Abstract
Background:
Response to inhaled corticosteroids is highly variable, and the association between DNA methylation and treatment response is not known.
Objective:
To examine the association between peripheral blood DNA methylation and inhaled corticosteroid response in children with persistent asthma.
Methods:
Epigenome-wide DNA methylation was analysed in individuals on inhaled corticosteroids in three independent and ethnically diverse cohorts—Childhood Asthma Management Program (CAMP); Children, Allergy, Milieu, Stockholm, Epidemiology (BAMSE); and Genetic Epidemiology of Asthma in Costa Rica Study (GACRS). Treatment response was evaluated using two definitions, the absence of emergency department visits and/or hospitalizations and the absence oral corticosteroid use while on inhaled corticosteroid therapy. CpG sites meeting nominal significance (P < 0.05) for each outcome were combined in a three-cohort meta-analysis with adjustment for multiple testing. DNA methylation was correlated with gene expression using Pearson and partial correlations.
Results:
In 154 subjects from CAMP, 72 from BAMSE, and 168 from GACRS, relative hypomethylation of cg00066816 (171 bases upstream of IL12B) was associated with the absence of emergency department visits and/or hospitalizations (Q = 0.03) in all cohorts and lower IL12B expression (ρ = 0.34, P = 0.01) in BAMSE. Relative hypermethylation of cg04256470 (688 bases upstream of CORT) was associated with the absence of oral corticosteroid use (Q = 0.04) in all cohorts and higher CORT expression (ρ = 0.20, P = 0.045) in CAMP.
Conclusion and Clinical Relevance:
Differential DNA methylation of IL12B and CORT are associated with inhaled corticosteroid treatment response in persistent childhood asthmatics. Pharmaco-methylation can identify novel markers of treatment sensitivity in asthma.
Clinical trial registration:
Childhood Asthma Management Program (CAMP) Phases I (Trial), II (CAMPCS), III (CAMPCS/2) and IV (CAMPCS/3) NCT00000575.
Keywords: asthma, DNA methylation and gene expression, paediatrics, pharmacogenetics
1 |. INTRODUCTION
Asthma continues to be the leading cause of emergency room visits, hospitalizations and school absenteeism in children.1 Inhaled corticosteroids (ICS) are the most effective long-term medication for asthma, yet up to one-third of patients have a poor response to treatment.2 Several genes have been associated with ICS treatment response, but none so far have been translated into reliable prognostic tools or therapeutic targets.3,4
Epigenetics can play a role in drug sensitivity and resistance.5–10 DNA methylation is an epigenetic mechanism that regulates both the activation and silencing of DNA transcription. Although DNA methylation has been implicated as a modifier of both asthma susceptibility and severity, the role of DNA methylation as a modifier of ICS treatment response in asthma has not been studied.11–13 In vitro studies have demonstrated that site-specific CpG methylation regulates dexamethasone sensitivity and resistance in human endothelial cells, supporting a role for DNA methylation in ICS pharmacogenetics.14 A small pilot study examined DNA methylation patterns in 33 paediatric asthmatics during asthma exacerbations and detected an association between methylation of the OTX2 promoter in nasal epithelial cells and oral corticosteroid (OCS) response.15 However, this association was not significant after adjustment for multiple testing, and the DNA methylation mark was not associated with OTX2 expression.15 Given the known complexity of ICS responsiveness in asthma, studies of substantially large sample size are needed to confidently explore a pharmaco-epigenetic hypothesis.
To address this, we now explore the relationship between DNA methylation and ICS response in children with persistent asthma. DNA methylation was studied in three independent and ethnically diverse cohorts—the Childhood Asthma Management Program (CAMP); the Children, Allergy, Milieu, Stockholm, Epidemiology (BAMSE); and the Genetic Epidemiology of Asthma in Costa Rica Study (GACRS). We further characterized the potential functional role of DNA methylation by correlation with gene expression.
2 |. METHODS
2.1 |. Study populations
Epigenome-wide DNA methylation analysis was performed in three independent cohorts—CAMP, BAMSE, and GACRS. A total of 394 subjects were included, 154 from CAMP, 72 from BAMSE, and 168 from GACRS. CAMP was a 4-year randomized, placebo-controlled trial of inhaled treatments for mild-to-moderate persistent childhood asthma in North America.16 In CAMP, asthma was defined as having at least one of the following findings for at least 6 months in the prior year: (a) asthma symptoms at least two times per week; (b) at least two usages per week of an inhaled bronchodilator; (c) daily asthma medication. Written informed consent was provided by all parents, with subject assent, and the institutional review boards (IRBs) of the Brigham and Women’s Hospital and each of the CAMP study centres approved the study (1999P0015492). BAMSE was a prospective, population-based cohort study on the risk factors for childhood asthma and allergic diseases in Stockholm, Sweden.17 In BAMSE, asthma was defined as having a positive history to two of the following three criteria: (a) doctor’s diagnosis of asthma; (b) asthma medication use in the past 12 months; (c) asthma symptoms (ie, wheezing, chest tightness, dyspnoea) in the past 12 months. BAMSE was approved by the Regional Ethical Review Board of the Karolinska Institutet (dnr 02–420) and written informed consent was obtained from the parents of all participating children. The GACRS was a cross-sectional study focused on an isolated Hispanic population with high asthma prevalence in the Central Valley of Costa Rica.18 In GACRS, asthma was defined as having a history of physician-diagnosed asthma and at least two respiratory symptoms or a history of asthma attacks in the previous year. Written parental consent and written subject assent were obtained for all GACRS participants. This study was approved by the IRBs of the Hospital Nacional de Niños (San José, Costa Rica) and Brigham and Women’s Hospital (2000P001130). Details on the study populations can be found in the Supporting information.
2.2 |. Inhaled corticosteroid response
The primary outcome was the absence of severe exacerbations while on ICS treatment, as defined by the absence of asthma-related emergency department (ED) visits and/or hospitalizations in the prior year. This outcome was measured in all three cohorts. The secondary outcome was the absence of asthma-related OCS use in the prior year. The secondary outcome was measured in the CAMP and GACRS cohorts but not in the BAMSE cohort.
2.3. DNA methylation
Peripheral blood DNA methylation analysis was performed using the Infinium HumanMethylation27 BeadChip assay (Illumina) in CAMP and the Infinium HumanMethylation450 BeadChip assay (Illumina) in BAMSE and GACRS. Standard quality control processing was performed in each cohort and described in the Supporting information. R software (version 3.4.0) was used for data analyses. In CAMP, the percentages of white blood cell counts measured from whole blood were used for adjustment of white blood cell composition in sub-sequent statistical models. The reference-based Houseman method was used to estimate the composition of white blood cells in BAMSE and GACRS.19
2.4 |. Gene expression
The association between DNA methylation and gene expression was investigated for the statistically significant CpG sites with the same direction of effect in the CAMP and BAMSE cohorts. Gene expression data from CAMP and BAMSE are described in prior publications and in the Supporting information.20,21 Only subjects with both DNA methylation and gene expression data were analysed. In CAMP, 109 subjects had both DNA methylation and gene expression data, and in BAMSE, 58 subjects had both DNA methylation and gene expression data.
2.5. Statistical analyses
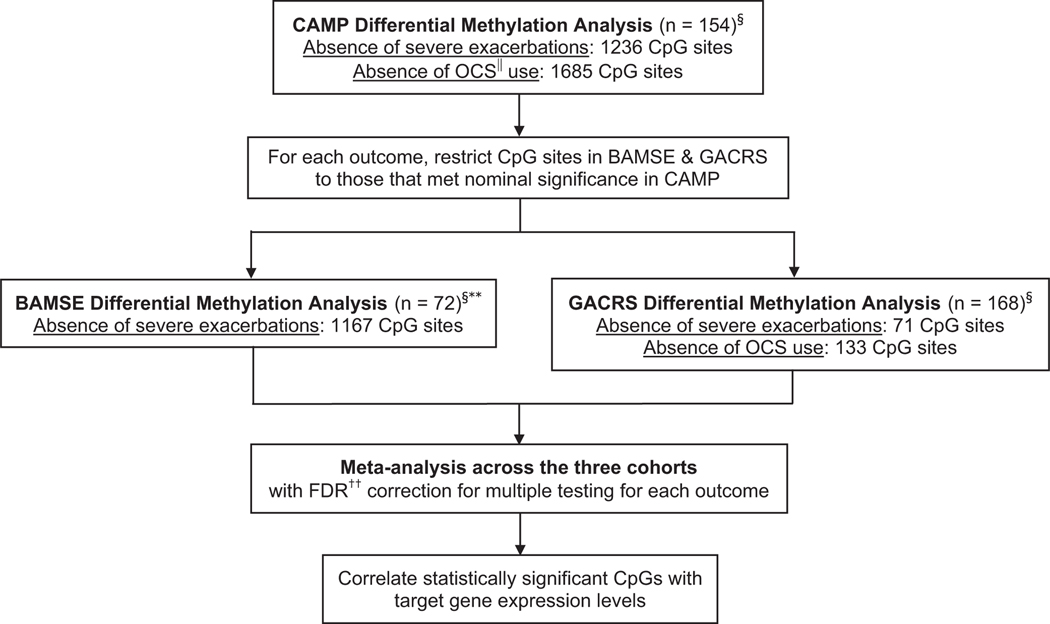
For each outcome, multivariable linear regression models were first generated in the CAMP DNA methylation dataset using the iCheck package (Figure 1).22 CAMP was used as the discovery cohort because it was the most phenotypically well characterized and had both ICS response outcomes. Next, the BAMSE and GACRS datasets were restricted to the CpG sites that met nominal significance in CAMP (P < 0.05) for each outcome and multivariable linear regression modelling was performed. A sequential analysis was performed because CAMP by design interrogated less CpG sites than BAMSE or GACRS. Prior to model building, DNA methylation beta values were converted to M values to correct for heteroscedasticity.23 The models are described in detail in the Supporting information. The CpG sites that met nominal significance in each cohort were combined in a meta-analysis using Fisher’s method for each respective outcome, and correction for multiple testing was performed by controlling the false discovery rate (FDR).24
FIGURE 1.
DNA methylation analysis of inhaled corticosteroid response in the CAMPa, BAMSEb, and GACRSc. aChildhood Asthma Management Program. bSwedish abbreviation for Children, Allergy, Milieu, Stockholm, Epidemiology. cGenetic Epidemiology of Asthma in Costa Rica Study. §The number of differentially methylated CpG sites for each outcome that met nominal P < 0.05 are shown. ||Oral corticosteroid. **The absence of oral corticosteroid use outcome was not measured in BAMSE. ††False discovery rate
For the statistically significant CpG sites on meta-analyses, normalized DNA methylation M values were correlated with log2 transformed and normalized gene expression using Pearson correlation. In addition, partial correlations between gene expression and DNA methylation controlling for the respective ICS treatment response outcome were performed. The partial correlation models are described in the Supporting information.
To determine if the effect of differential methylation on asthma exacerbation was specific to subjects on ICS treatment, we performed multivariable linear regression interaction modelling. Interaction models were generated for only the CpG sites that correlated with gene expression. These models were generated in CAMP because unlike BAMSE and GACRS, CAMP has DNA methylation data for subjects on ICS and placebo treatment (Table S1), and the primary interest was the interaction between treatment group and the respective asthma exacerbation outcome. Each model controlled for age, sex, batch effect, treatment group, and white blood cell count.
3 |. RESULTS
The mean age was 9.8 (±2.0) years in CAMP, 8.3 (±0.4) years in BAMSE, and 9.2 (±2.0) years in GACRS (Table 1). All three cohorts were predominantly male, consistent with other childhood asthma cohorts. In CAMP, BAMSE, and GACRS, 89.0%, 59.7%, and 23.8% of the respective subjects did not have ED visits and/or hospitalizations in the prior year while on ICS therapy. In CAMP and GACRS, 54.4% and 35.7% of the respective subjects did not have OCS use in the prior year while on ICS therapy.
TABLE 1.
| CAMP (n = 154) | BAMSE (n = 72) | GACRS (n = 168) | |
|---|---|---|---|
| Age (mean ± SD) | 9.8 (±2.0) | 8.3 (±0.4) | 9.2 (±2.0) |
| Age at asthma onset (median ± IQR) | 2.0 (±3.0) | 4.0 (±2.7) | 2.0 (±3.2) |
| Gender | |||
| Female | 64 (41.6%) | 26 (36.1%) | 69 (41.1%) |
| Male | 90 (58.4%) | 46 (63.9%) | 99 (58.9%) |
| EDd visit and/or hospitalization in the past year | |||
| No | 137 (89.0%) | 43 (59.7%) | 40 (23.8%) |
| Yes | 17 (11.0%) | 29 (40.3%) | 128 (76.2%) |
| Oral corticosteroid use in the past year | |||
| No | 80 (54.4%) | - | 60 (35.7%) |
| Yes | 67 (45.6%) | - | 108 (64.3%) |
| History of atopy | |||
| No | 113 (73.4%) | 25 (34.7%) | 18 (14.2%) |
| Yes | 41 (26.6%) | 47 (65.3%) | 109 (85.8%) |
| History of parental smokinge | |||
| No | 101 (66.0%) | 63 (87.5%) | 132 (79.5%) |
| Yes | 52 (34.0%) | 9 (12.5%) | 34 (20.5%) |
| History of parental asthma | |||
| No | 78 (53.1%) | 41 (60.3%) | 90 (53.9%) |
| Yes | 69 (46.9%) | 27 (39.7%) | 77 (46.1%) |
| IgE sensitizationf | 133 (86.4%) | 52 (72.2%) | 123 (73.7%) |
Childhood Asthma Management Program.
Swedish abbreviation for Children, Allergy, Milieu, Stockholm, Epidemiology.
Genetic Epidemiology of Asthma in Costa Rica Study.
Emergency department.
In BAMSE, history of parental smoking was defined as positive if the mother smoked at least one cigarette per day at the time of the baseline questionnaire and/or smoked at least one cigarette per day at any point of time during the pregnancy.
In CAMP, IgE sensitization was defined as any positive aeroallergen skin prick test. In BAMSE and GACRS, IgE sensitization was defined as any positive aeroallergen and/or food specific IgE (≥0.35 kU/L).
3.1 |. Absence of severe exacerbations on ICS treatment
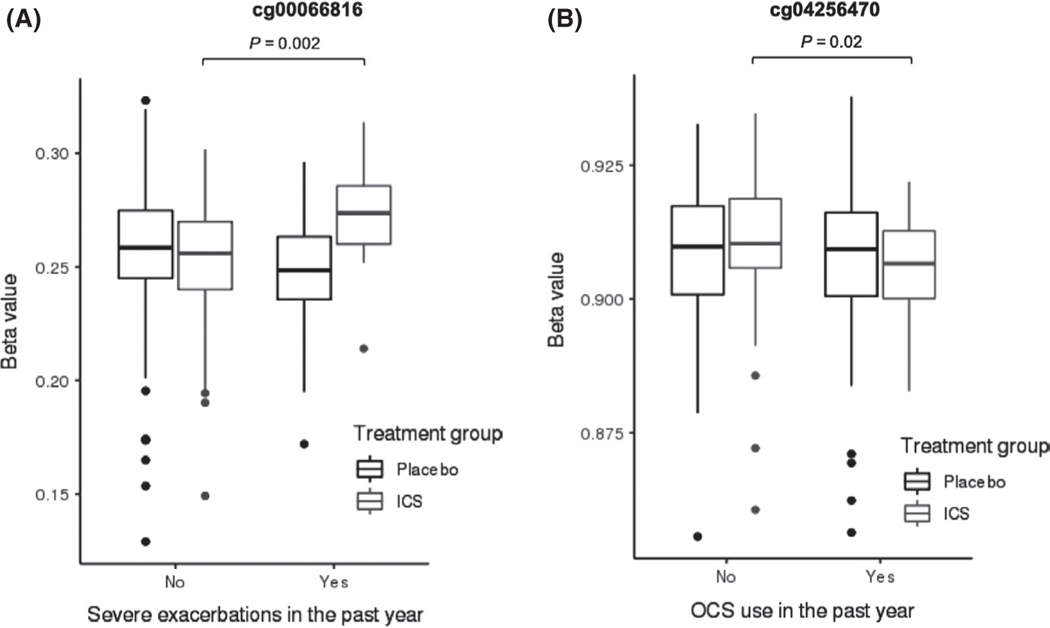
For the primary outcome of the absence of severe exacerbations, 1236 CpG sites in CAMP, 1167 CpG sites in BAMSE, and 71 CpG sites in GACRS were differentially methylated (nominal P < 0.05). On meta-analysis of the three cohorts, 42 CpG sites had the same direction of effect (Table S2), and one CpG site was significant after FDR adjustment (Table 2). Relative hypomethylation of cg00066816 was associated with the absence of ED visits and/or hospitalizations in the past year (standardized coefficient in CAMP –3.101, BAMSE –2.953, and GACRS −2.310; Q = 0.03) (Table 2, Figure S1). Interaction analysis demonstrated that the association between hypomethylation of cg00066816 and the absence of severe exacerbations was specific to the subjects on ICS (standardized coefficient –3.051, P = 0.002) (Figure 2A).
TABLE 2.
Differentially methylated CpG sites associated with inhaled corticosteroid response in the CAMP, BAMSE, and GACRS cohortsa
| CpG | Gene(s) | CAMP standardized coefficient | BAMSE standardized coefficient | GACRS standardized coefficient | CAMP P value (nominal) | BAMSE P value (nominal) | GACRS P value (nominal) | Meta-analysis Q value (FDR-adjusted) |
|---|---|---|---|---|---|---|---|---|
| Absence of severe exacerbationsb,c: | ||||||||
| cg00066816 | IL12B | −3.101 | −2.953 | −2.310 | 0.002 | 0.003 | 0.021 | 0.028 |
| Absence of asthma-related oral corticosteroid used: | ||||||||
| cg00557354 | ARHGEF7 | −3.490 | - | −2.238 | 0.001 | - | 0.027 | 0.035 |
| cg04256470 | CORT, CENPS | 3.620 | - | 2.329 | <0.001 | - | 0.021 | 0.035 |
| cg09495977 | HTRA3 | −2.420 | - | −3.466 | 0.017 | - | 0.001 | 0.035 |
| cgl2333095 | ANKRD13A | −3.485 | - | −2.336 | 0.001 | - | 0.021 | 0.035 |
| cgl3818573 | C1QL1 | -3.596 | - | −2.163 | <0.001 | - | 0.032 | 0.035 |
| cg21589280 | DDAH1 | −3.063 | - | −2.786 | 0.003 | - | 0.006 | 0.035 |
| cg03080985 | SH3BGRL2 | −3.077 | - | −2.458 | 0.003 | - | 0.015 | 0.039 |
| cg04330449 | NEUROG1 | −2.646 | - | −2.921 | 0.009 | - | 0.004 | 0.039 |
| cg05307923 | ADARB2 | −2.577 | - | −2.959 | 0.011 | - | 0.004 | 0.039 |
| cg08724517 | MAP9 | 2.951 | - | 2.653 | 0.004 | - | 0.009 | 0.039 |
| cgll665562 | PSMC1 | −3.250 | - | −2.275 | 0.001 | - | 0.024 | 0.039 |
| cgl4269514 | OAZ3, MRPL9 | −3.112 | - | −2.246 | 0.002 | - | 0.026 | 0.043 |
| cg24322623 | MYOD1 | −2.964 | - | −2.386 | 0.004 | - | 0.018 | 0.044 |
The binary outcome of interest was the absence of exacerbations. The presence of exacerbations was the reference level, measure respectively as severe exacerbations or oral corticosteroid use. All models controlled for age, sex, batch effect, and white blood cell composition.
CAMP analysis for the absence of asthma-related emergency department visits and/or hospitalizations included adjustment for history of atopy.
BAMSE analysis for the absence of asthma-related emergency department visits and/or hospitalizations included adjustment for history of atopy and IgE sensitization.
CAMP analysis for the absence of asthma-related oral corticosteroid use included adjustment for history of parental asthma.
FIGURE 2.
DNA methylation is a pharmaco-epigenetic marker of inhaled corticosteroid treatment responseab. A, Cg00066816 hypomethylation was associated with the absence of severe exacerbations only in subjects on inhaled corticosteroid treatment compared to placebo (standardized coefficient −3.051, P = 0.002). B, Cg04256470 hypermethylation was associated with the absence of oral corticosteroid only in subjects on inhaled corticosteroid treatment compared to placebo (standardized coefficient 2.322, P = 0.02). aThe analyses were performed using DNA methylation M values. β values are displayed for easier biologic interpretability. bInteraction analyses were performed only in CAMP because BAMSE and GACRS did not have DNA methylation data available on subjects on inhaled corticosteroids and placebo
3.2 |. Absence of OCS use on ICS treatment
For the secondary outcome of the absence of OCS use, 1685 CpG sites in CAMP and 133 CpG sites in GACRS were differentially methylated (nominal P < 0.05). The BAMSE cohort did not have data on OCS use. On meta-analysis of the CAMP and GACRS cohorts, 62 CpG sites had the same direction of effect (Table S3), and 13 CpG sites were significant after FDR adjustment (Table 2, Figure S2). Relative hypermethylation of cg04256470 was associated with the absence of OCS use while on ICS (standardized coefficient in CAMP 3.620 and GACRS 2.329; Q = 0.04), and this effect was specific to the subjects on ICS treatment on interaction analysis (standardized coefficient 2.322, P = 0.02) (Figure 2B).
3.3 |. Functional annotation of the identified CpG sites
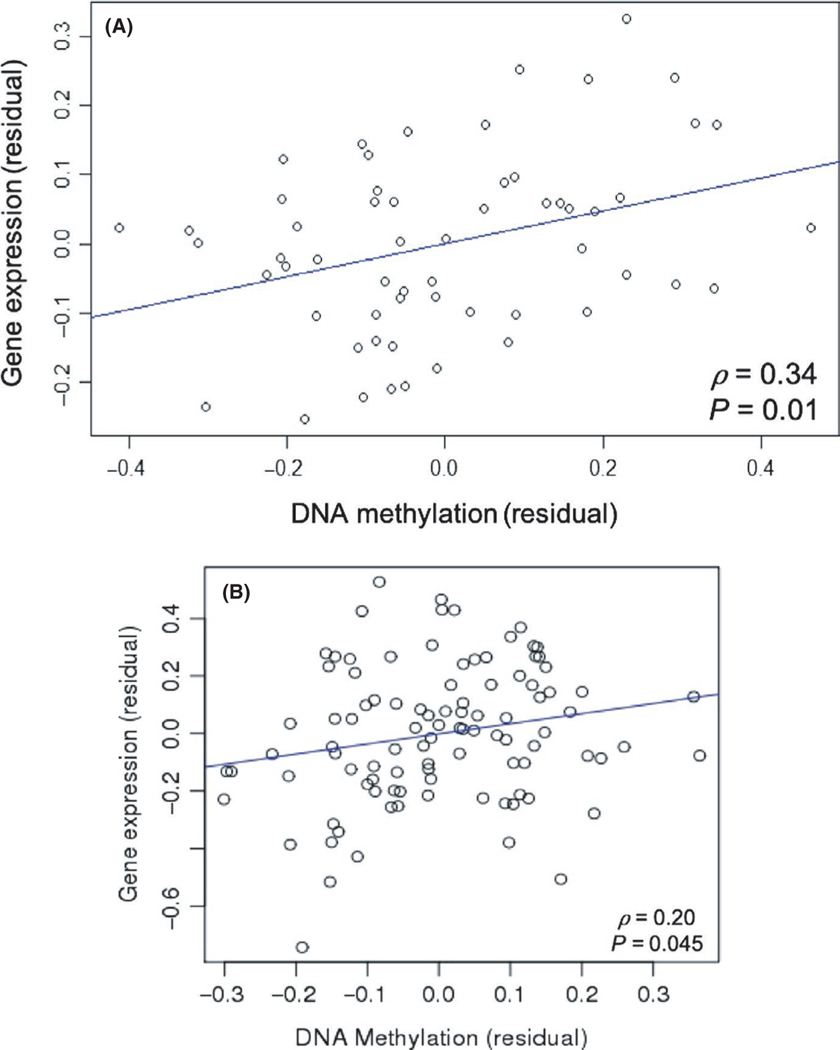
The CpG site associated with severe exacerbations (cg00066816) maps to two genes, the interleukin 12B gene (IL12B) and the long non-coding RNA LOC285626 on chromosome five. Cg00066816 is located 171 bases upstream of the IL12B transcription start site. Relative hypomethylation of cg00066816 was associated with lower IL12B expression in BAMSE (ρ = 0.34, P = 0.01) (Figure 3A). No association between cg00066816 methylation and IL12B expression was detected in CAMP.
FIGURE 3.
DNA methylation is associated with gene expression in the BAMSE and CAMP cohorts. A, Partial correlation between DNA methylation and gene expression controlling for emergency department visits and/or hospitalizations in the prior year while on inhaled corticosteroid therapy, in addition to age, sex, batch effect, and cell type composition, in BAMSE (ρ = 0.34, P = 0.01). B, Partial correlation between DNA methylation and gene expression controlling for oral corticosteroid use in the prior year while on inhaled corticosteroid therapy, in addition to age, sex, and batch effect, in CAMP (ρ = 0.20, P = 0.045)
Of the 13 CpG sites significantly associated with the absence of OCS use while on ICS treatment, only one CpG site (cg04256470) was associated with gene expression. Cg04256470 was positively correlated with CORT expression (ρ = 0.20, P = 0.045) in CAMP (Figure 3B). Only a small number of subjects in CAMP had gene expression cell type composition data available. Therefore, after adjustment for cell type composition, the partial correlation analysis was underpowered and not statistically significant (Figure S3). Cg04256470 methylation was not associated with gene expression in BAMSE. Cg04256470 is located 688 bases upstream of the CENPS-CORT transcription start site on chromosome one. The CENPS-CORT locus has read-through transcription of the centromere protein S (CENPS) gene and cortistatin (CORT) gene, and alternative splicing produces transcript variants of the genes.
4 |. DISCUSSION
We found DNA methylation patterns to be associated with response to ICS treatment in three independent and ethnically diverse cohorts of children with mild-to-moderate persistent asthma. Response to ICS was investigated using two definitions, the absence of severe exacerbations as measured by the absence of asthma-related ED visits and/or hospitalizations and the absence of OCS use for asthma while on ICS treatment. Hypomethylation of cg00066816 (IL12B) and hypermethylation of cg04256470 (CORT) were associated with response to ICS using the severe exacerbation and OCS use definitions, respectively. In addition, hypomethylation of cg00066816 was associated with lower IL12B expression, and hypermethylation of cg04256470 was associated with higher CORT expression. IL12B is involved in asthma pathogenesis and CORT regulates endogenous corticosteroids.25,26 Our results provide novel evidence on the association of pharmaco-methylation with ICS treatment resistance and response. The pharmaco-epigenetic aspect of these associations is further highlighted by the interaction analyses demonstrating differential DNA methylation in the ICS but not the placebo treatment groups (Figure 2).
Hypomethylation of IL12B was associated with the absence of severe exacerbations and decreased IL12B expression among children with asthma on ICS IL12B encodes the common beta subunit of interleukin (IL) 12 and IL-23. Depletion of IL-12 attenuates T helper (TH) 1 immune response and enhances TH2 antigen-induced airway hyperresponsiveness and pulmonary eosinophilia.27,28 In bronchial biopsies, asthmatics sensitive to prednisone have upregulation of IL-12 and downregulation of IL-13 expression, whereas prednisone-resistant asthmatics have no change in expression of either cytokine.29 These findings in lung tissue contrast with our findings in peripheral blood, and this difference may be due to the effects of different steroid concentrations in the lung versus the periphery after different routes of glucocorticoid exposure (oral versus inhaled), in addition to baseline differences in tissue-specific gene expression. IL-23 regulates TH17 differentiation, and stimulation with IL-23 in vitro protects human lung fibroblasts and endothelial cells from dexamethasone-induced apoptosis.30 IL-23 also weakens dexamethasone-induced inhibition of peripheral blood mononuclear cell proliferation.31 Therefore, IL-23 may have a role in corticosteroid resistance, and the reduction of IL-23 expression via hypomethylation of the IL12B subunit may be a mechanism for corticosteroid sensitivity.
Hypermethylation of CORT was associated with the absence of OCS use while on ICS and increased CORT expression. CORT encodes the neuropeptide cortistatin, which has high structural homology to somatostatin.25,26 Cortistatin regulates the hypothalamic-pituitary-adrenal axis and exerts antiinflammatory actions.25,26 In the human immune system, cortistatin production is upregulated in activated monocytes, macrophages, and dendritic cells, along with expression of its receptor sst2, suggesting an autocrine regulatory pathway.32 Cortistatin is also expressed by peripheral neurons, which in turn may have a paracrine role in regulating immune cells.32 In a mouse model of allogeneic skin transplantation, cortistatin treatment increased the numbers of regulatory T cells and prolonged survival of transplanted skin grafts.33 In experimental models of sepsis, cortistatin inhibited the release of local and systemic inflammatory cytokines (TNFα, IL-6, IL-1β, IFNγ, IL-12) and chemokines (MIP-2, RANTES), increased production of antiinflammatory IL-10, and protected against mortality in endotoxemic mice.34 Finally, cortistatin deficient mice are in a systemically immunosuppressed state with elevated basal levels of the glucocorticoid corticosterone and increased susceptibility to infection.35 Our data suggest that regulation of cortistatin by DNA methylation may modulate ICS sensitivity in childhood asthmatics.
In nasal epithelial cells, hypomethylation of OTX2 has been nominally associated with response to OCS in paediatric asthmatics during exacerbations, and interestingly, in the GACRS cohort, relative hypomethylation of OTX2 in peripheral blood was nominally associated with the absence of OCS use while on ICS therapy (standardized coefficient −2.120, P = 0.04).15 Hence, differential methylation of OTX2 is a potential marker of both OCS and ICS response, and larger studies with tissue-specific analyses are needed to confirm this association. In a prior study, differential methylation of the glucocorticoid receptor gene (NR3C1) was associated with dexamethasone sensitivity in endothelial cells, but we did not find an association of NR3C1 methylation with ICS response in peripheral blood.14 In addition, differential methylation of genetic loci previously associated with asthma exacerbations on ICS treatment (ST13, FCER2, P2RX7, CMTR1) was not detected.36 This absence of association may be due to phenotypic heterogeneity of the measured outcomes or a lack of methylation quantitative trait association at these genetic loci. Further research is needed to determine the association between genetic variation and quantitative changes in DNA methylation in asthma pharmacogenomics.
In this study, we demonstrate the first association between DNA methylation and response to ICS treatment in asthma across three independent populations of different ethnicities. Paediatric cohorts with drug treatment response outcomes and DNA methylation are few and with small sample sizes. The discovery of CpG sites in this study was limited by the small cohort sizes, but we were adequately powered to identify novel CpG sites by meta-analysing our results across cohorts. The variation in the rate of ED visits and/or hospitalizations and OCS use across the cohorts may reflect healthcare system differences and/or compliance, which are factors that may have influenced our results. Nonetheless, BAMSE and GACRS have successfully been used as replication cohorts for CAMP.37,38 CAMP and BAMSE are white cohorts, whereas GACRS is a Hispanic cohort. Racial differences in DNA methylation may explain the low numbers of replicated CpG sites in GACRS.39 However, the inclusion of replication in GACRS increases the global generalizability of our results. The number of CpG sites investigated was limited by the use of the Illumina 27 k methylation array in CAMP, which is enriched at CpG island promoters, and replication was performed in the corresponding subset of CpGs from BAMSE and GACRS. Therefore, CpGs involved in ICS response located outside of CpG island promoters may have been missed, and additional DNA methylation marks would likely have been found had more CpG sites been examined using newer technologies. Future studies interrogating a larger number of genome-wide CpGs are needed to identify the full extent of how DNA methylation is associated with ICS response.
DNA methylation and gene expression are tissue-specific, and peripheral blood DNA methylation and gene expression were examined because it is non-invasive and asthma inflammation and ICS absorption are detected systemically. Cell type deconvolution was performed to reduce false-positive results due to cell type heterogeneity in whole blood.40 Each identified CpG site was located far upstream of the respective gene, suggesting that methylation may be acting on an upstream gene modifier or that collaborating epigenetic modifications, such as histone modifications, may be contributing to the regulation of gene expression of both IL12B and CORT.41–46 The statistically significant CpG sites each mapped to more than one gene, but there is little known about these additional genes (LOC285626 and CENPS), including no known association with asthma. Gene expression data were collected after DNA methylation data in both CAMP and BAMSE, which may explain why the associations between DNA methylation and gene expression were not replicated across both cohorts. Lastly, as with most pharmacogenomic studies, we were not able to determine if DNA methylation preceded or resulted from ICS treatment. There is evidence that high-dose systemic glucocorticoids can induce DNA methylation changes in the buccal mucosal and peripheral blood, but it is not known if ICS exposure can mediate systemic changes in DNA methylation.47,48 Nonetheless, our results demonstrate that DNA methylation is a marker of treatment response in subjects on ICS. Future longitudinal studies are needed to understand the temporality and underlying mechanisms linking DNA methylation and ICS response.
There is an urgent need to understand the biologic basis for ICS response and resistance in order to develop new biomarkers and therapies for ICS resistant asthmatics. We identified DNA methylation marks in IL12B and CORT to be consistently associated with ICS response in persistent childhood asthmatics across three independent cohorts. Pharmaco-methylation is a novel approach in asthma research with promising applications across medicine. Our findings demonstrate its potential to identify treatment sensitivity in asthmatics. Epigenetic therapeutics that selectively target DNA methylation have been shown to increase the effectiveness of existing treatments in the field of cancer biology, and these therapeutics may have applications in other fields and diseases, including asthma.7,8,10 Further research into the identified DNA methylation marks may lead to the discovery of epigenetically mediated pathways of drug resistance and novel epigenetic therapeutics to increase sensitivity to ICS treatment.
Supplementary Material
ACKNOWLEDGEMENTS
The BAMSE team would like to thank Professors Jean Bousquet and Josep Maria Antó for coordinating the MeDALL project and Professor Charles Auffray and his team for generating the transcriptomics data in BAMSE.
Funding information:
NIH T32 AI007306, T32 HL007427, RC2 HL101543, R01 HL127332, R01 HL129935, U01 HL65899, P01 HL132825. BAMSE: The Swedish Heart-Lung Foundation, The Swedish Research Council, Stockholm County Council (ALF), MeDALL (Mechanisms of the Development of ALLergy; European Union grant agreement No. 261357) and the Swedish foundation for strategic research (SSF) (RBc08–0027).
Footnotes
DATA ACCESSIBILITY
Phenotypes for the CAMP cohort are available through the EVE Asthma Genetics Consortium (dbGaP Study Accession: phs001156. v2.p1). In addition, phenotype and DNA methylation data for CAMP and GACRS are available through the NHLBI Trans-Omics for Precision Medicine (TOPMed).49 Gene expression data for CAMP is available through the Asthma BioRepository for Integrative Genomic Exploration (Asthma BRIDGE).20 DNA methylation data for BAMSE are available through the European Genome-phenome Archive (EGA Study Accession: EGAS00001002746).
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
REFERENCES
- 1.Akinbami LJ, Moorman JE, Liu X. Asthma prevalence, health care use, and mortality: United States, 2005–2009. Natl Health Stat Report. 2011;32:1–14. [PubMed] [Google Scholar]
- 2.Szefler SJ, Martin RJ, King TS, et al. Significant variability in response to inhaled corticosteroids for persistent asthma. J Allergy Clin Immunol. 2002;109(3):410–418. [DOI] [PubMed] [Google Scholar]
- 3.Tantisira KG, Lake S, Silverman ES, et al. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum Mol Genet. 2004;13(13):1353–1359. [DOI] [PubMed] [Google Scholar]
- 4.Tantisira KG, Lasky-Su J, Harada M, et al. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N Engl J Med. 2011;365(13):1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powell TR, Smith RG, Hackinger S, et al. DNA methylation in interleukin-11 predicts clinical response to antidepressants in GENDEP. Transl Psychiatry. 2013;3:e300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeuchi N, Nonen S, Kato M, et al. Therapeutic response to paroxetine in major depressive disorder predicted by DNA methylation. Neuropsychobiology. 2017;75(2):81–88. [DOI] [PubMed] [Google Scholar]
- 7.Plumb JA, Strathdee G, Sludden J, Kaye SB, Brown R. Reversal of drug resistance in human tumor xenografts by 2’-deoxy-5-azacytidine-induced demethylation of the hMLH1 gene promoter. Cancer Res. 2000;60(21):6039–6044. [PubMed] [Google Scholar]
- 8.Shen L, Kondo Y, Ahmed S, et al. Drug sensitivity prediction by CpG island methylation profile in the NCI-60 cancer cell line panel. Cancer Res. 2007;67(23):11335–11343. [DOI] [PubMed] [Google Scholar]
- 9.Steele N, Finn P, Brown R, Plumb JA. Combined inhibition of DNA methylation and histone acetylation enhances gene re-expression and drug sensitivity in vivo. Br J Cancer. 2009;100(5): 758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha YN, Sung HY, Yang SD, Chae YJ, Ju W, Ahn JH. Epigenetic modification of alpha-N-acetylgalactosaminidase enhances cisplatin resistance in ovarian cancer. Korean J Physiol Pharmacol. 2018;22(1):43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brook PO, Perry MM, Adcock IM, Durham AL. Epigenome-modifying tools in asthma. Epigenomics. 2015;7(6):1017–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu C-J, Söderhäll C, Bustamante M, et al. DNA methylation in childhood asthma: an epigenome-wide meta-analysis. Lancet Respir Med. 2018;6(5):379–388. [DOI] [PubMed] [Google Scholar]
- 13.Gaffin JM, Raby BA, Petty CR, et al. beta-2 adrenergic receptor gene methylation is associated with decreased asthma severity in inner-city schoolchildren: asthma and rhinitis. Clin Exp Allergy. 2014;44(5):681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mata-Greenwood E, Jackson PN, Pearce WJ, Zhang L. Endothelial glucocorticoid receptor promoter methylation according to dexamethasone sensitivity. J Mol Endocrinol. 2015;55(2):133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Biagini Myers JM, Yadagiri VK, et al. Nasal DNA methylation differentiates corticosteroid treatment response in pediatric asthma: a pilot study. PLoS ONE. 2017;12(10):e0186150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The childhood asthma management program (CAMP): design, rationale, and methods. Childhood asthma management program research group. Control Clin Trials. 1999;20(1):91–120. [PubMed] [Google Scholar]
- 17.Thacher JD, Gruzieva O, Pershagen G, et al. Pre- and postnatal exposure to parental smoking and allergic disease through adolescence. Pediatrics. 2014;134(3):428–434. [DOI] [PubMed] [Google Scholar]
- 18.Hunninghake GM, Soto-Quiros ME, Avila L, et al. Sensitization to Ascaris lumbricoides and severity of childhood asthma in Costa Rica. J Allergy Clin Immunol. 2007;119(3):654–661 (0091–6749 (Print)). [DOI] [PubMed] [Google Scholar]
- 19.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Croteau-Chonka DC, Qiu W, Martinez FD, et al. Gene expression profiling in blood provides reproducible molecular insights into asthma control. Am J Respir Crit Care Med. 2017;195(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gref A, Merid SK, Gruzieva O, et al. Genome-wide interaction analysis of air pollution exposure and childhood asthma with functional follow-up. Am J Respir Crit Care Med. 2017;195(10):1373–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu W, Guo B, Anderson C, Klanderman B, Carey V, Raby B. QC pipeline and data analysis tools for high-dimensional Illumina mRNA expression data. 2016:R package version 1.6.0. [Google Scholar]
- 23.Du P, Zhang X, Huang C-C, et al. Comparison of beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc: Ser B (Methodol). 1995;57(1):289–300. [Google Scholar]
- 25.Giordano R, Picu A, Bonelli L, et al. The activation of somatostatinergic receptors by either somatostatin-14 or cortistatin-17 often inhibits ACTH hypersecretion in patients with Cushing’s disease. Eur J Endocrinol. 2007;157(4):393–398. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez-Rey E, Pedreno M, Delgado-Maroto V, Souza-Moreira L, Delgado M. Lulling immunity, pain, and stress to sleep with cortistatin. Ann N Y Acad Sci. 2015;1351:89–98. [DOI] [PubMed] [Google Scholar]
- 27.Szabo SJ, Jacobson NG, Dighe AS, Gubler U, Murphy KM. Developmental commitment to the Th2 lineage by extinction of IL-12 signaling. Immunity. 1995;2(6):665–675. [DOI] [PubMed] [Google Scholar]
- 28.Keane-Myers A, Wysocka M, Trinchieri G, Wills-Karp M. Resistance to antigen-induced airway hyperresponsiveness requires endogenous production of IL-12. J Immunol. 1998;161(2):919–926. [PubMed] [Google Scholar]
- 29.Naseer T, Minshall EM, Leung DY, et al. Expression of IL-12 and IL-13 mRNA in asthma and their modulation in response to steroid therapy. Am J Respir Crit Care Med. 1997;155(3):845–851. [DOI] [PubMed] [Google Scholar]
- 30.Halwani R , Sultana A , Al-Kufaidy R , Jamhawi A , Vazquez-Tello A , Al-Muhsen S . Th-17 regulatory cytokines inhibit corticosteroid induced airway structural cells apoptosis. Respir Res. 2016;17:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vazquez-Tello A, Halwani R, Hamid Q, Al-Muhsen S. Glucocorticoid receptor-beta upregulation and steroid resistance induction by IL-17 and IL-23 cytokine stimulation in peripheral mononuclear cells. J Clin Immunol. 2013;33(2):466–478. [DOI] [PubMed] [Google Scholar]
- 32.Dalm V, van Hagen PM, van Koetsveld PM, et al. Expression of somatostatin, cortistatin, and somatostatin receptors in human monocytes, macrophages, and dendritic cells. Am J Physiol Endocrinol Metab. 2003;285(2):E344–E353. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Zhao R, Zhang F, et al. Control of allograft rejection in mice by applying a novel neuropeptide, cortistatin. Adv Ther. 2008;25(12):1331–1341. [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez-Rey E, Chorny A, Robledo G, Delgado M. Cortistatin, a new antiinflammatory peptide with therapeutic effect on lethal endotoxemia. J Exp Med. 2006;203(3):563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Souza-Moreira L, Morell M, Delgado-Maroto V, et al. Paradoxical effect of cortistatin treatment and its deficiency on experimental autoimmune encephalomyelitis. J Immunol. 2013;191(5):2144–2154. [DOI] [PubMed] [Google Scholar]
- 36.Wang AL, Tantisira KG. Personalized management of asthma exacerbations: lessons from genetic studies. Expert Rev Precis Med Drug Dev. 2016;1(6):487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bunyavanich S, Melen E, Wilk JB, et al. Thymic stromal lymphopoietin (TSLP) is associated with allergic rhinitis in children with asthma. Clin Mol Allergy. 2011;9(1). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3032752/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forno E, Lasky-Su J, Himes B, et al. Genome-wide association study of the age of onset of childhood asthma. J Allergy Clin Immunol. 2012;130(1):83–90.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adkins RM, Krushkal J, Tylavsky FA, Thomas F. Racial differences in gene-specific DNA methylation levels are present at birth. Birth Defects Res A. 2011;91(8):728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Potaczek DP, Harb H, Michel S, Alhamwe BA, Renz H, Tost J. Epigenetics and allergy: from basic mechanisms to clinical applications. Epigenomics. 2017;9(4):539–571. [DOI] [PubMed] [Google Scholar]
- 41.Yu H-B, Yurieva M, Balachander A, et al. NFATc2 mediates epigenetic modification of dendritic cell cytokine and chemokine responses to dectin-1 stimulation. Nucleic Acids Res. 2015;43(2):836–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chandran A, Antony C, Jose L, Mundayoor S, Natarajan K, Kumar RA. Mycobacterium tuberculosis Infection Induces HDACl-Mediated Suppression of IL-12B Gene Expression in Macrophages. Front Cell Infect Microbiol. 2015;5:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tong Q, Gong AY, Zhang XT, et al. LincRNA-Cox2 modulates TNF-alpha-induced transcription of Il12b gene in intestinal epithelial cells through regulation of Mi-2/NuRD-mediated epigenetic histone modifications. FASEB J. 2016;30(3):1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin J, Xie X, Xiao Y, et al. Epigenetic regulation of the expression of Il 12 and Il23 and autoimmune inflammation by the deubiquitinase Trabid. Nat Immunol. 2016;17(3):259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu J, Xu X, Wang B, et al. Nuclear carbonic anhydrase 6B associates with PRMT5 to epigenetically promote IL-12 expression in innate response. Proc Natl Acad Sci U S A. 2017;114(32):8620–8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rubio A, Sanchez-Mut JV, Garcia E, et al. Epigenetic control of somatostatin and cortistatin expression by beta amyloid peptide. J Neurosci Res. 2012;90(1):13–20. [DOI] [PubMed] [Google Scholar]
- 47.Wan ES, Qiu W, Baccarelli A, et al. Systemic steroid exposure is associated with differential methylation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012;186(12):1248–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Braun PR, Tanaka-Sahker M, Chan AC et al. Genome-wide DNA methylation investigation of glucocorticoid exposure within buccal samples. Psychiatry Clin Neurosci. 2019;73(6):323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. TOPMed whole genome sequencing project. 2017 https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/GetPdf.cgi?xml:id=phd006969.2.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.