (CK5OE) or empty vector (EV). Cell lysates were collected and analyzed by immunoblot for CK5 expression using ⍺-tubulin as a loading control. Normalized CK5/β-actin levels are indicated as fold change over EV control within each cell line. E. T47D (left) and MCF7 (right) EV and CK5OE cells were analyzed for tumorsphere formation as described in 1B. Number of tumorspheres/well is indicated ± s.e.m. EV vs. CK5OE groups were compared by Student’s T-test. *P<0.05, **P<0.01. All experiments were repeated 3 times.
To study if exogenous CK5 expression would impact ER+ breast cancer cells, T47D, MCF7, and ZR75–1 cells were transduced with a viral construct overexpressing CK5 (CK5OE) or empty vector (EV) control. CK5 expression was confirmed by immunoblot (Figure 1D) and immunocytochemistry (ICC) (Supplemental Figure 1D). CK5OE T47D and MCF7 cells formed significantly more tumorspheres than EV cells (Figure 1E, Supplemental Figure 1E). ZR75–1 cells failed to form tumorspheres under any conditions. Taken together, these data support that CK5 is necessary for progestin-induced large tumorspheres and sufficient to increase tumorsphere formation in the absence of hormones in breast cancer cell lines. Thus, self-renewal dynamics of breast cancer cells may be regulated by CK5.
CK5 interacts with β-catenin
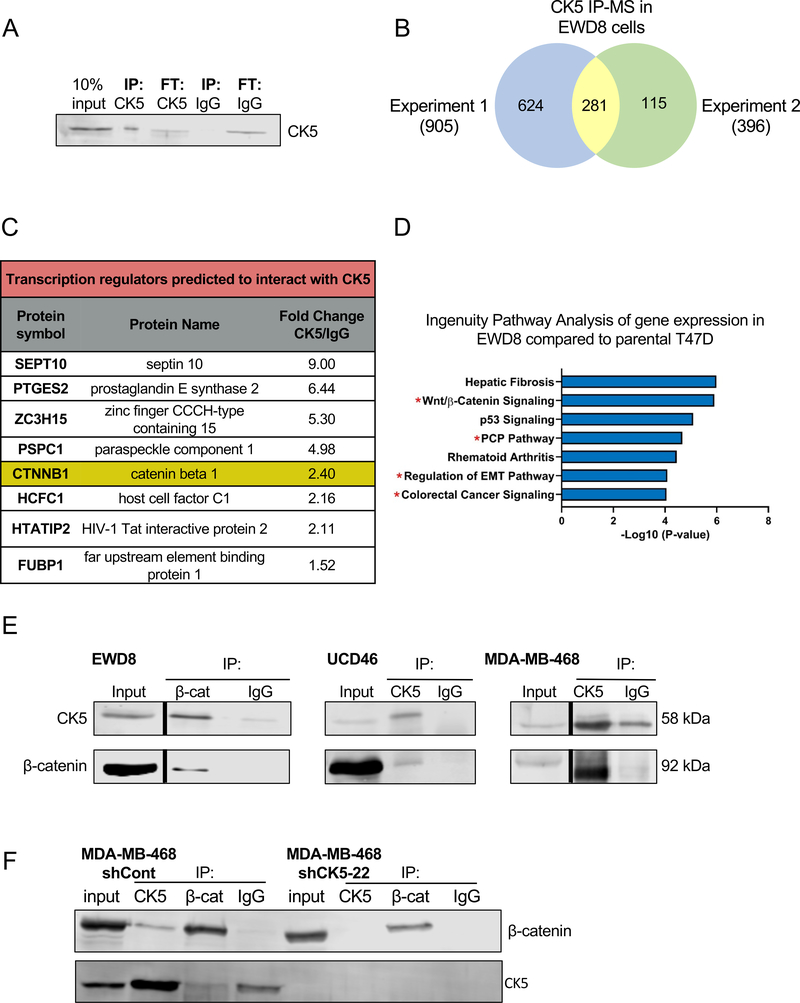
To interrogate mechanisms by which CK5 alters cell behavior and, based on published reports of CK interactions impacting cell signaling, we performed IP-MS to identify CK5 interacting proteins. For this experiment we utilized a syngeneic T47D cell line, EWD8, that has high basal levels of CK5 (5) (Supplemental Figure 2A). CK5 is efficiently immunoprecipitated with high sequence coverage in EWD8 cells (Figure 2A, Supplemental Figure 2B). Two independent IP-MS experiments were performed in EWD8 cells with 281 common proteins identified (Figure 2B). Several proteins with established CK5 interaction were identified including CK17 and desmosomal components such as plectin (Supplemental Figure 2C). We validated CK5 interaction with CK17, a heterodimeric binding partner of CK5, by co-IP (Supplemental Figure 2D).
Figure 2. CK5 interacts with β-catenin in luminal and basal breast cancer cells.
A. Immunoprecipitation for CK5 in the constitutive CK5+ T47D subline EWD8. 10% input, IP and flow-through (FT) fractions for CK5 vs IgG control antibodies were analyzed by immunoblot. B. IP-MS for CK5 was performed in EWD8 cells to identify prospective CK5 interacting proteins. The experiment was performed independently twice. Venn diagram depicts the number of CK5 interacting proteins identified in each experiment with 281 common proteins. C. Ingenuity Pathway Analysis was used to identify proteins classified as transcriptional regulators among the putative CK5 interacting proteins. Table shows protein symbol, protein name, and fold change of CK5 to IgG spectral counts. D. Ingenuity Pathway Analysis was used to identify the top enriched signaling pathways in EWD8 compared to parental T47D cells (5). Red asterisks indicate Wnt/β-catenin signaling and related pathways. E. Co-IP was performed with β-catenin and IgG antibodies in EWD8 cells, or CK5 and IgG antibodies in UCD46 PDX lysates and MDA-MB-468 cells. Immunoblots of input (10%) and indicated IP fractions were probed with CK5 and β-catenin antibodies. Black bars indicate omitted lanes on the same gel. Full blots are available in Supplemental Figure 3. Co-IPs were repeated for each model 2–3 times. F. Co-IP was performed with CK5, β-catenin, and IgG antibodies in control (shCont) or MDA-MB-468 cells containing a CK5 shRNA construct (shCK5–22). Immunoblots of input (10%) and indicated IP fractions were probed with CK5 and β-catenin antibodies. Co-IP was repeated twice.
To identify candidate interactors most likely to influence a CSC phenotype in conjunction with CK5, we performed several stratification steps. First, we filtered the list of 281 proteins identified by IP-MS through the CRAPome to remove proteins commonly found in >20% of affinity purification experiments (33). This reduced the number of proteins to 149. Second, we ran the 149 proteins through Ingenuity Pathway Analysis (IPA) and identified 8 transcription regulators (Figure 2C) including β-catenin, a key transcription co-factor of the Wnt signaling pathway and essential component of adherens junctions (34), and a well-known regulator of both normal and CSCs (35, 36). Third, IPA analysis on gene expression data comparing EWD8 to wild-type T47D cells (5), identified the Wnt signaling pathway among the top enriched pathways in EWD8 cells (Figure 2D, Supplemental Figure 2E). We confirmed that CK5 and β-catenin interact by co-IP in EWD8 cells and two additional CK5+ breast cancer models: patient-derived xenograft (PDX) UCD46 and BLBC cell line MDA-MB-468 (Figure 2E, Supplemental Figure 3). As an additional control, we confirmed the CK5/β-catenin interaction is lost in stable CK5 knockdown MDA-MB-468 cells (Figure 2F). Finally, knockdown of β-catenin via siRNA in both T47D-EV and CK5OE cells decreased tumorsphere formation (Supplemental Figure 4A&B). Thus, we pursued β-catenin as a CK5 associated protein that could mediate CSC properties.
CK5 enhances β-catenin transcriptional activity
Since canonical Wnt signaling is important for CSC maintenance, we first investigated how CK5 affects β-catenin activity. To test this, we employed the TOPFlash reporter assay which contains seven TCF/LEF binding sites upstream of the luciferase cDNA sequence to measure β-catenin transcriptional activity. We used lithium chloride (LiCl) as a positive control for all TOPFlash experiments, which inhibits GSK3-β, the key kinase in the β-catenin destruction complex (37). Since progestins increase several Wnt proteins, we first tested their effect on β-catenin transcriptional activity. In T47D and ZR75–1 cells transfected with TOPFlash, progestins stimulated β-catenin activity (Figure 3A&B), an effect eliminated in T47D CK5KO vs. control cells (Figure 3C). To test if Wnt expression is affected by CK5 status, we measured mRNA expression of Wnt1, a reported PR target gene (38), and a non-Wnt related PR target gene, MAFB. We found that Wnt1 expression was induced by R5020 in CRISPRcont cells but not CK5KO cells (Figure 3D, top). By contrast, MAFB expression was induced in both CRISPRcont and CK5KO cells (Figure 3D, bottom). These data support that CK5 is important for progestin-induced β-catenin transcriptional activity in ER+ luminal breast cancer cells, and imply that CK5+ cells may be the targets of P-induced Wnt expression.
Figure 3. CK5 enhances β-catenin transcriptional activity.
A. T47D cells were transfected with TOPFlash plasmid and treated with vehicle (EtOH), 100 nM progesterone (P4), 10 nM R5020, or 50 mM LiCl (positive control for TOPFlash reporter activity) for 24 h. Relative luciferase units were measured and are indicated as fold change over vehicle ± s.e.m. Treatment groups were compared by ANOVA/Tukey. *P<0.05. B. ZR75–1 cells were transfected with TOPFlash plasmid and treated with vehicle (EtOH), 10 nM 17β-estradiol (E2; to induce PR expression), E2 + 10 nM R5020, or 50 mM LiCl for 24 h. TOPFlash activity was measured as described in 3A.**P<0.01, ***P<0.001. C. T47D parental, CRISPRcont and CK5KO cell lines were transfected with TOPFlash plasmid and treated with vehicle, 10 nM R5020, or 50 mM LiCl for 24 h. TOPFlash activity was measured as described in 1A. Vehicle and R5020 groups within each cell line were compared using Student’s T-test. **P<0.01. D. T47D CRISPRcont-62 and CK5KO-44 cells were treated with vehicle or 10nM R5020 for 4h. mRNA expression of Wnt1 (top) and MAFB (bottom) were measured by qPCR. Experiment was repeated 5 times. Relative mRNA level is indicated as fold change normalized to vehicle treated CRISPRcont-62 ± s.e.m. Treatment groups were compared by ANOVA/Tukey *P<0.05. E. MDA-MB-468 cells with stable shRNA knockdown of CK5 (shCK5) were created using two different shRNAs. Knockdown efficiency was assessed by immunoblot compared to scrambled control (shCont). Relative change in CK5 levels was normalized to α-tubulin and compared to shCont. F. TOPFlash reporter assay was performed in shCont, shCK5–22 (left), and shCK5–88 (right) MDA-MB-468 cells treated with 200 ng/mL Wnt3a or 50 mM LiCl or for 24 h. Fold change is normalized to vehicle in each group ± s.e.m. Within each shCK5 cell line all groups were compared by ANOVA/Tukey. *P<0.05, **P<0.01,***P<0.001. All TOPFlash experiments were repeated 3 times.
Since we noted a CK5/β-catenin interaction in BLBC in addition to ER+PR+ luminal breast cancer cells, we next determined whether CK5 was necessary for β-catenin activity in BLBC. To test this, we created MDA-MB-468 BLBC cells with stable shRNA knockdown of CK5 using two constructs. One construct produced a near complete loss of CK5 (shCK5–22, >90%) while the other produced a partial knockdown (shCK5–88, 60%) (Figure 3E). Since MDA-MB-468 cells are PR negative, we used the canonical ligand Wnt3a as a stimulus. β-catenin activity was induced by Wnt3 in control (shCont) cells which was attenuated in the complete, but not the partial, CK5 knockdown cells (Figure 3E&F). Interestingly, β-catenin activity in response to LiCl positive control was highly and moderately reduced in the complete and partial CK5 knockdown cells, respectively (Figure 3E&F), an effect not observed with CK5 loss in luminal breast cancer cells. This implies β-catenin stability is highly dependent on CK5 in BLBC, even with inhibition of the destruction complex. In fact, LiCl treatment of EWD8 cells enhanced the β-catenin/CK5 interaction (Supplemental Figure 4C), suggesting luminal and BLBC differentially regulate β-catenin pools, as previously reported (39, 40).
CK5 promotes loss of β-catenin at the cell membrane
β-catenin is an essential component of cell adhesions through its interaction with E-cadherin on the cytoplasmic side of the cell membrane (34). Loss of β-catenin at the membrane leads to cytoplasmic accumulation where it is poised for nuclear translocation and activity upon Wnt stimulus, or otherwise degraded (41). To determine whether CK5 affects β-catenin localization, we performed dual ICC and confocal microscopy in our CK5 overexpressing cell lines. In CK5OE T47D and ZR75–1 cells, CK5+ cells show notable loss of membrane β-catenin compared to EV cells (Figure 4A&B). Quantitation of low (0–25%), medium (25–75%) and high (75–100%) membrane β-catenin found a significant shift in the proportion of CK5+ cells with low membrane coverage (16% to 68% in T47D and 27% to 52% in ZR75–1 EV and CK5OE cells, respectively) (Figure 4A&B). Note only a subpopulation of CK5OE T47D cells express CK5, similar to our published observations with progestin treatment (6–8), and the intra-culture CK5− cells retain membrane β-catenin (Figure 4A). We also assessed β-catenin localization by subcellular fractionation followed by immunoblot and found that membrane β-catenin decreased in T47D-CK5OE vs. control EV cells (Supplemental Figure 5). These data suggest that CK5 expression is sufficient to reduce membrane β-catenin.
Figure 4. CK5 promotes loss of β-catenin at the cell membrane.
A-E. Immunocytochemistry and confocal microscopy was performed for β-catenin (green), CK5 (red), and DAPI (blue). Representative fields are shown for T47D-EV and T47D-CK5OE cells (A), ZR75–1-EV and ZR75–1-CK5OE cells (B), T47D and EWD8 cells (C), T47D cells treated with vehicle or 10 nM R5020 24 h (D), and MDA-MB-468 shCont and shCK5–22 cells (E). Scale bars, 20 μm. Accompanying graphs in A-E depict quantification of membrane β-catenin coverage for each cell line or treatment group. 42–212 cells from each group were analyzed for membrane β-catenin in a blinded manner. Cells were quantified as low (0–25%), medium (26–50%) or high (76–100%) membrane β-catenin. In A and D, CK5+ cells (red arrows) and CK5− cells (green arrows) were quantified separately. A chi-square test was used to determine statistical significance.
We next assessed β-catenin localization in luminal breast cancer cells with CK5 induced by estrogen depletion (EWD8 cells) or progestin treatment. EWD8 cells had a fewer cells with high membrane β-catenin compared to parental T47D cells (Figure 4C). Loss of membranous β-catenin also occurred in CK5+ cells induced by progestin treatment (Figure 4E). However, a similar reduction in membrane β-catenin was also found in CK5− cells (Figure 4E). This suggests progestins may promote shuttling of β-catenin in breast cancer cells, regardless of CK5 status, although the ability to induce CK5 is necessary for β-catenin activity (Figure 3C).
In the CK5+ BLBC cell line MDA-MB-468, baseline membranous β-catenin is low and is instead concentrated around the nucleus (Figure 4D). Furthermore, CK5 knockdown (shCK5 #22) compared to control cells showed a complete loss of membrane β-catenin, marked lower β-catenin staining overall, and a transition to a more elongated mesenchymal-like morphology. This implies a strong dependency of β-catenin stability on CK5 in BLBC cells and explains the drastic loss of β-catenin transcriptional activity with CK5 loss (Figure 3F).
CK5 reduces membrane and total E-cadherin
Trafficking of membrane β-catenin is associated with loss of membrane E-cadherin (34). Given the profound effect of CK5 on β-catenin localization, we investigated its impact on E-cadherin localization. T47D and ZR75–1 CK5OE cells displayed loss of E-cadherin at the cell membrane, with the proportion of cells with low (0–25%) membrane E-cadherin staining shifting from 28 to 66% and 2 to 42% in T47D and ZR75–1 CK5OE vs. EV cells, respectively (Figure 5A&B). Immunoblot analysis showed that total E-cadherin, but not β-catenin, was decreased in T47D, ZR75–1, and MCF7 CK5OE compared to EV cells and EWD8 compared to parental T47D cells (Figure 5C). These results suggest that CK5 stabilizes cytosolic β-catenin and in the process destabilizes E-cadherin. Interestingly, MDA-MB-468 shCK5–22 compared to shCont cells had near total loss of both β-catenin and E-cadherin protein levels (80% and >95% decreases, respectively, Figure 4D). β-catenin, but not E-cadherin, could be partially rescued upon treatment with proteasome inhibitor MG-132 in shCK5–22 cells (Figure 5D). These data highlight differences in β-catenin dynamics between CK5+ luminal and BLBC cells, and that the CK5+ luminal cells should be considered a distinct cell type.
Figure 5. CK5+ cells lose E-cadherin.
A-B. Top: Immunocytochemistry and confocal microscopy was performed for E-cadherin (green), CK5 (red), and DAPI (blue) in T47D-EV and T47D-CK5OE cells (A), ZR75–1-EV and ZR75–1-CK5OE cells (B). Bottom: Membrane E-cadherin coverage was quantified for each comparison in a blinded manner as low (0–25%), medium (26–75%), or high (76–100%). 59–170 cells from each group were analyzed and a chi-square test was used to determine statistical significance. C. Cell lysates were harvested from EV and CK5OE T47D, ZR75–1, and MCF7 cells and T47D parental (non-genetically modified) and EWD8 cells. Lysates were analyzed by immunoblot for CK5, E-cadherin, and β-catenin expression using β-actin as a loading control and quantified as fold change of CK5OE vs. EV or EWD8 vs T47D. D. MDA-MB-468 shCont and shCK5–22 cells were treated with vehicle (DMSO) or 10 uM of the proteasome inhibitor MG132 for 4 h. Cell lysates were collected and analyzed by immunoblot for CK5, β-catenin, and E-cadherin expression using ⍺-tubulin as a loading control. Normalized protein levels are shown as fold change over vehicle. All immunoblots repeated 3 times.
CK5+ cells in ER+ patient-derived tumor models have altered adherens junctions
To assess whether the observed alterations in β-catenin and E-cadherin adherens junctions are present in solid tumor models, we analyzed PDX UCD15, which contains a mosaic of intratumoral CK5+ and ER+ cells (Figure 6A). Dual fluorescent IHC for CK5 and either β-catenin or E-cadherin found CK5+ UCD15 cells have reduced membrane β-catenin and E-cadherin compared to intratumoral CK5− cells (Figure 6B). To further interrogate this relationship, we analyzed single cell RNA sequencing data from PDX UCD15 and performed unbiased clustering analysis. UCD15 partitioned into 7 transcriptomic clusters, with CK5 (KRT5) being a defining gene for cluster #5. IPA analysis of all cluster #5 genes found the top functions are remodeling of adherens junctions and associated endocytosis pathways. These results confirm our cell line observations that CK5 expression is associated with loss of adherens junctions, a cell morphology associated with poor clinical outcome (39, 42, 43). Figure 7 depicts our proposed model of transition from a luminal (CK5−) to luminobasal (CK5+) state in ER+ breast cancer with respect to β-catenin and E-cadherin adherens junctions.
Figure 6. CK5+ cells in an ER+ patient-derived tumor model have altered adherens junctions.
A. Dual immunohistochemistry for CK5 (red, cytoplasmic) and ER (brown, nuclear) in PDX UCD15 was performed. Scale bar, 200 μM. B. Left: Dual fluorescent immunohistochemistry and confocal microscopy for CK5 (red) and either β-catenin (green, top) or E-cadherin (green, bottom) in UCD15. Individual and merged images are sown. Red arrows indicate CK5+ cells. 2–3 fields were acquired per tumor; representative images are shown. Right: Membrane β-catenin (59 cells, top) and E-cadherin (62 cells, bottom) coverage per cell were quantified as low (0–25%), medium (26–75%) or high (76–100%). C. Single-cell RNA-seq data from UCD15 tumors was analyzed by UMAP which identified 7 transcriptomic clusters. KRT5 is concentrated in and a defining gene for cluster 5. D. Top 8 pathways identified by Ingenuity Pathway Analysis of all transcriptomic cluster 5 genes. Pathways involved in cell junction signaling and remodeling are marked with a red asterisk.
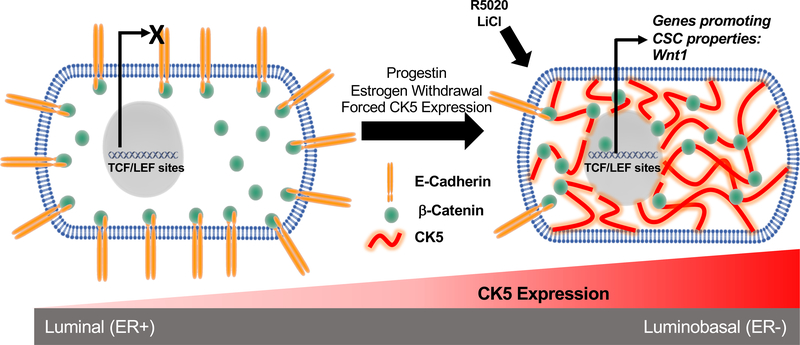
Figure 7. Proposed model of CK5 remodeling of ER+ breast cancer cells.
In ER+CK5− cells the β-catenin pool is largely localized to the inner cell membrane where it is bound to E-cadherin. Upon CK5 expression (via progestins, estrogen withdrawal, or forced CK5 expression) β-catenin translocates from the cell membrane to the cytoplasm where it prospectively interacts with CK5 which protects it from proteasomal degradation. Upon Wnt stimuli (R5020, LiCl, or Wnt3a), the cytosolic pool of β-catenin is poised for nuclear translocation and activation of TCF/LEF target genes, including Wnt1, sustaining a CSC phenotype. E-cadherin protein is also destabilized and lost in transitioned CK5+ cells, potentially enhancing invasive potential.
DISCUSSION
Cytokeratins are the major intermediate filament structural proteins of epithelial cells and are frequently used to stratify carcinomas into site of origin, tumor subtype, and to predict clinical course (44). CK5 is a Type II high molecular weight keratin of particular significance in the biology of normal and malignant breast tissue. In the normal human breast, while CK5 is ubiquitously expressed in the basal epithelial layer and has controversially been labeled a “basal cytokeratin” (16, 45), it is also expressed in some luminal epithelial cells in the TDLU (16). CK5 is a lineage marker of mammary stem and luminal progenitor cells(17); the latter are implicated as the cell of origin of BRCA1 mutant BLBC (17). Accordingly, CK5 is a signature marker of BLBC along with EGFR, and predicts quicker disease progression and worse overall survival compared to CK5− triple negative breast cancers (19, 46, 47). Likewise, ER+ and HER2 amplified/ER− breast cancers with positive vs. negative CK5 immunostaining have inferior outcome (20). CK5+ER− cells within ER+ breast cancers have CSC properties (6–10). The presence of CK5 in normal tissue regenerative cells, cancer initiating cells, and in detrimental cells across all breast cancer subtypes denotes its biological importance. Yet to our knowledge the functional contribution of CK5 to the ominous biology of such breast cancer cells has not been studied directly.
In the current study, we describe a direct relationship between CK5 expression and β-catenin dynamics in ER+ breast cancer in comparison to a typical CK5+ BLBC. Our previous and current data support that CK5, through genetic manipulation, is important for tumorsphere formation in ER+ breast cancer cell lines, which prompted us to explore unique CK5 protein interactors using unbiased IP-MS. Among several interesting candidates, the multifunctional protein β-catenin was pursued based on its role as a Wnt signaling effector and component of the cadherin cell adhesion complex (34). The Wnt/β-catenin signaling pathway regulates normal and cancer cell stemness (48), and has particular relevance to breast cancer, as over thirty years ago, Wnt1 overexpression was discovered as sufficient for murine mammary tumorigenesis (49). Progesterone upregulates Wnt4 during mammary morphogenesis (50) which acts in a paracrine manner to stimulate mammary stem cell expansion (51). In ER+PR+ breast cancer cells Wnt1 is upregulated by progestins (38). Here we show that progestins increase β-catenin transcriptional activity (Figure 3A&B) and that CK5KO blocks this activation without loss of total β-catenin protein levels (Figure 3C). Likewise, Wnt1 mRNA expression is induced by progestins in control cells and attenuated upon CK5KO (Figure 3D). This implies Wnt/β-catenin signaling is particularly activated in and important to progestin-induced CK5+ breast cancer cells. BLBC have heightened Wnt/β-catenin pathway activation (52) and display greater nuclear β-catenin staining than ER+ breast cancers (40). Curiously, CK5 knockdown in MDA-MB-468 cells impaired Wnt3 stimulated β-catenin activity. However, this occurs by concurrent reduction in β-catenin levels, which could be partially rescued by proteosome inhibition (Figure 3F&5D), suggesting CK5 shields β-catenin from degradation in BLBC, in contrast to luminal breast cancer cells, where β-catenin is anchored to E-cadherin at the membrane in the absence of CK5. Collectively these data support that CK5 stabilizes and primes cytosolic β-catenin for transcriptional activation.
β-catenin is an essential component of epithelial cell adherens junctions, linking the cytoplasmic portion of the transmembrane glycoprotein E-cadherin to the actin cytoskeleton through α-catenin (34, 53, 54). Disruption of adherens junctions is linked to increased motility and worse prognosis in breast cancer (42, 43, 55, 56). Our data show a striking loss of β-catenin at the cell membrane in CK5+ cells, whether CK5 is induced by progestins or long term estrogen withdrawal, exogenously expressed in ER+ breast cancer cells, or co-existing with ER+ cells in PDX models (Figure 4&6), with no change in total β-catenin levels. By contrast, we found that E-cadherin is not only lost at the membrane in each of these models, but its protein levels are reduced by CK5 expression (Figure 5&6). It is likely that cytosolic sequestering of β-catenin by CK5 destabilizes E-cadherin. Previous studies found E-cadherin mRNA levels were unchanged in CK5+ vs. CK5− EWD8 luminal cells (5), suggesting E-cadherin protein stability is altered. BLBC are reported to have predominant cytoplasmic and nuclear as opposed to membrane β-catenin staining, a phenotype associated with poor prognosis (40). We indeed observed MDA-MB-468 cells have a perinuclear β-catenin staining pattern (Figure 4E). However, in contrast to ER+ cells, knockdown of CK5 led to drastic loss of β-catenin and E-cadherin and a spindle-like cell morphology (Figure 4E & 5D). Remodeling of adherens junctions and trafficking of β-catenin is tightly regulated by the process of endocytosis (55, 57), which controls cell signaling by movement of proteins through endosomes (58). Several Rab GTPase proteins involved in membrane trafficking are predicted to interact with CK5 by our IP-MS data (Supplemental Table 1), suggesting a potential mechanism by which CK5 facilitates endocytic recycling of β-catenin, and perhaps other proteins. Regulation of endocytosis is important for stem cell asymmetric cell division and deregulation of this process is prospectively linked to cancer initiation (59). Thus, we speculate this may be an intriguing mechanism and target in CK5+ breast cancer cells.
Loss of adherens junctions, particularly E-cadherin, is a signature marker of epithelial-mesenchymal transition (EMT), a natural biological process that in cancer cells, leads to loss of intercellular adhesion and enhanced migration and invasion (60). EMT is classically associated with a transition from cytokeratin to vimentin intermediate filaments. In our studies, upon progestin treatment or estrogen withdrawal, ER+ cells transition from expression of simple (CKs 8, 18, 19) to stratified (CKs 5, 17) CKs (61), a state we previously referred to as luminobasal (5). Cells expressing stratified CKs (CK5, CK14) are reported to lead collective invasion of breast tumors and organoids (62). CK17 is the likely dimeric partner of CK5 in luminobasal cells that are void of CK14 and we demonstrate strong co-IP of CK17 with CK5 in EWD8 (Supplemental Figure 2D). Thus, cooperative scaffolding by CK5/CK17 dimers may facilitate cell junction remodeling, and CK17 has been implicated in cell signaling and oncogenesis (31, 63–65). Drugs such as retinoids that restrict expression of CK5, CK17 and other stratified CKs could be useful in luminobasal breast cancers (7, 66, 67).
In conclusion, our results suggest CK5 actively remodels ER+ breast cancer cells from a luminal to basal epithelial/CSC state, in part through recycling of membrane β-catenin to the cytoplasm and degradation of E-cadherin (Figure 7). Our data reinforce that CKs are an interesting group of molecules increasingly recognized as having cell regulator and signaling functions in addition to their central structural role as cell-stress protectors. CKs depend on protein-protein interactions for their function; thus, disruption of key interactions such as between CK5 and β-catenin or processes that facilitate cell remodeling such as endocytosis may be a unique way to target these molecules and their associated adverse cell behavior. Furthermore, this could be applicable to both ER+ disease and BLBC, which has a dire clinical course with limited targeted therapies.
MATERIALS AND METHODS
Cell culture and reagents
A table of breast cancer models used and their ER, PR, and CK5 status is included in Supplemental Figure 6. Breast cancer cell lines were obtained from the University of Colorado Cancer Center Tissue Culture core. T47D, ZR75–1, and MCF7 cells were maintained as previously described (7, 68). MDA-MB-468 cells were maintained in Dulbecco’s Modified Eagle Media supplemented with 10% fetal bovine serum. EWD8 cells were derived from T47D cells by serial passage through mice (5) in the absence of estrogen and are maintained in phenol red-free minimal Eagle’s medium, 5% charcoal stripped fetal bovine serum, 1x NEAA, 1 × 10−9 M insulin, 0.1 mg/mL sodium pyruvate and 2mM L-glutamine. Cell lines were authenticated by short tandem repeat analysis and routinely tested for mycoplasma using the MycoAlert mycoplasma detection kit (Lonza, Basel, Switzerland). Sigma Mission shRNAs targeting CK5 (shCK5–22 [TRCN0000425222], shCK5–88 [TRCN0000426388]) and non-targeting clone (SHC0002) were previously described (7). Cells were transduced with shRNA virus and stable pools selected with puromycin. Wnt3a was purchased from R&D Systems (Minneapolis, MN, USA) and MG-132 from Sigma (St. Louis, MO, USA).
Generation of CK5 knockout and overexpression cell lines
For CRISPR-Cas9 targeting, sgRNA was designed to target the first 250 bp of KRT5 exon 1 using crispr.mit.edu (no longer available). sgRNA sequences are as follows:
Forward: CACCGAGGATATCCATCAGCACTAG
Reverse: AAACCTAGTGCTGATGGATATCCTC
sgRNA was cloned into the pX458 plasmid which contains the cas9 sequence and a GFP reporter gene (48138, Addgene). The plasmid was transiently transfected into T47D cells using Lipofectamine 3000 (Invitrogen, Grand Island, NY, USA). FACS was used to single cell sort GFP positive cells into 96 well plates. Clones were grown and sequenced for KRT5 knockout by Sanger sequencing. CRISPRcont cell lines are clones that underwent the transfection, sorting, and selection process but have an intact KRT5 gene by Sanger sequencing and immunoblot.
For generation of CK5 overexpressing cells, a 1783 bp fragment of the KRT5 cDNA sequence was PCR amplified from pBabe RFP1-KRT5 hygro plasmid (58493, Addgene) using the Phusion High Fidelity Polymerase (New England Biolabs, Ipswich, MA, USA) and ligated into the pCDH1 retroviral vector (System Biosciences, Palo Alto, CA, USA). Cells were transduced with virus containing the pCDH1-KRT5 or empty vector and stable pools selected with puromycin.
Tumorsphere and colony formation assays
Tumorsphere assays were as previously described (7). For colony formation assays, 400 cells/well were seeded in 6 well plates in complete media, grown for 2 weeks, then fixed with 10% formalin and stained with Crystal Violet (Sigma Aldrich). Whole wells were imaged using a Cannon Power Shot camera and total colonies per plate counted.
Immunoblotting
All cell lysates for immunoblotting were harvested in RIPA buffer. Primary antibodies were as follows: CK5 (mouse NCL-L-CK5, Leica Biosystems, Buffalo Grove, IL, USA, 1:1500), α-tubulin (ST1568, Sigma, 1:1000), β-catenin (mouse 2698, or rabbit 9587, Cell Signaling Technologies, Danvers, MA, USA, 1:1000), E-cadherin (14472, Cell Signaling, 1:1000), Lamin B1 (12586, Cell Signaling, 1:1000), CK17 (NBP2–29421, Novus Biologicals, 1:1000), or β-actin (A5441, Sigma, 1:1000). Secondary antibodies were IRDye800CW Goat-Anti-Mouse IgG (926–32210, Li-Cor Biosciences, Lincoln, NE, USA) and IRDye 680LT Goat-Anti-Rabbit IgG (926–68021, Li-Cor Biosciences) both at 1:10,000. Immunoblots were imaged and analyzed with the Odyssey Infrared Imaging System and Image Studio Lite (Li-Cor Biosciences).
Immunoprecipitation and IP-MS
For immunoprecipitation, 15 cm plates of cells or 100 mg of frozen tumor as described (68) were suspended in NP-40 lysis buffer (1% NP-40, 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA) supplemented with a protease/phosphatase inhibitor (Halt™ Protease Inhibitor Cocktail, ThermoFisher Scientific, Waltham, MA). Lysates (500–1000 μg) were precleared with magnetic protein G beads (10004D, Fisher Scientific, Hampton, NH, USA). 5 ug of antibody to CK5 (905501, Biolegend, Dedham, MA), β-catenin (9587, Cell Signaling), or rabbit IgG isotype control (02–6102, ThermoFisher Scientific or sc-2027, Santa Cruz Biotechnology, Santa Cruz, CA, USA) was prebound and crosslinked to magnetic protein G beads with BS3. Precleared lysate was added to beads and incubated at 4C with rotary agitation for 2 h. Beads were washed four times with wash buffer (PBS + 0.05% Tween-20), then boiled in 1X SDS loading buffer for 10 min. Input, flow-through, and immunoprecipitation fractions were analyzed by immunoblot as above.
For IP-MS, protein input, amount of antibody, and all wash and incubation volumes were scaled up 5X. CHAPS buffer was used for cell lysis (0.3% CHAPS, 40mM HEPES pH 7.5, 120 mM NaCl, 1 mM EDTA). On-bead peptide digestion was performed as described (69). Nano UHPLC-MS/MS was performed using the LTQ Orbitrap Velos Pro Mass Spectrometer (Thermo Scientific) to identify proteins using peptide fingerprints and relative MS2 fragments. Data were analyzed using Mascot (Matrix Science) against a human database and a report generated with Scaffold (Proteome Software, Inc, Portland, OR, USA) using a protein threshold of 99%, minimum number of 2 peptides, and peptide threshold of 95% (70). Proteins with a spectral count difference ≥1 for CK5 versus IgG samples underwent further stratification steps.
Immunocytochemistry and immunohistochemistry
For ICC, 2–3 × 105 cells were seeded onto glass coverslips in 6-well plates. After treatments, cells were washed twice wish PBS and fixed with ice cold 70% Acetone/30% methanol for 10 min. Fixed cells were blocked with 10% normal goat serum (Vector Lab, Burlingame, CA, USA) in 0.05% TBS-T for 30 minutes followed by addition of primary antibodies (CK5, mouse NCL-L-CK5, Leica Biosystems; β-catenin, 9587, Cell Signaling Technologies, 1:200) for 2 h, secondary antibodies (A11029, A11037, Invitrogen, 1:200) for 1 h, and counterstained with DAPI. Cells were imaged using the Olympus BX40 fluorescent microscope or Olympus FV1000 laser scanning confocal microscope. Individual cells were scored for membrane β-catenin coverage of 0–25% (low), 25–75% (medium), or 75–100% (high) in a blinded manner. IHC was performed as previously described (71) using the antibodies described above and imaged using the Olympus FV1000 laser scanning confocal microscope.
TOP FLASH reporter assay
5 × 103 cells were seeded into 96 well plates in sextuplicate and transiently co-transfected with 90 ng TOPFlash (12456, Addgene) and 10 ng SV40-Renilla plasmids using Lipofectamine 3000 reagents (Invitrogen). All treatments were administered immediately. After 24 h, luciferase activity was measured with the Dual Luciferase Reporter Assay System (Promega).
Quantitative reverse transcription PCR
RNA was isolated using QIAzol lysis reagent (Qiagen, Venlo, Netherlands). cDNA synthesis was performed using the Verso cDNA Synthesis Kit (Thermo Scientific). Absolute Blue Sybr Green (Thermo Fisher) was used to perform qRT-PCR. mRNA expression was analyzed using the delta-delta CT method and normalizing to housekeeping gene gapdh. The following primers sequences were used:
Wnt1: Forward: AAAATCCGGGGATCCTGCAC; Reverse: AGCCTCGGTTGACGATCTTG
MAFB: Forward: GACGCAGCTCATTCAGCAG; Reverse: A CCGGAGTTGGCGAGTTTCT
GAPDH: Forward: GGTATCGTGGAAGGACTC; Reverse: GGATGATGTTCTGGAGAGC
Statistical methods
Data are represented as mean ± s.e.m. unless otherwise noted and analyzed by two-tailed Student’s t-test or one-way analysis of the variance followed by a Tukey post hoc test as noted. Prism 8.0 (Graphpad Software, La Jolla, CA, USA) was used for analyses when samples met variance and normality tests. P<0.05 were considered significant.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the University of Colorado School of Medicine Biological Mass Spectrometry Facility and Advanced Light Microscopy Core for their assistance. This work was supported by National Institutes of Health grants NIH F31 CA232456 (OM), NIH 2R01 CA140985 (CAS), and the Breast Cancer Research Foundation (CAS).
Support: This work was supported by grants from the National Institutes of Health grants NIH F31 CA232456 (OM), NIH 2R01 CA140985 (CAS), and the Breast Cancer Research Foundation (CAS).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest
REFERENCES
- 1.Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Annu Rev Med. 2011;62:233–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28(16):2784–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan H, Gray R, Braybrooke J, Davies C, Taylor C, McGale P, et al. 20-Year Risks of Breast-Cancer Recurrence after Stopping Endocrine Therapy at 5 Years. N Engl J Med. 2017;377(19):1836–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindstrom LS, Yau C, Czene K, Thompson CK, Hoadley KA, Van’t Veer LJ, et al. Intratumor Heterogeneity of the Estrogen Receptor and the Long-term Risk of Fatal Breast Cancer. J Natl Cancer Inst. 2018;110(7):726–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haughian JM, Pinto MP, Harrell JC, Bliesner BS, Joensuu KM, Dye WW, et al. Maintenance of hormone responsiveness in luminal breast cancers by suppression of Notch. Proc Natl Acad Sci U S A. 2012;109(8):2742–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Axlund SD, Yoo BH, Rosen RB, Schaack J, Kabos P, Labarbera DV, et al. Progesterone-inducible cytokeratin 5-positive cells in luminal breast cancer exhibit progenitor properties. Horm Cancer. 2013;4(1):36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fettig LM, McGinn O, Finlay-Schultz J, LaBarbera DV, Nordeen SK, Sartorius CA. Cross talk between progesterone receptors and retinoic acid receptors in regulation of cytokeratin 5-positive breast cancer cells. Oncogene. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horwitz KBD WW; Harrell JC; Kabos P; Sartorius CA Rare steroid receptor-negative basal-like tumorigenic cells in luminal subtype human breast cancer xenografts. PNAS. 2008;105(15):5774–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kabos P, Haughian JM, Wang X, Dye WW, Finlayson C, Elias A, et al. Cytokeratin 5 positive cells represent a steroid receptor negative and therapy resistant subpopulation in luminal breast cancers. Breast Cancer Res Treat. 2011;128(1):45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodman CR, Sato T, Peck AR, Girondo MA, Yang N, Liu C, et al. Steroid induction of therapy-resistant cytokeratin-5-positive cells in estrogen receptor-positive breast cancer through a BCL6-dependent mechanism. Oncogene. 2016;35(11):1373–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Truong TH, Dwyer AR, Diep CH, Hu H, Hagen KM, Lange CA. Phosphorylated Progesterone Receptor Isoforms Mediate Opposing Stem Cell and Proliferative Breast Cancer Cell Fates. Endocrinology. 2019;160(2):430–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stingl J, Raouf A, Emerman JT, Eaves CJ. Epithelial progenitors in the normal human mammary gland. J Mammary Gland Biol Neoplasia. 2005;10(1):49–59. [DOI] [PubMed] [Google Scholar]
- 13.Villadsen R, Fridriksdottir AJ, Ronnov-Jessen L, Gudjonsson T, Rank F, LaBarge MA, et al. Evidence for a stem cell hierarchy in the adult human breast. J Cell Biol. 2007;177(1):87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boecker WB H Evidence of progenitor cells of glandular and myoepithelial cell lineages in the human adult female breast epithelium: a new progenitor (adult stem) cell concept. Cell Proliferation. 2003;36:73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Böcker W, Moll R, Poremba C, Holland R, van Diest PJ, Dervan P, et al. Common Adult Stem Cells in the Human Breast Give Rise to Glandular and Myoepithelial Cell Lineages: A New Cell Biological Concept. Laboratory Investigation. 2002;82(6):737–46. [DOI] [PubMed] [Google Scholar]
- 16.Gusterson BA, Ross DT, Heath VJ, Stein T. Basal cytokeratins and their relationship to the cellular origin and functional classification of breast cancer. Breast Cancer Res. 2005;7(4):143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15(8):907–13. [DOI] [PubMed] [Google Scholar]
- 18.Molyneux G, Geyer FC, Magnay F-A, McCarthy A, Kendrick H, Natrajan R, et al. BRCA1 Basal-like Breast Cancers Originate from Luminal Epithelial Progenitors and Not from Basal Stem Cells. Cell Stem Cell. 2010;7(3):403–17. [DOI] [PubMed] [Google Scholar]
- 19.Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14(5):1368–76. [DOI] [PubMed] [Google Scholar]
- 20.Laakso M, Tanner M, Nilsson J, Wiklund T, Erikstein B, Kellokumpu-Lehtinen P, et al. Basoluminal carcinoma: a new biologically and prognostically distinct entity between basal and luminal breast cancer. Clin Cancer Res. 2006;12(14 Pt 1):4185–91. [DOI] [PubMed] [Google Scholar]
- 21.Malzahn K MM, Thoenes M, Moll R. Biological and prognostic significance of stratified epithelial cytokeratins in infiltrating ductal breast carcinomas. Virchows Arch. 1998;433(2):119–29. [DOI] [PubMed] [Google Scholar]
- 22.van de Rijn M, Perou CM, Tibshirani R, Haas P, Kallioniemi O, Kononen J, et al. Expression of Cytokeratins 17 and 5 Identifies a Group of Breast Carcinomas with Poor Clinical Outcome. The American Journal of Pathology. 2002;161(6):1991–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato T, Tran TH, Peck AR, Girondo MA, Liu C, Goodman CR, et al. Prolactin suppresses a progestin-induced CK5-positive cell population in luminal breast cancer through inhibition of progestin-driven BCL6 expression. Oncogene. 2014;33(17):2215–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coulombe PAO, M.B. ‘Hard’ and ‘soft’ principles defining the structure, function, and regulation of keratin intermediate filaments. Current Opinion in Cell Biology. 2002;14:110–22. [DOI] [PubMed] [Google Scholar]
- 25.Toivola DM, Strnad P, Habtezion A, Omary MB. Intermediate filaments take the heat as stress proteins. Trends Cell Biol. 2010;20(2):79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrmann H, Strelkov SV, Burkhard P, Aebi U. Intermediate filaments: primary determinants of cell architecture and plasticity. J Clin Invest. 2009;119(7):1772–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kreplak L, Aebi U, Herrmann H. Molecular mechanisms underlying the assembly of intermediate filaments. Exp Cell Res. 2004;301(1):77–83. [DOI] [PubMed] [Google Scholar]
- 28.Kim S, Wong P, Coulombe PA. A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature. 2006;441(7091):362–5. [DOI] [PubMed] [Google Scholar]
- 29.Ku NO, Fu H, Omary MB. Raf-1 activation disrupts its binding to keratins during cell stress. J Cell Biol. 2004;166(4):479–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mikami T, Maruyama S, Abe T, Kobayashi T, Yamazaki M, Funayama A, et al. Keratin 17 is co-expressed with 14–3-3 sigma in oral carcinoma in situ and squamous cell carcinoma and modulates cell proliferation and size but not cell migration. Virchows Arch. 2015;466(5):559–69. [DOI] [PubMed] [Google Scholar]
- 31.Escobar-Hoyos LF, Shah R, Roa-Pena L, Vanner EA, Najafian N, Banach A, et al. Keratin-17 Promotes p27KIP1 Nuclear Export and Degradation and Offers Potential Prognostic Utility. Cancer Res. 2015;75(17):3650–62. [DOI] [PubMed] [Google Scholar]
- 32.Boudreau A, Tanner K, Wang D, Geyer FC, Reis-Filho JS, Bissell MJ. 14–3-3sigma stabilizes a complex of soluble actin and intermediate filament to enable breast tumor invasion. Proc Natl Acad Sci U S A. 2013;110(41):E3937–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10(8):730–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303(5663):1483–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149(6):1192–205. [DOI] [PubMed] [Google Scholar]
- 36.de Sousa EMF, Vermeulen L. Wnt Signaling in Cancer Stem Cell Biology. Cancers (Basel). 2016;8(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klein P, Melton, DA. A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci. 1996;93:8455–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faivre EJ, Lange CA. Progesterone receptors upregulate Wnt-1 to induce epidermal growth factor receptor transactivation and c-Src-dependent sustained activation of Erk1/2 mitogen-activated protein kinase in breast cancer cells. Mol Cell Biol. 2007;27(2):466–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dolled-Filhart M, McCabe A, Giltnane J, Cregger M, Camp RL, Rimm DL. Quantitative in situ analysis of beta-catenin expression in breast cancer shows decreased expression is associated with poor outcome. Cancer Res. 2006;66(10):5487–94. [DOI] [PubMed] [Google Scholar]
- 40.Khramtsov AI, Khramtsova GF, Tretiakova M, Huo D, Olopade OI, Goss KH. Wnt/beta-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am J Pathol. 2010;176(6):2911–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farahani E, Patra HK, Jangamreddy JR, Rashedi I, Kawalec M, Rao Pariti RK, et al. Cell adhesion molecules and their relation to (cancer) cell stemness. Carcinogenesis. 2014;35(4):747–59. [DOI] [PubMed] [Google Scholar]
- 42.Kowalski PJ, Rubin MA, Kleer CG. E-cadherin expression in primary carcinomas of the breast and its distant metastases. Breast Cancer Res. 2003;5(6):R217–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siitonen SM KJ, Helin HJ, Rantala IS, Holli KA, Isola JJ. Reduced E-cadherin expression is associated with invasiveness and unfavorable prognosis in breast cancer. Anatomic Pathology. 1995;105(4). [DOI] [PubMed] [Google Scholar]
- 44.Moll R, Divo M, Langbein L. The human keratins: biology and pathology. Histochem Cell Biol. 2008;129(6):705–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gusterson B Do ‘basal-like’ breast cancers really exist? Nature Reviews Cancer. 2009;9:128–34. [DOI] [PubMed] [Google Scholar]
- 46.Abdelrahman AE, Rashed HE, Abdelgawad M, Abdelhamid MI. Prognostic impact of EGFR and cytokeratin 5/6 immunohistochemical expression in triple-negative breast cancer. Ann Diagn Pathol. 2017;28:43–53. [DOI] [PubMed] [Google Scholar]
- 47.Sutton LM, Han JS, Molberg KH, Sarode VR, Cao D, Rakheja D, et al. Intratumoral expression level of epidermal growth factor receptor and cytokeratin 5/6 is significantly associated with nodal and distant metastases in patients with basal-like triple-negative breast carcinoma. Am J Clin Pathol. 2010;134(5):782–7. [DOI] [PubMed] [Google Scholar]
- 48.Holland JD, Klaus A, Garratt AN, Birchmeier W. Wnt signaling in stem and cancer stem cells. Curr Opin Cell Biol. 2013;25(2):254–64. [DOI] [PubMed] [Google Scholar]
- 49.Tsukamoto AS GR, Guzman RC, Parslow T, Varmus HE. Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell. 1988;55:619–25. [DOI] [PubMed] [Google Scholar]
- 50.Brisken C HA, Chavarria T, Elenbaas B, Tan J, Dey SK, McMahon JA, McMahon AP, Weinberg RA. Essential function of Wnt-4 in mammary gland development downstream of progesterone signaling. Genes and Development. 2000;14:650–4. [PMC free article] [PubMed] [Google Scholar]
- 51.Joshi PA, Jackson HW, Beristain AG, Di Grappa MA, Mote PA, Clarke CL, et al. Progesterone induces adult mammary stem cell expansion. Nature. 2010;465(7299):803–7. [DOI] [PubMed] [Google Scholar]
- 52.Roarty K, Rosen JM. Wnt and mammary stem cells: hormones cannot fly wingless. Curr Opin Pharmacol. 2010;10(6):643–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wheelock MJ SA, Knudsen KA. Cadherin junctions in mammary tumors.. J Mammary Gland Biol Neoplasia. 2001;6:275–85. [DOI] [PubMed] [Google Scholar]
- 54.Valenta T, Hausmann G, Basler K. The many faces and functions of beta-catenin. EMBO J. 2012;31(12):2714–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kam Y, Quaranta V. Cadherin-bound beta-catenin feeds into the Wnt pathway upon adherens junctions dissociation: evidence for an intersection between beta-catenin pools. PLoS One. 2009;4(2):e4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Howard S, Deroo T, Fujita Y, Itasaki N. A positive role of cadherin in Wnt/beta-catenin signalling during epithelial-mesenchymal transition. PLoS One. 2011;6(8):e23899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hartwell KA, Miller PG, Mukherjee S, Kahn AR, Stewart AL, Logan DJ, et al. Niche-based screening identifies small-molecule inhibitors of leukemia stem cells. Nat Chem Biol. 2013;9(12):840–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mosesson Y, Mills GB, Yarden Y. Derailed endocytosis: an emerging feature of cancer. Nat Rev Cancer. 2008;8(11):835–50. [DOI] [PubMed] [Google Scholar]
- 59.Furthauer M, Gonzalez-Gaitan M. Endocytosis, asymmetric cell division, stem cells and cancer: unus pro omnibus, omnes pro uno. Mol Oncol. 2009;3(4):339–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20(2):69–84. [DOI] [PubMed] [Google Scholar]
- 61.Sartorius CA, Harvell DM, Shen T, Horwitz KB. Progestins initiate a luminal to myoepithelial switch in estrogen-dependent human breast tumors without altering growth. Cancer Res. 2005;65(21):9779–88. [DOI] [PubMed] [Google Scholar]
- 62.Cheung KJ, Gabrielson E, Werb Z, Ewald AJ. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 2013;155(7):1639–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Depianto D, Kerns ML, Dlugosz AA, Coulombe PA. Keratin 17 promotes epithelial proliferation and tumor growth by polarizing the immune response in skin. Nat Genet. 2010;42(10):910–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khanom R, Nguyen CT, Kayamori K, Zhao X, Morita K, Miki Y, et al. Keratin 17 Is Induced in Oral Cancer and Facilitates Tumor Growth. PLoS One. 2016;11(8):e0161163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sankar S, Tanner JM, Bell R, Chaturvedi A, Randall RL, Beckerle MC, et al. A novel role for keratin 17 in coordinating oncogenic transformation and cellular adhesion in Ewing sarcoma. Mol Cell Biol. 2013;33(22):4448–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jho SH, Radoja N, Im MJ, Tomic-Canic M. Negative response elements in keratin genes mediate transcriptional repression and the cross-talk among nuclear receptors. J Biol Chem. 2001;276(49):45914–20. [DOI] [PubMed] [Google Scholar]
- 67.Radoja N DD, Minars TJ, Freedberg IM, Blumenberg M, Tomic-Canic M. Specific organization of the negative response elements for retinoic acid and thyroid hormone receptors in keratin gene family. J Invest Dermatol. 1997;109:566–72. [DOI] [PubMed] [Google Scholar]
- 68.Finlay-Schultz J, Gillen AE, Brechbuhl HM, Ivie JJ, Matthews SB, Jacobsen BM, et al. Breast Cancer Suppression by Progesterone Receptors Is Mediated by Their Modulation of Estrogen Receptors and RNA Polymerase III. Cancer Res. 2017;77(18):4934–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Callipo L, Caruso G, Foglia P, Gubbiotti R, Samperi R, Lagana A. Immunoprecipitation on magnetic beads and liquid chromatography-tandem mass spectrometry for carbonic anhydrase II quantification in human serum. Anal Biochem. 2010;400(2):195–202. [DOI] [PubMed] [Google Scholar]
- 70.Searle BC. Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics. 2010;10(6):1265–9. [DOI] [PubMed] [Google Scholar]
- 71.Kabos P, Finlay-Schultz J, Li C, Kline E, Finlayson C, Wisell J, et al. Patient-derived luminal breast cancer xenografts retain hormone receptor heterogeneity and help define unique estrogen-dependent gene signatures. Breast Cancer Res Treat. 2012;135(2):415–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.