Abstract
Non-small cell lung cancer (NSCLC) accounts for about 85% of all lung cancer cases. The pathogenesis of NSCLC involves complex gene networks that include different types of non-coding RNAs, such as long non-coding RNAs (lncRNAs). The role of lncRNAs in NSCLC is gaining an increasing interest as their function is being explored in various human cancers. Recently, a new oncogenic lncRNA, LINC00152 (cytoskeleton regulator RNA (CYTOR)), has been identified in different tumor types. In NSCLC, the high expression of LINC00152 in tumor tissue and peripheral blood samples has been shown to be associated with worse prognoses of NSCLC patients. Overexpression of LINC00152 has been confirmed to promote the proliferation, invasion, and migration of NSCLC cells in vitro, as well as increase tumor growth in vivo. This review discusses the role of LINC00152 in NSCLC.
Keywords: Long non-coding RNA, LINC00152, Non-small cell lung cancer, Proliferation, Prognosis
1. Introduction
Lung cancer is one of the most common malignancies and is the leading cause of cancer-related mortality worldwide (Fidler and Bray, 2018). Around 85% of all lung cancer cases have been attributed to non-small cell lung cancer (NSCLC), which includes different histological types such as lung adenocarcinoma, squamous cell carcinoma, and large cell lung cancer. Among these subtypes, lung adenocarcinoma is the most common NSCLC (Ettinger et al., 2015). Despite the gratifying progress in the understanding of its molecular mechanisms and in the discovery of potential clinical treatments, the prognosis of lung cancer patients remains unsatisfactory. Indeed, most patients are diagnosed after reaching advanced stages, which hinders their chances to receive optimal treatment. Consequently, the five-year overall survival rate of NSCLC is only about 15% (Torre et al., 2016).
The role of long non-coding RNAs (lncRNAs) in the pathogenesis and progression of NSCLC is gaining increasing attention with the rapid development of high-throughput sequencing and various omics technologies (Esfandi et al., 2019). Some lncRNAs, such as HOX transcript antisense RNA (HOTAIR) (Liu et al., 2013), maternally expressed 3 (MEG3) (Liu et al., 2015), and colon cancer-associated transcript 1 (CCAT1) (Chen J et al., 2016), have been shown to participate in the etiology and deterioration of NSCLC, which can affect NSCLC diagnosis and treatment. Therefore, illuminating the function of lncRNAs would provide new insights to explore the molecular characteristics of NSCLC and would offer new possibilities to develop more effective therapeutic strategies (Bhan et al., 2017).
A newly discovered lncRNA, LINC00152 (cytoskeleton regulator RNA (CYTOR)), has been recently shown to exert various carcinogenic effects in a variety of tumors and has been demonstrated to serve as a potential diagnostic and prognostic biomarker (Bian et al., 2017; Deng et al., 2017; Cai et al., 2018; Chen PX et al., 2018). This review focuses on the pivotal role of LINC00152 in NSCLC.
2. Characteristics of lncRNA
While having little or no protein-coding capacity, lncRNAs are usually more than 200 nucleotides in length and participate in multiple biological processes (Dey et al., 2014). During tumor progression, lncRNAs play vital regulatory roles at epigenetic, transcriptional, and post-transcriptional levels (Orom et al., 2010). The function of lncRNAs is highly associated with their localization within the cells. In the nucleus, lncRNAs regulate gene expression by binding to transcription factors, chromatin modifiers, and heterogeneous nuclear ribonucleoproteins (hnRNPs). They can also regulate splicing, stabilization, and translation of host messenger RNAs (mRNAs) through post-transcriptional mechanism (Orom et al., 2010). On the other hand, lncRNAs in cytoplasm cannot only regulate the stability and translation of mRNAs, but also are involved in cellular signaling cascades. They can also bind specific microRNAs (miRNAs) as competing endogenous RNA (ceRNA), thus acting as “miRNA sponges” to protect target mRNAs from inhibition (Fatica and Bozzoni, 2014). In addition, some lncRNAs in the cytoplasm that contain small open reading frame (ORF) can be translated into bioactive short peptides (Choi et al., 2019).
LncRNAs act in various cancers either as tumor suppressors or oncogenes (Wang Y et al., 2018). In some classical tumor-related signaling pathways such as p53, nuclear factor-κB (NF-κB), and phosphoinositide-3-kinase (PI3K)/AKT, lncRNAs can serve as the scaffold for receptors, protein kinases, and transcription factors in signaling cascades (Peng et al., 2017).
3. Relation of overexpression of LINC00152 with worse prognosis of NSCLC patients
The LINC00152 gene is located on chromosome 2p11.2, with a transcript length of 828 nucleotides. The localization of LINC00152 differs between tumor cells from different origins (Table 1). Nevertheless, LINC00152 acts as an oncogene regardless of its localization in tumor cells. Yu Y et al. (2017) reviewed the pivotal oncogenic effect of LINC00152 in different human cancers, including gastric cancer, hepatocellular carcinoma, colon cancer, gallbladder cancer, and renal cell carcinoma. While the carcinogenic function of LINC00152 has been confirmed in multiple cancers (Table 2), only one study on colon cancer showed contradictory findings about the expression and mechanism of action of LINC00152 (Zhang et al., 2016). This discrepancy could be related to different factors such as the sample size, demographic characteristics, polymerase chain reaction (PCR) primers, experimental protocols, and laboratory conditions.
Table 1.
Predominant distribution of LINC00152 in tumor cells from different origins
| Type of cancer | Nucleus | Cytoplasm | Reference |
| Hepatoma | Yes | Ji et al., 2015 | |
| Gastric cancer | Yes | Zhou et al., 2015; Chen WM et al., 2016 | |
| Osteosarcoma | Yes | Zheng et al., 2019 | |
| Ovarian cancer | Yes | Chen PX et al., 2018 | |
| Glioma | Yes | Reon et al., 2018 | |
| Breast cancer | Yes | Yes | Wu et al., 2018; Shen et al., 2019 |
| Kidney cancer | Yes | Yes | Wang et al., 2017 |
| Lung cancer | Yes | Yes | Chen et al., 2017; Feng et al., 2017; Zhang and Li, 2018 |
| Colon cancer | Yes | Yes | Bian et al., 2017; Nishizawa et al., 2018; Wang X et al., 2018 |
Table 2.
Expression of LINC00152 in tumor tissues from different origins
| Type of cancer | Expression level | Reference |
| Hepatoma | ↑ | Ji et al., 2015; Ma et al., 2018 |
| Gastric cancer | ↑ | Zhao et al., 2015; Zhou et al., 2015; Chen WM et al., 2016; Yang et al., 2016; Huang et al., 2018; Wang HF et al., 2019 |
| Osteosarcoma | ↑ | Zheng et al., 2019 |
| Ovarian cancer | ↑ | Chen PX et al., 2018 |
| Glioma | ↑ | Yu MJ et al., 2017; Cai et al., 2018; Chen X et al., 2018; Liu et al., 2018; Reon et al., 2018; Zhu et al., 2018 |
| Breast cancer | ↑ | Wu et al., 2018; Shen et al., 2019 |
| Kidney cancer | ↑ | Wu et al., 2016; Wang et al., 2017 |
| Gallbladder cancer | ↑ | Cai et al., 2016, 2017 |
| Tongue squamous cell carcinoma | ↑ | Yu JJ et al., 2017 |
| Oral cancer | ↑ | Li MH et al., 2018; Chen et al., 2019 |
| Esophageal squamous cell carcinoma | ↑ | Hu HB et al., 2016; Liu DL et al., 2019 |
| Hemangioma | ↑ | Wang YL et al., 2019 |
| Acute myeloid leukemia | ↑ | Zhang and Tao, 2019 |
| Multiple myeloma | ↑ | Yu et al., 2018 |
| Retinoblastoma | ↑ | Li SH et al., 2018 |
| Papillary thyroid cancer | ↑ | Sun et al., 2019 |
| Lung cancer | ↑ | Chen et al., 2017; Feng et al., 2017; Li et al., 2017; Ma et al., 2017; Nötzold et al., 2017; Xu et al., 2017; Zhang PP et al., 2017; Reon et al., 2018; Zhang and Li, 2018; Zhao et al., 2018; Liu ZZ et al., 2019 |
| Colon cancer | ↑ | Qiu and Yan, 2015; Bian et al., 2017; Nishizawa et al., 2018; Wang X et al., 2018; Yue et al., 2018 |
| ↓ | Zhang et al., 2016 |
LINC00152 is overexpressed in lung adenocarcinoma tissues and can be used as a predictor of reduced patient survival (Table 3). It serves as an independent risk factor for disease-free survival (DFS) and overall survival (OS) (Zhang PP et al., 2017). This conclusion has been also confirmed by other reports (Feng et al., 2017; Reon et al., 2018) on the Gene Expression Omnibus (GEO) (Seo et al., 2012) and The Cancer Genome Atlas (TCGA) databases (Collisson et al., 2014). Zhao et al. (2018) analyzed the expression profiles of lncRNAs in three pairs of lung adenocarcinoma tissues and adjacent normal tissues. They found that LINC00152, LINC00691, and LINC00578 are the most significantly up-regulated lncRNAs in lung adenocarcinoma tissues, compared with adjacent tissues. The results were further identified by quantitative real-time polymerase chain reaction (qRT-PCR) in 90 lung adenocarcinoma tissue samples. Unexpectedly, LINC00152 showed no significant correlation with the clinicopathologic characteristics including age, gender, tumor stage, or metastasis (Zhao et al., 2018). These inconsistent results indicate that future studies warrant more accurate experimental designs and standard operating procedures with larger sample sizes.
Table 3.
LINC00152 expression in NSCLC tissues
| Tissue sample (n) | Expression level | Correlation | No correlation | Reference |
| 101 | ↑ | Poor prognosis | Age, gender, smoking history, tumor differentiation, tumor stage, lymph node status, KRAS mutation | Feng et al., 2017 |
| 60 | ↑ | TNM stage, tumor size, lymph node metastasis, poor survival | Age, gender, smoking history | Chen et al., 2017 |
| 64 | ↑ | Tumor size, advanced TNM stage, lymph node metastasis, shorter overall survival | Gender, age, smoking status, differentiation grade | Zhang and Li, 2018 |
| 110 | ↑ | Lymph node metastasis, distant metastasis, TNM stage, decreased survival | T stage of lung adenocarcinoma | Zhang PP et al., 2017 |
| 27 | ↑ | Nötzold et al., 2017 | ||
| 30 | ↑ | EGFR expression | Zhang Y et al., 2017 | |
| 80 | ↑ | Smoking history, tumor size, differentiation, metastasis | Liu ZZ et al., 2019 | |
| 90 | ↑ | Age, gender, tumor stage, metastasis | Zhao et al., 2018 | |
| 72 | Tumor size, tumor stage | Age, gender, histology subtype, lymph node metastasis | Li et al., 2017 |
EGFR: epidermal growth factor receptor; TNM: tumor-node-metastasis
4. Circulating LINC00152 as a biomarker for NSCLC
Tumor cells can release a large number of RNAs into the peripheral blood (Rapisuwon et al., 2016). These RNAs, such as lncRNAs, can be used as biomarkers to quantitatively measure the progression of pathological disorders in tumor patients (Hu HB et al., 2016; Hu XD et al., 2016; Yang et al., 2016; Mao et al., 2019). Some lncRNAs, such as metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), HOTAIR, and growth arrest specific 5 (GAS5), have been found to be abnormally expressed in the serum of patients with NSCLC (Xie et al., 2018).
Li et al. (2017) measured the expression level of LINC00152 in the plasma of 100 NSCLC patients. They found that LINC00152 expression is markedly up-regulated and significantly associated with tumor size and stage. They also observed that LINC00152 was stable in whole blood and plasma samples by environmental intervention through the inappropriate storage condition of plasma samples. In addition, LINC00152 level was able to effectively discriminate NSCLC patients from healthy controls and patients with benign lung disease, although there was no apparent variance in plasma LINC00152 level between the latter two groups. Moreover, there was a positive relationship between plasma LINC00152 level and a traditional NSCLC marker, carcinoembryonic antigen (CEA). The combined measurement of LINC00152 and CEA showed a higher diagnostic ability, compared with LINC00152 or CEA alone. Even for NSCLC patients in stage I, the plasma level of LINC00152 showed higher diagnostic accuracy than that of CEA. Li et al. (2017) also observed a significant positive correlation between LINC00152 level in tumor tissue and that in plasma samples. Furthermore, post-operative plasma LINC00152 levels in 47 patients one month after surgery were lower than the preoperative levels in the same patients. In relapsed patients, plasma LINC00152 level rapidly rebounded, suggesting that circulating lncRNAs could be mainly originated from tumor cells.
Smoking is one of the causes of lung cancer. A recent report demonstrated that LINC00152 was present at higher levels in the serum of smokers and was positively correlated with the exposure dose (Liu ZZ et al., 2019). Li J et al. (2015) also reported that lncRNAs in circulating peripheral blood can be used as biomarkers of diagnosis and prognosis. One explanation of the potential mechanism is that lncRNAs can be selectively encapsulated into membrane-covered vesicles including exosomes, particles, and apoptotic bodies. LncRNAs can also be folded into complex secondary and tertiary structures, or can bind to proteins to avoid degradation. Another possible explanation is that circulating lncRNAs resist RNase digestion through methylation, adenylation, and uridinylation (Li QE et al., 2015).
5. Effect of LINC00152 on NSCLC cells
LINC00152 is highly expressed in lung cancer cells (Liu ZZ et al., 2019). Chen et al. (2017) examined LINC00152 level in six lung adenocarcinoma cell lines: A549, SPCA1, PC-9, H1299, H1975, and H226. They found that A549 and SPCA1 cells had the highest expression level of LINC00152, while H1299 cells expressed the lowest level. The heterogeneity of basal LINC00152 expression within cell lines of various options can be exploited to perform both knockdown and overexpression experiments to elucidate in vitro function (Chen et al., 2017).
Overexpression of LINC00152 in NSCLC cells facilitates cell proliferation, invasion, and migration. In contrast, down-regulation of LINC00152 significantly inhibits lung cancer cells’ proliferation, invasion, and migration in vitro. It also restrains lung tumor growth and distant metastasis in vivo (Zhang PP et al., 2017; Zhang Y et al., 2017). Zhang Y et al. (2017) speculated that the oncogenic effect of LINC00152 depends on epidermal growth factor receptor (EGFR)/PI3K/AKT signaling pathway. However, their study lacked direct evidence. Interestingly, studies on gastric cancer and vascular endothelial cells confirmed that LINC00152 can activate PI3K/AKT signaling pathway through direct binding to EGFR (Zhou et al., 2015; Teng et al., 2017).
Epithelial mesenchymal transition (EMT) plays a critical role in the process of tumor metastasis. Ectopic expression of LINC00152 expedites EMT program. Some EMT markers (for example, epithelial marker E-cadherin) were negatively correlated with LINC00152 level. Other EMT markers, such as mesenchymal markers N-cadherin and vimentin, were positively correlated with LINC00152 level in various types of tumors (Zhang Y et al., 2017; Hu et al., 2018; Li MH et al., 2018).
LINC00152 regulates EMT process through several canonical molecular pathways. For example, LINC00152 was shown to mediate EMT process through Wnt/β-catenin signaling pathway in colon cancer (Yue et al., 2018). LINC00152 blocks casein kinase 1 (CK1)–β-catenin interaction by competitively binding non-phospho-β-cateninSer45. Then it drives β-catenin nuclear translocation. After entering nucleus, β-catenin binds to T-cell factor (TCF). The β-catenin/TCF complex then induces nuclear LINC00152 transcription and directly trans-activates downstream target genes via TCF-binding elements (TBEs) that are presented in their cis-regulatory regions, thus forming a positive feedback loop in colon cancer cells to promote EMT process. Another report in colon cancer cells found a direct interaction between LINC00152 and two RNA binding proteins, nucleolin (NCL) and Sam68 (Src-associated in mitosis, 68 kDa). The NCL-LINC00152-Sam68 complex was shown to promote colon cancer progression and EMT process by activating NF-κB signaling pathway. In this progress, EXON1 of LINC00152 is indispensable for its interaction with NCL and Sam68 (Wang X et al., 2018). These results suggest that the regulatory network by LINC00152 is complex and that future studies to analyze the mechanism of LINC00152 in EMT process of lung cancer cells will require referral to the current evidence.
Through analyzing RNA-Seq data, Feng et al. (2017) confirmed that LINC00152, with histone acetylation, was highly expressed in 30 NSCLC cell lines. However, the DNA copy number or promoter DNA methylation of LINC00152 genomic locus was not changed. After silencing LINC00152 in 12 lung cancer cell lines, the authors observed that cell proliferation was repressed in nine cell lines. Interestingly, while the growth of PC-9 (EGFR mutant) and H838 (EGFR wild type) cells was both most sensitive to LINC00152 knockdown, the invasion abilities of these two cell lines were not affected. In addition, in contrast to the results of Zhang PP et al. (2017), LINC00152 knockdown did not change the growth of H1650 lung cancer cells. Hence, the underlying reason behind these findings warrants further investigation.
The p38 mitogen-activated protein kinase (MAPK) pathway is known to respond to various extracellular stresses (Cuadrado and Nebreda, 2010). Cyclic adenosine monophosphate (cAMP)-responsive element binding protein 1 (CREB1), signal transducer and activator of transcription 1 (STAT1), and signal transducer and activator of transcription 3 (STAT3) are all key elements in this signaling pathway. CREB1 modulates cell cycle and apoptosis by up-regulating the expression of cyclin E1 (CCNE1). P38 knockdown can lead to increased expression of phosphorylated STAT3 (p-STAT3), which in turn up-regulates other genes involved in tumor cell proliferation and survival, such as MYC proto-oncogene (c-MYC), cyclin D1 (CCND1), cyclin D2, B-cell lymphoma-extra large (BCL-XL), and myeloid cell leukemia 1 (MCL1). Feng et al. (2017) showed that small interfering RNA (siRNA)-based knockdown of LINC00152 reduced the protein expression of p38a, STAT1, STAT3, CREB1, CCNE1, and c-MYC. However, the mRNA expression of the genes did not change, except for STAT3 and CCNE1, which suggests that LINC00152 might regulate STAT3 and CCNE1 expression at transcriptional level, while it can regulate the expression of the other genes at post-transcriptional level. It is worth noticing that the protein expression of EGFR, AKT serine/threonine kinase 1 (AKT1), or extracellular signal-regulated kinase 1/2 (ERK1/2) did not change in this study, which suggests that cell growth would be affected if LINC00152 reaches a certain level and that the genomic background might be a key element for LINC00152 to exert its effects on cell growth. These results are very encouraging to further conduct follow-up studies on the activity of LINC00152 in NSCLC.
6. LINC00152 involved in cell cycle arrest and apoptosis in lung adenocarcinoma
Zhang Y et al. (2017) reported that LINC00152 knockdown impaired cell proliferation, invasion, and metastasis in lung adenocarcinoma. LINC00152 knockdown also promoted cell apoptosis and induced cell cycle arrest in G1 phase. Their results showed that the protein expression levels of EGFR, PI3K, AKT, fibronectin, and vimentin in tumor cells significantly decreased after LINC00152 down-regulation. In contrast, the protein level of cyclin-dependent kinase inhibitor 1A (p21), associated with cell cycle arrest and apoptosis (El-Deiry, 2016), was obviously increased (Zhang Y et al., 2017). The enhancer of zeste homolog 2 (EZH2) is a core catalytic element of polycomb repressive complex 2 (PRC2), which inhibits the transcription of target gene and affects cancer progression by modulating histone H3 lysine 27 trimethylation (H3K27me3). A study on gastric cancer demonstrated that LINC00152 can inhibit target gene expression at epigenetic level by recruiting EZH2 to cyclin-dependent kinase inhibitor 2B (p15) and p21 promoters through H3K27me3, thereby promoting cell proliferation and cell cycle progression (Chen WM et al., 2016).
Chen et al. (2017) demonstrated that LINC00152 knockdown can promote apoptosis and induce cell cycle at G1 phase. In contrary to the results reported by Feng et al. (2017), they observed that LINC00152 was predominantly distributed in the nucleus of A549 and SPCA1 cells, and it can directly bind to EZH2 and lysine demethylase 1A (LSD1) in lung cancer cells. Their result of chromatin immunoprecipitation (ChIP) further demonstrated that EZH2, but not LSD1, was bound to the promoter locus of interleukin 24 (IL-24). Correspondingly, LINC00152 knockdown reduced H3K27me3 occupation in IL24 promoter region. These results indicated that, at least partially, LINC00152 can repress IL24 expression at epigenetic level by recruiting EZH2 to IL24 promoter, thereby promoting the proliferation of lung adenocarcinoma cells. A study on renal cancer also showed that LINC00152 can inhibit cyclin-dependent kinase inhibitor 2A (p16) expression through interaction with EZH2 and LSD1 at epigenetic level by regulating H3K27me3 (Wang et al., 2017). These findings revealed that the mechanism of LINC00152 is heterogeneous, even in homologous tumors, which should be considered in future studies.
Liu ZZ et al. (2019) established a malignant transformed cell model (16HBE-M) by treating human bronchial epithelial cell lines with cigarette smoke extract (CSE). During this process, LINC00152 promoted cell malignant transformation by binding to miR-193b, leading to the up-regulated CCND1. Silencing LINC00152 inhibited proliferation, invasion, and metastasis of 16HBE-M and lung cancer cells along with cell cycle arrest at G0/G1 phase. In contrast, Reon et al. (2018) reported that the interaction with PRC2 or the M8 hairpin RNA is not necessary for LINC00152-mediated invasion. They also ruled out the “miRNA sponge” mechanism proposed by Zhang and Li (2018). M8 is a protein-binding site that constitutes nucleotides 280‒401 of the stem-loop structure at the 3' end of LINC00152, and its overexpression has been shown to stimulate glioma cells invasion.
While LINC00152 and CCND1 were up-regulated with increased dose and duration, miR-193b level was down-regulated in 16HBE-M cells acutely exposed to CSE. Similarly, the expression of E-cadherin decreased, while that of N-cadherin and vimentin increased after CSE treatment. In addition, the proportion of cells in G0/G1 phase was remarkably reduced, while that in S and G2/M phases was obviously elevated. LINC00152 or CCND1 knockdown lowered the mRNA and protein expression of cyclin-dependent kinase 4 (CDK4), CDK6, retinoblastoma protein (Rb), and E2F transcription factor 1 (E2F1), which indicates that LINC00152 can control cell cycle and proliferation through CCND1 and its downstream CDK4/CDK6/Rb/E2F1 pathway (Liu ZZ et al., 2019).
It has been shown that LINC00152 plays a crucial role in cell cycle progression through the regulation of mitosis. LINC00152 depletion induced mitotic arrest in about 75% HeLa cells, of which 42% underwent apoptosis, 28% exited mitosis, and 5% experienced cytokinesis failure. The remaining 25% of cells died directly at an earlier time point (12–24 h). Therefore, apoptosis may not be a direct consequence of LINC00152 knockdown, and it might be a follow-up event that occurs after cell cycle arrest (Nötzold et al., 2017).
The mechanism of LINC00152 in NSCLC is still far from being elucidated. However, clues can be obtained from other diseases to identify potential targets of LINC00152 regulatory network. The cellular distribution and the expression level of LINC00152 followed by manual intervention should be taken into account in in vitro experiments. Until now, in vivo studies using lncRNA knockdown strategies to reduce NSCLC tumor growth are very limited (Gutschner et al., 2013). This could be mainly related to the challenging nature of the experimental strategies, such as RNA interference (RNAi), antisense oligonucleotide (ASO), morpholinos, and clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 9 nuclease (Cas9) (Arun et al., 2018). ASO is an ideal approach for achieving effective lncRNA knockdown that can exert noteworthy inhibitory effects on tumor growth and progression (Crooke, 2017). One of the major advances in ASO chemistry is the development of locked nucleic acid (LNA) (Vester and Wengel, 2004) that is synthesized using modified RNA nucleotides. Recently, the antimetastatic effect of MALAT1 targeting by ASO was reported in a lung cancer xenograft model, highlighting the potential use of ASO as a promising therapeutic approach for targeting lncRNA (Gong et al., 2019). On the other hand, transcriptional silencing of lncRNA using CRISPR-based approaches is also feasible. Indeed, the recently identified CRISPR-Cas13 system represents another promising approach for lncRNA knockdown (Abudayyeh et al., 2017). The good news is that sequence-based nucleic acid therapeutics are evolving at a rapid pace and more potential approaches could be identified in the future (Gudenas et al., 2019).
7. Effect of LINC00152 on the radiosensitivity of NSCLC cells
Irradiation reduces the viability of lung cancer cells and induces apoptosis in a dose-dependent manner. Overexpression of LINC00152 can restore the decrease of cell viability and inhibit irradiation-mediated cell apoptosis through direct binding to miR-195 (Zhang and Li, 2018). Indeed, the expression levels of LINC00152 in NSCLC tissues and cells were up-regulated after radiation and were negatively correlated with miR-195 levels. Consequently, some target genes of miR-195, such as coactivator-associated arginine methyltransferase 1 (CARM1), Yes-associated protein 1 (YAP), glycerophosphodiester phosphodiesterase domain containing 5 (GDPD5), and Wnt family member 3A (WNT3A), were significantly increased. In contrast, LINC00152 suppression in A549 and 95D lung cancer cells induced the expression of miR-195 and reduced the expression of its target genes. Additionally, transfection of miR-195 inhibitor reversed the favorable changes on cell biological behavior caused by LINC00152 knockdown, such as radiosensitivity. These results further support the oncogenic behavior of LINC00152 in NSCLC.
Chemotherapeutic and targeted drug resistance has always been the dilemma facing clinical treatment. It has been reported that LINC00152/miR-193a-3p/ERBB4/AKT axis can regulate the sensitivity of colon cancer cells to oxaliplatin (L-OHP) treatment (Yue et al., 2016). On the other hand, LINC00152/miR-139-5p/NOTCH1 was shown to influence 5-fluorouracil resistance in colon cancer (Bian et al., 2017). Interestingly, LINC00152 expression in doxorubicin-resistant MCF-7 breast cancer cells was significantly higher than that in the parental cells (Hu et al., 2018). Decreasing LINC00152 expression promoted chemosensitivity to temozolomide and enhanced survival of patients with high-grade glioma (Wang W et al., 2018). These results indicate that LINC00152 can serve as a predictor of chemoresistance. However, there is currently no report on the effect of LINC00152 on lung cancer chemotherapy or molecule-targeted therapy. Whether LINC00152 plays a regulatory role in the drug resistance of lung cancer is to be determined.
8. Bioinformatics analysis on LINC00152 in lung cancer
Xu et al. (2017) analyzed data from the TCGA and GEO databases. They found that LINC00152 is highly expressed and associated with poor prognoses in bladder, cervical, colon, renal, liver, lung, and other cancers. They explored the relationship between LINC00152 expression and histone modification using the encyclopedia of DNA elements (ENCODE) database (Davis et al., 2018). Among 15 tumor cell lines, including lung cancer, LINC00152 was regarded as a promoter-like lncRNA because its transcription initiation site had an H3K4me3/H3K4me1 value of >1.2. Furthermore, they demonstrated that LINC00152 promoted tumor cell proliferation, invasion, and apoptosis by binding to EZH2 in vitro and in vivo. Reon et al. (2018) analyzed the expression of LINC00152 in all tumor samples in the TCGA database, with their matched normal controls, which showed that LINC00152 was highly expressed in almost all tumor samples. High LINC00152 expression was associated with poor prognosis of head and neck squamous cell carcinoma, lung adenocarcinoma, renal clear cell carcinoma, etc. Feng et al. (2017) extended the analysis of LINC00152 expression using RNA-Seq data of 6220 cancer cases from the MiTranscriptome database. High expression of LINC00152 was observed in multiple types of tumors, including lung squamous cell carcinoma and large cell lung cancer, which further confirms that LINC00152 can serve as a tumor biomarker or potential therapeutic target in the future.
9. Conclusions and perspectives
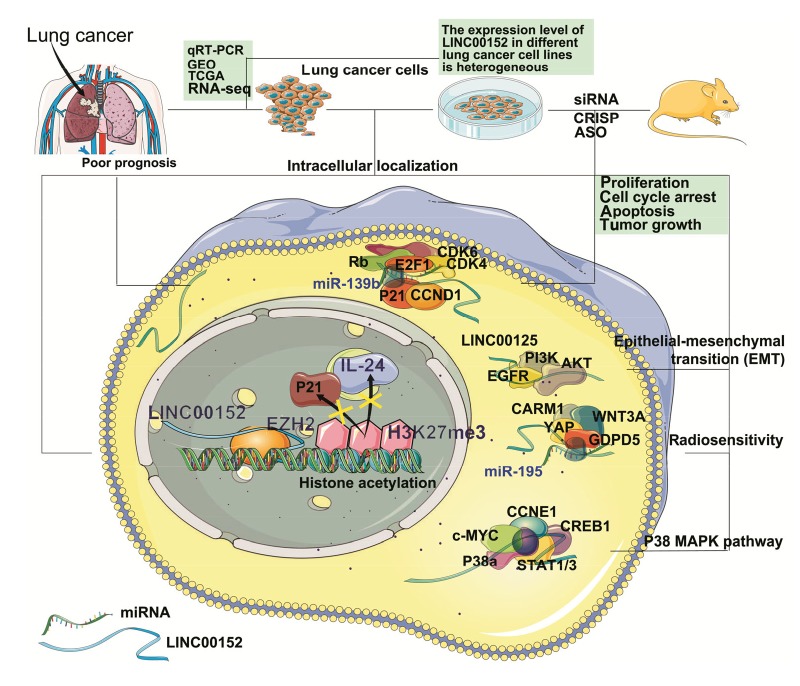
The experimental research on LINC00152 in NSCLC is still scarce and the effect of LINC00152 in NSCLC has not been fully elucidated (Fig. 1). However, the long-established function of LINC00152 in other cancers suggests a potential role for LINC00152 in NSCLC (Zhou et al., 2015; Teng et al., 2017; Su et al., 2018).
Fig. 1.
Potential mechanisms of LINC00152 in NSCLC
LINC00152 has been associated with poor prognosis of non-small cell lung cancer (NSCLC) patients. It has been shown to be highly expressed in tumor tissues and peripheral blood samples of NSCLC patients through analysis of the data from quantitative real-time polymerase chain reaction (qRT-PCR) assay, RNA sequencing (RNA-seq), The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. In vitro, the expression level of LINC00152 in different NSCLC cell lines is heterogenous. It is located in both cytoplasm and the nucleus, and it can exert carcinogenic effects through different molecular mechanisms. In the nucleus, LINC00152 regulates gene expression by binding to enhancer of zeste homolog 2 (EZH2), while it affects cellular signaling cascades in the cytoplasm. It can also bind specific microRNAs (miRNAs) as competing endogenous RNA (ceRNA) to regulate the expression of target messenger RNAs (mRNAs). On the other hand, the mechanisms of LINC00152 can be studied in vivo by regulating its expression using small interfering RNA (siRNA), clustered regularly interspaced short palindromic repeat (CRISPR), and antisense oligonucleotide (ASO) approaches in xenograft models. CDK: cyclin-dependent kinase; E2F1: E2F transcription factor 1; CCND1: cyclin D1; PI3K: phosphoinositide-3-kinase; EGFR: epidermal growth factor receptor; CARM1: coactivator-associated arginine methyltransferase 1; WNT3A: Wnt family member 3A; YAP: yes-associated protein 1; GDPD5: glycerophosphodiester phosphodiesterase domain containing 5; CCNE1: cyclin E1; CREB1: cyclic adenosine monophosphate (cAMP)-responsive element binding protein 1; STAT1/3: signal transducer and activator of transcription 1/3
Chen et al. (2017) showed that, even in the same type of tumor cell lines, the subcellular localization and expression of LINC00152 are heterogeneous. Thus, it is difficult to give a clear conclusion on the sub-cellular location of LINC00152. On the one hand, the heterogeneous LINC00152 expression can be used as a profitable condition to study the effect of LINC00152 up-or down-regulation in the corresponding cell lines. On the other hand, it can also reveal the complexity of LINC00152 mechanisms, which greatly influences research effort.
It has been shown that LINC00152 is adjacent to another lncRNA on chromosome 2 (chr2), MIR4435-2 host gene (MIR4435-2HG); LINC00152 is located on chr2 at 87 455 476‒87 606 739, while MIR4435-2HG is located on chr2 at 111 196 350‒111 495 115 (Nötzold et al., 2017; Reon et al., 2018). Interestingly, the sequences of the two lncRNA strands differ only by 6 bases, and they both contain the M8 sequence. In glioma cells, the expression levels of LINC00152 and MIR4435-2HG were identical. Targeting LINC00152 by siRNA simultaneously down-regulated the expression of LINC00152 and MIR4435-2HG, which suggests that these two lncRNAs might have common regulatory effects. Meanwhile, MIR4435-2HG is a host gene for miR-4435 that is transcribed from a MIR4435-2HG intron (Reon et al., 2018). In fact, this raises concerns about finding from multiple previous studies. For example, it raises concerns about the sequence specificity and efficiency of LINC00152 PCR and siRNA primers. Such problems should be fully considered in future studies.
At present, almost all studies on LINC00152 have been performed in China. Verification on tumor tissue and blood samples also poses some problems such as relatively small sample size and lack of consensus on detection standard (Yang et al., 2016). In the future, it would be necessary to develop uniform testing procedure, expand the sample size, and construct multi-center cooperations. Nevertheless, the current attempts offer the opportunity to study lung cancer pathogenesis and progression. In this regard, interpreting the orchestrated regulatory network of lncRNAs would be helpful in elucidating novel diagnostic and therapeutic targets to improve clinical outcomes.
Footnotes
Project supported by the Health Technology Innovation Project of Jilin Province (No. 2017J025), China
Contributors: Hong YU and Shu-bin LI wrote, edited, and revised the manuscript. Both authors have read and approved the final manuscript.
Compliance with ethics guidelines: Hong YU and Shu-bin LI declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by either of the authors.
References
- 1.Abudayyeh OO, Gootenberg JS, Essletzbichler P, et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550(7675):280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol Med. 2018;24(3):257–277. doi: 10.1016/j.molmed.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bian ZH, Zhang JW, Li M, et al. Long non-coding RNA LINC00152 promotes cell proliferation, metastasis, and confers 5-FU resistance in colorectal cancer by inhibiting miR-139-5p. Oncogenesis. 2017;6(11):395. doi: 10.1038/s41389-017-0008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai JQ, Zhang JW, Wu PF, et al. Blocking LINC00152 suppresses glioblastoma malignancy by impairing mesenchymal phenotype through the miR-612/AKT2/NF-κB pathway. J Neuro-Oncol. 2018;140(2):225–236. doi: 10.1007/s11060-018-2951-0. [DOI] [PubMed] [Google Scholar]
- 6.Cai Q, Wang ZQ, Wang SH, et al. Upregulation of long non-coding RNA LINC00152 by SP1 contributes to gallbladder cancer cell growth and tumor metastasis via PI3K/AKT pathway. Am J Transl Res. 2016;8(10):4068–4081. [PMC free article] [PubMed] [Google Scholar]
- 7.Cai Q, Wang ZQ, Wang SH, et al. Long non-coding RNA LINC00152 promotes gallbladder cancer metastasis and epithelial-mesenchymal transition by regulating HIF-1α via miR-138. Open Biol. 2017;7(1):160247. doi: 10.1098/rsob.160247. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Chen J, Zhang K, Song HZ, et al. Long noncoding RNA CCAT1 acts as an oncogene and promotes chemoresistance in docetaxel-resistant lung adenocarcinoma cells. Oncotarget. 2016;7(38):62474–62489. doi: 10.18632/oncotarget.11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen MW, Xu XN, Ma HQ. Identification of oncogenic long noncoding RNAs CASC9 and LINC00152 in oral carcinoma through genome-wide comprehensive analysis. Anti-Cancer Drugs. 2019;30(4):356–362. doi: 10.1097/cad.0000000000000725. [DOI] [PubMed] [Google Scholar]
- 10.Chen PX, Fang XL, Xia B, et al. Long noncoding RNA LINC00152 promotes cell proliferation through competitively binding endogenous miR-125b with MCL-1 by regulating mitochondrial apoptosis pathways in ovarian cancer. Cancer Med. 2018;7(9):4530–4541. doi: 10.1002/cam4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen QN, Chen X, Chen ZY, et al. Long intergenic non-coding RNA 00152 promotes lung adenocarcinoma proliferation via interacting with EZH2 and repressing IL24 expression. Mol Cancer. 2017;16(1):17. doi: 10.1186/s12943-017-0581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen WM, Huang MD, Sun DP, et al. Long intergenic non-coding RNA 00152 promotes tumor cell cycle progression by binding to EZH2 and repressing p15 and p21 in gastric cancer. Oncotarget. 2016;7(9):9773–9787. doi: 10.18632/oncotarget.6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Li DH, Gao Y, et al. Long intergenic noncoding RNA 00152 promotes glioma cell proliferation and invasion by interacting with miR-16. Cell Physiol Biochem. 2018;46(3):1055–1064. doi: 10.1159/000488836. [DOI] [PubMed] [Google Scholar]
- 14.Choi SW, Kim HW, Nam JW. The small peptide world in long noncoding RNAs. Brief Bioinf. 2019;20(5):1853–1864. doi: 10.1093/bib/bby055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collisson EA, Campbell JD, Brooks AN, et al. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crooke ST. Molecular mechanisms of antisense oligonucleotides. Nucleic Acid Ther. 2017;27(2):70–77. doi: 10.1089/nat.2016.0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuadrado A, Nebreda AR. Mechanisms and functions of p38 MAPK signalling. Biochem J. 2010;429(3):403–417. doi: 10.1042/bj20100323. [DOI] [PubMed] [Google Scholar]
- 18.Davis CA, Hitz BC, Sloan CA, et al. The encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46(D1):D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng X, Zhao XF, Liang XQ, et al. Linc00152 promotes cancer progression in hepatitis B virus-associated hepatocellular carcinoma. Biomed Pharmacother. 2017;90:100–108. doi: 10.1016/j.biopha.2017.03.031. [DOI] [PubMed] [Google Scholar]
- 20.Dey BK, Mueller AC, Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5(4):e944014. doi: 10.4161/21541272.2014.944014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El-Deiry WS. p21(WAF1) mediates cell-cycle inhibition, relevant to cancer suppression and therapy. Cancer Res. 2016;76(18):5189–5191. doi: 10.1158/0008-5472.CAN-16-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esfandi F, Taheri M, Omrani MD, et al. Expression of long non-coding RNAs (lncRNAs) has been dysregulated in non-small cell lung cancer tissues. BMC Cancer, 19: 222. 2019 doi: 10.1186/s12885-019-5435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ettinger DS, Wood D, Akerley W, et al. Non-small cell lung cancer, version 6.2015. J Natl Compr Canc Netw. 2015;13(5):515–524. doi: 10.6004/jnccn.2015.0071. [DOI] [PubMed] [Google Scholar]
- 24.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 25.Feng SM, Zhang J, Su WM, et al. Overexpression of LINC00152 correlates with poor patient survival and knockdown impairs cell proliferation in lung cancer. Sci Rep. 2017;7(1):2982. doi: 10.1038/s41598-017-03043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fidler MM, Bray F. Global cancer inequalities. Front Oncol, 8:293. 2018 doi: 10.3389/fonc.2018.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong NQ, Teng XC, Li JH, et al. Antisense oligonucleotide-conjugated nanostructure-targeting lncRNA MALAT1 inhibits cancer metastasis. ACS Appl Mater Interfaces. 2019;11(1):37–42. doi: 10.1021/acsami.8b18288. [DOI] [PubMed] [Google Scholar]
- 28.Gudenas BL, Wang J, Kuang SZ, et al. Genomic data mining for functional annotation of human long noncoding RNAs. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2019;20(6):476–487. doi: 10.1631/jzus.B1900162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutschner T, Hämmerle M, Eißmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu HB, Jie HY, Zheng XX. Three circulating lncRNA predict early progress of esophageal squamous cell carcinoma. Cell Physiol Biochem. 2016;40(1-2):117–125. doi: 10.1159/000452529. [DOI] [PubMed] [Google Scholar]
- 31.Hu XD, Bao JT, Wang Z, et al. The plasma lncRNA acting as fingerprint in non-small-cell lung cancer. Tumour Biol. 2016;37(3):3497–3504. doi: 10.1007/s13277-015-4023-9. [DOI] [PubMed] [Google Scholar]
- 32.Hu XL, Wang J, He W, et al. Down-regulation of lncRNA Linc00152 suppressed cell viability, invasion, migration, and epithelial to mesenchymal transition, and reversed chemo-resistance in breast cancer cells. Eur Rev Med Pharmacol Sci. 2018;22(10):3074–3084. doi: 10.26355/eurrev_201805_15067. [DOI] [PubMed] [Google Scholar]
- 33.Huang Y, Luo H, Li F, et al. LINC00152 down-regulated miR-193a-3p to enhance MCL1 expression and promote gastric cancer cells proliferation. Biosci Rep. 2018;38(3):BSR20171607. doi: 10.1042/bsr20171607. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Ji J, Tang JW, Deng L, et al. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget. 2015;6(40):42813–42824. doi: 10.18632/oncotarget.5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, Wang X, Tang J, et al. HULC and Linc00152 act as novel biomarkers in predicting diagnosis of hepatocellular carcinoma. Cell Physiol Biochem. 2015;37(2):687–696. doi: 10.1159/000430387. [DOI] [PubMed] [Google Scholar]
- 36.Li MH, Ning J, Li ZH, et al. LINC00152 promotes the growth and invasion of oral squamous cell carcinoma by regulating miR-139-5p. OncoTargets Ther. 2018;11:6295–6304. doi: 10.2147/ott.s168807. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Li ND, Feng XB, Tan Q, et al. Identification of circulating long noncoding RNA Linc00152 as a novel biomarker for diagnosis and monitoring of non-small-cell lung cancer. Disease markers, 2017:7439698. 2017 doi: 10.1155/2017/7439698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li QE, Shao YF, Zhang XJ, et al. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumor Biol. 2015;36(3):2007–2012. doi: 10.1007/s13277-014-2807-y. [DOI] [PubMed] [Google Scholar]
- 39.Li SH, Wen DC, Che ST, et al. Knockdown of long noncoding RNA 00152 (LINC00152) inhibits human retinoblastoma progression. OncoTargets Ther. 2018;11:3215–3223. doi: 10.2147/ott.s160428. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Liu DL, Gao M, Wu K, et al. LINC00152 facilitates tumorigenesis in esophageal squamous cell carcinoma via miR-153-3p/FYN axis. Biomed Pharmacother, 112:108654. 2019 doi: 10.1016/j.biopha.2019.108654. [DOI] [PubMed] [Google Scholar]
- 41.Liu J, Wan L, Lu KH, et al. The long noncoding RNA MEG3 contributes to cisplatin resistance of human lung adenocarcinoma. PLoS ONE. 2015;10(5):e0114586. doi: 10.1371/journal.pone.0114586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu XZ, Yidayitula Y, Zhao H, et al. LncRNA LINC00152 promoted glioblastoma progression through targeting the miR-107 expression. Environ Sci Pollut Res. 2018;25(18):17674–17681. doi: 10.1007/s11356-018-1784-x. [DOI] [PubMed] [Google Scholar]
- 43.Liu ZL, Sun M, Lu KH, et al. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21WAF1/CIP1 expression. PLoS ONE. 2013;8(10):e77293. doi: 10.1371/journal.pone.0077293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu ZZ, Liu AF, Nan A, et al. The linc00152 controls cell cycle progression by regulating CCND1 in 16HBE cells malignantly transformed by cigarette smoke extract. Toxicol Sci. 2019;167(2):496–508. doi: 10.1093/toxsci/kfy254. [DOI] [PubMed] [Google Scholar]
- 45.Ma P, Zhang ML, Nie FQ, et al. Transcriptome analysis of EGFR tyrosine kinase inhibitors resistance associated long noncoding RNA in non-small cell lung cancer. Biomed Pharmacother. 2017;87:20–26. doi: 10.1016/j.biopha.2016.12.079. [DOI] [PubMed] [Google Scholar]
- 46.Ma P, Wang HT, Sun JY, et al. LINC00152 promotes cell cycle progression in hepatocellular carcinoma via miR-193a/b-3p/CCND1 axis. Cell Cycle. 2018;17(8):974–984. doi: 10.1080/15384101.2018.1464834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mao Y, Tie Y, Du J, et al. LINC00152 promotes the proliferation of gastric cancer cells by regulating B-cell lymphoma-2. J Cell Biochem. 2019;120(3):3747–3756. doi: 10.1002/jcb.27655. [DOI] [PubMed] [Google Scholar]
- 48.Nishizawa Y, Konno M, Asai A, et al. Hypoxia stimulates the cytoplasmic localization of oncogenic long noncoding RNA LINC00152 in colorectal cancer. Int J Oncol. 2018;52(2):453–460. doi: 10.3892/ijo.2017.4218. [DOI] [PubMed] [Google Scholar]
- 49.Nötzold L, Frank L, Gandhi M, et al. The long non-coding RNA LINC00152 is essential for cell cycle progression through mitosis in HeLa cells. Sci Rep. 2017;7(1):2265. doi: 10.1038/s41598-017-02357-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orom UA, Derrien T, Beringer M, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qiu JJ, Yan JB. Long non-coding RNA LINC01296 is a potential prognostic biomarker in patients with colorectal cancer. Tumor Biol. 2015;36(9):7175–7183. doi: 10.1007/s13277-015-3448-5. [DOI] [PubMed] [Google Scholar]
- 53.Rapisuwon S, Vietsch EE, Wellstein A. Circulating biomarkers to monitor cancer progression and treatment. Comput Struct Biotechnol J. 2016;14:211–222. doi: 10.1016/j.csbj.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reon BJ, Karia BTR, Kiran M, et al. LINC00152 promotes invasion through a 3'-hairpin structure and associates with prognosis in glioblastoma. Mol Cancer Res. 2018;16(10):1470–1482. doi: 10.1158/1541-7786.mcr-18-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seo JS, Ju YS, Lee WC, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012;22(11):2109–2119. doi: 10.1101/gr.145144.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen X, Zhong JX, Yu P, et al. YY1-regulated LINC00152 promotes triple negative breast cancer progression by affecting on stability of PTEN protein. Biochem Biophys Res Commun. 2019;509(2):448–454. doi: 10.1016/j.bbrc.2018.12.074. [DOI] [PubMed] [Google Scholar]
- 57.Su M, Xiao YH, Tang JM, et al. Role of lncRNA and EZH2 interaction/regulatory network in lung cancer. J Cancer. 2018;9(22):4156–4165. doi: 10.7150/jca.27098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun ZH, Guo X, Zang MC, et al. Long non-coding RNA LINC00152 promotes cell growth and invasion of papillary thyroid carcinoma by regulating the miR-497/BDNF axis. J Cell Physiol. 2019;234(2):1336–1345. doi: 10.1002/jcp.26928. [DOI] [PubMed] [Google Scholar]
- 59.Teng W, Qiu CG, He ZH, et al. Linc00152 suppresses apoptosis and promotes migration by sponging miR-4767 in vascular endothelial cells. Oncotarget. 2017;8(49):85014–85023. doi: 10.18632/oncotarget.18777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Torre LA, Siegel RL, Jemal A. Lung cancer statistics. In: Ahmad A Gadgeel S., editor. Lung Cancer and Personalized Medicine: Current Knowledge and Therapies. Springer, Cham; 2016. pp. 1–19. [DOI] [Google Scholar]
- 61.Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43(42):13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 62.Wang HF, Chen WX, Yang P, et al. Knockdown of linc00152 inhibits the progression of gastric cancer by regulating microRNA-193b-3p/ETS1 axis. Cancer Biol Ther. 2019;20(4):461–473. doi: 10.1080/15384047.2018.1529124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang W, Wu F, Zhao Z, et al. Long noncoding RNA LINC00152 is a potential prognostic biomarker in patients with high-grade glioma. CNS Neurosci Ther. 2018;24(10):957–966. doi: 10.1111/cns.12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X, Yu HF, Sun WJ, et al. The long non-coding RNA CYTOR drives colorectal cancer progression by interacting with NCL and Sam68. Mol Cancer. 2018;17(1):110. doi: 10.1186/s12943-018-0860-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y, Zhou J, Xu YJ, et al. Long non-coding RNA LINC00968 acts as oncogene in NSCLC by activating the Wnt signaling pathway. J Cell Physiol. 2018;233(4):3397–3406. doi: 10.1002/jcp.26186. [DOI] [PubMed] [Google Scholar]
- 66.Wang YJ, Liu JZ, Bai HZ, et al. Long intergenic non-coding RNA 00152 promotes renal cell carcinoma progression by epigenetically suppressing P16 and negatively regulates miR-205. Am J Cancer Res. 2017;7(2):312–322. [PMC free article] [PubMed] [Google Scholar]
- 67.Wang YL, Li MM, Dong CX, et al. Linc00152 knockdown inactivates the Akt/mTOR and Notch1 pathways to exert its anti-hemangioma effect. Life Sci. 2019;223:22–28. doi: 10.1016/j.lfs.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 68.Wu JL, Shuang ZY, Zhao JF, et al. Linc00152 promotes tumorigenesis by regulating DNMTs in triple-negative breast cancer. Biomed Pharmacother. 2018;97:1275–1281. doi: 10.1016/j.biopha.2017.11.055. [DOI] [PubMed] [Google Scholar]
- 69.Wu Y, Tan C, Weng WW, et al. Long non-coding RNA Linc00152 is a positive prognostic factor for and demonstrates malignant biological behavior in clear cell renal cell carcinoma. Am J Cancer Res. 2016;6(2):285–299. [PMC free article] [PubMed] [Google Scholar]
- 70.Xie YJ, Zhang Y, Du LT, et al. Circulating long noncoding RNA act as potential novel biomarkers for diagnosis and prognosis of non-small cell lung cancer. Mol Oncol. 2018;12(5):648–658. doi: 10.1002/1878-0261.12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu SP, Wan L, Yin HZ, et al. Long noncoding RNA Linc00152 functions as a tumor propellant in pan-cancer. Cell Physiol Biochem. 2017;44(6):2476–2490. doi: 10.1159/000486170. [DOI] [PubMed] [Google Scholar]
- 72.Yang T, Zeng HM, Chen WQ, et al. Helicobacter pylori infection, H19 and LINC00152 expression in serum and risk of gastric cancer in a Chinese population. Cancer Epidemiol. 2016;44:147–153. doi: 10.1016/j.canep.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 73.Yu JJ, Liu Y, Guo C, et al. Upregulated long non-coding RNA LINC00152 expression is associated with progression and poor prognosis of tongue squamous cell carcinoma. J Cancer. 2017;8(4):523–530. doi: 10.7150/jca.17510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu MJ, Xue YX, Zheng J, et al. Linc00152 promotes malignant progression of glioma stem cells by regulating miR-103a-3p/FEZF1/CDC25A pathway. Mol Cancer. 2017;16(1):110. doi: 10.1186/s12943-017-0677-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75.Yu TH, Xu ZH, Zhang XH, et al. Long intergenic non-protein coding RNA 152 promotes multiple myeloma progression by negatively regulating microRNA-497. Oncol Rep. 2018;40(6):3763–3771. doi: 10.3892/or.2018.6721. [DOI] [PubMed] [Google Scholar]
- 76.Yu Y, Yang J, Li QP, et al. LINC00152: a pivotal oncogenic long non-coding RNA in human cancers. Cell Prolif. 2017;50(4):e12349. doi: 10.1111/cpr.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yue B, Cai DL, Liu CC, et al. Linc00152 functions as a competing endogenous RNA to confer oxaliplatin resistance and holds prognostic values in colon cancer. Mol Ther. 2016;24(12):2064–2077. doi: 10.1038/mt.2016.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yue B, Liu CC, Sun HM, et al. A positive feed-forward loop between lncRNA-CYTOR and Wnt/β-catenin signaling promotes metastasis of colon cancer. Mol Ther. 2018;26(5):1287–1298. doi: 10.1016/j.ymthe.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang J, Li WQ. Long noncoding RNA CYTOR sponges miR-195 to modulate proliferation, migration, invasion and radiosensitivity in nonsmall cell lung cancer cells. Biosci Rep. 2018;38(6):BSR20181599. doi: 10.1042/BSR20181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang PP, Wang YQ, Weng WW, et al. Linc00152 promotes cancer cell proliferation and invasion and predicts poor prognosis in lung adenocarcinoma. J Cancer. 2017;8(11):2042–2050. doi: 10.7150/jca.18852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang XX, Tao WG. Long noncoding RNA LINC00152 facilitates the leukemogenesis of acute myeloid leukemia by promoting CDK9 through miR-193a. DNA Cell Biol. 2019;38(3):236–242. doi: 10.1089/dna.2018.4482. [DOI] [PubMed] [Google Scholar]
- 82.Zhang Y, Xiang C, Wang YL, et al. LncRNA LINC00152 knockdown had effects to suppress biological activity of lung cancer via EGFR/PI3K/AKT pathway. Biomed Pharmacother. 2017;94:644–651. doi: 10.1016/j.biopha.2017.07.120. [DOI] [PubMed] [Google Scholar]
- 83.Zhang YH, Fu J, Zhang ZJ, et al. LncRNA-LINC00152 down-regulated by miR-376c-3p restricts viability and promotes apoptosis of colorectal cancer cells. Am J Transl Res. 2016;8(12):5286–5297. [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao B, Xu H, Ai X, et al. Expression profiles of long noncoding RNAs in lung adenocarcinoma. OncoTargets Ther. 2018;11:5383–5390. doi: 10.2147/ott.s167633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao J, Liu YC, Zhang WH, et al. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14(19):3112–3123. doi: 10.1080/15384101.2015.1078034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zheng LL, Hu N, Zhou XZ. TCF3-activated LINC00152 exerts oncogenic role in osteosarcoma through regulating miR-1182/CDK14 axis. Pathol Res Pract. 2019;215(2):373–380. doi: 10.1016/j.prp.2018.12.031. [DOI] [PubMed] [Google Scholar]
- 87.Zhou JP, Zhi XF, Wang LJ, et al. Linc00152 promotes proliferation in gastric cancer through the EGFR-dependent pathway. J Exp Clin Cancer Res. 2015;34(1):135. doi: 10.1186/s13046-015-0250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhu ZK, Dai JH, Liao YF, et al. Knockdown of long noncoding RNA LINC00152 suppresses cellular proliferation and invasion in glioma cells by regulating miR-4775. Oncol Res. 2018;26(6):857–867. doi: 10.3727/096504017x15016337254597. [DOI] [PMC free article] [PubMed] [Google Scholar]