Abstract
Recombinase polymerase amplification (RPA) is a quick, specific, sensitive molecular tool carried out at a constant temperature for pathogen detection. In the present study, RPA and reverse transcription (RT) RPA assays were optimized for the detection of piper yellow mottle virus (PYMoV) infecting black pepper. Out of the eight primer pairs targeted to amplify open reading frames (ORFs) 2 and 3 of the virus, the primer pair targeted to ORF2 gave specific amplification only with DNA isolated from infected plant but not with healthy plant. A magnesium acetate concentration of 18 mM, 40 min of incubation time and a temperature of 37–42 °C was found optimum for detection of the virus in RPA assay. Comparison of sensitivity of detection revealed that RPA could detect the virus up to 10−5 dilution of the total DNA while PCR could detect the virus up to 10−4 dilution indicating that RPA is 10 times more sensitive than PCR. RPA was further simplified using crude extract as template which could detect the virus up to 10−3 dilution. RT-RPA was optimized for the detection of PYMoV using total RNA isolated from infected plants as the template. Both RT-RPA and RPA assays were validated using field samples of black pepper representing different varieties and geographical regions by using CTAB isolated DNA, crude DNA extract and cDNA. Our study showed that RPA and RT-RPA can be successfully adopted as a substitute to PCR for detection of PYMoV infecting black pepper.
Electronic supplementary material
The online version of this article (10.1007/s13337-019-00566-x) contains supplementary material, which is available to authorized users.
Keywords: Isothermal amplification, Reverse transcription-RPA, Diagnosis, Sensitivity, Polymerase chain reaction
Introduction
Piper yellow mottle virus (PYMoV), a pararetrovirus belonging to the genus, Badnavirus infects black pepper and related species in all black pepper growing countries of the world including India. PYMoV infection is identified by different symptoms such as yellow mottles and deformation of leaves, shortened internodes leading to stunting of plants [2, 8, 13]. PYMoV has a double stranded circular DNA genome of about 7.5 kb in size containing four open reading frames (ORFs) [2, 9]. As black pepper is propagated by vegetative means, use of virus-free plants is necessary to reduce spread of the virus. Currently available assays for detection of PYMoV include: polymerase chain reaction (PCR), real-time PCR and loop-mediated isothermal amplification (LAMP) [3–5, 8, 13]. Being a pararetrovirus, PYMoV replicates via a RNA intermediate, hence it can also be detected using RNA as template by reverse transcription (RT) PCR [2, 6].
Recombinase polymerase amplification (RPA) which was introduced in the year 2006 is extensively used for detection of pathogens that gives results within 5–20 min [7, 10]. RPA uses recombinase, single-strand DNA binding protein (SSB) and DNA polymerase with strand-displacement activity for amplification. Recombinase forms complex with primers and then scans the target sequence for homology and invades this region to form limited separation of DNA strands. The SSB stabilizes the separated strands and recombinase extends the primer by DNA polymerase. RPA cycle repeats that leads to an exponential increase of target DNA. RNA virus can be amplified by the addition of the enzyme, reverse transcriptase to RPA components known as reverse transcription (RT)-RPA [7, 19].
RPA was successfully used for detection of many DNA viruses infecting different crops such as banana [12], beans, and tomato [14]. Similarly, RT-RPA is reported for the diagnosis of RNA viruses infecting different crops such as yam, rose, plum and sweet cherry [1, 15, 18, 20]. In the current study we report optimization and validation of RPA and RT-RPA assays for quick diagnosis of the PYMoV infecting black pepper.
Materials and methods
DNA isolation by CTAB method
Total DNA was isolated from 50 mg tissues of black pepper using CTAB (cetyltrimethyl ammonium bromide) method [5]. The method involved grinding the sample in 10 volumes (w/v) of CTAB buffer [100 mM Tris–HCl (pH 8), 4 mM EDTA (pH 8), 1.4 M NaCl, 2% CTAB, 1% PVP] containing 2.5 µl β-mercaptoethanol. The resultant mixture was kept at 65 °C for 30 min, added with equal volume of phenol: chloroform: isoamyl alcohol (25:24:1) and subjected to centrifugation at 2500 g for 10 min. The supernatant was added with 50 µl of 10% CTAB and equal volume of chloroform: isoamyl alcohol (24:1). The resultant mixture was subjected to centrifugation at 2500 g for 10 min at room temperature. The aqueous phase was mixed with 50 µl of 3 M sodium acetate (pH 5.2) and an equivalent volume of isopropanol. After incubating the mixture in ice for 30 min, the contents were subjected to centrifugation at 10,000 rpm for 15 min at 4 °C to pelletize DNA. The DNA pellet was washed with 70% ethanol, dried and dissolved in 50 μl sterile water.
Crude DNA extraction
Tissue (50 mg) was ground in 500 µl of 0.5 N NaOH with mortar and pestle [16]. The contents were subjected to centrifugation at 5000 g for 30 s. The supernatant obtained was then diluted (1:10) using 0.1 M Tris (pH 8) and used as template for RPA.
Polymerase chain reaction (PCR)
PCR was done with CTAB isolated DNA and crude DNA extract as template using, forward (5′-TTTGTCAAGCCAAGAGACCAC-3′) and reverse (5′-TTGAGTGATTTGGTCCTCCAC-3′) primers designed based on ORF 2 sequence of PYMoV. The 25 μl PCR reaction mix contained 12.5 μl of master mix (Emerald Amp GT PCR, Takara), 0.25 μl (10 μM) each forward and reverse primers, 1 μl template DNA and 11 μl water. DNA was denatured at 98 °C for 1 min, then the reactions were cycled 35 times at 98 °C for 10 s, annealed at 60 °C for 1 min and extended at 72 °C for 30 s, finally an extension step at 72 °C for 10 min. PCR products were analysed on 0.8% agarose gel electrophoresis.
Recombinase polymerase amplification (RPA)
For RPA eight pairs of primers were designed (Supplementary Table 1) based on the conserved sequences in the ORF 2 and ORF 3 of available PYMoV sequences in the GenBank according to the TwistDx guidelines (https://www.twistdx.co.uk). Initially to determine the suitable primer pair for the specific detection of PYMoV, RPA was carried out using eight pairs of primers using total DNA isolated by using CTAB method from the PCR tested known infected and healthy plants as template. Water control without template was also used to check for any non-specific amplification in the RPA. RPA was carried out using TwistDx, Twist Amp® basic kit, reaction was set up by reconstituting supplied freeze dried reaction pellets with rehydration buffer (29.5 µl) and 8.2 μl nuclease free water. The resultant mixture was equally divided into five fresh PCR tubes (7.54 μl each) to make five individual reactions of 10 μl each. For each PCR tubes, primers (both forward and reverse 0.48 µl each (480 nM each) and template (1 µl) were added. RPA reaction was initiated by the adding of 0.5 μl magnesium acetate solution (14 mM/10 µl). Reaction mix was incubated at 39 °C for 4 min, after which samples were taken out of incubator and mixed by pipetting, spun and returned back to the incubator. Incubation was continued for a total of 40 min. The RPA reaction was then stopped by placing the tubes for 10 min at 65 °C. RPA samples were analyzed through electrophoresis on 2% agarose gel.
Optimization of RPA
Optimization of the RPA assay for the short listed primer pair was done by changing concentration of magnesium acetate (12–20 mM per 10 µl reaction mix), reaction incubation time (10–50 min) and incubation temperature (37–42 °C) using DNA isolated from identified virus-infected and virus-free black pepper plant as template.
RPA with crude DNA extract
RPA with crude DNA extract was carried out using 1 μl of undiluted and diluted (1:10 with 0.1 M Tris pH 8) crude DNA extract using the method optimized above.
Total RNA extraction
Total RNA was extracted from 50 mg of leaf tissue ground in 500 μl trizol reagent containing 5 μl of β-mercaptoethanol and 50 μl of 0.5% sodium sulphite [17]. To this, 50 μl of 2 M sodium acetate (pH 4), 500 μl of water saturated phenol and 100 μl of chloroform: isoamyl alcohol (24:1) was added. Contents were shaken vigorously for 10 s, incubated for 15 min on ice and centrifuged at 12,000 rpm at 4 °C for 15 min. Supernatant was aspirated and DNase treated for 30 min at 37 °C. The mixture was added with chloroform: isoamyl alcohol (24:1) and centrifuged for 15 min at 12,000 rpm. Aqueous phase was collected, added with equal volume isopropanol, mixed well and incubated in − 80 °C for 1 h followed by centrifugation for 15 min at 12,000 rpm at 4 °C. Pellet obtained was washed in 75% ethanol, dried and re-suspended in 50 μl nuclease free water.
cDNA synthesis
Total RNA isolated above was denatured at 80 °C in a water bath for 10 min and snap cooled on ice for 5 min was used for cDNA synthesis. cDNA synthesis was done using 4 μl of 5 × reaction buffer, 20U RNase inhibitor, 10 mM dNTP mix,10.5 μl of denatured RNA, 1 μM reverse primer (5′-TTGAGTGATTTGGTCCTCCAC-3′) corresponding to ORF 2 region of PYMoV and 200U of reverse transcriptase in a final volume of 20 μl. The reaction tube was kept at 42 °C for 60 min followed by 70 °C for 10 min.
Reverse transcription (RT)-RPA
One micro litre of cDNA served as template for the detection of PYMoV through RT-RPA. The components and reaction condition of RT-RPA remained same as described in RPA.
Comparison of sensitivity of RPA, RT-RPA and PCR
The optimized RPA and RT-RPA assays were compared for their sensitivity of detection of PYMoV in comparison to PCR using serial dilutions (100–10−8) of CTAB isolated DNA, diluted crude DNA extract and cDNA (100–10−8). Each dilution was subjected to PCR, RPA and RT-RPA as explained above. The CTAB isolated DNA and cDNA dilutions were made using nuclease free water while dilutions of crude DNA extract was made using 0.1 M Tris (pH 8).
Validation of RPA and RT-RPA assays
The optimized RPA and RT-RPA assays were validated along with PCR assay using 1 μl each of CTAB isolated DNA, diluted crude DNA extract and cDNA from test samples of black pepper representing different varieties namely, IISR Girimunda, IISR Malabar Excel, IISR Shakthi, Panchami, Panniyur 1, Panniyur 2, Panniyur 3, Panniyur 4, Panniyur 5, Panniyur 6, PLD 2, Pournami, Thekken and Vijay.
Results
Development of RPA assay
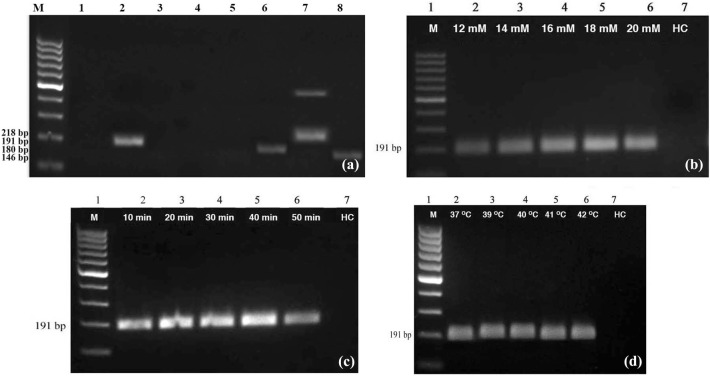
PCR using ORF 2 specific primer gave expected band at 352 bp for positive control while negative control and water control did not give any product. The DNA of positive control plant was used to determine primer pair suitable for RPA assay using eight pairs of primers synthesized. Results showed that primer pair, AIB 453–454 gave expected specific band at 191 bp indicating that it is suitable for RPA assay while remaining seven pair of primers were found unsuitable as they either did not give any product or produced non-specific bands (Fig. 1a). Primers AIB 452–453, AIB 454–455, AIB 456–455 and AIB 452–455 did not give any band while AIB 483–484 and AIB 487–488 gave very faint band. The primer pair AIB 485–486 gave non-specific band in addition to the specific band. Optimization of RPA assay, using the primers AIB 453–454 showed that 18 mM magnesium acetate (Fig. 1b) and 40 min incubation time was optimum (Fig. 1c). All the temperatures tested (37 °C, 39 °C, 40 °C, 41 °C and 42 °C) gave similar results indicating that any temperature from 39 to 42 °C can be used in the RPA assay (Fig. 1d). Optimized RPA assay using the primer pair, AIB 453–454 gave expected band at 191 bp for positive control while negative control and water control did not give any product (Fig. 2a). Further, identity of the RPA product was established by directly sequencing the product.
Fig. 1.
Optimization of recombinase polymerase amplification (RPA) assay for the detection of piper yellow mottle virus (PYMoV). a Identification of suitable primer pair. Lane M: 100 bp DNA ladder, Lane 1–8: Loaded with RPA product obtained with primers AIB 452–453, AIB 453–454, AIB 454–455, AIB 456–455, AIB 452–455, AIB 483–484, AIB 485–486, AIB 487–488. Details of the primers and expected product size are provided in supplementary Table 1. b determination of optimum concentration of magnesium acetate, c incubation time and d temperature. Lane HC: Healthy control
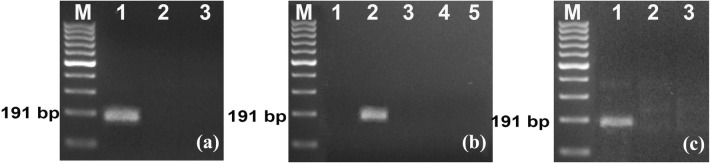
Fig. 2.
a Detection of PYMoV through optimized RPA assay using CTAB isolated DNA as template. Lane M: 100 bp DNA ladder; Lane 1: DNA from infected plant; Lane 2: DNA from healthy plant; Lane 3: Water control. b Detection of PYMoV through optimized RPA assay using crude DNA extract. Lane 1: crude DNA extract from infected plant; Lane 2: 1:10 diluted DNA crude extract from infected plant; Lane 3: crude DNA extract from healthy plant; Lane 4: 1:10 diluted crude DNA extract from healthy plant; Lane 5: Water control. c Detection of PYMoV through reverse transcription (RT)-RPA assay using cDNA as template. Lane 1: cDNA from infected plant; Lane 2: cDNA from healthy plant; Lane 3: Water control
RPA with crude extract
Optimized RPA assay was successful in the detection of PYMoV using diluted crude DNA extract as template (Fig. 2b). Use of crude extract directly as template failed to detect the PYMoV. However, 1:10 diluted crude DNA extract from infected plant showed positive reaction while healthy plant and water control did not give any product. Similar results were obtained in PCR too.
RT-RPA assay
PYMoV was also detected successfully by RT-RPA using 1 μl of cDNA as template. Amplification was obtained for positive control while negative control and water control did not give any product (Fig. 2c).
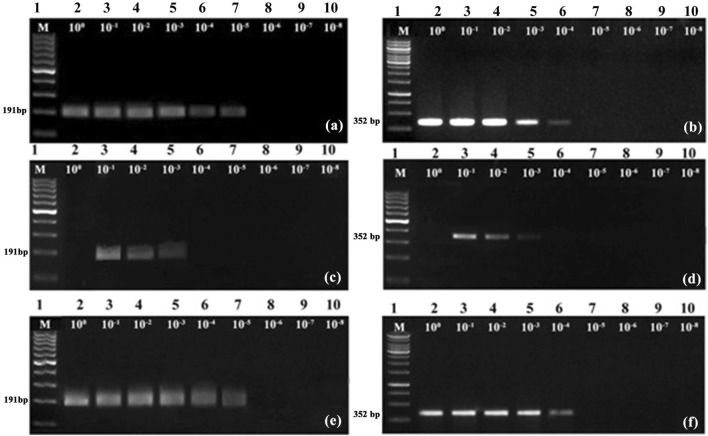
Comparison of sensitivity of RPA and PCR assays
When CTAB isolated total DNA was used as template, sensitivity of detection of PYMoV was up to 10−5 in RPA (Fig. 3a) while it was upto10−4 in PCR (Fig. 3b) indicating that RPA assay is ten times more sensitive compared to PCR for the detection of PYMoV. Sensitivity of detection of PYMoV using crude DNA extract was up to 10−3 for both RPA (Fig. 3c) and PCR (Fig. 3d). Sensitivity of detection of PYMoV using cDNA template was similar to the results of RPA and PCR obtained with CTAB isolated DNA (Fig. 3e, f).
Fig. 3.
Comparison of sensitivity of detection of PYMoV by RPA (a, c, e) and PCR (b, d, f) assays. Lane 2–10 loaded with RPA/PCR products obtained with template DNA dilutions from 100–10−8 using template DNA isolated by CTAB (a, b), crude DNA extract (c, d) and cDNA (e, f). Lane M in a, c, d, e are loaded with 100 bp DNA ladder while lane M in b and f are loaded with 1 kb DNA ladder
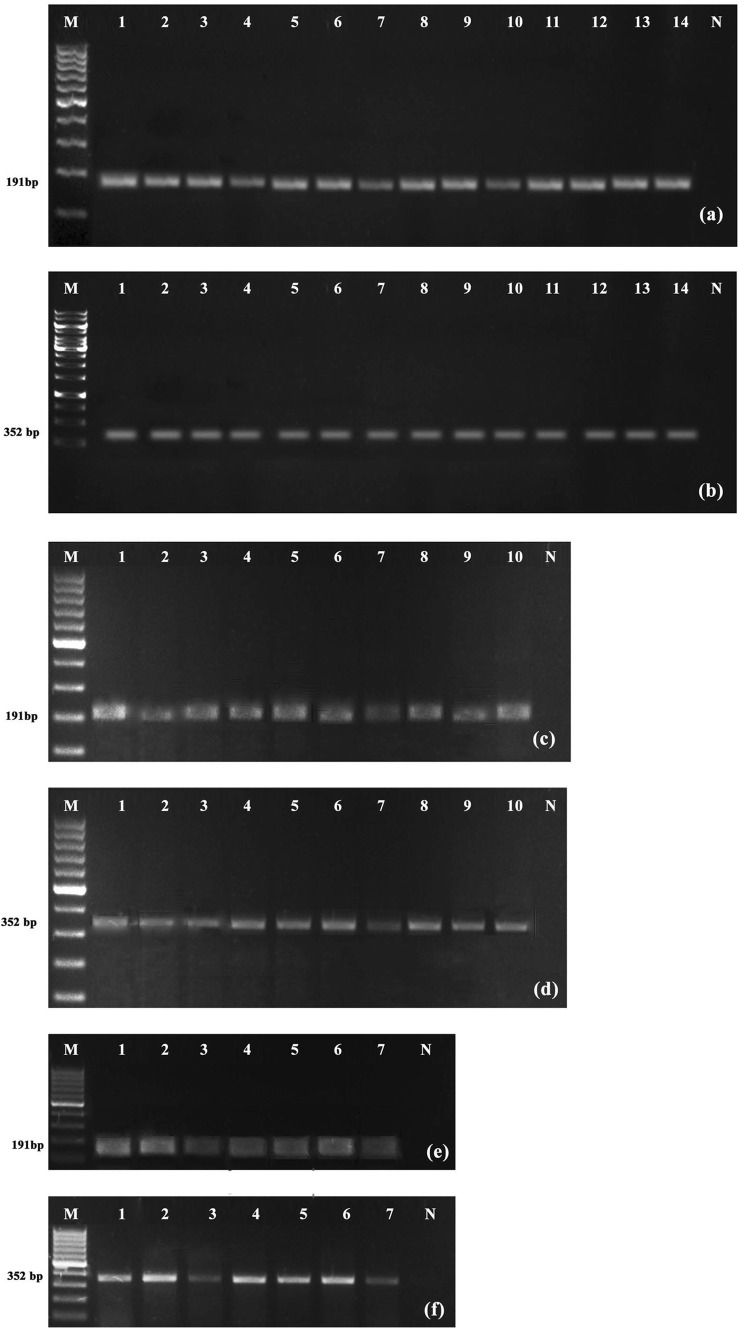
Validation of RPA and RT-RPA assays
Validation of RPA and RT-RPA assay using CTAB isolated DNA, diluted crude DNA extract and cDNA from different varieties of black pepper showed detection of PYMoV in all varieties tested (Fig. 4a, c, e). Similar results were also obtained with PCR (Fig. 4b, d, f).
Fig. 4.
Validation of RPA (a, c, e) and PCR (b, d, f) assays for the detection of PYMoV using template DNA isolated by CTAB (a, b), crude extract (c, d) and cDNA (e, f) respectively. Lane M in a, c, d, e, f are loaded with 100 bp DNA ladder while lane M in b is loaded with kb DNA ladder; Lane N is DNA from known healthy plant while rest of the lanes are loaded with DNA/cDNA from test plants
Discussion
The present study optimized a successful RPA and RT-RPA assays for the rapid detection of PYMoV infecting black pepper. Black pepper being a vegetatively propagated crop, identification and production of virus-free plants is necessary to control virus spread. Though PCR and real-time PCR assays are available for PYMoV detection, these methods require sophisticated laboratory and takes more time for the test [4, 5, 8]. Other isothermal methods like LAMP assay is also reported for the detection of PYMoV. It requires high temperature, longer incubation time and about six primers for amplification [3]. On the other hand, RPA is a very simple assay that can be performed in a resource poor set up with less than 40 min time without using thermal cycler.
The present study showed that RPA using CTAB isolated DNA is ten times additional sensitive than PCR as described earlier for the detection of cucumber green mottle mosaic virus [11]. Further, in the present study we simplified RPA assay successfully by using diluted crude DNA extract as template, as reported for detection of BBTV in banana [12]. Sensitivity of detection of PYMoV by both RPA and PCR were on par when crude DNA extract was used as template in contrast to some of the earlier studies that reported RPA as more sensitive compared to PCR [12, 14]. The diluted crude DNA extract as template for detection of PYMoV will lead to quick large scale detection of PYMoV at the field level. Nucleic acid extraction is a laborious process that requires laboratory and skill. To make the detection technique simpler we need to simplify nucleic acid extraction method. The detection of PYMoV by RPA and PCR using crude DNA extract from black pepper in the present study is a right step in this direction. This would help to take diagnostics to rural areas where lab facilities do not exist. Another outcome of the present study is the reduced reaction volume of just 10 μl (instead of 50 μl) to successfully detect the virus thus reducing the cost of detection per sample.
Pararetroviruses like PYMoV replicate via an RNA intermediate, thus they have both DNA and RNA phase in their replication cycle. Hence these viruses can also be detected using cDNA as template. Thus the present study also developed RT-RPA assay for the detection of PYMoV using cDNA as template. The sensitivity of detection of PYMoV through RT-RPA was up to 10−5 using cDNA template that was on par with the RPA assay using CTAB isolated DNA and 10 times additional sensitive compared to PCR assay [13]. Many RNA viruses such as yam mosaic virus, rose rosette virus, plum pox virus and little cherry virus 2 were successfully detected using RT-RPA [1, 15, 18–20]. Both RPA and RT-RPA assays developed in the present study were validated using field samples confirming its utility in testing the virus in field samples. This would help in identifying virus-free plants for propagation and further distribution to farmers that would ultimately check the spread of the virus.
Thus the present study developed a simple, rapid and sensitive RPA assay for the detection of the PYMoV infecting black pepper. RPA assay was further simplified using crude DNA extract as template. As PYMoV has both DNA and RNA phases in its replication cycle, an RT-RPA assay was also developed for detection of the virus. Both RPA and RT-RPA assays were validated using field samples representing different varieties of black pepper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Authors express their thanks to the Department of Biotechnology, Government of India for the funding (BT/PR15202/BPA/118/156/2015), Head (Division of Crop Protection) and Director, ICAR-Indian Institute of Spices Research, Kozhikode, Kerala, India for facilities. First author is also thankful to the Kerala State Council for Science Technology and Environment, Trivandrum, Kerala for the research fellowship.
Compliance with ethical standards
Conflict of interest
Both authors declare that they have no conflict of interest.
Ethical approval
This research did not involve any experimentation on humans or animals.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Babu B, Washburn BK, Ertek TS, Miller SH, Riddle CB, Knox GW, Ochoa-Corona FM, Olson J, Katırcıoğlu YZ, Paret ML. A field based detection method for Rose rosette virus using isothermal probe based reverse transcription-recombinase polymerase amplification assay. J Virol Methods. 2017;247:81–90. doi: 10.1016/j.jviromet.2017.05.019. [DOI] [PubMed] [Google Scholar]
- 2.Bhat AI, Hohn T, Selvarajan R. Badnaviruses: the current global scenario. Viruses. 2016;8:177. doi: 10.3390/v8060177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhat AI, Siljo A, Deeshma KP. Rapid detection of Piper yellow mottle virus and Cucumber mosaic virus infecting black pepper (Piper nigrum) by loop-mediated isothermal amplification (LAMP) J Virol Methods. 2013;193:190–196. doi: 10.1016/j.jviromet.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Bhat AI, Siljo A. Detection of viruses infecting black pepper by SYBR Green based real-time PCR assay. Plant Pathol J. 2014;96:105–109. [Google Scholar]
- 5.Bhat AI, Siljo A, Jiby MV, Thankamani CK, Mathew PA. Polymerase chain reaction (PCR) based indexing for screening black pepper (Piper nigrum L.) plants against Piper yellow mottle virus. J Spices Aromat Crop. 2009;18:28–32. [Google Scholar]
- 6.Chabannes M, Iskra L, Caruana ML. Endogenous pararetroviruses-a reservoir of virus infection in plants. Curr Opin Virol. 2013;3:615–620. doi: 10.1016/j.coviro.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 7.Daher RK, Stewart G, Boissinot M, Bergeron MG. Recombinase polymerase amplification for diagnostic applications. Clin Chem. 2016;62:947–956. doi: 10.1373/clinchem.2015.245829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Silva DPP, Jones P, Shaw MW. Identification and transmission of Piper yellow mottle virus and Cucumber mosaic virus infecting black pepper (Piper nigrum L.) in Sri Lanka. Plant Pathol. 2002;51:537–545. doi: 10.1046/j.0032-0862.2002.00757.x. [DOI] [Google Scholar]
- 9.Hany U, Adams IP, Glover R, Bhat AI, Boonham N. The complete genome sequence of Piper yellow mottle virus (PYMoV) Arch Virol. 2014;159:385–388. doi: 10.1007/s00705-013-1824-2. [DOI] [PubMed] [Google Scholar]
- 10.Jauset-Rubio M, Svobodovà T, Mairal C, McNeil N, Keegan A, Saeed MN, Abbas MS, El-Shahawi AS, Bashammakh AO, Alyoubi CK, Sullivan O. Ultrasensitive, rapid and inexpensive detection of DNA using paper based lateral flow assay. Sci Rep. 2016;6:1–10. doi: 10.1038/srep37732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiao Y, Jiang J, Wu Y, Xia Z. Rapid detection of Cucumber green mottle mosaic virus in watermelon through a recombinase polymerase amplification assay. J Virol Methods. 2019;270:146–149. doi: 10.1016/j.jviromet.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 12.Kapoor R, Srivastava N, Kumar S, Saritha RK, Sharma SK, Baranwal VK. Development of a recombinase polymerase amplification assay for the diagnosis of banana bunchy top virus in different banana cultivars. Arch Virol. 2017;162:2791–2796. doi: 10.1007/s00705-017-3399-9. [DOI] [PubMed] [Google Scholar]
- 13.Lockhart BEL, Kirtisak KA, Jones P, Padmini DD, Olszieewski NE, Lockhart N, Nuarchan D, Sangalang J. Identification of Piper yellow mottle virus, a mealy bug transmitted badnavirus infecting Piper spp. in South East Asia. Eur J Plant Pathol. 1997;103:303–311. doi: 10.1023/A:1008699414536. [DOI] [Google Scholar]
- 14.Londono MA, Harmon CL, Polston JE. Evaluation of recombinase polymerase amplification for detection of begomoviruses by plant diagnostic clinics. J Virol. 2016;13:1–9. doi: 10.1186/s12985-016-0504-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mekuria TA, Zhang S, Eastwell KC. Rapid and sensitive detection of Little cherry virus 2 using isothermal reverse transcription-recombinase polymerase amplification. J Virol Methods. 2014;205:24–30. doi: 10.1016/j.jviromet.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 16.Satya P, Mitra S, Ray DP, Mahapatra BS, Karan M, Jana S, Sharma AK. Rapid and inexpensive NaOH based direct PCR for amplification of nuclear and organelle DNA from ramie (Boehmeria nivea), a bast fibre crop containing complex polysaccharides. Ind Crop Prod. 2013;50:532–536. doi: 10.1016/j.indcrop.2013.07.049. [DOI] [Google Scholar]
- 17.Siju S, Madhubala R, Bhat AI. Sodium sulphite enhances RNA isolation and sensitivity of cucumber mosaic virus detection by RT-PCR in black pepper. J Virol Methods. 2007;141:107–110. doi: 10.1016/j.jviromet.2006.11.032. [DOI] [PubMed] [Google Scholar]
- 18.Silva G, Bömer M, Nkere C, Kumar PL, Seal SE. Rapid and specific detection of Yam mosaic virus by reverse-transcription recombinase polymerase amplification. J Virol Methods. 2015;222:138–144. doi: 10.1016/j.jviromet.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y, Qin X, Song Y, Zhang W, Hu G, Dou Y, Li Y, Zhang Z. Development of real-time and lateral flow strip reverse transcription recombinase polymerase amplification assays for rapid detection of peste des petits ruminants virus. J Virol. 2017;14:1–10. doi: 10.1186/s12985-016-0669-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S, Ravelonandro M, Russell P, McOwen N, Briard P, Bohannon S, Vrient A. Rapid diagnostic detection of plum pox virus in prunus plants by isothermal amplify RP® using reverse transcription-recombinase polymerase amplification. J Virol Methods. 2014;207:114–120. doi: 10.1016/j.jviromet.2014.06.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.