Abstract
Purpose:
To validate a machine learning model trained on an open source dataset and subsequently optimize it to chest X-rays with large pneumothoraces from our institution.
Methods:
The study was retrospective in nature. The open-source chest X-ray (CXR8) dataset was dichotomized to cases with pneumothorax (PTX) and all other cases (non-PTX), resulting in 41946 non-PTX and 4696 PTX cases for the training set and 11120 non-PTX and 541 PTX cases for the validation set. A limited supervision machine learning model was constructed to incorporate both localized and unlocalized pathology. Cases were then queried from our health system from 2013 to 2017. A total of 159 pneumothorax and 682 non-pneumothorax cases were available for the training set. For the validation set, 48 pneumothorax and 1287 non-pneumothorax cases were available. The model was trained, a receiver operator curve (ROC) was created, and output metrics, including area under the curve (AUC), sensitivity and specificity were calculated.
Results:
Initial training of the model using the CXR8 dataset resulted in an AUC of 0.90 for pneumothorax detection. Naively inferring our own validation dataset on the CXR8 trained model output an AUC of 0.59. After re-training the model with our own training dataset, the validation dataset inference output an AUC of 0.90.
Conclusion:
Our study showed that even though you may get great results on open-source datasets, those models may not translate well to real world data without an intervening retraining process.
Keywords: Chest X-ray, machine learning, artificial intelligence, pneumothorax, neural network
1. Introduction:
Using machine learning to evaluate radiologic studies and pathology have demonstrated utility in several fields such as neuroimaging [1,2], breast imaging [3], and orthopedic imaging [4–6]. In the realm of thoracic radiology, machine learning had previously been implemented for prognosis evaluation in smokers using CT images [7] and pulmonary tuberculosis classification using chest X-rays [8]. Thoracic imaging evaluation using machine learning took a big step forward when Want et al. released the largest open-source chest X-ray dataset to date containing 14 common thoracic diseases (denoted CXR8 with 112,120 images) [9]. Using this CXR8 dataset, several studies constructed machine learning models to detect thoracic diseases using weak supervision without incorporation of localization information [9,10]. Li et al. formulated a limited supervision algorithm incorporating both localized and unlocalized pathology, improving performance on the CXR8 dataset compared to prior studies [11]. Using limited supervision was valuable since having a trained Radiologist localize pathology on every single study quickly becomes resource exhaustive.
While several studies have been published utilizing the CXR8 open source dataset [9–11], there is a paucity of studies validating these models against real medical images. In addition, while being able to detect many of the common pathologies is valuable, certain pathologies, such as large pneumothoraces, require emergent management to prevent adverse outcomes [12].
Therefore, we decided to validate a model trained on the CXR8 dataset and subsequently optimize it to chest X-rays with large pneumothoraces from our institution.
2. Materials and Methods
There are no conflicts of interest. There is no overlap in subjects from prior publications. Study was approved by the local institutional review board (IRB) with waiver of consent.
The study was retrospective in nature. The CXR8 dataset [9] was dichotomized to cases with pneumothorax (PTX) and all other cases (non-PTX). Because there were a significantly higher number of non-PTX cases compared to PTX cases, only half of the non-PTX cases were utilized. The same training and validation data split was utilized as the reference study [9], resulting in 4696 PTX and 41946 non-PTX cases for the training set and 541 PTX and 11120 non-PTX cases for the validation set. The machine learning model was constructed based on the algorithm by Li et al [11], which was built to incorporate both localized and unlocalized imaging. Unlike the original study, we performed augmentation with random brightness, random contrast, and random horizontal flip when training with the CXR8 dataset. Images were resized to 300 × 300 and pixel values were scaled between −1 to 1. L2 regularization was performed at the final convolutional layer with an alpha value of 0.05. The learning rate was initially set to .0002 and decayed based on the Adam algorithm [13]. The machine learning models were built with Tensorflow version 1.7 [14]; additional packages utilized included Scikit-learn version 0.19.1 [15] and SciPy version 1.1.0 [16]. Imaging manipulation was done using Scikit-image version 0.14.0 [17] and OpenCV version 3.4.2.17 [18]. Using a batch size of 6, the model was initially trained using the CXR8 dataset over a course of approximately 1,200,000 batch-iterations.
Our data for this study was collected from the Vendor Neutral Archive across the years 2013 to 2017. Positive and negative cases were collected concurrently with the same sex and age distribution by searching radiology reports for the term “pneumothorax.” There was no patient overlap between the two datasets, and only 1 chest X-ray study was obtained from each unique patient. Laterality of the pneumothorax was extracted from the radiology report. The Digital Imaging and Communications in Medicine (DICOM) files were de-identified for The Health Insurance Portability and Accountability Act of 1996 (HIPAA) compliance. Data was stored behind the institution firewall.
The DICOM files were parsed with Pydicom version 1.1.0 [19] and processed step-wise. The size description of the pneumothorax was extracted from the radiology report. Only cases which explicitly stated that the imaging demonstrated a large or tension pneumothorax was included in the study, and we selected cases with large and tension pneumothoraces as they need to be treated expeditiously. To remove the derivative views, the DerivationDescription DICOM header was checked. Similarly, to remove lateral views, the ViewPosition DICOM header was examined. If the PhotometricInterpretation DICOM header was equal to MONOCHROME1, the image was inverted. The images were visually inspected by a board-certified radiologist (corresponding author) to confirm the underlying diagnosis. The pathology was not equivocal as a large pneumothorax was readily appreciable when present. Simultaneously, lateral chest radiographs that escaped filtering were manually removed. Once the images were processed and inspected, a total of 159 pneumothorax and 682 non-pneumothorax cases were available for the training set. For the validation set, 48 pneumothorax and 1287 non-pneumothorax cases were available.
The machine learning model, pre-trained with the CXR8 dataset, was retrained using our training dataset, which was resized to 315 × 315 and augmented with random brightness, random contrast, random horizontal flip, and randomly cropped to the final size of 300 × 300 during each training step. For comparison purposes, retraining was also performed without augmentation using images resized to 300 × 300. Pixel values were initially downscaled to between 0 and 255 before resizing and scaled between −1 to 1 at the time of resizing. Using a batch size of 6, the training was completed after approximately 280,000 batch-iterations.
Augmentation was performed by randomly perturbing each image and rescaling the pixels values as a batch rather than individually. This technique allowed the images to retain the random changes to brightness and contrast, which would have been otherwise lost. Localization of our data was done by using the laterality of the pneumothorax extracted from the radiology report. We applied a mask to the unaffected side and directed the model to focus on the side with the pathology.
A receiver operator curve (ROC) was created and the area under the curve (AUC) was calculated using Scikit-learn [15]. Sensitivity and specificity values were calculated from the upper-left corner threshold of the ROC. For all cases, the output metrics were calculated using the held-out validation dataset, which was not exposed to the training process. Model training was performed on a Linux cluster using the Nvidia GeForce GTX 1080 graphical processing unit (GPU). All code is available at the GitHub repository of the corresponding author.
3. Results:
Initial model training using the CXR8 dataset resulted in an AUC of 0.90 for pneumothorax detection with the open-source held-out validation dataset, which was in line with previously published pneumothorax detection AUC values between 0.87 and 0.88 [10,11]. Naively inferring our held-out validation dataset without re-training the model output an AUC of 0.59. After retraining the model with our own training dataset using both localization and augmentation, our validation dataset inference output an AUC of 0.90. Retraining the model with only augmentation or localization resulted in degradation of AUC in each case (Figure 1). Similarly, sensitivity and specificity were suboptimal when naively inferring our own validation dataset on the CXR8 model, and the best output metrics were obtained utilizing the model retrained with both localization and augmentation (Table 1). Examples of correctly and incorrectly classified cases from our validation dataset on the final model are shown in Figure 2 and 3; the superimposed heatmaps were generated by overlaying the final activation tensor onto the original image after resizing.
Fig1.
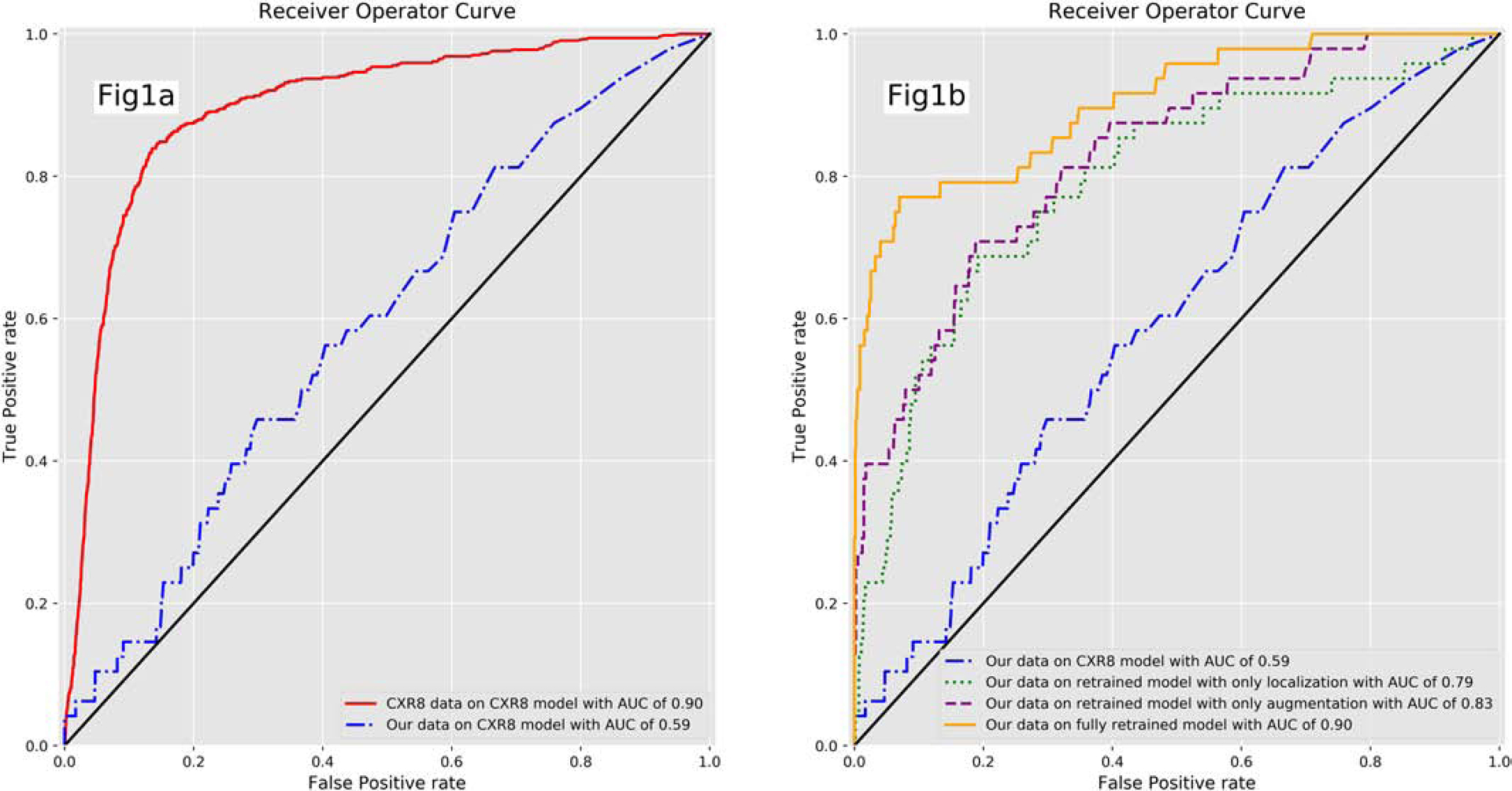
Receiver operator curve (ROC) for the CXR8 model and retrained model. Fig1a. ROC for the CXR8 model evaluating the CXR8 and our validation dataset. The ROC is significantly degraded when evaluating our own validation dataset with the CXR8 trained model. Fig1b. There is improvement in the ROC as the model is retrained with our dataset, particularly with both augmentation and localization.
Table 1:
Output metrics for models trained using the CXR8 dataset and models retrained with our dataset. There is a significant degradation in the output metrics when our own validation dataset is evaluated with the CXR8 trained model. The outputs metrics improve once the models are retrained, particularly with both localization and augmentation.
| Input data | Model | Area under the curve (AUC) | Sensitivity | Specificity |
|---|---|---|---|---|
| CXR8 data | CXR8 model | 0.90 | 0.84 | 0.86 |
| Our data | CXR8 model | 0.59 | 0.46 | 0.70 |
| Our data | Retrained model with only localization | 0.79 | 0.67 | 0.81 |
| Our data | Retrained model with only augmentation | 0.83 | 0.71 | 0.81 |
| Our data | Retrained model with both localization and augmentation | 0.90 | 0.77 | 0.93 |
Fig2.
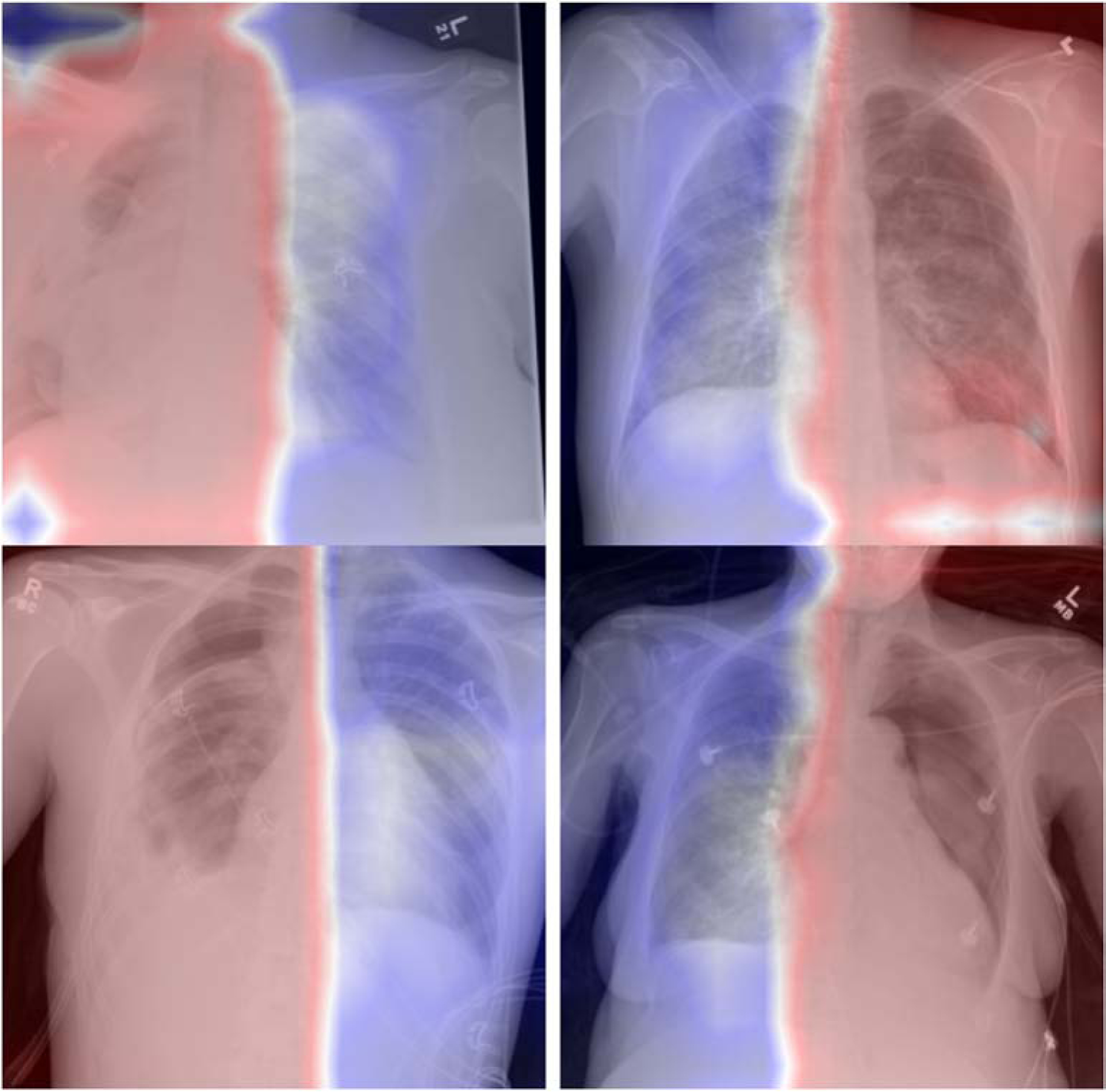
Sample of correctly classified positive pneumothorax cases from our validation dataset. The retrained model is correctly detecting the presence and laterality of the pneumothorax. The board localization is a result of the retraining method, where laterality of the pneumothorax was extracted from the radiology report.
Fig3.
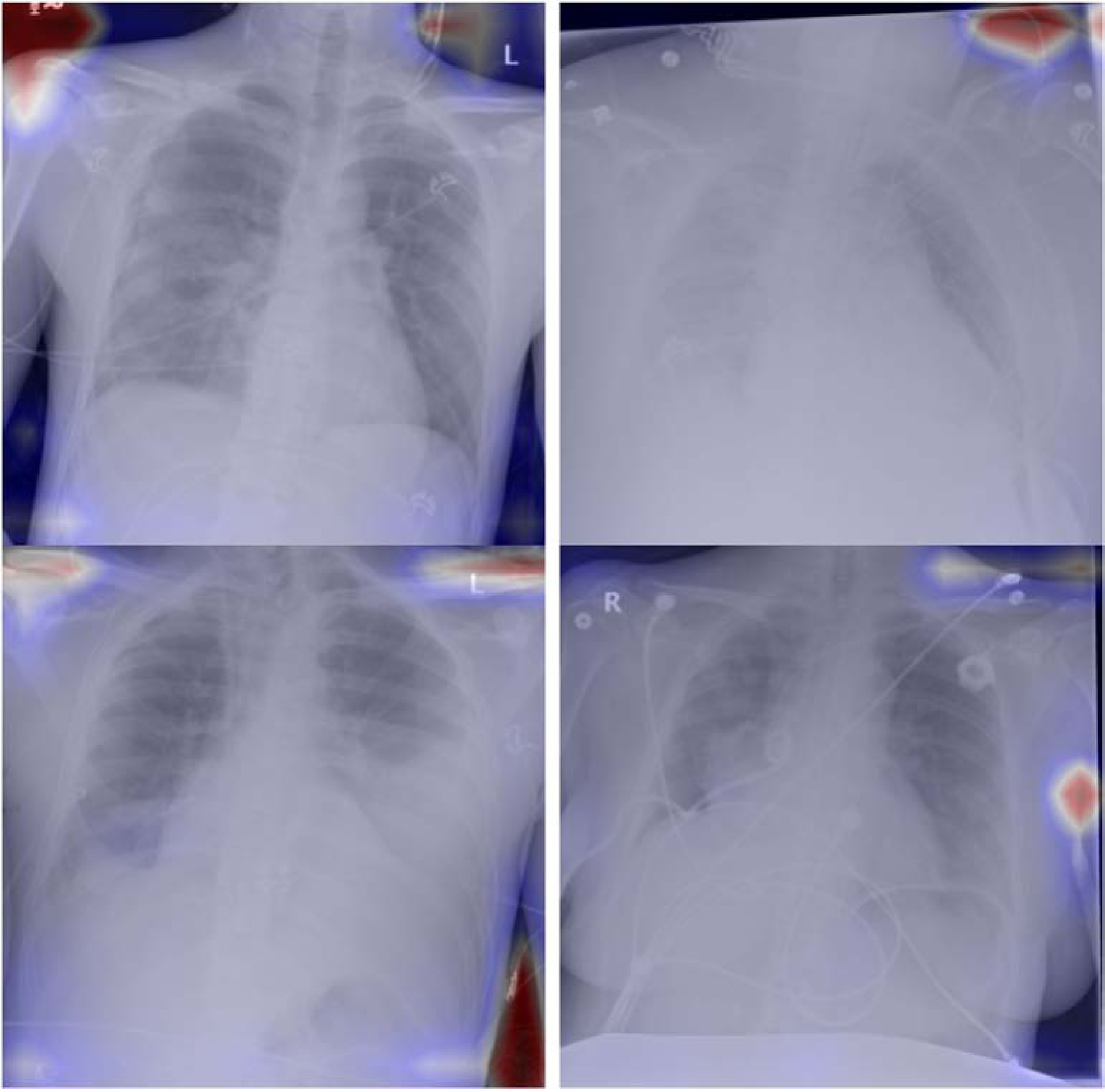
Sample of incorrectly classified positive pneumothorax cases from our validation dataset. The model fails to identify and localize the pneumothorax.
4. Discussion:
We initially demonstrated that the machine learning algorithm was properly functioning by training and testing on the open source CXR8 dataset, obtaining an AUC in line with reference values [11]. However, simply attempting to classify our validation dataset with the CXR8 trained model demonstrated suboptimal performance. There are several possibilities for this lack of model transference; the first consideration is a difference in pixel range. The CXR8 dataset is saved in a Portable Network Graphic (PNG) format with a pixel range between 0 to 255 (bit size of 8), while medical images in DICOM format often have a pixel range of 0 to 4096 (bit size of 12) [20]. While our images were initially scaled down to the range of 0 to 255, some degree of data loss is inevitable with downscaling. Second, while another study did show good transference of a model trained on the CXR8 dataset for airspace disease detection [10], their model was a relatively simple DenseNet [21] without localization information incorporation. Our model based on Li et al. [11] incorporated both unlocalized and localized cases. Unlocalized cases have board labels assigned to each image (e.g. Chest X-ray with pneumothorax) while localized cases have a label and location assigned to each image (e.g. Chest X-ray with pneumothorax on the left upper lung). So we speculate that more complex models may not transfer as well between datasets. Thirdly, it is known that there is some labeling discrepancy with the open source CXR8 dataset, and this may be another confounding factor affecting the performance with our dataset. Finally, there may simply be some inherent difference between the open source dataset and our own dataset, which may be secondary to different support lines overlying the patient, acquisition technique (mA, kVp, field-of-view, etc.) or display format (window/level, etc.).
Once our model was retrained, it showed promising results with an AUC value of 0.90, which was in line with the open source dataset output. Because we used our own verified dataset, we can now be more confident that the model is optimized to our institutional images and pathology of interest without concerns of labeling discrepancies. In addition, while the reported sensitivity and specificity values are the best, balanced values at the upper left corner of the ROC, the threshold can be moved to increase one value at the expense of the other depending on workflow needs.
There were 4 limitations to the study. Firstly, the sample size, while adequate, could have been larger. The open-source dataset contains approximately 112,120 images, while we worked with just 2176 images. Secondly, we decided to focus only on detecting the pathology with the highest clinical impact, which was large pneumothoraces. Thirdly, we only collected imaging data from our Health Plan, and although we encompass many distinct hospitals, incorporating imaging data from outside institutions may have made our study more robust. Finally, we decided not to perform any manual, explicit localization of pathology, but instead worked with broad localization based on the radiology report. However, even with broad localization, we showed there were performance improvements.
5. Conclusion
While various studies have shown promising results on the open source CXR8 dataset, these trained models must perform well on real medical images for practical applications. Our study showed that even though you may get great results on open-source datasets, those models may not translate well to real world data without an intervening retraining process. There is still significant value in these open source datasets, as they can be used to evaluate model performances before they are retrained to real medical images. As the number of datasets, models and companies that tout machine learning grow in the coming years, it is important to keep in mind that not only do they need validation to your institutional data, they may require some degree of re-training for optimal performance.
Acknowledgement:
This research was supported in part by the University of Pittsburgh Center for Research Computing through the resources provided.
Funding:
The project described was supported by the National Institutes of Health through Grant Number UL1 TR001857.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Mathotaarachchi S, Pascoal TA, Shin M, Benedet AL, Kang MS, Beaudry T, et al. Identifying incipient dementia individuals using machine learning and amyloid imaging. Neurobiol Aging 2017;59:80–90. [DOI] [PubMed] [Google Scholar]
- [2].Schnyer DM, Clasen PC, Gonzalez C, Beevers CG. Evaluating the diagnostic utility of applying a machine learning algorithm to diffusion tensor MRI measures in individuals with major depressive disorder. Psychiatry Res 2017;264:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bahl M, Barzilay R, Yedidia AB, Locascio NJ, Yu L, Lehman CD. High-Risk Breast Lesions: A Machine Learning Model to Predict Pathologic Upgrade and Reduce Unnecessary Surgical Excision. Radiology 2017:170549. [DOI] [PubMed] [Google Scholar]
- [4].Gale W, Oakden-Rayner L, Carneiro G, Bradley AP, Palmer LJ. Detecting hip fractures with radiologist-level performance using deep neural networks. arXiv [csCV] 2017. [Google Scholar]
- [5].Cao Y, Wang H, Moradi M, Prasanna P, Syeda-Mahmood TF. Fracture detection in x-ray images through stacked random forests feature fusion. 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), 2015, p. 801–5. [Google Scholar]
- [6].Olczak J, Fahlberg N, Maki A, Razavian AS, Jilert A, Stark A, et al. Artificial intelligence for analyzing orthopedic trauma radiographs. Acta Orthop 2017;88:581–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].González G, Ash SY, Vegas Sanchez-Ferrero G, Onieva Onieva J, Rahaghi FN, Ross JC, et al. Disease Staging and Prognosis in Smokers Using Deep Learning in Chest Computed Tomography. Am J Respir Crit Care Med 2017. doi: 10.1164/rccm.201705-0860OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lakhani P, Sundaram B. Deep Learning at Chest Radiography: Automated Classification of Pulmonary Tuberculosis by Using Convolutional Neural Networks. Radiology 2017;284:574–82. [DOI] [PubMed] [Google Scholar]
- [9].Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. ChestX-ray8: Hospital-scale Chest X-ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. arXiv [csCV] 2017. [Google Scholar]
- [10].Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, et al. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. arXiv [csCV] 2017. [Google Scholar]
- [11].Li Z, Wang C, Han M, Xue Y, Wei W, Li L-J, et al. Thoracic Disease Identification and Localization with Limited Supervision. arXiv [csCV] 2017. [Google Scholar]
- [12].Parlak M, Uil SM, van den Berg JWK. A prospective, randomised trial of pneumothorax therapy: manual aspiration versus conventional chest tube drainage. Respir Med 2012;106:1600–5. [DOI] [PubMed] [Google Scholar]
- [13].Kingma D, Ba - arXiv preprint arXiv:1412.6980 J, 2014 Adam: A method for stochastic optimization. Arxiv.org 1412.
- [14].Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. arXiv [csDC] 2016. [Google Scholar]
- [15].Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res 2011;12:2825–30. [Google Scholar]
- [16].Jones E, Oliphant T, Peterson - URL http://scipy.org P, 2011 SciPy: Open source scientific tools for Python, 2009 2011.
- [17].van der Walt S, Schönberger JL, Nunez-Iglesias J, Boulogne F, Warner JD, Yager N, et al. scikit-image: image processing in Python. PeerJ 2014;2:e453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bradski G The OpenCV Library. Dr Dobb’s Journal of Software Tools 2000. [Google Scholar]
- [19].Mason - Medical Physics D, 2011. SU- E- T- 33: Pydicom: An Open Source DICOM Library Wiley Online Library; 2011. [Google Scholar]
- [20].Mahesh M The Essential Physics of Medical Imaging, Third Edition Medical Physics; 2013;40:077301. doi: 10.1118/1.4811156. [DOI] [PubMed] [Google Scholar]
- [21].Huang G, Liu Z, van der Maaten L, Weinberger KQ. Densely Connected Convolutional Networks. arXiv [csCV] 2016. [Google Scholar]


