Abstract
DNA double-stranded breaks (DSBs) are strongly associated with active transcription, and promoter-proximal pausing of RNA polymerase II (Pol II) is a critical step in transcriptional regulation. Mapping the distribution of DSBs along actively expressed genes and identifying the location of DSBs relative to pausing sites can provide mechanistic insights into transcriptional regulation. Using genome-wide DNA break mapping/sequencing techniques at single-nucleotide resolution in human cells, we found that DSBs are preferentially located around transcription start sites of highly transcribed and paused genes and that Pol II promoter-proximal pausing sites are enriched in DSBs. We observed that DSB frequency at pausing sites increases as the strength of pausing increases, regardless of whether the pausing sites are near or far from annotated transcription start sites. Inhibition of topoisomerase I and II by camptothecin and etoposide treatment, respectively, increased DSBs at the pausing sites as the concentrations of drugs increased, demonstrating the involvement of topoisomerases in DSB generation at the pausing sites. DNA breaks generated by topoisomerases are short-lived because of the religation activity of these enzymes, which these drugs inhibit; therefore, the observation of increased DSBs with increasing drug doses at pausing sites indicated active recruitment of topoisomerases to these sites. Furthermore, the enrichment and locations of DSBs at pausing sites were shared among different cell types, suggesting that Pol II promoter-proximal pausing is a common regulatory mechanism. Our findings support a model in which topoisomerases participate in Pol II promoter-proximal pausing and indicated that DSBs at pausing sites contribute to transcriptional activation.
Keywords: RNA polymerase II, DNA topoisomerase, DNA transcription, transcription regulation, DNA sequencing, DNA damage, gene expression, DNA double-stranded breaks, HeLa cells, RNA polymerase II promoter-proximal pausing, topoisomerase inhibitors, topoisomerase I and II
Introduction
DNA double-stranded breaks (DSBs)3 are strongly associated with active transcription and regulate the expression of highly expressed genes (1–3). DNA topoisomerases I and II (TOP1 and TOP2, respectively) have been shown to participate in this regulation by relieving torsional stress of the DNA duplex (4–6). Lensing et al. (7) applied DSBCapture to in situ capture and sequencing of DNA breaks and found that DSBs are enriched at promoters and 5′ UTRs and that the number of DSBs correlates with the expression level of genes. Recently, Gothe et al. (8) demonstrated a dependence of DNA fragility on the direction of active transcription, and Canela et al. (9) showed that TOP2-mediated DNA breaks are enhanced in actively transcribed regions and contribute to gene translocations. In activated B cells and primary neural stem/progenitor cells, analysis of the junctions derived from translocation events showed that DSBs were clustered around the transcription start sites (TSSs) of actively expressed genes and shared in these two cell types (2).
RNA polymerase II (Pol II) promoter-proximal pausing is a common but poorly understood step in the regulation of actively expressed genes across cell types (10, 11). Although it was hypothesized that DNA torsional stress could cause Pol II pausing and recruit DNA topoisomerases at pausing sites, this has not been explicitly shown (6, 12). Recently, Dellino et al. (13) demonstrated that Pol II pausing signal (using Pol II–pSer5 ChIP-seq) is enriched at TSSs of fragile promoters (those having DSB hot spots) compared with TSSs of control promoters. However, the study was focused on the characterization of a small subset of 627 fragile promoters. The distribution of DSBs with respect to genome-wide Pol II pausing sites and whether there is any correlation of DSBs with strength of pausing was not explored.
Previously we have used pausing-relevant datasets, including Pol II ChIP-seq, GRO-seq, NET-seq, and mNET-seq data and established, at nearly single-nucleotide resolution, a set of pausing sites ranked on robust criteria for Pol II pausing in HeLa cells independent of annotations (14). Here we performed genome-wide DSB mapping/sequencing in HeLa cells and analyzed the distribution of DSBs around TSSs and the location of DSBs relative to the refined Pol II pausing sites (n = 13,910). We found a strong association between DSBs and Pol II pausing strength. Additionally, using camptothecin and etoposide, inhibitors of TOP1 and TOP2 religation activity, respectively, we directly detected regions where TOP1 and TOP2 cause DNA breaks. Following analysis of TOP1 and TOP2 ChIP-seq data, we found that TOP2B (and, to a lesser extent, TOP1) displays a strong binding peak at and around pausing sites and that the peak overlaps with the DSB peak. In TOP2B knockout cells, the break peak at the pausing site (observed in WT cells) diminished. Therefore, our data elucidate a direct role of TOP1 and TOP2 in the generation of DNA breaks at pausing sites. Furthermore, we showed that the degree of pausing and the enrichment of DSBs at Pol II pausing sites are shared among different cell types, suggesting that DNA breaks play a ubiquitous role in the process of Pol II pausing.
Results
DSBs are preferentially located at the TSS of paused genes compared with nonpaused or no-Pol II genes
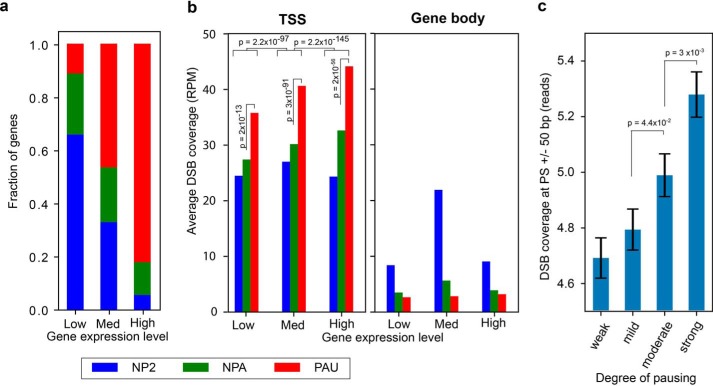
Actively expressed genes are commonly regulated by Pol II promoter-proximal pausing (10). About 80% of highly expressed genes in HeLa cells are paused (Fig. 1a), which is consistent with observations in other cells (14). Here we stratified RefSeq-annotated genes into three groups: paused (PAU), nonpaused (NPA), and no-Pol II (NP2) genes based on the traveling ratio derived from Pol II ChIP-seq of HeLa cells (15). We performed genome-wide DNA DSB mapping/sequencing in HeLa cells adapted from the DSBCapture protocol (7) with two biological replicates (Table S1, untreated N1 and N2), which showed very high reproducibility of genomic coverage (Pearson's correlation r = 0.986, p ∼ 0, Fig. S1). Combining these two DSB mapping/sequencing data, we found that, in RefSeq-annotated genes, increased DSB frequency at TSSs ± 500 bp is correlated with increasing gene expression. This trend is not detected, however, within gene bodies. More importantly, TSSs of paused genes, but not the rest of their gene bodies, possessed a significantly higher amount of DSBs than that of the nonpaused and no-Pol II groups, regardless of the gene expression level (Fig. 1b, p ∼ 0, two-sided Kolmogorov–Smirnov test). This demonstrates an association between DSBs and Pol II pausing and suggests that DSBs could play a role in Pol II pausing.
Figure 1.
DSBs are clustered at TSSs of highly expressed paused genes in HeLa cells. a, fractions of PAU (red), NPA (green), and NP2 (blue) genes based on the traveling ratio derived from Pol II ChIP-seq of HeLa cells are indicated for low, medium (Med), and highly expressed genes based on RNA-seq data of HeLa cells. b, number of DSBs (RPM, reads per million reads) detected at TSSs ± 500 bp (left panel) or within the rest of the gene bodies (right panel) for the three gene expression groups. The p values were determined by two-sided Kolmogorov–Smirnov tests. c, DSB reads at pausing sites (n = 13,910) are stratified into four equal size groups (quartiles) based on pausing site ranks, established previously using multiple pausing-relevant data sets from HeLa cells (14). Error bars denote the standard error of the mean. The p values were calculated using a two-sided Mann–Whitney U test.
To further investigate whether the enrichment of DSBs at TSSs of paused genes is associated with the degree of Pol II pausing, we utilized a set of ranked pausing sites (PSs, n = 13,910) we established previously in HeLa cells (14). This set of PSs provides comprehensive information based on the following features. First, it was derived from a combination of Pol II pausing-related datasets (Pol II ChIP-seq, GRO-seq (16), NET-seq (17), and mNET-seq (18)) and not limited to just one dataset. Second, it was entirely based on measurements rather than gene annotations. Third, it provided the location of pausing sites at single-nucleotide resolution, as determined by mNET-seq (18). The mNET-seq data contained single-nucleotide resolution genome-wide sequence data of nascent RNA for each Pol II–bound region in HeLa cells (Fig. S2a). This allowed us to investigate the relationship between pausing strength and DSBs at single-nucleotide resolution. These PSs are stratified into four equal size groups (quartiles) based on pausing site ranks, as weak, mild, moderate, and strong pausing strength (Fig. S2b). Fig. 1c shows that a significant increase in DNA break levels at PS ± 50 bp corresponds to increasing pausing strength (p = 0.044 for mild versus moderate, p = 0.003 for moderate versus strong, two-sided Mann–Whitney U test). This suggests that DNA breaks at pausing sites are associated with molecular activities facilitated by Pol II pausing.
DSBs are enriched at pausing sites independent of a proximal annotated TSS
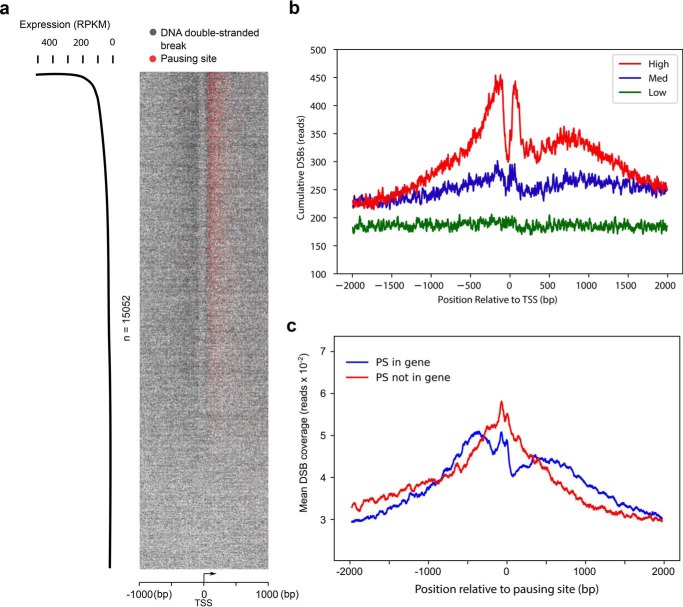
Because the set of ranked PSs provides the location of pausing at single-nucleotide resolution, we mapped the distribution of DSBs and PSs around the TSSs of transcribed genes (expression > 0 reads per kilobase per million reads, RPKM). The heatmap (Fig. 2a) and the average plot (Fig. 2b) show that DNA breaks accumulate immediately upstream and downstream of the TSS of highly transcribed genes, whereas, at low and moderately expressed genes, the amount of DNA breaks was lower and evenly distributed across the entire region. The locations of PSs are clearly superimposed on the break sites just downstream of TSSs (Fig. 2a). Furthermore, the colocalization and enrichment of DNA breaks and PSs are more prominent as gene expression activity increases (Fig. 2a).
Figure 2.
DSBs are enriched at pausing sites. a, heatmap representation of DSBs (one gray dot per site) and PSs (one red dot per site) at transcription start sites of actively transcribed genes (RPKM > 0, n = 15,052). Pileups of breaks or PSs result in a darker color from overlaid dots. Genomic regions are ordered by gene expression based on RNA-Seq in HeLa cells. b, cumulative, single-nucleotide-resolution profiles of measured DSBs at TSSs of genes stratified by their levels of transcription: low, medium (Med), and high. c, DSBs are enriched at pausing sites regardless of the presence of annotated TSSs. Shown are average cumulative profiles of DSBs at pausing sites located within RefSeq-annotated genes (blue, n = 7,941) and not within genes (red, n = 5,969). The pausing sites within RefSeq genes were located about 100 nt downstream of TSSs, whereas the pausing sites in the latter group were located either in intergenic regions or enhancer/promoter regions of genes and farther away from the nearest TSS.
The observation of two break cluster peaks immediately flanking each side of TSSs prompted us to investigate whether DNA breaks located at PSs are influenced by the proximity to annotated TSSs or affected by Pol II pausing activity. Among the list of 13,910 ranked PSs we established (14), 7,941 sites are located within RefSeq-annotated genes, and 5,969 sites are located in either intergenic regions or enhancer/promoter regions of genes. The pausing sites within RefSeq genes were located about 100 nt downstream of TSSs (14), whereas the pausing sites in the latter group are farther away from the nearest TSS (the majority are more than 10 kb away). Therefore, to examine whether DNA breaks at PSs can occur far from annotated TSSs, DNA break frequency around the PSs of these two groups were analyzed. We found that both groups share a bimodal distribution of DNA breaks immediately upstream of and at the PS (Fig. 2c). This demonstrates that DNA breaks are enriched at PSs regardless of the presence of an annotated TSS and that the DSB enrichment is beyond the RefSeq-annotated genes.
TOP1 and TOP2 act at pausing sites, resulting in DSBs
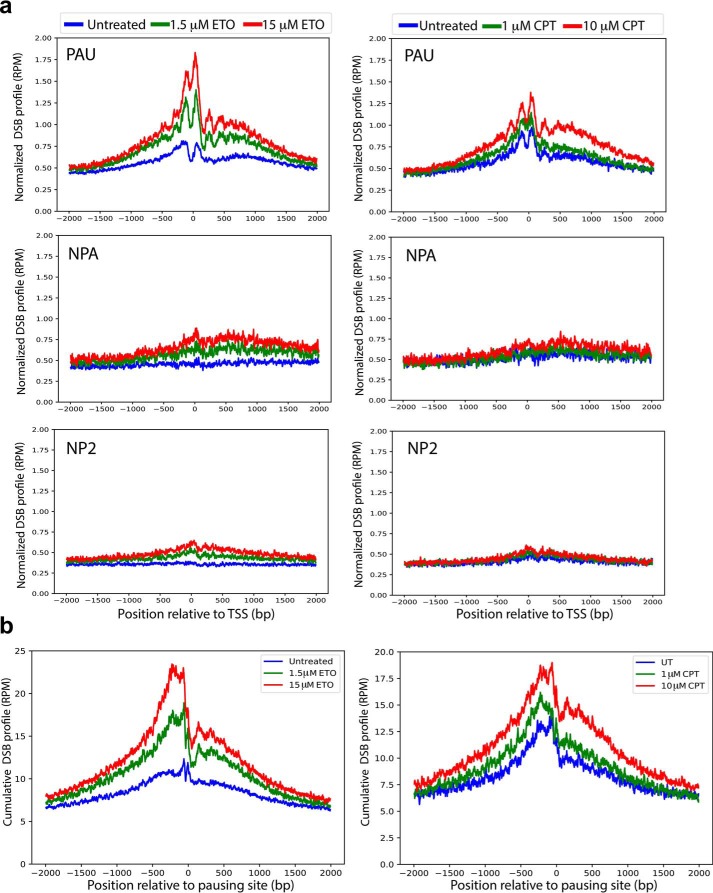
Several studies have suggested involvement of TOP1 and TOP2 in DNA breaks at highly expressed genes (6, 9, 19). A recent study using ChIP-seq of TOP1, TOP2B, and Pol II–pSer5 showed recruitment of the three proteins to the regions around the TSSs of 627 fragile promoters, with a lesser extent for TOP1 (13), but it did not address the location and the level of DNA breaks relative to pausing sites. To investigate whether the presence of TOP1 and TOP2 contributes to the generation of DNA breaks at pausing sites, we analyzed genome-wide break mapping/sequencing data from HeLa cells treated with camptothecin and etoposide, which inhibit the DNA ligation activity but not the DNA cleavage activity of TOP1 and TOP2, respectively. Upon treatment with camptothecin or etoposide, the TSS regions of paused genes display significant DSB enrichment over the background of untreated cells (p ∼ 0, untreated versus each chemical concentration and between concentrations of etoposide or camptothecin, two-sided Wilcoxon signed-rank test), and the DNA break increase corresponds to the increased concentrations of etoposide or camptothecin (Fig. 3a). In contrast, the DSB enrichment was not observed at the TSS regions of nonpaused and no-Pol II genes. These results suggest that DSBs at TSSs of paused genes are generated by the action of TOP1 and TOP2. Next we analyzed DSBs at pausing sites of HeLa cells and found a significant increase in DNA breaks at the pausing sites of HeLa cells irrespective of gene annotations (using the 13,910 ranked pausing sites, p ∼ 0, untreated versus each chemical concentration and between concentrations of etoposide or camptothecin, two-sided Wilcoxon signed-rank test). Notably, this increase is proportional to the increase in each drug concentration (Fig. 3b). These observations indicate that DNA breaks at pausing sites are caused by topoisomerase DNA cleavage activity, and both TOP1 and TOP2 activity contribute to DNA breaks at pausing sites. Examples of three individual genes displaying the general trend of colocalization of DSBs with pausing sites are shown in Fig. 4. At or immediately upstream of the pausing site location (mNET-seq spikes), there are sharp increases in DSBs with increasing doses of etoposide, along with a shift in DSBs in the 3′ direction toward the site of pausing. This is in agreement with the observation from the average plot (Fig. 3, a and b, left panels). The action of topoisomerases (cleavage, DNA strand passage, and religation) generates primarily transient breaks, whereas higher concentrations of etoposide increasingly trap these TOP2-mediated breaks. Therefore, increasing DSBs exclusively at pausing sites indicates that TOP2 is recruited to these sites with high efficiency. We also noticed that endogenous DSBs in untreated cells showed an enrichment at and around pausing sites (Fig. 3), suggesting that DNA breaks at pausing sites are a general phenomenon associated with transcriptional activation.
Figure 3.
Etoposide and camptothecin induce DSBs preferentially in paused genes and at pausing sites. a, cumulative, read-normalized, single-nucleotide-resolution profiles of DSBs induced by etoposide (ETO, left panels) and camptothecin (CPT, right panels) at TSSs of PAU (top panels), NPA (center panels), and NP2 (bottom panels) genes. The values are also normalized for the different gene numbers in PAU, NPA, and NP2 to make comparisons among groups. For paused genes, p ∼ 0; p values were calculated using a two-sided Wilcoxon signed-rank test for untreated versus each chemical concentration and between concentrations of etoposide and camptothecin among paused genes. b, cumulative, read-normalized, single-nucleotide-resolution profiles of DSBs at PSs (n = 13,910) in response to etoposide (left panel) and camptothecin (right panel) treatment. These PSs are derived from the combination of Pol II pausing-related datasets and provide the location of the sites at single-nucleotide resolution. p ∼ 0; p values were calculated using a two-sided Wilcoxon signed-rank test for untreated versus each chemical concentration and between concentrations of etoposide and camptothecin.
Figure 4.
Gene tracks display enrichment of DSBs at pausing sites at three genes (RTEL1, PRPF19, and LRRC47) upon etoposide treatment. Each gene example shows read-normalized break mapping in untreated and two concentrations of etoposide-treated HeLa cells. The HeLa mNET-seq spikes mark the location of the pausing sites. ETO, etoposide.
Evidence of direct involvement of TOP2 and TOP1 at pausing sites
Although the DNA breaks induced by etoposide treatment at the pausing sites suggested involvement of TOP2, it does not provide evidence of direct involvement at these sites. Therefore, we analyzed TOP2B ChIP-seq data from MCF10A cell lines from Dellino et al. (13) at pausing sites. TOP2B displays a strong binding peak at pausing sites, in agreement with our observation, and it supports our conclusion that TOP2B is directly involved in DNA breakage at pausing sites (Fig. S3a). We also analyzed the TOP1 ChIP-seq data from the same study and observed that TOP1 is also directly involved, although to a lesser extent, at the pausing sites and associated with DSBs (Fig. S3b). Furthermore, we examined the effect of TOP2B knockout in the generation of DNA breaks at pausing sites using the recently published cleavage complexes (CC)-seq data on RPE-1 cells by Gittens et al. (20). In these experiments, G1-arrested TOP2B knockout cells were treated with etoposide and compared with G1-arrested etoposide-treated WT cells. The TOP2 transient covalent complex was trapped, and the DNA breaks associated with the complex were sequenced at single-nucleotide resolution. We found that the break peak immediately upstream of the pausing site (observed in WT cells) disappeared in TOP2B knockout cells (Fig. S3c), supporting our conclusion that TOP2B directly acts at pausing sites, resulting in DSBs. These results demonstrated that TOP2 and TOP1 bind at pausing sites and that TOP2 is enzymatically active at these sites. Combining our etoposide-induced DSB data, we conclude that TOP1 and TOP2 act directly to induce breaks at these pausing sites.
Location of pausing sites and distribution of DSBs at these pausing sites are shared among different cell types
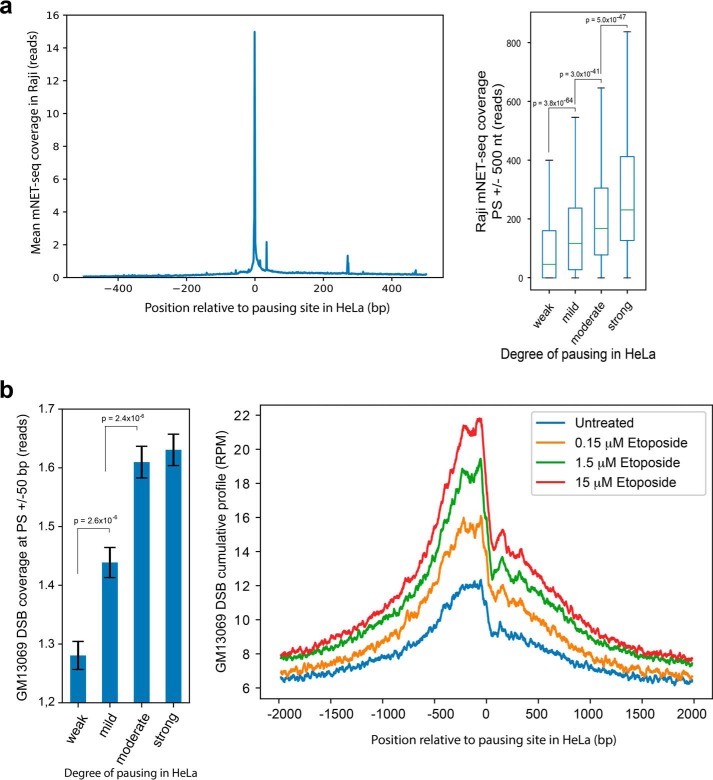
If DSBs at pausing sites serve as a common regulatory step for transcription, then strong overlaps of pausing sites with similar DSB enrichment will be observed among different cell types. Therefore, we next examined the extent to which these associations described above in HeLa cells occur in other cell types. Notably, the degree of pausing is generally comparable among different cell types (10, 14). To test whether the locations of PSs at nucleotide resolution in HeLa were similar in other cells, we used mNET-seq data derived from Raji cells (21), a Burkitt's lymphoma cell line, and plotted it against the position and strength of PSs defined from HeLa cells. We found that the majority of pausing site locations in HeLa cells are strong pausing sites in Raji cells, as indicated by the high mNET-seq reads (Fig. 5a, left panel). This demonstrates that the positions of strong PSs in HeLa cells (Fig. S2a) are shared with Raji cells. Furthermore, the intensity of pausing in Raji cells around these HeLa cell–defined sites is very similar to that of HeLa cells, as pausing intensity measured in Raji cells using mNET-seq read coverage matched that of HeLa cells using the degree of pausing defined in HeLa cells (Fig. 5a, right panel, compared with Fig. S2b). The four groups of pausing sites (weak, mild, moderate, and strong) based on the degree of pausing defined in HeLa cells showed significant increases from weak to strong in pausing intensity measured in Raji cells (Fig. 5a, right panel, p ∼ 0, two-sided Mann–Whitney U test).
Figure 5.
Distribution of pausing sites and DSBs across different cell types. a, pausing intensity in Raji cells relative to pausing sites defined from HeLa cells. Left panel, average mNET-seq coverage measured in Raji cells over pausing sites determined in HeLa cells. Right panel, mNET-seq coverage measured in Raji cells over the pausing sites determined in HeLa cells and divided into quartiles based on pausing site ranks from HeLa cells. Boxes denote 25th and 75th percentiles, center bars show median, and whiskers span from 5% to 95%. The p values were determined by two-sided Mann–Whitney U test. b, DNA breaks mapped in GM13069 at pausing sites established in HeLa cells. Left panel, DSBs measured in untreated GM13069 cells over HeLa-defined pausing sites stratified into four groups based on pausing sites rank from HeLa cells. The p values were calculated using a two-sided Mann–Whitney U test. Right panel, average cumulative profiles of DSBs in GM13069 induced by etoposide treatment at pausing sites established in HeLa cells. p ∼ 0; p values were calculated using a two-sided Wilcoxon signed-rank test for untreated versus each chemical concentration and between concentrations of etoposide.
Next we analyzed DSBs from genome-wide break mapping/sequencing of GM13069 cells (22), a nonmalignant lymphoblastoid cell line, and found that DNA break frequency is positively correlated with the degree of pausing defined from HeLa cells (Fig. 5b, left panel), similar to DNA break data from HeLa cells (Fig. 1c). We then analyzed genome-wide break mapping/sequencing data from etoposide-treated GM13069 cells and found that treatment with the TOP2 inhibitor etoposide increased DNA breaks at the PSs identified in HeLa cells (Fig. 5b, right panel), resembling the break pattern induced by etoposide treatment of HeLa cells (Fig. 3b, left panel). A similar trend was observed when single-nucleotide resolution DSBs from RPE-1 cell lines and TOP2B ChIP-seq data from MCF10A cell lines were plotted on HeLa pausing sites (Fig. S3, a and b). These results indicate that Pol II promoter-proximal pausing could be a common regulatory step shared among different cell types and that TOP2 participates in generation of DNA breaks at PSs.
Discussion
Utilizing a genome-wide DNA break mapping/sequencing technique and a set of ranked pausing sites, we determined exact locations of DNA breaks around TSS regions in HeLa cells. A subset of those breaks can be attributed to pausing sites, with the DNA break frequency increasing as the strength of pausing increases. This relationship is also observed in other cell types. The involvement of TOP1 and TOP2 in the generation of DNA breaks at pausing sites suggests that TOP1 and TOP2 activity could influence RNA Pol II pausing and/or that Pol II pausing could affect TOP1 and TOP2 activity at pausing sites.
Bunch et al. (12) have shown the involvement of TOP2 in DNA break–induced signaling to promote transcription elongation and demonstrated that, upon transcriptional activation, a DNA break event became intensified at the PS of the HSPA1B gene. Recently, Dellino et al. (13), employing the BLISS protocol reported that Pol II pausing (defined by the presence of Pol II–pSer5 ChIP-seq) is enriched at fragile promoters (subsets of promoters having DSB hot spots). TOP2 and, to a lesser extent, TOP1 are present at these promoters, and non-homologous end joining repair proteins, such as XRCC4 and PARP1, are recruited to these sites. However, they also suggested that transcription might not favor break formation, as they observed that ∼86% of high and moderately transcribed genes do not have fragile promoters. Here we directly mapped DNA breaks at PSs in a genome-wide manner regardless of the presence of an annotated TSS and found evidence in support of the idea that DNA breaks at PSs could contribute to transcriptional activation. Gittens et al. (20) observed direct overlaps of TOP2 cleavage complex sites with the GRO-seq signal peaks (measuring Pol II pausing). When we analyzed their data relative to the set of pausing sites we identified, we found the same results (Fig. S3c). We also explored the ChIP-seq data of a commonly used DSB marker, γH2AX (23), at pausing sites and observed a dip in the immediately upstream region of the pausing site (Fig. S4), suggesting that the region is free of nucleosomes because it is occupied by the RNA Pol II and topoisomerase cleavage complex. The same pattern was observed by Dellino et al. (13), where the regions enriched in TOP2B were deprived of γH2AX marks. This is consistent with the observation that γH2AX surrounds sites of DNA damage propagating megabases from these sites but is not at the break sites themselves (24).
We also observed DNA break enrichment just upstream of the TSSs of highly expressed genes (Fig. 2b), and these breaks also increase upon etoposide and camptothecin treatment (Fig. 3a). The presence of DNA breaks at the promoter regions of highly expressed genes have been suggested based on enrichment of elevated mutations (25–27) and translocation junctions (2) at promoters. In this work, we show direct evidence of the presence of DNA breaks and involvement of TOP1 and TOP2 in break formation at TSSs. The results of several genome-wide break mapping approaches are in agreement with our findings. Lensing et al. (7), using DSBCapture, demonstrated that DSBs are enriched at TSSs of highly expressed genes. Employing the BLISS techniques, Gothe et al. (8) showed that transcription is a major contributor to DSBs, with more than 75% of DSB hot spots occurring within transcriptionally active regions. Furthermore, Canela et al. (9), using the END-seq protocol, reported that etoposide-induced chromosomal translocations are also dependent on transcriptional activity.
The two break cluster peaks immediately flanking each side of TSSs emphasize a sharp dip in DNA breaks at the TSSs among highly expressed genes (Fig. 2b). The absence of detectable DNA breaks is likely due to the regions occupied by the RNA Pol II complex. In support of this, the position of the dip in DNA breaks matches the exclusion of the H3K4me3 signal in highly expressed genes (Fig. S5).
Furthermore, we showed previously that, immediately upstream of PSs, DNA has a high propensity to form stable secondary structures (14), which can affect RNA Pol II promoter-proximal pausing. The location of these structures corresponds to the peaks of DNA breaks and the peaks of topoisomerases binding at PSs (Fig. S3, a and b), suggesting a possible role of these structures in DNA breaks at PSs. In addition, we found that all of the pausing sites located within RefSeq-annotated genes that are highly expressed (n = 5533) contain a folding free energy favorable for formation of DNA secondary structures (lower than three standard deviations of the genome average) and that 99.5% of them have a free energy lower than four standard deviations of the genome average, indicating the potential presence of energetically favorable DNA secondary structures (the genome average free energy is −1.26 kcal/mol with a standard deviation of 1.42). Interestingly, several studies demonstrated that a property of TOP1 and TOP2 is to recognize and preferentially cleave DNA at regions capable of forming stable DNA secondary structures (28–32). Site-specific cleavage by TOP2 at centromeric DNA with dyad symmetries (potential to form hairpins and four-way junctions) is found in yeast, fruit fly, chicken, and human (32, 33). Moreover, mismatched bases, which are often present in the multiple stem-loop type of DNA secondary structures, when in the proximity of TOP2 cleavage sites, can greatly stimulate TOP2 cleavage activity and hinder DNA end religation (34, 35). This provides a possible notion that TOP1 and TOP2 could recognize and cleave DNA at pausing sites via the presence of DNA secondary structures and that supercoiling can promote the formation of DNA secondary structures (Fig. S6).
Our study directly demonstrates the common presence of enriched DSBs at pausing sites of highly expressed genes and involvement of topoisomerases in the generation of break enrichment. Further studies to investigate how DNA breaks at pausing sites influence transcriptional activation will provide critical insights into transcriptional regulation.
Experimental procedures
Cell culture and treatments
HeLa cells (ATCC) and GM13069 cells (ATCC) were grown in DMEM (Gibco, 11965) and RPMI 1640 medium (Gibco, 11875), respectively, supplemented with 10% fetal bovine serum and plated at 2 × 106 cells/100-mm cell culture dish. Cells were treated 18 h later with etoposide (1.5 or 15 μm, Sigma) or camptothecin (1 or 10 μm, Sigma) for 24 h, along with untreated controls. HeLa cells were trypsinized, and cells were washed twice with cold PBS containing the treatment dose of etoposide or camptothecin and collected by centrifugation at 4 °C.
Genome-wide break mapping and sequencing
Detection of DNA breaks was adapted from DSBCapture and performed as described previously (7). Briefly, fixed nuclei were subjected to blunting/A-tailing reactions and Illumina P5 adaptor ligation to capture broken DNA ends. Genomic DNA was then purified and fragmented by sonication and subsequently ligated to the Illumina P7 adaptor, and the libraries were PCR-amplified for 15 cycles. Prepared libraries were then subjected to whole-genome 75-bp and 150-bp paired-end sequencing with the Illumina NextSeq 500 and HiSeq X Ten platforms, respectively.
DSB read processing
Sequencing reads were aligned to the human genome (GRCh38/hg38) with the bowtie2 (v.2.3.4.1) aligner running in high sensitivity mode (--very-sensitive). Restriction of the fragment length from 100 to 2000 nt (-X 2000 -I 100 options) was imposed. Unmapped, nonprimary, supplementary, and low-quality reads were filtered out with SAMtools (v. 1.7, −F 2820). Furthermore, PCR duplicates were marked with picard-tools (v. 1.95) MarkDuplicates, and finally, the first mate of nonduplicated pairs (−f 67 −F 1024) were filtered with SAMtools for continued analysis. For each detected break, the 5′-most nucleotide of the first mate defined the DNA break position. Sequencing and alignment statistics for the DSB mapping/sequencing libraries prepared from HeLa cells are listed in Table S1. Biological duplicates of each sample (untreated N1 and N2, Table S1), which showed very high reproducibility of genomic coverage (Pearson's correlation r = 0.986, p ∼ 0, Fig. S1), were combined for downstream data analysis. This strong correlation confirms that the break mapping procedure does not introduce significant amounts of random DNA breaks that could convert single-stranded nicks into DSBs.
Downstream data analysis
Downstream data analysis following DSB read processing was performed with BEDtools (v. 2.27.1) and standard Linux (Ubuntu 18) tools to compute coverages and annotation densities and generate heatmaps. Results were visualized in Python3 (v. 3.6.5) with matplotlib (v. 2.2.2), numpy (v. 1.15.0), and pandas (v. 0.23.3). Statistical tests were performed using Python3 (v. 3.6.5) with scipy (v. 0.19.1). RNA-Seq and mNET-seq were aligned with TopHat (v. 2.1.1). ChIP-seq data of TOP1 and TOP2 were aligned using bowtie2 (v.2.3.4.1).
Pol II pausing analysis
Traveling ratio (TR) was calculated as described previously (14). In brief, RefSeq genes (build GRCh38/hg38) were first stratified into two groups: intersecting and not intersecting with Pol II ChIP-seq peaks of HeLa cells. For Pol II–bound genes, we calculated the TR using ChIP-seq read coverage in two regions: −30 to +300 nt from the TSS and in the rest of the gene body. We then determined the Pol II ChIP-seq read density by calculating the read coverage and dividing this by the length of the region. TR was calculated as a ratio of the density of reads in the −30 to +300 nt from the TSS region over the read density within the rest of the gene. Based on the definitions above, all genes were divided into three groups: NP2, NPA (TR ≤ 2, and PAU (TR > 2).
PS ranks in HeLa cells were established based on Pol II ChIP-seq, GRO-seq, NET-seq, and mNET-seq data as described previously (14). Using the combination of Pol II pausing–related data sets, we identified 13,910 pausing sites genome-wide, based on measurements rather than gene annotations as in previously proposed methods such as TR (36–38), and it provides the location of pausing sites at single-nucleotide resolution. Among them, 7941 sites are located within a RefSeq annotated gene, and 5969 sites are located in either intergenic regions or enhancer/promoter regions of genes. PSs, determined previously for the GRCh37/hg19 assembly of the human genome, were converted to GRCh38/hg38 with LiftOver from the UCSC Genome Browser. Twenty-two PSs were overlapping, and the PSs with the higher rank were kept (Table S2).
Data availability
Pausing sites in HeLa cells (GRCh37/hg19) can be accessed as Additional File 2 and Additional File 3 in Szlachta et al. (14). The high-throughput sequencing data used in this study were downloaded from the Gene Expression Omnibus (GSE numbers) or from the ENCODE project (15) through the UCSC Genome Browser (GSE and wgEncode numbers) (39). For HeLa-S3 cells, we downloaded Pol II ChIP-seq (GSM935395, wgEncodeEH000613) (15), mNET-seq (GSE60358) (18), H3K4me3 ChIP-seq (GSM733682, wgEncode001013) (15), and RNA-Seq in HeLa (GSE95452, WT) (40) data. For Raji cells, we downloaded mNET-seq (GSE96056) (21) data. TOP1 and TOP2 ChIP-seq data for MCF10A cell lines were downloaded from study GSE93038 (13). TOP2B knockout and the corresponding WT DSB data in RPE-1 cell lines were downloaded from study GSE136943 (20). ChIP-Seq data of γH2AX in Jurkat cells was obtained from study GSE25577. The DSB mapping data can be accessed at the Sequence Read Archive (SRA) under the accession number PRJNA497476 for GM13069 cells (22).
Author contributions
S. S., K. S., and H. M. R. formal analysis; S. S., K. S., and H. M. R. validation; S. S., K. S., H. M. R., and Y.-H. W. visualization; S. S., H. M. R., M. D., S. B., and Y.-H. W. writing-review and editing; K. S. and Y.-H. W. conceptualization; K. S. data curation; K. S. and Y.-H. W. writing-original draft; A. M., M. D., and Y.-H. W. investigation; H. M. R. and Y.-H. W. funding acquisition; S. B. and Y.-H. W. supervision; Y.-H. W. project administration.
Supplementary Material
This work was supported by NIGMS, National Institutes of Health Grants RO1GM101192 (to Y.-H. W.) and T32GM008136 (to H. M. R.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S6 and Tables S1 and S2.
The DSB mapping data were deposited into the Sequence Read Archive under accession number PRJNA579071.
- DSB
- double-stranded break
- TOP1
- topoisomerase I
- TOP2
- topoisomerase II
- TSS
- transcription start site
- Pol II
- RNA polymerase II
- PAU
- paused
- NPA
- nonpaused
- NP2
- no-Pol II
- PS
- pausing site
- RPKM
- reads per kilobase per million reads
- CC
- cleavage complexes
- TR
- traveling ratio.
References
- 1. Madabhushi R., Gao F., Pfenning A. R., Pan L., Yamakawa S., Seo J., Rueda R., Phan T. X., Yamakawa H., Pao P. C., Stott R. T., Gjoneska E., Nott A., Cho S., Kellis M., and Tsai L. H. (2015) Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell 161, 1592–1605 10.1016/j.cell.2015.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schwer B., Wei P. C., Chang A. N., Kao J., Du Z., Meyers R. M., and Alt F. W. (2016) Transcription-associated processes cause DNA double-strand breaks and translocations in neural stem/progenitor cells. Proc. Natl. Acad. Sci. U.S.A. 113, 2258–2263 10.1073/pnas.1525564113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. D'Alessandro G., and d'Adda di Fagagna F. (2017) Transcription and DNA damage: holding hands or crossing swords? J. Mol. Biol. 429, 3215–3229 10.1016/j.jmb.2016.11.002 [DOI] [PubMed] [Google Scholar]
- 4. Haffner M. C., Aryee M. J., Toubaji A., Esopi D. M., Albadine R., Gurel B., Isaacs W. B., Bova G. S., Liu W., Xu J., Meeker A. K., Netto G., De Marzo A. M., Nelson W. G., and Yegnasubramanian S. (2010) Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat. Genet. 42, 668–675 10.1038/ng.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. King I. F., Yandava C. N., Mabb A. M., Hsiao J. S., Huang H. S., Pearson B. L., Calabrese J. M., Starmer J., Parker J. S., Magnuson T., Chamberlain S. J., Philpot B. D., and Zylka M. J. (2013) Topoisomerases facilitate transcription of long genes linked to autism. Nature 501, 58–62 10.1038/nature12504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pommier Y., Sun Y., Huang S. N., and Nitiss J. L. (2016) Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 17, 703–721 10.1038/nrm.2016.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lensing S. V., Marsico G., Hänsel-Hertsch R., Lam E. Y., Tannahill D., and Balasubramanian S. (2016) DSBCapture: in situ capture and sequencing of DNA breaks. Nat. Methods 13, 855–857 10.1038/nmeth.3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gothe H. J., Bouwman B. A. M., Gusmao E. G., Piccinno R., Petrosino G., Sayols S., Drechsel O., Minneker V., Josipovic N., Mizi A., Nielsen C. F., Wagner E. M., Takeda S., Sasanuma H., Hudson D. F., et al. (2019) Spatial chromosome folding and active transcription drive DNA fragility and formation of oncogenic MLL translocations. Mol. Cell 75, 267–283.e12 10.1016/j.molcel.2019.05.015 [DOI] [PubMed] [Google Scholar]
- 9. Canela A., Maman Y., Huang S. N., Wutz G., Tang W., Zagnoli-Vieira G., Callen E., Wong N., Day A., Peters J. M., Caldecott K. W., Pommier Y., and Nussenzweig A. (2019) Topoisomerase II-induced chromosome breakage and translocation is determined by chromosome architecture and transcriptional activity. Mol. Cell 75, 252–266.e8 10.1016/j.molcel.2019.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jonkers I., and Lis J. T. (2015) Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 167–177 10.1038/nrm3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Core L., and Adelman K. (2019) Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev. 33, 960–982 10.1101/gad.325142.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bunch H., Lawney B. P., Lin Y. F., Asaithamby A., Murshid A., Wang Y. E., Chen B. P., and Calderwood S. K. (2015) Transcriptional elongation requires DNA break-induced signalling. Nat. Commun. 6, 10191 10.1038/ncomms10191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dellino G. I., Palluzzi F., Chiariello A. M., Piccioni R., Bianco S., Furia L., De Conti G., Bouwman B. A. M., Melloni G., Guido D., Giacò L., Luzi L., Cittaro D., Faretta M., Nicodemi M., et al. (2019) Release of paused RNA polymerase II at specific loci favors DNA double-strand-break formation and promotes cancer translocations. Nat. Genet. 51, 1011–1023 10.1038/s41588-019-0421-z [DOI] [PubMed] [Google Scholar]
- 14. Szlachta K., Thys R. G., Atkin N. D., Pierce L. C. T., Bekiranov S., and Wang Y. H. (2018) Alternative DNA secondary structure formation affects RNA polymerase II promoter-proximal pausing in human. Genome Biol. 19, 89 10.1186/s13059-018-1463-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Core L. J., Waterfall J. J., and Lis J. T. (2008) Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 10.1126/science.1162228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mayer A., di Iulio J., Maleri S., Eser U., Vierstra J., Reynolds A., Sandstrom R., Stamatoyannopoulos J. A., and Churchman L. S. (2015) Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161, 541–554 10.1016/j.cell.2015.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nojima T., Gomes T., Grosso A. R. F., Kimura H., Dye M. J., Dhir S., Carmo-Fonseca M., and Proudfoot N. J. (2015) Mammalian NET-Seq reveals genome-wide nascent transcription coupled to RNA processing. Cell 161, 526–540 10.1016/j.cell.2015.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bunch H., Lawney B. P., Burkholder A., Ma D., Zheng X., Motola S., Fargo D. C., Levine S. S., Wang Y. E., and Hu G. (2016) RNA polymerase II promoter-proximal pausing in mammalian long non-coding genes. Genomics 108, 64–77 10.1016/j.ygeno.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gittens W. H., Johnson D. J., Allison R. M., Cooper T. J., Thomas H., and Neale M. J. (2019) A nucleotide resolution map of Top2-linked DNA breaks in the yeast and human genome. Nat. Commun. 10, 4846 10.1038/s41467-019-12802-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gressel S., Schwalb B., Decker T. M., Qin W., Leonhardt H., Eick D., and Cramer P. (2017) CDK9-dependent RNA polymerase II pausing controls transcription initiation. Elife 10.7554/eLife.29736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Szlachta K., Raimer H. M., Comeau L. D., and Wang Y. H. (2020) CNCC: an analysis tool to determine genome-wide DNA break end structure at single-nucleotide resolution. BMC Genomics 21, 25 10.1186/s12864-019-6436-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seo J., Kim S. C., Lee H. S., Kim J. K., Shon H. J., Salleh N. L., Desai K. V., Lee J. H., Kang E. S., Kim J. S., and Choi J. K. (2012) Genome-wide profiles of H2AX and gamma-H2AX differentiate endogenous and exogenous DNA damage hotspots in human cells. Nucleic Acids Res. 40, 5965–5974 10.1093/nar/gks287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tanwar V. S., Jose C. C., and Cuddapah S. (2019) Role of CTCF in DNA damage response. Mutat. Res. 780, 61–68 10.1016/j.mrrev.2018.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Katainen R., Dave K., Pitkänen E., Palin K., Kivioja T., Välimäki N., Gylfe A. E., Ristolainen H., Hänninen U. A., Cajuso T., Kondelin J., Tanskanen T., Mecklin J. P., Järvinen H., Renkonen-Sinisalo L., et al. (2015) CTCF/cohesin-binding sites are frequently mutated in cancer. Nat. Genet. 47, 818–821 10.1038/ng.3335 [DOI] [PubMed] [Google Scholar]
- 26. Kaiser V. B., Taylor M. S., and Semple C. A. (2016) Mutational biases drive elevated rates of substitution at regulatory sites across cancer types. PLoS Genet. 12, e1006207 10.1371/journal.pgen.1006207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Perera D., Poulos R. C., Shah A., Beck D., Pimanda J. E., and Wong J. W. (2016) Differential DNA repair underlies mutation hotspots at active promoters in cancer genomes. Nature 532, 259–263 10.1038/nature17437 [DOI] [PubMed] [Google Scholar]
- 28. Been M. D., and Champoux J. J. (1984) Breakage of single-stranded DNA by eukaryotic type 1 topoisomerase occurs only at regions with the potential for base-pairing. J. Mol. Biol. 180, 515–531 10.1016/0022-2836(84)90025-1 [DOI] [PubMed] [Google Scholar]
- 29. Froelich-Ammon S. J., Gale K. C., and Osheroff N. (1994) Site-specific cleavage of a DNA hairpin by topoisomerase II: DNA secondary structure as a determinant of enzyme recognition/cleavage. J. Biol. Chem. 269, 7719–7725 [PubMed] [Google Scholar]
- 30. Jonstrup A. T., Thomsen T., Wang Y., Knudsen B. R., Koch J., and Andersen A. H. (2008) Hairpin structures formed by α satellite DNA of human centromeres are cleaved by human topoisomerase IIα. Nucleic Acids Res. 36, 6165–6174 10.1093/nar/gkn640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. West K. L., and Austin C. A. (1999) Human DNA topoisomerase IIβ binds and cleaves four-way junction DNA in vitro. Nucleic Acids Res. 27, 984–992 10.1093/nar/27.4.984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mills W., Spence J., Fukagawa T., and Farr C. (2018) Site-specific cleavage by topoisomerase 2: a mark of the core centromere. Int. J. Mol. Sci. 10.3390/ijms19020534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Spence J. M., Fournier R. E., Oshimura M., Regnier V., and Farr C. J. (2005) Topoisomerase II cleavage activity within the human D11Z1 and DXZ1 α-satellite arrays. Chromosome Res. 13, 637–648 10.1007/s10577-005-1003-8 [DOI] [PubMed] [Google Scholar]
- 34. Bigioni M., Zunino F., Tinelli S., Austin C. A., Willmore E., and Capranico G. (1996) Position-specific effects of base mismatch on mammalian topoisomerase II DNA cleaving activity. Biochemistry 35, 153–159 10.1021/bi951736p [DOI] [PubMed] [Google Scholar]
- 35. Kingma P. S., and Osheroff N. (1998) The response of eukaryotic topoisomerases to DNA damage. Biochim. Biophys. Acta 1400, 223–232 10.1016/S0167-4781(98)00138-9 [DOI] [PubMed] [Google Scholar]
- 36. Rahl P. B., Lin C. Y., Seila A. C., Flynn R. A., McCuine S., Burge C. B., Sharp P. A., and Young R. A. (2010) c-Myc regulates transcriptional pause release. Cell 141, 432–445 10.1016/j.cell.2010.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eddy J., Vallur A. C., Varma S., Liu H., Reinhold W. C., Pommier Y., and Maizels N. (2011) G4 motifs correlate with promoter-proximal transcriptional pausing in human genes. Nucleic Acids Res. 39, 4975–4983 10.1093/nar/gkr079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kellner W. A., Bell J. S., and Vertino P. M. (2015) GC skew defines distinct RNA polymerase pause sites in CpG island promoters. Genome Res. 25, 1600–1609 10.1101/gr.189068.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rosenbloom K. R., Sloan C. A., Malladi V. S., Dreszer T. R., Learned K., Kirkup V. M., Wong M. C., Maddren M., Fang R., Heitner S. G., Lee B. T., Barber G. P., Harte R. A., Diekhans M., Long J. C., et al. (2013) ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 41, D56–D63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tchasovnikarova I. A., Timms R. T., Douse C. H., Roberts R. C., Dougan G., Kingston R. E., Modis Y., and Lehner P. J. (2017) Hyperactivation of HUSH complex function by Charcot-Marie-Tooth disease mutation in MORC2. Nat. Genet. 49, 1035–1044 10.1038/ng.3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Pausing sites in HeLa cells (GRCh37/hg19) can be accessed as Additional File 2 and Additional File 3 in Szlachta et al. (14). The high-throughput sequencing data used in this study were downloaded from the Gene Expression Omnibus (GSE numbers) or from the ENCODE project (15) through the UCSC Genome Browser (GSE and wgEncode numbers) (39). For HeLa-S3 cells, we downloaded Pol II ChIP-seq (GSM935395, wgEncodeEH000613) (15), mNET-seq (GSE60358) (18), H3K4me3 ChIP-seq (GSM733682, wgEncode001013) (15), and RNA-Seq in HeLa (GSE95452, WT) (40) data. For Raji cells, we downloaded mNET-seq (GSE96056) (21) data. TOP1 and TOP2 ChIP-seq data for MCF10A cell lines were downloaded from study GSE93038 (13). TOP2B knockout and the corresponding WT DSB data in RPE-1 cell lines were downloaded from study GSE136943 (20). ChIP-Seq data of γH2AX in Jurkat cells was obtained from study GSE25577. The DSB mapping data can be accessed at the Sequence Read Archive (SRA) under the accession number PRJNA497476 for GM13069 cells (22).