Abstract
Deubiquitinases deconjugate ubiquitin modifications from target proteins and are involved in many cellular processes in eukaryotes. The functions of deubiquitinases are regulated by post-translational modifications, mainly phosphorylation and ubiquitination. Post-translational modifications can result in subtle changes in structural and dynamic properties, which are difficult to identify but functionally important. In this work, we used NMR spectroscopy to characterize the conformational properties of the human deubiquitinase A (DUBA), a negative regulator of type I interferon. DUBA activity is regulated by phosphorylation at a single serine residue, Ser-177. We found that the catalytic rate constant of DUBA is enhanced by phosphorylation. By comparing NMR and enzyme kinetics data among different forms of DUBA with low and high activities, we concluded that a two-state equilibrium that was present only in phosphorylated DUBA is important for DUBA activity. Our results highlight the importance of defining conformational dynamics in understanding the mechanism of DUBA activation.
Keywords: nuclear magnetic resonance (NMR), deubiquitylation (deubiquitination), protein conformation, post-translational modification (PTM), enzyme kinetics, conformational dynamics, deubiquitinase A (DUBA), enzyme activation, protein homeostasis
Introduction
Ubiquitination is a prevalent form of post-translational modification (PTM)2 that controls the fate and activity of many target proteins in a wide variety of cellular processes in eukaryotes, such as protein degradation, protein trafficking, DNA damage repair, and immune responses (1–3). Deubiquitinases (DUBs) are proteases that cleave monoubiquitin or polyubiquitin chains from target proteins and can also disassemble free polyubiquitin chains. Human genome encodes ∼100 DUBs, which can be divided into seven families (4–6). The abundance, subcellular localization, activity, and interaction specificity of DUBs are tightly regulated by a variety of mechanisms (7, 8). Deregulation is associated with human diseases, including cancer, neurodegeneration, inflammation, and autoimmunity (9, 10). DUBs are potential therapeutic targets for immune disorders and cancers (9, 11). PTM is an important mechanism for regulating both the activity and substrate specificity of DUBs in a reversible manner (12). For example, phosphorylation can enhance or suppress the catalytic activities of DUBs (12). Substrate specificity can also be modified by phosphorylation, as recently demonstrated on OTUD4 (13) and A20 (14). A large number of DUBs are subject to PTMs, including phosphorylation, ubiquitination, SUMOylation, lipidation, and oxidation (15). However, the structural and mechanistic basis of regulation by PTMs is not known for most of the DUBs. Modulation of conformational dynamics has been proposed as a plausible mechanism when structural changes are minimal (16).
Deubiquitinase A (DUBA), also named OTUD5, is a deubiquitinase whose catalytic activity is regulated by phosphorylation at a single serine residue. It is a cysteine protease that belongs to the ovarian tumor (OTU) family of DUBs, which is second to the largest family (4). The site of phosphorylation, Ser-177, is close to the N terminus of the catalytic domain and ∼13 Å away from the catalytic triad (17). DUBA was first identified as a negative regulator of type I interferon induction in macrophages by cleaving Lys-63–linked polyubiquitin chains from TRAF3 (tumor necrosis factor receptor-associated factor 3) (18). It also negatively regulates the production of cytokines in other innate immune cells (19) and T cells (20). A new role of DUBA as a regulator of DNA damage response has recently been identified (21). DUBA cleaves Lys-48– and Lys-63–linked diubiquitins in vitro, and the catalytic domain retains the substrate specificity and activity compared with the full-length protein, which consists of a ubiquitin-interacting motif at the C terminus (17).
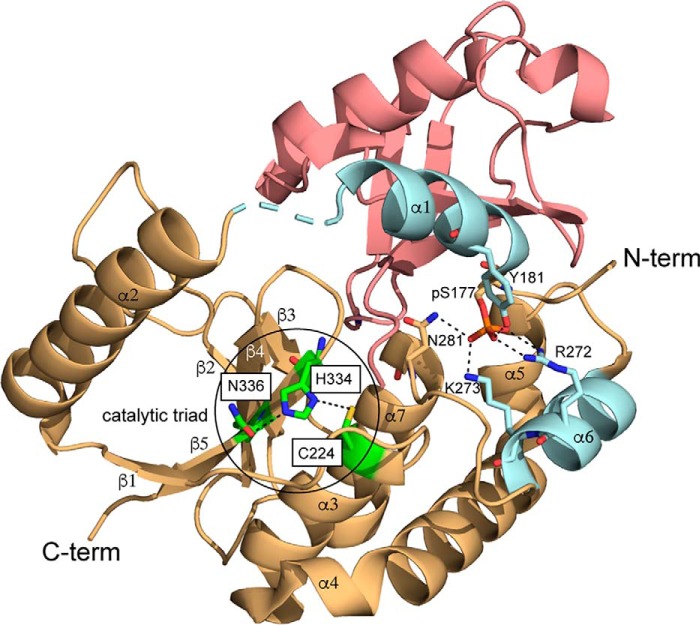
According to previous studies on the catalytic domain of DUBA, using both X-ray crystallography and NMR, phosphorylation induces minimal structural changes, including the active site (Fig. S1) (17). Crystal structure of phosphorylated DUBA (p-DUBA) conjugated to ubiquitin–aldehyde reveals interactions between the phosphate group and the surrounding residues (Fig. 1). These interactions presumably lead to structural and/or dynamic changes of DUBA, which are important for the catalytic cycle. However, the conformational properties unique to the active form of DUBA have not been identified because of the minimal structural changes upon phosphorylation and the electron density missing for the functionally important parts of DUBA (Table S1). In this work, we combined NMR spectroscopy and enzyme activity assays to characterize WT DUBA and several mutants that display low activities, similar to nonphosphorylated DUBA (np-DUBA). We observed a two-state conformational equilibrium unique to the p-DUBA. Our results highlight the importance of defining conformational dynamics in understanding the activation mechanism of DUBA.
Figure 1.
Crystal structure of p-DUBA conjugated to ubiquitin–aldehyde (PDB code 3TMP). p-DUBA is displayed in gold and cyan; ubiquitin–aldehyde is displayed in salmon. In the crystal structure of DUBA alone (PDB code 3PFY), helices α1 and α6 (cyan) are not visible.
Results and discussion
In a previous report, the activity of p-DUBA has been quantified using fluorescently labeled substrates under steady-state conditions (17), although the measured Km and kcat likely have large uncertainties because of the limited concentrations of substrates with respect to Km. Because no activity data were available on the nonphosphorylated form, it was not clear whether the activation of DUBA is due to a higher catalytic rate constant or the higher affinity to substrates. To answer this question, we performed activity assays using ubiquitin–7-amino-4-methylcoumarin (Ub-AMC) as the substrate under single-turnover conditions ([enzyme]/[substrate] > 10) and varied the enzyme concentration to circumvent the problem of insufficiently high substrate concentration. We were able to detect activity and quantify the enzyme kinetics on both forms of DUBA. Treating the np-DUBA samples with λ-protein phosphatase does not change the measured rate constant, confirming that the detected activity is not due to a small amount of phosphorylated protein. We also measured the activity of a phosphomimetic mutant, S177E, which was known to show no activity in the steady-state experiment from the previous report (17). Our data indicate that the increase in activity upon phosphorylation mainly results from ∼300-fold enhancement in the catalytic rate constant, although the substrate-binding affinity also becomes slightly higher (Fig. 2 and Table 1).
Figure 2.
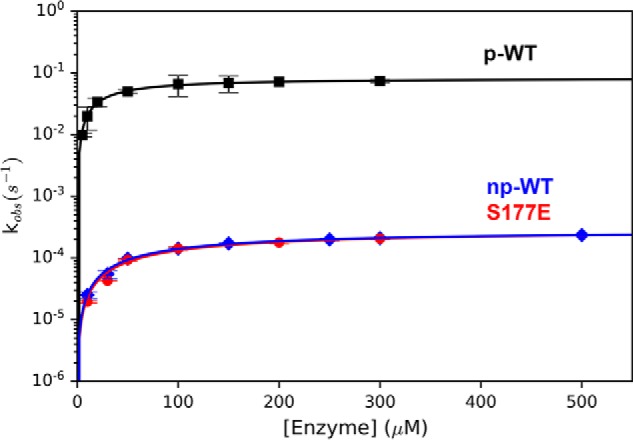
DUBA activity assays. The average kobs values from triplicated measurements at variable enzyme concentrations are displayed. The concentrations of the substrate, Ub-AMC, were 25 nm (p-WT), 1 μm (np-WT), and 0.5 μm (S177E), respectively.
Table 1.
Single-turnover kinetic parameters of the wildtype DUBA and the S177E mutant
| Enzyme | k2 | Kd |
|---|---|---|
| 10−3 s−1 | μm | |
| np-WT | 0.283 ± 0.007 | 102 ± 7 |
| p-WT | 83 ± 15 | 31 ± 2 |
| S177E | 0.29 ± 0.03 | 118 ± 25 |
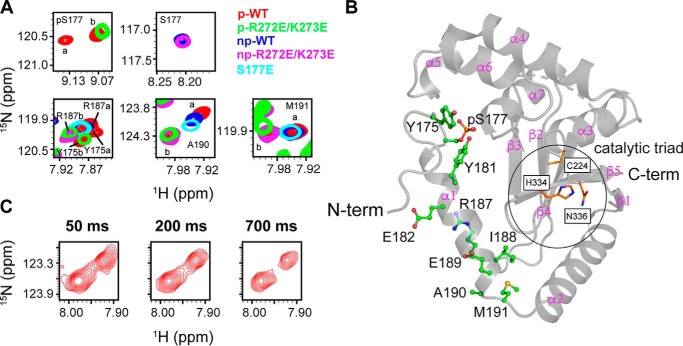
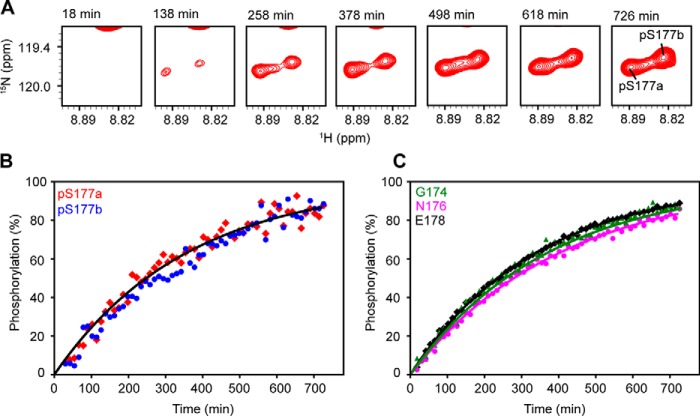
Previous NMR studies (17) indicate that phosphorylation does not induce significant structural changes because chemical shift perturbations were only observed in several residues (Gly-174–Tyr-181) close to the phosphorylation site, Ser-177 (Fig. S2A and Table S2). It was suggested that the extensive interactions between helices α1 and α6 mediated by phosphorylated Ser177 (pSer-177) are important for DUBA activity, but the differences in the conformational properties of p-DUBA and np-DUBA were not well-defined. The phosphomimetic mutation, S177E, in principle, can mimic these interactions, but this mutant was previously reported to show no activity in the steady-state experiment (17) and displayed low activity similar to the np-DUBA in our single-turnover experiment. In p-DUBA, we have identified two sets of cross-peaks in the 15N TROSY spectrum for a subset of residues within the helix α1 and the immediately neighboring residues, including pSer-177, and the loop connecting helices α1 and α2, whereas the np-DUBA displays only one set of cross-peaks (Fig. 3 and Table S2) (22). We hypothesized that the presence of two conformers in slow exchange on the NMR time scales is functionally important. To test this hypothesis, we examined two mutants, S177E and R272E/K273E, which displayed low or nondetectable activity. The charge reversal mutations, R272E and K273E, were designed to disrupt the interactions between phosphate group and these two positively charged residues (Fig. 1). Indeed, the R272E/K273E mutant in both the nonphosphorylated (np-R272E/K273E) and the phosphorylated (p-R272E/K273E) forms displayed activity too low to be quantified in the single-turnover kinetic assays that we performed. We recorded 15N TROSY spectra of these two mutants (Fig. S2A) and the HNCACB spectra to confirm the assignments. Fig. 3A shows that both mutants yield one set of cross-peaks, supporting our hypothesis. Unlike the WT, phosphorylation of the R272E/K273E mutant did not result in two conformers distinguishable from each other by different chemical shifts (Fig. 3A and Table S2). This observation suggests that the presence of the phosphate group in pSer-177 is not sufficient to induce the formation of two conformers, which were detected only in the WT p-DUBA. Because the observed two conformers are similarly populated, each cross-peak cannot be easily assigned to a particular conformer based on either peak intensity or volume. Interestingly, the cross-peaks observed in the p-R272E/K273E mutant overlap with one of the two sets of cross-peaks observed in the WT p-DUBA. It is reasonable to assume that the single set of cross-peaks present in the R272E/K273E mutant belongs to one conformer, in which the interactions between pSer-177 and helix α6 are weak or nonexistent. Under this assumption, we assigned the cross-peaks from the WT p-DUBA to two conformers, which we named a and b, respectively. The b conformer is present in both the WT p-DUBA and the p-R272E/K273E mutant, whereas the formation of the a conformer requires pSer-177 to interact with the two positively charged residues in α6. We determined that the fractional population of the a conformer was 61% by quantifying the relative cross-peak volumes from two conformers across seven residues (Table S3). The a and b conformers share similar secondary structures because the differences in the 13C chemical shifts are negligibly small (Table S4), suggesting that the two conformers result from different modes of interactions between helix α1 and the other regions of DUBA, especially helix α6, rather than local folding and unfolding events. Helix α1 is flexible and does not become more rigid upon phosphorylation, based on the 15N transverse relaxation rates (Fig. S3), consistent with the relatively low helical content predicted by TALOS-N (23) (Fig. S4); low 15N heteronuclear NOE values have previously been reported (17). We performed 15N ZZ-exchange experiment at 303 K on p-DUBA to quantify the exchange rate, but no exchange cross-peaks were observed (Fig. 3C). The rate of exchange between the two conformers was therefore estimated to be slower than 0.1 s−1. We also monitored the phosphorylation of DUBA by casein kinase 2 using real-time NMR. The relative populations of the a and b conformers remain the same throughout the entire time course (Fig. 4), suggesting that the two conformers reach their equilibrium populations at a rate faster than the sampling rate (∼12 min each spectrum). The results of this experiment also ensure that the appearance of the second set of cross-peaks is not due to much slower processes such as sample degradation.
Figure 3.
Two conformational states of p-DUBA observed by NMR spectroscopy. A, the overlaid 15N TROSY spectra of p-WT DUBA, p-R272E/K273E mutant, np-WT DUBA, np-R272E/K273E mutant, and S177E mutant. B, structure of DUBA (PDB code 3TMP) with residues showing two conformers in p-WT DUBA highlighted by stick-and-ball representation. C, 15N ZZ-exchange spectra of p-WT DUBA at three mixing times, showing the cross-peaks from two conformers of Ala-190. The sample temperature was 300 K for all TROSY experiments and 303 K for the ZZ-exchange experiment. The protein concentrations were ∼500 μm for all samples. C-term, C terminus; N-term, N terminus.
Figure 4.
Time courses of DUBA phosphorylation. A, cross-peaks of pSer-177 in 15N TROSY spectra recorded during the time course. B and C, time courses of phosphorylation calculated from cross-peak intensities. The apparent rate constant of phosphorylation (kapp) was determined to be 2.8 × 10−3 min−1 (pSer-177a and pSer-177b), 2.7 × 10−3 min−1 (Gly-174), 2.5 × 10−3 min−1 (Asn-176), and 3.0 × 10−3 min−1 (Glu-178). The cross-peaks from other residues are not well-resolved between the phosphorylated and nonphosphorylated species and not analyzed.
Consistent with our hypothesis that the observed two-state equilibrium is essential for DUBA activity, the S177E mutant displays only one set of cross-peaks, whose positions are similar to those in the WT np-DUBA. The enzyme kinetic parameters of the S177E mutant are almost the same as the np-DUBA (Table 1). For both, the positions of cross-peaks lie between the two conformers observed in p-DUBA, raising the possibility of fast exchange between the two conformers on the NMR time scales. In principle, the relative populations of two conformers in fast exchange can be determined from the averaged peak positions relative to those in the two conformers, according to the formula ¯δ = paδa + pbδb (24), where δa and δb are the chemical shifts of the a and b conformers, and pa and pb are the fractional populations. However, we did not perform such an analysis because of the relatively small difference in chemical shifts between the two conformers; both conformers may also display slightly different chemical shifts across different forms of DUBA and invalidate the analysis. Combining NMR and activity data on all variants of DUBA that we studied, we conclude that the two conformers in p-DUBA and the slow exchange rate are unique features of p-DUBA. Absence of these features is associated with low activity.
Conformational dynamics of DUBA have been hypothesized to be critical for its activity according to the crystal structure and the NMR characterization of motions faster than the molecular tumbling. It was proposed that the motions of helices α1 and α6 allow DUBA to switch between the open and closed conformers to allow substrate binding (17). Our study provides NMR spectroscopic evidence that p-DUBA adopts two conformers distinguishable by chemical shifts. The a conformer can possibly resemble the crystal structure of p-DUBA covalently linked to ubiquitin–aldehyde (Fig. 1), although the low 15N R2 relaxation rates in the α1 (Fig. S3; methods detailed in supporting text) raise the possibility of dynamic averaging of many conformers on the fast time scale. To understand the structural differences between the two ensembles of conformers, we have performed the CLEANEX experiment (25) (supporting text) to measure solvent-exchange rates of the amide 1H, which report on the extent of exposure to solvent. The exchange rates are slower for the a conformer compared with the b conformer (Fig. S5). In a simplified model, the conformational ensemble a can be divided into two subensembles, named ae and au, where the subscript e stands for exposed, and the u stands for unexposed. au may largely resemble the crystal structure, where the pSer-177 is mostly buried (accessible surface area = ∼13 Å2). The lower limit of the fractional population of the subconformer au can be estimated from the ratio (kb − ka)/kb, where kb and ka are the solvent-exchange rates of the conformers a and b, respectively, by assuming that the b represents the completely exposed conformer. The estimated lower limit to the population of the au subconformer is ∼20% (Fig. S5B). In other words, if the a conformer results from the dynamic averaging between the exposed and buried conformers, at least 20% of the conformers need to be completely buried. Alternatively, a continuum of conformers ranging from exposed to buried can be sampled on the fast time scale. We also performed pH titration on p-DUBA samples (supporting text) within a limited range of pH values, as allowed by sample stability, to estimate the pKa value of the phosphate group associated with the transition between the −1 and −2 charge states. The two conformers display very similar pKa values according to the amide 1H chemical shifts (Fig. S6A), and both are close to previously reported pKa values of 6.0–6.1 of solvent-exposed phosphoserine in model peptides (26–28). The a conformer displays a slightly lower pKa, 0.08 pH unit lower than the b conformer. A previous study (29) has reported a ∼0.7-pH unit downward shift in the pKa value upon formation of salt bridges involving the −2 charge state. The lower pKa value of the phosphate in the a conformer is consistent with the formation of salt bridges within the subconformer au. Overall, both the solvent-exchange rates and the pKa values of the phosphate group suggest that the a and b conformers are structurally different, with the a conformer less exposed to the solvent. 15N NOESY–heteronuclear single quantum correlation (HSQC) experiment has been attempted on p-DUBA (supporting text) to obtain structural constraints. However, no NOE cross-peaks indicating interactions between the α1 and the other regions of DUBA were observed, most likely because of the presence of ∼45 residues undetectable by 15N TROSY, among which most of the surface residues are clustered in the regions interacting with helix α1 in the crystal structure of p-DUBA conjugated to the ubiquitin–aldehyde (Fig. S7). The significant line broadening in both the missing residues and approximately half of the assigned residues may be induced by the transient interactions between helix α1 and the well-folded regions of DUBA on the microsecond-to-millisecond time scales. For two residues in the helix α1 or the α1–α2 loop, Tyr-175 and Glu-189, the b conformer shows significantly elevated 15N transverse relaxation rates compared with the a conformer and also the average rate across all residues in the helix α1 (Fig. S3). Likewise, the 1H line widths are significantly different between the a and b conformers for a few residues (Table S5). These differences are not explained by the differential solvent-exchange rates and likely indicate different dynamic properties on the microsecond-to-millisecond time scales. Combining all evidence, the a and b conformers display detectable differences in both the structural and dynamic properties.
The two-state equilibrium of active and inactive states has been previously observed in kinases, such as Abl kinase (30), where the relative populations of the two states are highly correlated with activity. Fast exchange between the inactive and active states on the NMR time scales has also been observed in signaling proteins whose activities are regulated by phosphorylation (31, 32) or autoinhibition (33). In these systems, the effects of conformational dynamics are primarily thermodynamic, because the exchange between two states is fast compared with the catalytic rate constant and therefore not rate-limiting. By contrast, the exchange between two conformers in p-DUBA can be comparable or slower than the catalytic turnover rate, which is ∼0.08 s−1 in p-DUBA, and rate-limiting. The rate of conformational process can influence various steps of enzymatic cycle, including substrate binding (34) and product release (35). Alternatively, as previously reported on a scavenger mRNA-decapping enzyme, the increase in the rate of the observed opening and closing motions of the dimeric enzyme caused by the increase in substrate concentration is correlated with decrease in the catalytic turnover rate, although the motions are necessary for catalysis (36). In other words, excess motions can lower catalytic activity of enzymes, at least in this particular case. It is reasonable to hypothesize that the much slower interconversion between the two states in p-DUBA compared with np-DUBA is the underlying reason why the p-DUBA displays a much higher catalytic turnover rate. Future studies on DUBA mutants, in which the interconversion rates vary within the range defined by np- and p-DUBA, need to be carried out to understand the detailed mechanism of DUBA activation at the molecular level.
In addition to the two-state equilibrium observed from the NMR experiments, an important observation from our studies is that phosphorylation of DUBA primarily enhances the catalytic rate constant rather than the substrate affinity. The increase in the catalytic rate constant by binding of the accessory domains or regulatory proteins to the catalytic domain has been previously reported on members of the USP (ubiquitin-specific protease) family of DUBs (37). DUBs can also distinguish between polyubiquitin chains of different linkages based on distinct catalytic rate constants rather than substrate affinities, as shown for Cezanne (cellular zinc finger anti–NF-κB protein) (38), a member of the OTU family. Our results provide an interesting example of a deubiquitinase whose catalytic rate constant is enhanced by phosphorylation, and it remains to be seen whether other DUBs subject to PTMs are regulated by similar mechanisms.
Experimental procedures
Sample preparation
The plasmid OTUD5, which encodes residues 172–344 of DUBA in isoform 2 was a gift from Cheryl Arrowsmith (Addgene plasmid no. 28270). Mutants were generated using the WT construct as the template. The R272E/K273E mutant was generated using the mutagenesis service provided by GenScript. The S177E mutant was generated using QuikChange II site-directed mutagenesis kit (Agilent Technology) and the primer 5′-GCCTCATACTCGTCCTCCTCGTTGTAGCCTGCGCCGA-3′. Expression, purification, and phosphorylation of the 15N-labeled, [2H,15N]-labeled, and [2H,13C,15N]-labeled DUBA and DUBA variants were carried out using previously reported protocols (17, 22). All kinetic assays were performed on 15N-labeled DUBA. The purity and homogeneity of p-DUBA samples were verified using 15N TROSY spectra and the mass spectrometric analysis (22).
NMR spectroscopy
The NMR sample buffer contains 50 mm sodium phosphate, pH 7.0, 100 mm NaCl, 1 mm tris(2-carboxyethyl)phosphine, 0.03% (w/v) NaN3, and 90% H2O/10% D2O (v/v), unless stated otherwise. 0.2 mm 4,4-dimethyl-4-silapentane-1-sulfonic acid was used for chemical shift referencing.
NMR experiments were performed on a Varian 700-MHz spectrometer equipped with a cryoprobe unless stated otherwise. The sample temperature was calibrated using methanol-d4 (Cambridge Isotope Laboratories) (39). The NMR data were processed with NMRPipe (40) and visualized using SPARKY (41, 42). Peak integration was performed using PINT (43). The 15N ZZ-exchange experiment (44) was performed as previously described (45). The HNCACB experiment was performed using the TROSY-detected method (46).
Phosphorylation monitored by real-time NMR
The phosphorylation reactions were carried out at 303 K on samples containing 480 μm of DUBA and monitored by real-time NMR spectroscopy on a Bruker Avance NEO 600-MHz spectrometer equipped with a cryoprobe. The reactions were initiated by adding 2000 units of casein kinase 2 (New England Biolabs) to 200 μl of [2H,15N]-labeled DUBA in 20 mm Tris, pH 6.6, 50 mm KCl, 0.1 mm EGTA, 10 mm MgCl2, 5 mm ATP, 5 mm DTT, and 6% D2O. 15N TROSY spectra were acquired with 2 scans using an interscan delay of 1 s. 1024 (1H) × 150 (15N) complex points were recorded for each spectrum, which results in an acquisition time of ∼12 min. The rates of phosphorylation were obtained by fitting the cross-peak intensities to the first-order rate equation,
| (Eq. 1) |
where kapp is the apparent phosphorylation rate constant, and I∞ is the intensity that corresponds to 100% phosphorylated protein.
Enzyme kinetic assays
Kinetic assays were performed under the single-turnover conditions by ensuring that the enzyme concentration is at least 10 times that of the substrate. Increase in the fluorescent intensities resulting from cleavage of the substrate, Ub-AMC (Boston Biochem), was monitored using the SpectraMax Gemini XPS plate reader (Molecular Devices) or Fluoromax-4 fluorometer (HORIBA). The excitation and emission wavelengths were set to λex = 340 nm and λem = 440 nm, respectively. All activity assays were performed at 25 °C in the buffer containing 50 mm HEPES, pH 7.5, 100 mm NaCl, 5 mm DTT, and 0.1 mg/ml BSA. For assays performed on the plate reader, the reactions were carried out in a 20-μl volume on 384-well plates (Corning). For assays performed on the Fluoromax-4 fluorometer, the reaction volume was 60 μl. The reactions were initiated by mixing equal volumes of 2× substrate and 2× enzyme solutions. All measurements were performed in triplicate. To test whether np-DUBA samples might contain small amount of phosphorylated protein, the samples were treated with λ-protein phosphatase (New England Biolabs) according to the instruction manual and compared with the untreated samples.
The kinetic data were interpreted using the simplest kinetic model for acyl-enzyme (47).
| (Eq. 2) |
The change in the fluorescent intensity of AMC results from the second step of the reaction, where the AMC is detached from the substrate. Data fitting was performed using Mathematica (Wolfram). kobs was determined by fitting the time course to the equation,
| (Eq. 3) |
where I∞ and I0 are the final and initial intensities. kobs was measured at a range of enzyme concentrations to determine the k2 and Kd. The data were fit to the equation,
| (Eq. 4) |
where [E] is the enzyme concentration.
Author contributions
A.K., E. R., and Y. L. data curation; A.K. and Y. L. formal analysis; A.K. and Y. L. investigation; A.K. and Y. L. writing-review and editing; Y. L. conceptualization; Y. L. supervision; Y. L. funding acquisition; Y. L. writing-original draft.
Supplementary Material
This work was supported by National Institutes of Health Grant R15GM123391 (to Y. L.) and by startup funds from University of Louisville. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supporting text, Tables S1–S5, and Figs. S1–S7.
- PTM
- post-translational modification
- AMC
- 7-amino-4-methylcoumarin
- DUBA
- deubiquitinase A
- np
- nonphosphorylated
- p
- phosphorylated
- OTU
- ovarian tumor
- TROSY
- transverse relaxation optimized spectroscopy
- PDB
- Protein Data Bank
- Ub
- ubiquitin.
References
- 1. Komander D., and Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- 2. Swatek K. N., and Komander D. (2016) Ubiquitin modifications. Cell Res. 26, 399–422 10.1038/cr.2016.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yau R., and Rape M. (2016) The increasing complexity of the ubiquitin code. Nat. Cell Biol. 18, 579–586 10.1038/ncb3358 [DOI] [PubMed] [Google Scholar]
- 4. Clague M. J., Barsukov I., Coulson J. M., Liu H., Rigden D. J., and Urbé S. (2013) Deubiquitylases from genes to organism. Physiol. Rev. 93, 1289–1315 10.1152/physrev.00002.2013 [DOI] [PubMed] [Google Scholar]
- 5. Abdul Rehman S. A., Kristariyanto Y. A., Choi S. Y., Nkosi P. J., Weidlich S., Labib K., Hofmann K., and Kulathu Y. (2016) MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol. Cell 63, 146–155 10.1016/j.molcel.2016.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coleman K. E., and Huang T. T. (2018) In a class of its own: a new family of deubiquitinases promotes genome stability. Mol. Cell 70, 1–3 10.1016/j.molcel.2018.03.022 [DOI] [PubMed] [Google Scholar]
- 7. Sahtoe D. D., and Sixma T. K. (2015) Layers of DUB regulation. Trends Biochem. Sci. 40, 456–467 10.1016/j.tibs.2015.05.002 [DOI] [PubMed] [Google Scholar]
- 8. Reyes-Turcu F. E., Ventii K. H., and Wilkinson K. D. (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78, 363–397 10.1146/annurev.biochem.78.082307.091526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harrigan J. A., Jacq X., Martin N. M., and Jackson S. P. (2018) Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat. Rev. Drug Discov. 17, 57–78 10.1038/nrd.2017.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Popovic D., Vucic D., and Dikic I. (2014) Ubiquitination in disease pathogenesis and treatment. Nat. Med. 20, 1242–1253 10.1038/nm.3739 [DOI] [PubMed] [Google Scholar]
- 11. Bedford L., Lowe J., Dick L. R., Mayer R. J., and Brownell J. E. (2011) Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat. Rev. Drug Discov. 10, 29–46 10.1038/nrd3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leznicki P., and Kulathu Y. (2017) Mechanisms of regulation and diversification of deubiquitylating enzyme function. J. Cell Sci. 130, 1997–2006 10.1242/jcs.201855 [DOI] [PubMed] [Google Scholar]
- 13. Zhao Y., Mudge M. C., Soll J. M., Rodrigues R. B., Byrum A. K., Schwarzkopf E. A., Bradstreet T. R., Gygi S. P., Edelson B. T., and Mosammaparast N. (2018) OTUD4 is a phospho-activated K63 deubiquitinase that regulates MyD88-dependent signaling. Mol. Cell 69, 505–516.e5 10.1016/j.molcel.2018.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wertz I. E., Newton K., Seshasayee D., Kusam S., Lam C., Zhang J., Popovych N., Helgason E., Schoeffler A., Jeet S., Ramamoorthi N., Kategaya L., Newman R. J., Horikawa K., Dugger D., et al. (2015) Phosphorylation and linear ubiquitin direct A20 inhibition of inflammation. Nature 528, 370–375 10.1038/nature16165 [DOI] [PubMed] [Google Scholar]
- 15. Kessler B. M., and Edelmann M. J. (2011) PTMs in conversation: activity and function of deubiquitinating enzymes regulated via post-translational modifications. Cell Biochem. Biophys. 60, 21–38 10.1007/s12013-011-9176-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mevissen T. E. T., and Komander D. (2017) Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 86, 159–192 10.1146/annurev-biochem-061516-044916 [DOI] [PubMed] [Google Scholar]
- 17. Huang O. W., Ma X., Yin J., Flinders J., Maurer T., Kayagaki N., Phung Q., Bosanac I., Arnott D., Dixit V. M., Hymowitz S. G., Starovasnik M. A., and Cochran A. G. (2012) Phosphorylation-dependent activity of the deubiquitinase DUBA. Nat. Struct. Mol. Biol. 19, 171–175 10.1038/nsmb.2206 [DOI] [PubMed] [Google Scholar]
- 18. Kayagaki N., Phung Q., Chan S., Chaudhari R., Quan C., O'Rourke K. M., Eby M., Pietras E., Cheng G., Bazan J. F., Zhang Z., Arnott D., and Dixit V. M. (2007) DUBA: a deubiquitinase that regulates type I interferon production. Science 318, 1628–1632 10.1126/science.1145918 [DOI] [PubMed] [Google Scholar]
- 19. González-Navajas J. M., Law J., Nguyen K. P., Bhargava M., Corr M. P., Varki N., Eckmann L., Hoffman H. M., Lee J., and Raz E. (2010) Interleukin 1 receptor signaling regulates DUBA expression and facilitates Toll-like receptor 9-driven antiinflammatory cytokine production. J. Exp. Med. 207, 2799–2807 10.1084/jem.20101326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rutz S., Kayagaki N., Phung Q. T., Eidenschenk C., Noubade R., Wang X., Lesch J., Lu R., Newton K., Huang O. W., Cochran A. G., Vasser M., Fauber B. P., DeVoss J., Webster J., et al. (2015) Deubiquitinase DUBA is a post-translational brake on interleukin-17 production in T cells. Nature 518, 417–421 10.1038/nature13979 [DOI] [PubMed] [Google Scholar]
- 21. de Vivo A., Sanchez A., Yegres J., Kim J., Emly S., and Kee Y. (2019) The OTUD5-UBR5 complex regulates FACT-mediated transcription at damaged chromatin. Nucleic Acids Res. 47, 729–746 10.1093/nar/gky1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kabra A., Benson C. A., and Li Y. (2019) Backbone 1H, 13C, 15N resonance assignments of deubiquitinase A in non-phosphorylated and phosphorylated forms. Biomol. NMR Assign. 13, 37–42 10.1007/s12104-018-9847-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shen Y., and Bax A. (2013) Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 56, 227–241 10.1007/s10858-013-9741-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Loria J. P., Berlow R. B., and Watt E. D. (2008) Characterization of enzyme motions by solution NMR relaxation dispersion. Acc. Chem. Res. 41, 214–221 10.1021/ar700132n [DOI] [PubMed] [Google Scholar]
- 25. Hwang T. L., van Zijl P. C., and Mori S. (1998) Accurate quantitation of water-amide proton exchange rates using the phase-modulated CLEAN chemical EXchange (CLEANEX-PM) approach with a Fast-HSQC (FHSQC) detection scheme. J. Biomol. NMR 11, 221–226 10.1023/A:1008276004875 [DOI] [PubMed] [Google Scholar]
- 26. Vogel H. J., and Bridger W. A. (1982) Phosphorus-31 nuclear magnetic resonance studies of the two phosphoserine residues of hen egg white ovalbumin. Biochemistry 21, 5825–5831 10.1021/bi00266a016 [DOI] [PubMed] [Google Scholar]
- 27. Hoffmann R., Reichert I., Wachs W. O., Zeppezauer M., and Kalbitzer H. R. (1994) 1H and 31P NMR spectroscopy of phosphorylated model peptides. Int. J. Pept. Protein Res. 44, 193–198 [DOI] [PubMed] [Google Scholar]
- 28. Bienkiewicz E. A., and Lumb K. J. (1999) Random-coil chemical shifts of phosphorylated amino acids. J. Biomol. NMR 15, 203–206 10.1023/A:1008375029746 [DOI] [PubMed] [Google Scholar]
- 29. Kumar P., Chimenti M. S., Pemble H., Schönichen A., Thompson O., Jacobson M. P., and Wittmann T. (2012) Multisite phosphorylation disrupts arginine-glutamate salt bridge networks required for binding of cytoplasmic linker-associated protein 2 (CLASP2) to end-binding protein 1 (EB1). J. Biol. Chem. 287, 17050–17064 10.1074/jbc.M111.316661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saleh T., Rossi P., and Kalodimos C. G. (2017) Atomic view of the energy landscape in the allosteric regulation of Abl kinase. Nat. Struct. Mol. Biol. 24, 893–901 10.1038/nsmb.3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Volkman B. F., Lipson D., Wemmer D. E., and Kern D. (2001) Two-state allosteric behavior in a single-domain signaling protein. Science 291, 2429–2433 10.1126/science.291.5512.2429 [DOI] [PubMed] [Google Scholar]
- 32. Gardino A. K., Villali J., Kivenson A., Lei M., Liu C. F., Steindel P., Eisenmesser E. Z., Labeikovsky W., Wolf-Watz M., Clarkson M. W., and Kern D. (2009) Transient non-native hydrogen bonds promote activation of a signaling protein. Cell 139, 1109–1118 10.1016/j.cell.2009.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li P., Martins I. R., Amarasinghe G. K., and Rosen M. K. (2008) Internal dynamics control activation and activity of the autoinhibited Vav DH domain. Nat. Struct. Mol. Biol. 15, 613–618 10.1038/nsmb.1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boehr D. D., Dyson H. J., and Wright P. E. (2006) An NMR perspective on enzyme dynamics. Chem. Rev. 106, 3055–3079 10.1021/cr050312q [DOI] [PubMed] [Google Scholar]
- 35. Watt E. D., Shimada H., Kovrigin E. L., and Loria J. P. (2007) The mechanism of rate-limiting motions in enzyme function. Proc. Natl. Acad. Sci. U.S.A. 104, 11981–11986 10.1073/pnas.0702551104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Neu A., Neu U., Fuchs A. L., Schlager B., and Sprangers R. (2015) An excess of catalytically required motions inhibits the scavenger decapping enzyme. Nat. Chem. Biol. 11, 697–704 10.1038/nchembio.1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Faesen A. C., Luna-Vargas M. P., Geurink P. P., Clerici M., Merkx R., van Dijk W. J., Hameed D. S., El Oualid F., Ovaa H., and Sixma T. K. (2011) The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem. Biol. 18, 1550–1561 10.1016/j.chembiol.2011.10.017 [DOI] [PubMed] [Google Scholar]
- 38. Mevissen T. E. T., Kulathu Y., Mulder M. P. C., Geurink P. P., Maslen S. L., Gersch M., Elliott P. R., Burke J. E., van Tol B. D. M., Akutsu M., Oualid F. E., Kawasaki M., Freund S. M. V., Ovaa H., and Komander D. (2016) Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne. Nature 538, 402–405 10.1038/nature19836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Findeisen M., Brand T., and Berger S. (2007) A 1H-NMR thermometer suitable for cryoprobes. Magn. Reson. Chem. 45, 175–178 10.1002/mrc.1941 [DOI] [PubMed] [Google Scholar]
- 40. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 41. Goddard T. D., and Kneller D. G. (2004) SPARKY 3: NMR Assignment Program, University of California, San Francisco, CA [Google Scholar]
- 42. Lee W., Tonelli M., and Markley J. L. (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 10.1093/bioinformatics/btu830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ahlner A., Carlsson M., Jonsson B. H., and Lundström P. (2013) PINT: a software for integration of peak volumes and extraction of relaxation rates. J. Biomol. NMR 56, 191–202 10.1007/s10858-013-9737-7 [DOI] [PubMed] [Google Scholar]
- 44. Farrow N. A., Zhang O., Forman-Kay J. D., and Kay L. E. (1994) A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J. Biomol. NMR 4, 727–734 10.1007/BF00404280 [DOI] [PubMed] [Google Scholar]
- 45. Li Y., Altorelli N. L., Bahna F., Honig B., Shapiro L., and Palmer A. G. 3rd (2013) Mechanism of E-cadherin dimerization probed by NMR relaxation dispersion. Proc. Natl. Acad. Sci. U.S.A. 110, 16462–16467 10.1073/pnas.1314303110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhu G., Xia Y., Lin D., and Gao X. (2004) TROSY-based correlation and NOE spectroscopy for NMR structural studies of large proteins. Methods Mol. Biol. 278, 57–78 [DOI] [PubMed] [Google Scholar]
- 47. Bicknell R., and Waley S. G. (1985) Single-turnover and steady-state kinetics of hydrolysis of cephalosporins by β-lactamase I from Bacillus cereus. Biochem. J. 231, 83–88 10.1042/bj2310083 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.