Abstract
Functional protein-coding small open reading frames (smORFs) are emerging as an important class of genes. However, the number of translated smORFs in the human genome is unclear because proteogenomic methods are not sensitive enough, and, as we show, Ribo-Seq strategies require additional measures to ensure comprehensive and accurate smORF annotation. Here, we integrate de novo transcriptome assembly and Ribo-Seq into an improved workflow that overcomes obstacles with previous methods to more confidently annotate thousands of smORFs. Evolutionary conservation analyses suggest that hundreds of smORF-encoded microproteins are likely functional. Additionally, many smORFs are regulated during fundamental biological processes, such as cell stress. Peptides derived from smORFs are also detectable on human leukocyte antigen complexes, revealing smORFs as a source of antigens. Thus, by including additional validation into our smORF annotation workflow, we accurately identify thousands of unannotated translated smORFs that will provide a rich pool of unexplored, functional human genes.
Annotation of open reading frames (ORFs) from genome sequencing was initially carried out by locating in-frame start (AUG) and stop codons1–3. This approach resulted in unreasonably large numbers of ORFs smaller than 100 codons called small open reading frames (smORFs). A length cutoff was then introduced to remove smORFs4,5, which were largely presumed to be meaningless random occurrences1,2. With the advent of more sensitive detection methods, functional proteins encoded by smORFs, dubbed microproteins, have been characterized with more regularity6,7. In fruit flies, tal/pri was shown to encode three 11- and one 32-amino acid microproteins that control proper physiological development8,9. This example, and others, highlighted the importance of investigating smORFs, and paved the way for work in higher organisms. Recently, several mammalian microproteins have been characterized with fundamental roles ranging from DNA repair10, mitochondrial function11,12, RNA regulation13, and muscle development14. These studies demonstrated that genomes contain many functional smORFs and therefore annotating all protein-coding smORFs is important.
Advances in proteomics and next-generation sequencing (NGS) technologies provided the tools necessary to identify protein-coding smORFs. For example, the integration of RNA-Seq and proteomics approaches identified hundreds of novel microproteins in human cell lines15,16. While proteomics provides evidence that a smORF produces a microprotein of sufficient abundance for detection, it is limited in sensitivity and some microproteins do not have suitable tryptic peptides. With the development of ribosome profiling (Ribo-Seq), NGS can be utilized to identify ORFs that are undergoing active translation with high sensitivity and accuracy by revealing the position of elongating ribosomes throughout the transcriptome17. Ribo-Seq has been applied successfully to smORF discovery in fruit flies18 and zebra fish19, identifying hundreds of novel translated smORFs, which is significantly more than were detected by mass spectrometry in these organisms.
Ribo-Seq has also been used more recently to annotate novel protein-coding smORFs in human cell lines and tissues. SmProt20 and sORFs.org21 are two prominent smORF databases, containing >17,000 and >500,000 unique Ribo-Seq predicted human protein-coding smORFs, respectively. However, this order of magnitude difference, despite analyzing many of the same datasets, raised concerns, as accurate smORF annotations are critical for downstream biological studies. SmProt and sORFs.org employ different strategies for identifying and filtering protein-coding smORFs, which may contribute to the size disparity. Another possible contributor is that unannotated smORFs might be less reliably called translated than annotated ORFs using Ribo-Seq due to their low relative abundance, inherent small size, or other distinguishing properties. Thus, major questions about smORFs remain, including: (1) Is Ribo-Seq as robust at identifying translated unannotated smORFs as annotated ORFs? (2) How many bona fide protein-coding smORFs are in the human genome? (3) Is there evidence that protein-coding smORFs are regulated similarly to annotated genes? To answer these questions, we developed a top-down workflow that combines de novo transcriptome assembly and multiple Ribo-Seq experiments to rigorously annotate novel protein-coding smORFs.
We found that while detection of annotated ORFs is robust, smORF detection is noisier. Application of this workflow in HEK293T, HeLa-S3, and K562 cells, uncovered >2,500 confidently annotated protein-coding smORFs—our gold standard set—and >7,500 in total. We also demonstrated that while smORF-encoded microproteins have distinguishing properties from annotated proteins, their expression is similarly regulated during cell stress, and they are also presented as cell surface antigens. These results dramatically increase the coding potential of the genome and provide several strategies for finding potentially functional smORFs.
Results
Overview of top-down smORF annotation workflow
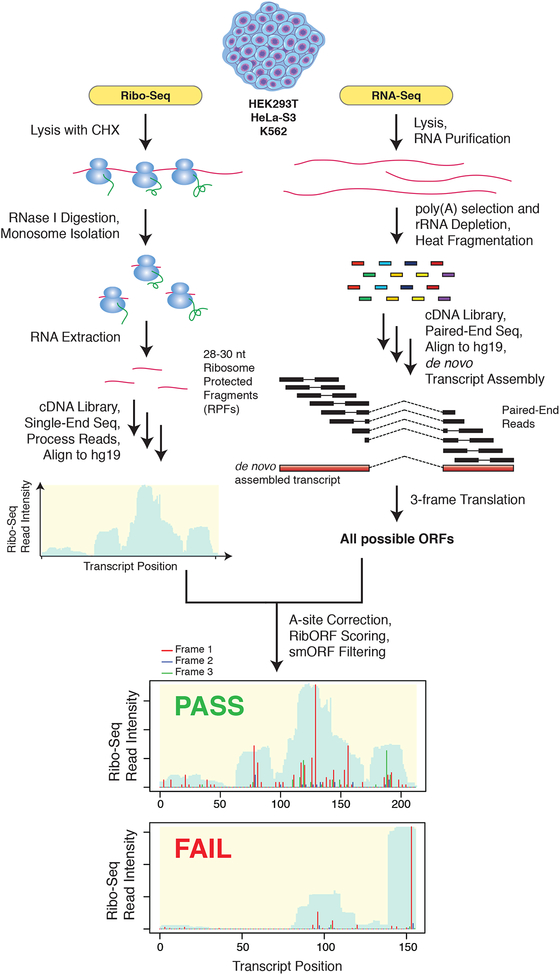
Ribo-Seq maps the position of elongating ribosomes throughout the transcriptome by first footprinting with RNase I (Fig. 1). The resulting 28–29 nt ribosome protected mRNA fragments (RPFs) are then sequenced and aligned to the transcriptome. Typically, Ribo-Seq reads are mapped onto reference transcriptome databases, such as RefSeq or Ensembl, which are not representative of every cell type. Our top-down workflow utilizes transcripts obtained by de novo assembly of RNA-Seq data. This approach identified entirely new transcripts as well as isoforms of annotated transcripts, allowing for more comprehensive smORF discovery. We then define ORFs across all three reading frames of the de novo assembled transcriptome to generate an ORF database that includes smORFs.
Figure 1. Outline of top-down smORF annotation workflow.
RNA-Seq and Ribo-Seq datasets were collected for HEK293T, HeLa-S3, and K562 cell lines and utilized for the prediction of novel translated smORFs. RNA-Seq reads were de novo assembled into a transcriptome using Cufflinks. The assembled transcriptome for each cell line was then in silico 3-frame translated to create a database of all possible ORFs. In parallel, multiple biological replicates of Ribo-Seq data were also collected for each cell line and utilized to assess translation of all smORFs in the accompanying 3-frame database. For each replicate, RibORF was used to define the A-site position of each ribosome protected fragment (RPF) and then score each smORF for translation. Those smORFs which passed RibORF scoring, did not overlap with annotated ORFs, and lacked significant similarity to RefSeq annotated proteins were retained. Shown at the bottom are examples of a smORF passing RibORF scoring with high coverage and in-frame ribosome A-site reads (Frame 1) and a smORF failing RibORF scoring due to poor read coverage.
After obtaining Ribo-Seq data, we scored all ORFs in the database for translation using RibORF (Fig. 1), a support vector machine-based classifier of translation22. RibORF uses the fraction of RPF reads aligned in-frame with the candidate ORF to calculate the overall probability of translation, which depends on the resolution of the dataset. Sub-codon- or high-resolution Ribo-Seq datasets can display >70% of RPFs aligned in-frame with annotated coding sequences (CDS)23 by metagene analysis, enabling more accurate identification of unannotated protein-coding smORFs. RibORF also scores the uniformity and distribution of RPF reads over the entire ORF to avoid possible artifacts22.
Following RibORF scoring, the list of predicted translated ORFs was filtered to remove ORFs less than 6 codons, which are not amenable to detection by mass spectrometry, and greater than 150 codons, as unannotated protein-coding smORFs larger than 100 codons have been discovered15. Next, translated smORFs found to overlap with annotated CDS regions in the UCSC database were removed to filter out both annotated genes and out-of-frame overlappers. Finally, encoded microproteins were analyzed for similarity to human RefSeq proteins by BLASTp. Only low scoring hits were retained, removing likely pseudogenes and any additional annotated genes. The remaining hits constitute the set of novel microprotein-encoding smORFs (Fig. 1).
Annotating protein-coding smORFs in HEK293T cells
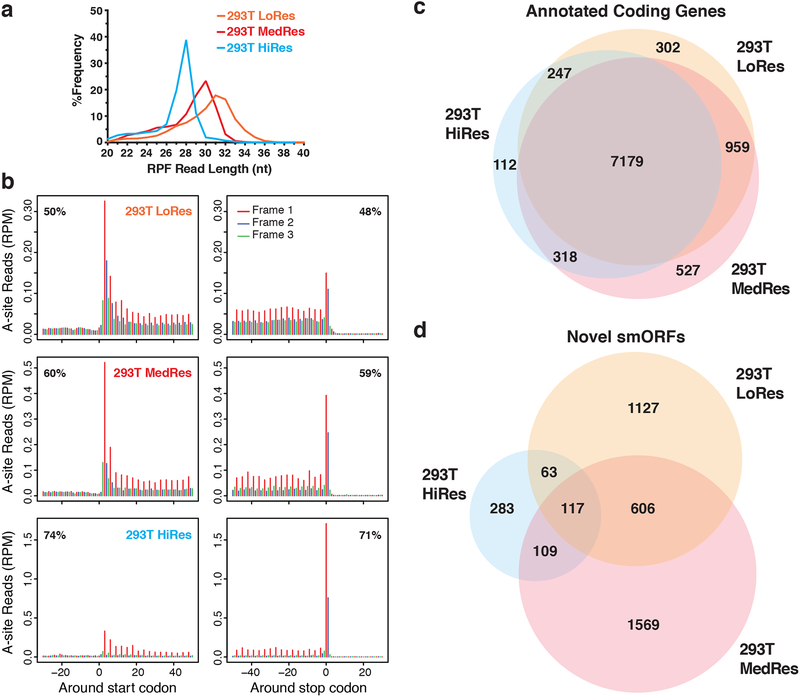
We first tested our workflow in HEK293T cells, which we previously identified dozens of microproteins in by proteomics15. Ribosome footprints were initially prepared using a protocol that afforded high resolution data in HEK293 cells24. However, only ~50% of reads aligned in-frame by metagene analysis, and RPF lengths peaked at 31-nt (Fig. 2a). While this resolution is comparable to several published datasets (Supplementary Fig. 1), we collected higher resolution datasets as well to ensure identification of translated smORFs that require greater accuracy. To gain finer control over nuclease digestion, we followed a reported strategy that normalizes the amount of nuclease added to the RNA concentration23. We generated two additional HEK293T Ribo-Seq datasets with ~60% and >70% of reads in-frame by metagene analysis and RPF lengths that peaked at 30 and 28-nt, respectively (Fig. 2a). Given that published datasets show a wide range of resolutions (Supplementary Fig. 1), we carried all three datasets forward for protein-coding smORF prediction.
Figure 2. Comparison of translation prediction for smORFs versus annotated ORFs.
a RPF read length distribution plot showing the differences in footprint sizes across HEK293T Ribo-Seq datasets. Biological replicates were subjected to increasing RNase I nuclease digestion resulting in a range of Ribo-Seq resolutions: low (LoRes), medium (MedRes), and high (HiRes). The expected RPF size is 28-nt. b Metagene plots showing RPF read alignment around the start site and stop site for each dataset. The 5’-position of each RPF read was shifted to the ribosomal A-site and then mapped to all hg19 RefSeq coding transcripts. The metagene coding region is in frame 1, while frame 2 and frame 3 are out-of-frame. The percentage of in-frame reads is noted in the top corner. 28–34 nt reads were used for LoRes, 29–33 nt for MedRes, and 25–29 nt for HiRes. c Venn diagram showing overlap of annotated RefSeq genes passing RibORF scoring between all three HEK293T Ribo-Seq datasets. d Venn diagram showing overlap of novel protein-coding smORFs passing RibORF scoring and our smORF filters.
Several previous studies combined reads from multiple Ribo-Seq experiments to increase the sensitivity of translation scoring22–25. However, this strategy can also allow for more false positives when the same thresholds are applied due to reads accumulating on an ORF because of non-productive ribosomal binding or noise inherent to the Ribo-Seq protocol26. Additionally, combining experiments does not allow one to assess the reproducibility of translation predictions, which is critical in other NGS-based assays27. When analyzed separately, novel smORFs scored as translated in every experiment regardless of noise and sequencing depth are more confidently protein-coding than those found in a single experiment, and also allows one to observe how differences in RPF preparation affect translation scoring. Therefore, to improve the confidence of smORF translation prediction, we analyzed each Ribo-Seq experiment separately.
To confirm the quality of our HEK293T Ribo-Seq datasets and determine the noise level for bona fide genes, we used RibORF to score RefSeq genes. Despite differences in resolution and sequencing depth, we observed high overlap among the 9,644 canonical genes called translated, with 74% found in all three experiments (Fig. 2b). For smORFs, however, we found that these differences had a strong influence on the total number called translated (Fig. 2c). We identified 1,913, 2,401, and 572 predicted translated smORFs, with 117 smORFs called translated in every experiment and 895 smORFs in at least two experiments. Interestingly, 606 smORFs were found in both lower resolution datasets but not the high-resolution dataset. Thus, translation prediction is noisier for smORFs than annotated ORFs, but the analysis of several Ribo-Seq experiments can improve confidence. The set of reproducibly detected smORFs is also greater than the 24 novel microproteins we identified in HEK293T cells by proteomics15,28, highlighting the value of Ribo-Seq for smORF discovery. Ribo-Seq data validated nine of our proteomics-detected smORFs (Supplementary Data 1), with the others missing due to overlap with annotated genes or insufficient read coverage.
Endoplasmic reticulum stress-regulated smORFs
Having identified thousands of novel protein-coding smORFs, we next searched for evidence of their regulation as a means to uncover possible biological roles. We chose to look for expression changes induced by endoplasmic reticulum (ER) stress, which leads to the accumulation of unfolded and mis-folded proteins and triggers a well-characterized signaling cascade dubbed the Unfolded Protein Response (UPR)29. To induce ER stress, HEK293T cells were treated with either thapsigargin (TG) or tunicamycin (TM), and RNA-Seq and Ribo-Seq data were collected for each sample (Supplementary Fig. 2a and 3). Applying our workflow, we identified 666 additional predicted translated smORFs, increasing the total to 4,540 in HEK293T (Supplementary Data 1). Confirming TG- and TM-induced activation of the UPR, HSPA5, HYOU1, DDIT3, and other known UPR genes were upregulated30 (Fig. 4a and Supplementary Data 2). Gene Ontology (GO) analysis also revealed enrichment in UPR and cell stress related genes (Supplementary Data 2).
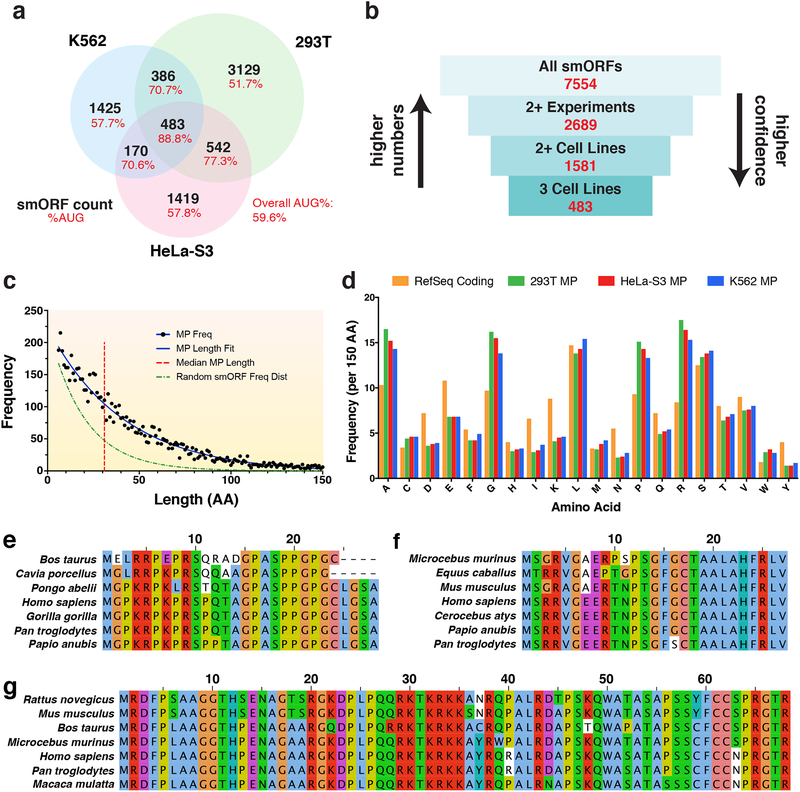
Figure 4. Characteristics of protein-coding smORFs.
a Venn diagram showing the overlap of predicted translated smORFs across HEK293T, HeLa-S3, and K562 cell lines. The percent of smORFs containing an AUG start codon for each sector is also shown. b Diagram showing the number of annotated smORFs in increasingly confident subsets. c Frequency distribution of smORF-encoded microprotein (MP) lengths in amino acids (aa). The median microprotein size is 32 aa. The MP length distribution can be fit with a decay curve of the formula N0e−λx, where N0 = 224 and λ = 0.024. This is a slower decay than the expected frequency distribution of randomly occurring MPs, where λ = 0.057. d Frequency of aa occurrence per 150 aa for annotated RefSeq proteins and novel microproteins identified in each cell line. e Sequence alignment for a novel microprotein encoded by the smORF found within the 5’-UTR of four jointed box 1 (FJX1). This smORF has an average PhyloCSF score of 3.49 using the 29-mammal alignment. f Sequence alignment for a novel microprotein encoded by a smORF found within the 5’-UTR of nuclear casein kinase and cyclin dependent kinase substrate 1 (NUCKS1). This smORF shows high similarity to translated regions in other mammalian species by tBLASTn and has a negative PhyloCSF score. g Sequence alignment for a novel microprotein encoded by a smORF within the 5’-UTR of B-cell CLL/lymphoma 9 (BCL9). This smORF shows high similarity to proteins in other mammalian species by BLASTp and has a negative PhyloCSF score.
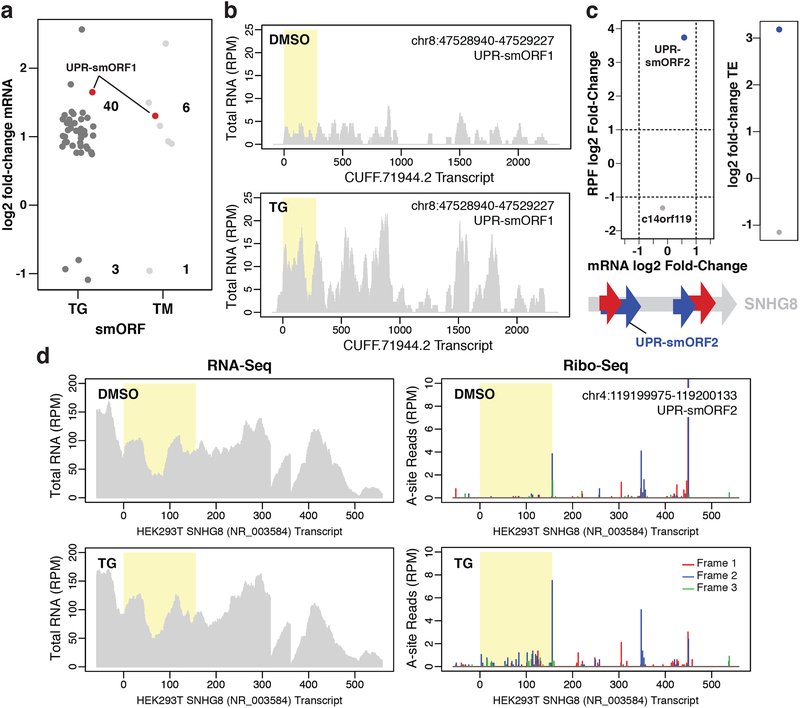
We then analyzed smORFs for transcriptional regulation under ER stress, focusing on reproducibly detected smORFs. TG and TM induced significant mRNA expression changes in 43 and 7 smORFs, respectively (Fig. 3a), suggesting that the encoded microproteins might function in the UPR. For instance, one upregulated smORF, UPR-smORF1, is found on a de novo assembled transcript isoform of asparagine synthase pseudogene 1 (ASNSP1) (Fig. 3b, Supplementary Data 2). While ASNSP1 is a predicted pseudogene, translated pseudogenes are being reassessed for functional importance31. In addition, asparagine synthase (ASNS) is a known UPR target gene32, supporting the possibility that the UPR-smORF1 microprotein might have a role in the UPR.
Figure 3. smORF regulation during ER stress.
a Changes in mRNA expression for novel smORFs induced by 1 μM TG or 5 μg/mL TM for 4 h relative to DMSO treatment (padj < 0.05). Only smORFs identified in at least two Ribo-Seq experiments were considered. The novel predicted translated smORF UPR-smORF1 is shown in red. Two biological replicates for each condition were analyzed. b Representative RNA-Seq read coverage plots for the unannotated de novo assembled transcript, CUFF.71994.2, with UPR-smORF1 highlighted in yellow. The y-axes show the intensity of read peaks in reads per million (RPM). c, top Changes in mRNA expression versus changes in RPF levels for a novel smORF found on SNHG8, UPR-smORF2, and the annotated smORF c14orf119 in response to TG treatment. Plot showing the resulting change in translational efficiency (TE, ΔRPF/ΔmRNA) is also shown (padj < 0.1). c, bottom Schematic showing the location of UPR-smORF2 on the SNHG8 transcript. Four novel smORFs were identified at least twice on SNHG8: two in frame 1 and two in frame 2. d Representative RNA-Seq read coverage and ribosomal A-site plots (Ribo-Seq) for SNHG8 with UPR-smORF2 highlighted in yellow.
Several UPR pathway genes have been shown to be translationally regulated during ER stress33,34. Therefore, we sought to identify any translationally regulated smORFs. Translational regulation was monitored by assessing changes in translational efficiency (TE) using Xtail35, which quantifies the changes in RPF densities relative to mRNA expression levels using Ribo-Seq and RNA-Seq data, respectively. Both TG and TM induced higher TE for ATF4, IFRD1, and SEC61G, which are known to be regulated during ER stress36–38, as well as many other genes (Supplementary Fig. 4b–c, Supplementary Data 2). Analysis of smORFs revealed a robust change in TE for a single smORF located on SNHG8, dubbed UPR-smORF2 (Fig. 3c). Increased TE for UPR-smORF2 is clearly visualized by comparing the Ribo-Seq and RNA-Seq read coverage plots for SNHG8 (Fig. 3d), which show an increase in ribosome occupancy and little change in transcript levels between vehicle- and TG-treated cells. In addition, the TE of an annotated but uncharacterized smORF, c14orf119, significantly decreased in response to TG.
Annotation of smORFs in additional cell lines
To determine whether the smORFs identified in HEK293T are unique, and test the generality of our observations, we profiled additional cell lines for protein-coding smORFs. Because they differ in their tissue of origin from HEK293T, we selected the chronic myeloid leukemia-derived cell line K562, and the cervical cancer-derived HeLa-S3 cell line. Both of these cell lines are also included in ENCODE, providing a wealth of high-quality genomic, transcriptomic, and functional data available for follow-up analyses39.
As with HEK293T, HeLa-S3 cell lysates were digested using different conditions to maximize the number and accuracy of smORFs identified. Metagene analysis showed a range of resolutions across the four datasets collected, from ~50–70% reads in-frame (Supplementary Fig. 3b and 5). Altogether, 2,614 novel smORFs were called translated, with 777 smORFs found in at least two experiments (Supplementary Data 1). Next, we collected three Ribo-Seq datasets from K562 using a range of digestion conditions. All digestion conditions tested in K562 resulted in >75% reads in-frame by metagene analysis (Supplementary Fig. 6). However, K562 HiRes3 displayed a broader footprint length distribution (Supplementary Fig. 2c). In total, 2,464 predicted protein-coding smORFs were identified in K562 cells, with 542 smORFs found in at least two experiments (Supplementary Data 1).
Across the three cell lines profiled, we identified 7,554 novel predicted protein-coding smORFs. The majority of these smORFs are only identified in a single experiment, but there are thousands of smORFs that overlap between cell lines or are found in multiple experiments from a single cell line. In total, 483 smORFs were detected in all three cell lines, 1,581 in at least two cell lines, and 2,689 in at least two experiments across any cell line (Fig. 4a,b). We define this last set of smORFs as our gold standard protein-coding smORF annotations given their reproducibility. These results reveal that smORFs, like larger annotated genes, can be ubiquitous and cell type specific. Notably, we also observed that smORFs called translated in two or more cell lines are more likely to utilize an AUG initiation codon than smORFs found in only one cell line (Fig. 4a), which supports their robust detection across different cell types.
Next, we quantified the abundance of smORF-containing transcripts to determine whether smORFs called translated in only a single experiment are relatively less expressed. We found that the median transcript FPKM values are significantly greater for smORFs called translated in multiple experiments than for singly identified smORFs (Supplementary Fig. 7). These results suggest that the ability to reproducibly detect translated smORFs by Ribo-Seq may be limited in part due to transcript abundance.
Protein-coding smORFs on annotated transcripts
Over half of all predicted translated smORFs are located on RefSeq transcripts. The majority of smORFs are found within the 5’-UTR of known genes (Supplementary Fig. 8), including ~76% of predicted translated smORFs identified in all three cell lines. These 5’-UTR smORFs are also called upstream open reading frames (uORFs), and often regulate translation of the downstream CDS through engagement with the ribosome40. While the microprotein products of uORFs are often assumed to be non-functional, there are examples with characterized functions, such as the 70 amino acid MIEF1 uORF microprotein11,41. We identified 597 uORFs containing >50 codons that are candidates for encoding functional microproteins (Supplementary Data 1).
Beyond uORFs, a small portion of predicted protein-coding smORFs were found within the 3’-UTR, on antisense transcripts, and on ncRNAs. Notably, 623 translated smORFs are located on RefSeq ncRNAs, and several more on UCSC ncRNAs, and many are high confidence identifications found in several experiments. For instance, translated smORFs on the ncRNAs, BC013229 and LOC100287015 (Supplementary Fig. 9a,b), were identified in every HeLa-S3 dataset. We also observed ncRNAs containing multiple protein-coding smORFs, such as LINC00534, which contains two novel smORFs in different reading frames (Supplementary Fig. 9c). Some ncRNAs even contained more than two predicted translated smORFs, such as the colon cancer-related gene CCAT1, which contains two confidently identified smORFs and several more called translated only once (Supplementary Fig. 9d and Supplementary Data 1).
Analyzing microprotein properties
We next sought to determine distinguishing properties of microproteins from annotated proteins. First, the median length of encoded microproteins is 32 amino acids (Fig. 4c, red line), whereas the median human protein length in the Pfam database is 416 amino acids42. The frequency distribution of microprotein lengths can be fit by a decay curve that has a slower decay than expected for randomly occurring microproteins, based on an ~5% chance of encountering a stop codon7 (Fig. 4c). Thus, predicted protein-coding smORFs occur at a higher frequency than expected by chance alone. In addition, comparing microprotein amino acid usage to that of annotated proteins revealed a clear difference in several amino acid frequencies, including increased usage of alanine, glycine, proline, and arginine, as well as depletion of aspartic acid, glutamic acid, isoleucine, lysine, and tyrosine (Fig. 4d). We also analyzed microproteins for common structural features, including transmembrane helices and conserved protein domains. Only 48 reproducibly detected smORF-encoded microproteins contain predicted transmembrane helix domains (Supplementary Data 3), and another 17 are predicted to contain conserved protein domains.
Given that uORFs might generally behave as regulators of downstream translation, we also checked whether their encoded microproteins differ from those of non-uORFs. We found that the median length of uORF microproteins is shorter than for non-uORF microproteins, at 24 versus 43 amino acids, respectively (Supplementary Fig. 10a,b). However, uORFs and non-uORFs show little difference in amino acid usage (Supplementary Fig. 10c). We also compared the transcript abundances and ribosome densities for uORFs versus non-uORFs. In each cell line, median transcript FPKM values and RPF RPKM values were greater in uORFs versus non-uORFs (Supplementary Fig. 10d,e). These results are consistent with uORFs sharing their transcripts with relatively well expressed annotated ORFs and their regulation of downstream translation.
Evidence of smORF conservation
Based on functionally characterized smORFs10,11,13,14, we hypothesized that some microproteins would show sequence conservation across other mammalian species. We first employed PhyloCSF, which uses a multi-species nucleotide alignment to examine sequences for signatures of conserved coding regions43. At least one exon with a positive average PhyloCSF score was found in 432 smORFs (Supplementary Data 1), such as the novel smORF within the 5’-UTR of FJX1 (Fig. 4e). We also searched for sequence similarities across other species using tBLASTn and BLASTp as evidence for possible protein conservation. Using tBLASTn, 4,687 microproteins were found to have high similarity to translated RNA sequences from at least one other species, including 273 to mouse sequences (Supplementary Data 1). Additionally, 476 microproteins with high similarity to known and predicted proteins were found in other species using BLASTp. In many instances, clear sequence similarity was observed across several species using tBLASTn and BLASTp despite having negative PhyloCSF scores (Fig. 4f,g). These data suggest that many novel microproteins show evidence of conservation, and therefore are likely to have cellular and physiological functions.
Identifying smORF translation initiation sites
Approximately 40% of the predicted protein-coding smORFs lack an in-frame canonical AUG start codon (Fig. 4a), making their translation initiation sites difficult to identify. Through treatment with initiation-specific inhibitors, such as harringtonine (Harr) and lactimidomycin (LTM)44, one can use Ribo-Seq to identify translation initiation sites. For example, Harr treatment induced RPF accumulation centered on the first AUG start codon in a novel METTL3 uORF (Supplementary Fig. 11a). Start site inhibitors also helped identify alternative initiation codons. LTM treatment enriched RPF coverage over the near cognate start codon UUG in a TMEM33 uORF (Supplementary Fig. 11b), supporting its translation despite the lack of an in-frame AUG start codon.
These inhibitors were also helpful in identifying the predominant codons for translation initiation when multiple canonical or near cognate start codons were present. For example, there are three in-frame AUG codons within a novel uORF on GTF2H1. Surprisingly, Harr treatment induced the highest RPF accumulation on the third AUG codon, with only a small peak present over the first AUG (Supplementary Fig. 11c), suggesting that both a long and predominant short form of the microprotein are made. Similarly, we observed mixed start site usage for the uORF on FBXO9, with translation initiation peaks on a CUG codon and a downstream AUG codon (Supplementary Fig. 11d). Interestingly, no initiation peak was observed over the most upstream in-frame AUG codon.
Protein-coding smORFs on unannotated transcripts
By including de novo transcriptome assembly, we were able to identify a large portion of predicted protein-coding smORFs on transcripts that are missing from the RefSeq assembly. For example, we observed a 5’-extension of c6orf62 which contains a translated smORF (Supplementary Fig. 12a). Other examples include novel exons, such as the smORF-containing EYA4 isoform found specifically in HeLa-S3 samples (Supplementary Fig. 12b) and the GGPS1 isoform with an alternative 5’-UTR containing a novel smORF (Supplementary Fig. 12c).
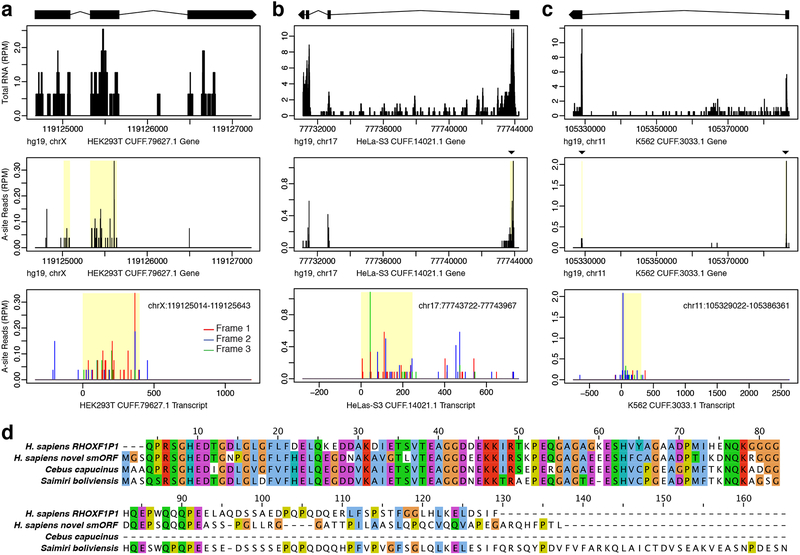
Several predicted protein-coding smORFs were also found on transcripts that do not overlap with any annotated gene, and many of these unannotated transcripts are cell type specific (Fig. 5a–c). BLAST sequence analysis can help identify the function of these unannotated genes. For instance, the HEK293T-specific smORF-encoded microprotein in Fig. 5a shows high similarity to a sequence on the X-linked reproductive homeobox (rhox) pseudogene RHOXF1P1, as well as two predicted rhox-like X-linked homeobox genes in other mammals (Fig. 5d, Supplementary Data 1). Moreover, this novel X-linked homeobox candidate is located within 90 kb of the rhox gene cluster and 20 kb of RHOXF1P1. Given that there are 33 known rhox genes in mouse but only 3 in humans45, it’s possible that this novel smORF is a missing rhox family gene.
Figure 5. Protein-coding smORFs identified on novel unannotated transcripts.
Novel protein-coding smORFs were identified on unannotated de novo assembled transcripts which had no overlap with annotated genes. Examples shown are specific to a HEK293T, b HeLa-S3, and c K562. The top plot shows RNA coverage at the genomic level with the exon model of the Cufflinks assembled transcript shown above. Black boxes represent the exons, connecting lines represent the introns, and the strand orientation is noted by the arrowhead. The middle A-site plot shows the Ribo-Seq coverage at the gene level with the smORF highlighted in yellow. The bottom A-site plot shows the Ribo-Seq coverage at the transcript level with reads colored by frame. The smORF coordinates are shown in the top corner. The smORFs in a and b are in frame 1, while the smORF in c is in frame 2. d Sequence alignment for the novel smORF in a shows high similarity to the human X-linked reproductive homeobox pseudogene RHOXF1P1, as well as predicted X-linked homeobox genes in C. capucinus and S. boliviensis.
Detection of microprotein peptides on HLA-I complexes
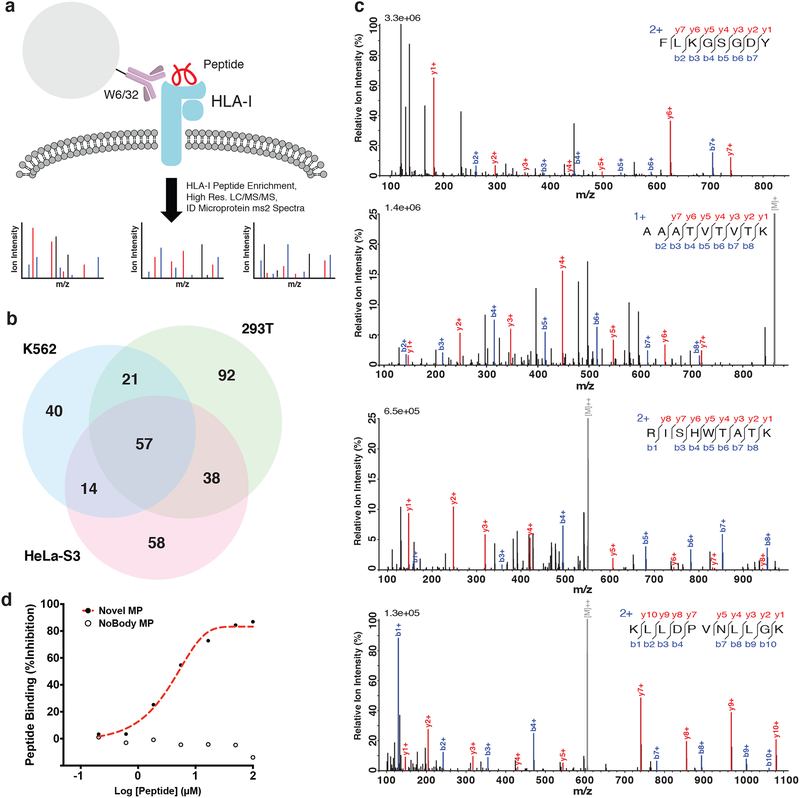
While Ribo-Seq is effective for identifying translated smORFs, it cannot determine whether the encoded microproteins are sufficiently long-lived to be functional. Mass spectrometry provides direct evidence of proteins that accumulate to a concentration above the limit of detection, offering important complementary data. We therefore re-analyzed published proteomics datasets to validate some of the Ribo-Seq predicted translated smORFs. Proteomic analysis of immnuoprecipitated HLA-I complexes has been used to identify antigenic peptides from annotated genes. We reasoned that HLA-I immunoprecipitation serves as an ideal enrichment step to enhance microprotein peptide detection, and simultaneously allow for identification of microprotein-derived antigens (Fig. 6a). Searching a published HLA-I proteomics dataset46 against the human Swiss-Prot database appended with the 7,554 novel predicted microproteins identified peptides from 320 microproteins (Fig. 6b). A previous study detected over 100 microprotein peptides in the same proteomics dataset, which is consistent with and expanded by these data47. Of the 320 microproteins identified, 192 (~60%) were from smORFs identified in at least two Ribo-Seq experiments, 131 (~41%) were found in at least two cell lines, and 279 (~87%) had an in-frame AUG start codon (Supplementary Data 3), which are all higher frequencies than in the total dataset (~36%, ~20%, and ~60%, respectively). Representative spectra demonstrate good fragment ion coverage, regardless of the number of times detected by Ribo-Seq (Fig. 6c). We also verified binding of three microprotein peptides to the HLA-I complex using a fluorescence-based competition assay (Fig. 6d and Supplementary Fig. 13). These results validated the translation of hundreds of smORFs at the protein level, and demonstrated that they are capable of being presented on HLA-I complexes.
Figure 6. Novel microproteins detected in HLA-I complexes.
a Schematic of HLA-I bound peptide enrichment experiment carried out in Bassani-Sternberg et al.46. The pan-HLA-I antibody, W6/32, was used to pull-down and enrich HLA-I complexes, and bound small peptides were further enriched by solid phase extraction. High resolution tandem mass spectrometry data of enriched HLA-I peptide samples (PXD000394) was then searched against a database containing human Swiss-Prot proteins and the 7,554 novel smORF-encoded microproteins. b 320 novel microproteins were identified across all three cell lines. c ms2 spectra examples of peptides from smORFs called translated in (top-bottom): three cell lines, two cell lines, one cell line (multiple experiments), and one cell line (single experiment). d Binding of a novel microprotein peptide, RMKDFLCLK (chr1:39875291–39875422), was validated by a competition-based fluorescence assay. The novel microprotein peptide was able to compete off the control peptide, indicating binding, while the negative control peptide, TPNGGSTTL, from the recently characterized microprotein NoBody13 was not.
Comparison to other smORF databases
Our Ribo-Seq-based workflow differs in several key ways from the SmProt20 and sORFs.org21 databases that improve the quality of our smORF annotations. First, we intentionally incorporated Ribo-Seq data of varying resolution and demonstrate that it affects smORF translation prediction. Neither SmProt nor sORFs.org shows the metagene analyses for the published datasets utilized in their workflows, making it impossible to tell whether the underlying data used is of sufficient quality for smORF annotation. Second, we define smORFs by the most upstream in-frame AUG start codon or stop codon and provide accompanying initiation drug treated samples to help identify the utilized start codon. SmProt does not include initiation data, and sORFs.org includes separate entries for smORFs defined by all possible in-frame AUG and non-AUG start codons, resulting in multiple entries for what is likely a single smORF. Third, neither database incorporates de novo transcript assembly in their workflows. Both other databases do, however, contain predicted translated smORFs that overlap with annotated ORFs, which we leave out due to the increased likelihood of being scored inaccurately with low-resolution data. Another key difference is that sORFs.org uses its own noise-filtering algorithm that does not incorporate 3-nt periodicity, leading to many smORFs called translated despite poor Ribo-Seq evidence. SmProt utilizes RiboTaper for its translation predictions, which incorporates 3-nt periodicity similarly to RibORF and several other translation scoring software packages48. Lastly, sORFs.org contains many smORF entries that overlap in-frame with annotated ORFs, which cannot be separated as unique translation products by Ribo-Seq.
As a result, our gold standard database contains fewer unique annotated smORFs than SmProt and sORFs.org. Despite having more protein-coding smORF entries, both other databases miss a substantial number of smORFs annotated in our datasets. The sORFs.org database contains 3,269 predicted translated smORFs in common with our annotations, only 1,574 of which overlap with our gold standard set (Supplementary Data 1). Similarly, SmProt shares just 1,169 Ribo-Seq annotated smORFs in common, 798 of which overlap with our gold standard set. SmProt and sORFs.org are also missing many smORFs for which we identified peptides in the HLA I proteomics data, including only 217 and 128 out of 320 smORFs, respectively. Thus, our database retains high confidence annotations without incorporating as many likely false positives. Still, both SmProt and sORFs.org include more Ribo-Seq datasets from additional cell lines, and thus likely include bona fide protein-coding smORFs that were not found in our datasets.
Discussion
This study serves three key purposes: the development of a reliable workflow for smORF annotation, the curation of a human protein-coding smORF database, and the demonstration of strategies for finding smORFs related to pathways of interest. Utilizing our workflow, we were able to rigorously annotate thousands of novel protein-coding smORFs in three human cell lines. By analyzing individual experiments, we showed that predicting smORF translation from Ribo-Seq data is noisier than for annotated genes. Differences in Ribo-Seq resolution, sequencing library construction, sequencing depth, as well as biological variations such as passage number and cell density can affect smORF translation analysis. However, given that annotated ORFs showed much greater overlap between the same experiments, it is most likely that overall lower transcription and translation levels explain why smORFs are more difficult to detect reproducibly. We also show that it is beneficial to use a range of RNase I digestion conditions to annotate smORFs, as there are several hundred reproducibly detected smORFs that were only identified in lower or higher resolution datasets. Based on these results, we suggest that there is an optimal range of digestion conditions for identifying translated smORFs, below which causes low accuracy translation predictions and above which causes overall reduced RPF coverage. This latter point is supported by previous studies which showed that monosome stability is particularly sensitive to digestion by RNase I compared to other RNases in some mammalian cell lines and tissues49,50. Importantly, we also demonstrate that de novo transcriptome assembly is necessary for comprehensive smORF annotation.
For many smORFs, these data provide the first evidence of translation. Therefore, we propose using our higher confidence gold standard set of >2,500 smORFs called translated in multiple experiments for follow-up functional studies. For a smaller library, one can use the even higher confidence set of smORFs found in multiple cell lines, which require both transcript assembly and sufficient Ribo-Seq evidence in each cell line. Supporting their higher confidence, our gold standard set is enriched among smORFs validated in the HLA-I proteomics data, though this could also suggest that microproteins found in multiple cell lines are more likely to have peptides presented on HLA-I complexes. Notwithstanding their lack of reproducibility, smORFs identified in a single experiment are worth including in large-scale studies, as many just failed the stringent RibORF scoring filter in other experiments and might pass with higher sequencing depth or in an additional replicate. In support of their inclusion, peptides from singly identified smORFs were also validated in the HLA-I proteomics data, and over 1,800 overlapped with the sORFs.org or SmProt databases.
Beyond reproducibility, useful methods for uncovering biologically functional smORFs include identifying those that are regulated, bound to protein complexes, or evolutionarily conserved. For example, smORFs that are regulated during ER stress, such as UPR-smORF1 and UPR-smORF2, might have functions in the UPR. Similarly, microprotein peptides presented on HLA-I complexes may be immunogenic or serve as useful biomarkers. Functional inferences can also be drawn from microprotein sequence similarity, such as the potential X-linked homeobox gene in HEK293T cells. Having identified thousands of smORFs, additional biological data can easily be mined to help elucidate their roles.
While our data represent a significant step in comprehensive protein-coding smORF annotation, we expect future studies to find additional novel smORFs. First, these numbers are an underestimation, because we chose to exclude smORFs that overlap with annotated ORFs in our analyses, though such smORFs are known16. By definition, overlapping smORFs have RPF reads aligned out-of-frame relative to another ORF, which limits the scoring of both. Our highest resolution datasets may be suitable for identifying abundant overlappers, however, we expect to find a significant number of artifacts using our lower resolution datasets due to the higher percentage of noisy out-of-frame reads. Second, we utilized ENCODE cell lines, which are valuable but likely different from primary cells or tissues. Finally, improvements to transcript assembly through long read sequencing, small RNA library construction, and to computational methods for short read alignment and analysis of Ribo-Seq for translation will be critical for complete annotation of protein-coding smORFs. There are currently several newer translation scoring software that could help identify additional smORFs missed by RibORF48.
Given the number of protein-coding smORFs annotated, their diversity of amino acid composition, and cell type specificity, we anticipate smORFs being involved in all facets of biology. In addition, new insights into translational regulation can be gained by studying polycistronic RNAs and how multiple start sites are employed for the same reading frame. These results also add to the growing evidence that some ncRNAs can operate as both a functional molecule and a coding template. In summary, smORFs offer a rich opportunity for uncovering new biology, and in the future perhaps a new avenue for therapeutic discovery.
Online Methods
Cell Culture
HeLa-S3 cells (CCL-2.2) were purchased from ATCC (Manassas, VA). HEK293T cells (HCL4517) were purchased from GE Life Sciences (Pittsburgh, PA). K562 cells (89121407) were purchased from MilliporeSigma (Carlsbad, CA). HEK293T, and HeLaS3 cells were maintained in DMEM (10–013-CV, Corning, San Diego, CA) supplemented with 10% Fetal Bovine Serum (FBS; Corning, 35–010-CV). K562 cells were maintained in RPMI 1640 (Corning, 10–040-CV) supplemented with 10% FBS. All cells were maintained at 37 °C with 5% CO2.
Paired-End RNA-Seq and de novo Transcriptome Assembly
The HEK293T Cufflinks assembled transcriptome was generated previously28, and used to create the ORF database for scoring translation with RibORF. For HeLaS3 and K562, total RNA was harvested and purified from two biological replicates using an RNeasy Kit (Qiagen, Germantown, MD) with gDNA eliminator columns. For each cell line, two separate cDNA libraries were prepared for each replicate: one using the TruSeq Stranded mRNA Kit (Illumina, San Diego CA) and the other using the TruSeq Total RNA Kit (Illumina). This allowed for representation from poly-A tailed mRNA and non-poly-A RNAs in the transcriptome assembly. Paired-end 125 or 150 base reads were collected for all 4 libraries on a single lane of an Illumina HiSeq 2500 or NextSeq 500, respectively. At least 250M reads were generated for each cell line. Aligned reads were assembled into transcripts by Cufflinks using default parameters, fragment bias correction, multi-read correction, fr-firststrand library construction, and the hg19 human genome sequence as a guide. Cufflinks was used to measure the FPKM values of the assembled transcripts.
Ribosome Footprinting
Preparation of ribosome footprints for Ribo-Seq experiments was performed as described24 with some modifications. For all ribosome footprinting experiments, adherent cells were grown to about 80% confluency in 10 cm or 15 cm diameter tissue culture dishes and suspension cells were grown to a density of approximately 500,000 cells/mL. Cells were washed with 5 mL ice-cold Phosphate Buffered Saline (PBS) with 100 μg/mL cycloheximide (CHX) added. Immediately after removing PBS, 400 μL of ice-cold lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1% Triton X-100, with 1 mM DTT, 25 U/mL Turbo DNase (AM2238, Thermo Fisher, Waltham, MA), and 100 μg/mL CHX added fresh) was dripped onto the plate or added to the cell pellet. Cells were incubated on ice in lysis buffer for 10 min with periodic vortexing and pipetting to disperse the cells. The lysate was then clarified by centrifugation at 15,000 g for 10 min. Cell lysates were flash frozen in liquid nitrogen and stored at −80°C for up to 5 d prior to ribosome footprinting. For experiments profiling translation initiation, the same procedure was followed except for the addition of either 2 μg/mL harringtonine (ab141941, Abcam, Cambridge, MA) for 2 min or 20 μg/mL lactimidomycin (506291, MilliporeSigma) for 30 min to media prior to PBS wash and lysis. A variety of digestion conditions were tested in this study and are summarized in Supplementary Data 1. Briefly, RNA digestions using 250 U RNase I (AM2294, Thermo Fisher) per 100 μL lysate were used in the low resolution 293T and HeLaS3 experiments. For high-resolution experiments, 15 to 30 U TruSeq Nuclease (Illumina) was used to digest 30 to 60 μg RNA in up to 300 μL lysate. Digestion reactions were run for 45 to 60 min at RT and quenched with 100 to 200 U Superase-In RNase I inhibitor (AM2694, Thermo Fisher) on ice. Following digestion, ribosome protected fragments (RPF) were purified from small RNA fragments using MicroSpin S-400 HR columns (GE Life Sciences) according to the TruSeq Ribo Profile Kit (Illumina). Low resolution experiments were cleaned up with Zymo RNA Clean & Concentrator-25 kit, while high resolution experiments were purified by acid phenol:chloroform extraction followed by isopropanol precipitation. Ribosomal RNAs were depleted from RPF fragments by Ribo-Zero Mammalian Kit (Illumina) following the manufacturer’s protocol. cDNA sequencing libraries were then prepared using the TruSeq Ribo Profile Kit (Illumina) following the manufacturer’s protocol. Single-end 50 base reads were collected for each library on an Illumina HiSeq2500 with no more than 4 samples sequenced on a single lane. Each Ribo-Seq experiment was prepared from a different biological replicate except for K562 HiRes1 & 2 which were prepared from the same lysate using different digestion conditions. For K562 HiRes3, CHX was added to the media prior to pelleting cells and washing with PBS.
Ribo-Seq and Short Read RNA-Seq Read Processing
Ribo-Seq and accompanying short fragment total RNA-Seq reads were first trimmed of excess 3’ adaptor sequences as in Calviello et al.24 using the FASTX-toolkit. Trimmed Ribo-Seq reads aligning to tRNA and rRNA sequences were then removed using STAR v2.5.2b51 as in Wang et al.52. Next, the remaining Ribo-Seq reads were aligned to the UCSC hg19 human genome assembly containing chromosomes 1–22, X, and Y with the hg19 refGene transcript annotation using STAR. Up to two mismatches were allowed during alignment, keeping only uniquely mapped reads. Ribo-Seq and RNA-Seq alignments were checked for overall quality using the CollectRnaSeqMetrics script from the Picard Tools software suite.
RibORF Scoring
Following Ribo-Seq read processing and quality control, the RibORF software package22 was used to score individual ORFs for translation. First, metagene analysis was conducted using coding genes from the hg19 refGene annotation included with RibORF. Metagene analysis is run for individual processed read lengths ranging from 25–34 nt. Using the metagene plots, the offset shift needed to align the 5’-most position with the A-site, or +3 position, for each read length is assessed. Next, the entire Ribo-Seq alignment is corrected by the offset shift for each length. For high-resolution data, reads ranging from 25–30 nt in length were included depending on the sample’s footprint length distribution. For lower resolution data, reads ranging from 28–35 nt were included. The offset-corrected read alignments were used for scoring individual ORFs as translated. Following the suggestions of the RibORF developers, only ORFs with RibORF scores ≥0.7 and at least 10 reads mapped to the ORF were considered translated in each individual Ribo-Seq dataset. Each Ribo-Seq dataset was analyzed individually for translated smORF predictions. RNA coverage and Ribo-Seq A-site plots for individual smORFs were plotted using R scripts.
Defining ORFs
RibORF does not define boundaries of putative ORFs based on Ribo-Seq coverage and thus requires a user-generated list of candidate ORFs. Generation of ORF databases from the de novo assembled transcriptome of each cell line was done using a custom java script, GTFtoFASTA (Supplementary Data 4). For each cell line’s de novo assembled transcriptome, ORFs were defined by identifying the most distal in-frame upstream AUG start codon for every stop codon across all three reading frames. Because Ribo-Seq evidence is expected to occur solely within a putative ORF, it is important to limit ORFs to AUG start codons, which are mostly likely to be initiation sites based on the scanning model of translation, when available instead of beginning at upstream stops. However, if no AUG start codon is found, the ORF was defined from stop codon to stop codon to allow for the identification of non-AUG initiated smORFs. The resulting millions of ORFs were then assembled into a database containing the exon coordinates for each ORF in refFlat format. In Ribo-Seq datasets, translation termination peaks are often overrepresented and have a higher fraction of reads aligned to the second position (out of frame) compared to non-stop codons, as observed by metagene analysis (Fig. 2a). Therefore, for RibORF scoring, only the first position of the stop codon was included in the ORF as opposed to the full stop codon. By only including the first position of the stop codon in the ORF definition, we limited the scoring penalty that frequently occurs due to the higher frequency of out of frame reads. A previous study dealt with the extreme nature of translation termination peaks by excluding the stop codon altogether from scoring19, while others include the entire stop codon and do not handle it differently22. While the majority of smORFs called translated do not change whether the stop codon is included or not, our strategy results in the highest number of predicted protein-coding smORFs and offers the best overlap with each alternative option across all different levels of overall Ribo-Seq resolution tested (Supplementary Fig. 14).
Differential Translation Analysis
Differential translation analysis was conducted using the R package Xtail v1.1.535. First, HTSeq-count53 in intersection-strict mode was used to calculate total RNA read counts for hg19 refGene annotations. For smORFs, HTSeq-count was run in union mode and allowed for non-unique reads to be counted. RPF read counts for the same annotations were calculated using the custom python script in Xiao et al.35, which retains only uniquely mapped reads occurring within the middle of the CDS region. For hg19 RefGene annotated genes, reads aligning after the first 15 codons and before the last 5 codons were counted. For novel protein-coding smORFs, reads aligning after the first and before the last codon were counted. Xtail was used to calculate the log2 fold-changes in TE between DMSO- and tunicamycin- or thapsigargin-treated cells from the read count tables. Genes not considered ‘stable’ by xtail and with a log2 fold-change ≥ 1 or ≤ −1 were assigned as either ‘homodirectional,’ ‘transcription-only,’ or ‘translation-only’ category of differential translation. DESeq254 was also run in parallel with Xtail to calculate differential mRNA expression for hg19 refGene annotations and smORFs. Plots summarizing the results from both analyses were generated using R.
PhyloCSF and BLAST Analyses of protein-coding smORFs
Smoothed PhyloCSF scores for the 29-mammals alignment were extracted for all smORFs from the UCSC genome browser’s PhyloCSF Track Hub using the bedtools map function. The scores represent the log-odds that codons in the smORF are in the coding state. The average smoothed PhyloCSF scores are shown for each protein-coding smORF by exon (Supplementary Data 1).
All smORFs were queried for similarity against the non-redundant database using tBLASTn and BLASTP under default parameters. BLAST alignments were considered significant if the BLAST score ≥ 80 or if ≥80% of the microprotein sequence matched ≥80% of the aligned subject sequence. This second condition allowed for the identification of short but high similarity sequence alignments, which otherwise have a low BLAST score under default parameters.
Microprotein domain predictions
Microprotein sequences were assessed for possible transmembrane helices using TMHMM2.055 under default parameters. Sequences were also analyzed for similarity to known protein domains using the CD-Search tool and the Conserved Domain Database56 v3.16.
Mass Spectrometry Data Analysis
Mass spectrometry data from PXD00039446 were downloaded from the PRIDE archive. Tandem mass spectra were extracted from RAW files using RawConverter 1.0.0.0. Next, the spectra were searched against a database containing human Swiss-Prot proteins, novel microproteins, and common contaminants using ProLuCID57. The enzyme specificity was set to none and no variable modifications were included. The false discovery rate was set to 1% for peptides. Identified spectra were then filtered and grouped into proteins using DTASelect58. Mass spectrometry analyses were separated by different cell lines from the study. We also utilized the pFind 3 Open-pFind59 search engine to identify microprotein-derived peptides by an open search strategy, which allows for many variable modifications, using the same database and false discovery rate.
HLA-I peptide binding assay
The affinities of novel microprotein-derived peptides for HLA-I were measured as previously described60. Briefly, SupB15 cells (HLA-I: A3, A11, B51, B52 serotype) were harvested and the cell surface HLA complex was disassembled by treating with citric acid elution buffer (pH 2.9) for 90 seconds. Then, cells were incubated with a high-affinity fluorescein-labeled reference peptide KVFPC(FITC)ALINK (1 μM) and increasing concentrations of a non-labeled microprotein-derived peptide for 20 hours at 4°C. A negative control peptide from the recently characterized microprotein NoBody13 (TPNGGSTTL, B7 serotype binder) was also tested for comparison. Fluorescence intensities were measured by flow cytometry. Binding of novel microprotein-derived peptides at each concentration was calculated as percentage inhibition of reference peptide binding relative to background (without reference peptide, MFbg) and the maximal response (reference peptide only, MFref) using the following equation:
The data were then plotted and fit for IC50 calculation using Prism 5.
Peptide synthesis
Peptides were purchased from Peptide 2.0. Fluorescein-labeled reference peptide KVFPC(FITC)ALINK was synthesized by covalently coupling of fluorescein to the cysteine residue with 5-(iodoacetamido)fluorescein (M0638, Marker Gene Technologies, Eugene, OR) for use in the HLA-binding assay. All peptides were purified by high-performance liquid chromatography and confirmed by mass spectrometry.
Supplementary Material
Acknowledgements
We thank the Saghatelian lab for helpful comments and suggestions throughout the study, and N. Ingolia for advice on RNase I digestion conditions. We also thank M. Ku, N. Hah, and the Salk Institute Next Generation Sequencing Core for preparation of RNA-Seq libraries and high-throughput sequencing of Ribo-Seq and RNA-Seq libraries. This research was supported by NIH/NIGMS (R01 GM102491, A.S.), Leona M. and Harry B. Helmsley Charitable Trust grant (A.S.), Dr. Frederick Paulsen Chair/Ferring Pharmaceuticals (A.S.), NIH/NIGMS postdoctoral fellowship (F32 GM123685, T.F.M.), George E. Hewitt Foundation for medical research (Q.C.), Pioneer Fellowship (D.T.). This work was also supported by the Razavi Newman Integrative Genomics and Bioinformatics Core and the Next Generation Sequencing Core Facilities of the Salk Institute with funding from the NIH-NCICCSG (P30 014195) and the Chapman Foundation.
Footnotes
Competing interests: The authors declare no competing interests.
Data Availability
All sequencing datasets generated in this study are available through GEO (GSE125218).
Code Availability
A custom java script used for 3-frame in silico translation of assembled transcripts is included as Supplementary Data 4.
References
- 1.Basrai MA, Hieter P & Boeke JD Small open reading frames: beautiful needles in the haystack. Genome Res 7, 768–71 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Ochman H Distinguishing the ORFs from the ELFs: short bacterial genes and the annotation of genomes. Trends Genet 18, 335–7 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Lawrence J When ELFs are ORFs, but don’t act like them. Trends Genet 19, 131–2 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Dujon B et al. Complete DNA sequence of yeast chromosome XI. Nature 369, 371–8 (1994). [DOI] [PubMed] [Google Scholar]
- 5.Goffeau A et al. Life with 6000 genes. Science 274, 546, 563–7 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Saghatelian A & Couso JP Discovery and characterization of smORF-encoded bioactive polypeptides. Nat Chem Biol 11, 909–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couso JP & Patraquim P Classification and function of small open reading frames. Nat Rev Mol Cell Biol 18, 575–589 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Galindo MI, Pueyo JI, Fouix S, Bishop SA & Couso JP Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol 5, e106 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondo T et al. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 9, 660–5 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Arnoult N et al. Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature 549, 548–552 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rathore A et al. MIEF1 Microprotein Regulates Mitochondrial Translation. Biochemistry 57, 5564–5575 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stein CS et al. Mitoregulin: A lncRNA-Encoded Microprotein that Supports Mitochondrial Supercomplexes and Respiratory Efficiency. Cell Rep 23, 3710–3720 e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D’Lima NG et al. A human microprotein that interacts with the mRNA decapping complex. Nat Chem Biol 13, 174–180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Q et al. The microprotein Minion controls cell fusion and muscle formation. Nat Commun 8, 15664 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma J et al. Improved Identification and Analysis of Small Open Reading Frame Encoded Polypeptides. Anal Chem 88, 3967–75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slavoff SA et al. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nature chemical biology 9, 59–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ingolia NT, Ghaemmaghami S, Newman JRS & Weissman JS Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–23 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aspden JL et al. Extensive translation of small open reading frames revealed by Poly-Ribo-Seq. Elife 3, e03528 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bazzini AA et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J 33, 981–93 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hao Y et al. SmProt: a database of small proteins encoded by annotated coding and non-coding RNA loci. Brief Bioinform 19, 636–643 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Olexiouk V, Van Criekinge W & Menschaert G An update on sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res 46, D497–D502 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji Z, Song R, Regev A & Struhl K Many lncRNAs, 5’UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife 4, e08890 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu PY et al. Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proc Natl Acad Sci U S A 113, E7126–E7135 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calviello L et al. Detecting actively translated open reading frames in ribosome profiling data. Nat Methods 13, 165–70 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Raj A et al. Thousands of novel translated open reading frames in humans inferred by ribosome footprint profiling. eLife 5, e13328 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diament A & Tuller T Estimation of ribosome profiling performance and reproducibility at various levels of resolution. Biol Direct 11, 24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robasky K, Lewis NE & Church GM The role of replicates for error mitigation in next-generation sequencing. Nat Rev Genet 15, 56–62 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma J, Saghatelian A & Shokhirev MN The influence of transcript assembly on the proteogenomics discovery of microproteins. PLoS One 13, e0194518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oslowski CM & Urano F Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol 490, 71–92 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C-L et al. Genome-wide analysis of tunicamycin-induced endoplasmic reticulum stress response and the protective effect of endoplasmic reticulum inhibitors in neonatal rat cardiomyocytes. Mol Cell Biochem 413, 57–67 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Xu J & Zhang J Are Human Translated Pseudogenes Functional? Mol Biol Evol 33, 755–60 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gjymishka A, Su N & Kilberg MS Transcriptional induction of the human asparagine synthetase gene during the unfolded protein response does not require the ATF6 and IRE1/XBP1 arms of the pathway. Biochem J 417, 695–703 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andreev DE et al. Translation of 5 leaders is pervasive in genes resistant to eIF2 repression. Elife 4, e03971 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sidrauski C, McGeachy AM, Ingolia NT & Walter P The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. Elife 4(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao Z, Zou Q, Liu Y & Yang X Genome-wide assessment of differential translations with ribosome profiling data. Nat Commun 7, 11194 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guan BJ et al. Translational control during endoplasmic reticulum stress beyond phosphorylation of the translation initiation factor eIF2alpha. J Biol Chem 289, 12593–611 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao C, Datta S, Mandal P, Xu S & Hamilton T Stress-sensitive regulation of IFRD1 mRNA decay is mediated by an upstream open reading frame. J Biol Chem 285, 8552–62 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sundaram A, Plumb R, Appathurai S & Mariappan M The Sec61 translocon limits IRE1alpha signaling during the unfolded protein response. Elife 6(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Consortium EP An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chew GL, Pauli A & Schier AF Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nat Commun 7, 11663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delcourt V et al. The Protein Coded by a Short Open Reading Frame, Not by the Annotated Coding Sequence, Is the Main Gene Product of the Dual-Coding Gene MIEF1. Mol Cell Proteomics 17, 2402–2411 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brocchieri L & Karlin S Protein length in eukaryotic and prokaryotic proteomes. Nucleic Acids Res 33, 3390–400 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin MF, Jungreis I & Kellis M PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27, i275–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ingolia NT, Brar GA, Rouskin S, McGeachy AM & Weissman JS Genome-wide annotation and quantitation of translation by ribosome profiling. Curr Protoc Mol Biol Chapter 4, Unit 4.18 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacLean JA 2nd, & Wilkinson MF The Rhox genes. Reproduction 140, 195–213 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bassani-Sternberg M, Pletscher-Frankild S, Jensen LJ & Mann M Mass Spectrometry of Human Leukocyte Antigen Class I Peptidomes Reveals Strong Effects of Protein Abundance and Turnover on Antigen Presentation. Molecular & Cellular Proteomics 14, 658–673 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Erhard F et al. Improved Ribo-seq enables identification of cryptic translation events. Nat Methods 15, 363–366 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calviello L & Ohler U Beyond Read-Counts: Ribo-seq Data Analysis to Understand the Functions of the Transcriptome. Trends Genet 33, 728–744 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Cenik C et al. Integrative analysis of RNA, translation, and protein levels reveals distinct regulatory variation across humans. Genome Res 25, 1610–21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerashchenko MV & Gladyshev VN Ribonuclease selection for ribosome profiling. Nucleic Acids Res 45, e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang H, McManus J & Kingsford C Isoform-level ribosome occupancy estimation guided by transcript abundance with Ribomap. Bioinformatics 32, 1880–2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anders S, Pyl PT & Huber W HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krogh A, Larsson B, von Heijne G & Sonnhammer EL Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305, 567–80 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Marchler-Bauer A et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45, D200–D203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu T et al. ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J Proteomics 129, 16–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cociorva D, L.T. D & Yates JR Validation of tandem mass spectrometry database search results using DTASelect. Curr Protoc Bioinformatics Chapter 13, Unit 13 4 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Chi H et al. Comprehensive identification of peptides in tandem mass spectra using an efficient open search engine. Nat Biotechnol 36, 1059–1061 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Kessler JH et al. Competition-based cellular peptide binding assay for HLA class I. Curr Protoc Immunol Chapter 18, Unit 18 12 (2004) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.