Abstract
Following endocytosis, receptors that are internalized to sorting endosomes are sorted to different pathways, in part by sorting nexin (SNX) proteins. Notably, SNX17 interacts with a multitude of receptors in a sequence-specific manner to regulate their recycling. However, the mechanisms by which SNX17-labeled vesicles that contain sorted receptors bud and undergo vesicular fission from the sorting endosomes remain elusive. Recent studies suggest that a dynamin-homolog, Eps15 homology domain protein 1, catalyzes fission and releases endosome-derived vesicles for recycling to the plasma membrane. However, the mechanism by which EHD1 is coupled to various receptors and regulates their recycling remains unknown. Here we sought to characterize the mechanism by which EHD1 couples with SNX17 to regulate recycling of SNX17-interacting receptors. We hypothesized that SNX17 couples receptors to the EHD1 fission machinery in mammalian cells. Coimmunoprecipitation experiments and in vitro assays provided evidence that EHD1 and SNX17 directly interact. We also found that inducing internalization of a SNX17 cargo receptor, low-density lipoprotein receptor–related protein 1 (LRP1), led to recruitment of cytoplasmic EHD1 to endosomal membranes. Moreover, surface rendering and quantification of overlap volumes indicated that SNX17 and EHD1 partially colocalize on endosomes and that this overlap further increases upon LRP1 internalization. Additionally, SNX17-containing endosomes were larger in EHD1-depleted cells than in WT cells, suggesting that EHD1 depletion impairs SNX17-mediated endosomal fission. Our findings help clarify our current understanding of endocytic trafficking, providing significant additional insight into the process of endosomal fission and connecting the sorting and fission machineries.
Keywords: endosome, sorting, sorting nexin (SNX), trafficking, vesicles, receptor recycling, receptor endocytosis, receptor, intracellular trafficking, endocytosis, endosomal fission, Eps15 homology domain protein 1 (EHD1), LDL receptor–related protein 1 (LRP1), retromer, sorting nexin 17 (SNX17)
Introduction
The early/sorting endosome (SE)2 serves as the initial intracellular focal site for fusion of receptor-laden vesicles that have internalized from the plasma membrane (1). Receptors are sorted at the SE and packaged into budding vesicles that are ultimately released for intracellular transport to a variety of destinations, including recycling endosomes, the plasma membrane, late endosomes and lysosomes, and other cellular organelles (1, 2). Until recently, the mechanisms by which receptors are sorted at the SE have been poorly understood, and little is known about the mechanisms by which budding vesicles undergo fission from SE. Moreover, the mode of coupling between the sorting and fission machineries at SE remains elusive.
The retromer is a key sorting complex at SE, comprised of a cargo selection complex trimer of VPS35, VPS29, and either a VPS26a or VPS26b isoform, along with a dimer of sorting nexins (SNX1 or SNX2 and SNX5, SNX6, or SNX32) involved in sorting of cargo receptors at the SE (3, 4). Although the cargo selection complex was initially thought to be the primary contact site for interactions with receptors, recent studies have demonstrated that sorting nexins, especially SNX27 and SNX17, directly interact with receptor tails (5–8). For example, SNX17, a sorting nexin that attaches to the SE via its Phox homology (PX) domain (9–12), interacts with an additional complex called Retriever at the SE to regulate recycling of receptors by interaction of the SNX17 FERM domain with canonical NPXY/NXXY motifs, thus controlling the recycling of the β1 integrin receptor, LRP1 receptor, and more than 100 additional receptors (9, 11, 13).
When receptors at SE have been packaged into budding transport vesicles to be trafficked to their next destination, the budding vesicles need to undergo fission. The mechanisms of endosomal fission are not well-defined, but evidence supports a role of the Wiskott-Aldrich syndrome protein (WASH) complex in this process. The WASH complex is comprised of WASH1 (also known as WASHC1), Strumpellin (WASHC5), CCDC53 (WASHC3), KIAA1033/SWIP (WASHC4), and Fam21 (also known as WASHC2) (61, 62), which binds to the Retromer and initiates filamentous actin nucleation on SE (63, 64), potentially facilitating fission and vesicle release. However, whether WASH complex–mediated actin nucleation is sufficient for fission at SE remains unknown, and how receptor cargo is coupled to sorting and the fission machinery is similarly not understood.
A growing body of literature suggests that the endocytic regulatory protein Eps15 homology domain protein 1 (EHD1) localizes to SE and recycling endosomes and is capable of inducing ATP-catalyzed fission of membranes (14–21). Indeed, it has been demonstrated that EHD1 function is required for normal recycling of a wide variety of receptors that traverse endocytic pathways (22, 23). However, to date, the mechanism of EHD1 recruitment and coupling to SE has not been elucidated. Here we demonstrate that EHD1 undergoes recruitment to endosomal membranes upon induction of receptor-mediated endocytosis. Recruitment of EHD1 to the SE leads to its direct interaction with SNX17 and depends on the presence of the SNX17 FERM B and FERM C domains. Indeed, EHD1 depletion leads to enlarged SE structures, supporting a role of EHD1 in SE fission. Our data are consistent with a new model of endosomal fission in which the interaction between SNX17 and EHD1 provides a molecular explanation to couple the endosomal sorting and fission machineries.
Results
Recent studies have highlighted a significant role of SNX17 in sorting of select cargo receptors at the SE and regulation of their recycling to the plasma membrane (9–12). However, the mechanisms of coupling SNX17 and its cargo receptors to specific fission machinery at the SE remain unknown. The endocytic regulatory protein EHD1 regulates receptor recycling (16, 19, 24–26), and increasing evidence suggests that its primary function is in the fission of endosomes (14–21). However, to date, direct interactions between EHD1 and the cytoplasmic tails of receptors have not been identified. Accordingly, we hypothesized that SNX17 may serve as a link to connect the sorting machinery and recycling receptors with EHD1 and the endosomal fission machinery.
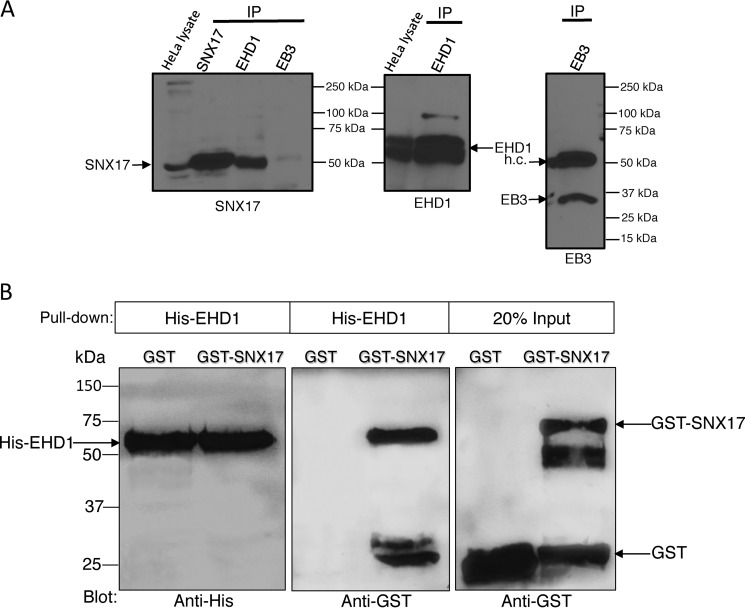
To first test whether SNX17 and EHD1 interact in cells (despite the lack of a NPXY/NXXY motif in EHD1), we performed coimmunoprecipitations. Antibodies specific for SNX17 detected an ∼53-kDa band in HeLa cell lysates and by immunoprecipitation (IP) with anti-SNX17 (IP SNX17) (Fig. 1A). In addition, IP with anti-EHD1 antibodies also led to detection of SNX17 (IP EHD1). As a control, an anti-EB3 antibody failed to precipitate SNX17 (IP EB3). These data demonstrate that EHD1 and SNX17 reside in a complex in cells.
Figure 1.
Interaction between EHD1 and SNX17. A, HeLa cell lysates were incubated at 4 °C overnight with either anti-SNX17, anti-EHD1, or anti-EB3 antibodies (from left to right). Protein G beads were then added to the lysate–antibody mixture at 4 °C for 4 h. Bound proteins were then eluted by boiling at 95 °C in β-mercaptoethanol–containing loading buffer, separated by SDS-PAGE, and immunoblotted with anti-SNX17 antibodies (left panel), anti-EHD1 antibodies (center panel), or anti-EB3 antibodies (right panel). Input lysates (20%) are depicted on the left of the immunoblots. h.c., immunoglobulin heavy chain. B, purified His-EHD1 was bound to Ni2+-NTA beads prior to incubation with either GST alone or GST-SNX17. Bound proteins were then eluted by boiling at 95 °C in β-mercaptoethanol–containing loading buffer, separated by SDS-PAGE, and immunoblotted with anti-His (left panel) or anti-GST antibodies (center and right panels). Input refers to the amounts of purified GST and GST-SNX17 used for incubation with His-EHD1. Data shown are representative of three independent experiments.
In addition to SNX17, it has been reported that SNX27 also associates with the retromer and plays a role in recycling receptors from endosomes to the plasma membrane (5, 7, 8, 27). Accordingly, we also tested whether EHD1 coimmunoprecipitates with SNX27. Although IP of SNX27 with an antibody to SNX27 led to detection of the anticipated ∼61-kDa band, IP with antibodies to EHD1 led to detection of a weak, ∼50-kDa band that was not observed in the lysate fraction for SNX27 (Fig. S1A, unknown). Although it remains possible that this is a different SNX27 species of lower molecular mass, given its absence in the lysate fraction, the most likely explanation is that it is a nonspecific band that is enriched in the course of the IP. Although MS and additional tests will be needed to ultimately determine whether there may be a weak interaction between EHD1 and SNX27, in this study we focused on the novel and well-defined interaction we identified between EHD1 and SNX17.
We next asked whether EHD1 and SNX17 directly interact with one another. Accordingly, we used purified GST-SNX17 (or purified GST as a control) together with purified His-tagged EHD1 (His-EHD1) to perform in vitro binding assays (Fig. 1B). Using equal concentrations of His-EHD1 (Fig. 1B, left panel), we showed that His-EHD1 pulled down significant levels of GST-SNX17 (roughly equivalent to the 20% observed in the input), whereas His-EHD1 pulled down no detectable GST (Fig. 1B, center panel, negative control). These data support the notion that EHD1 and SNX17 reside within a complex in cells and are capable of direct interaction.
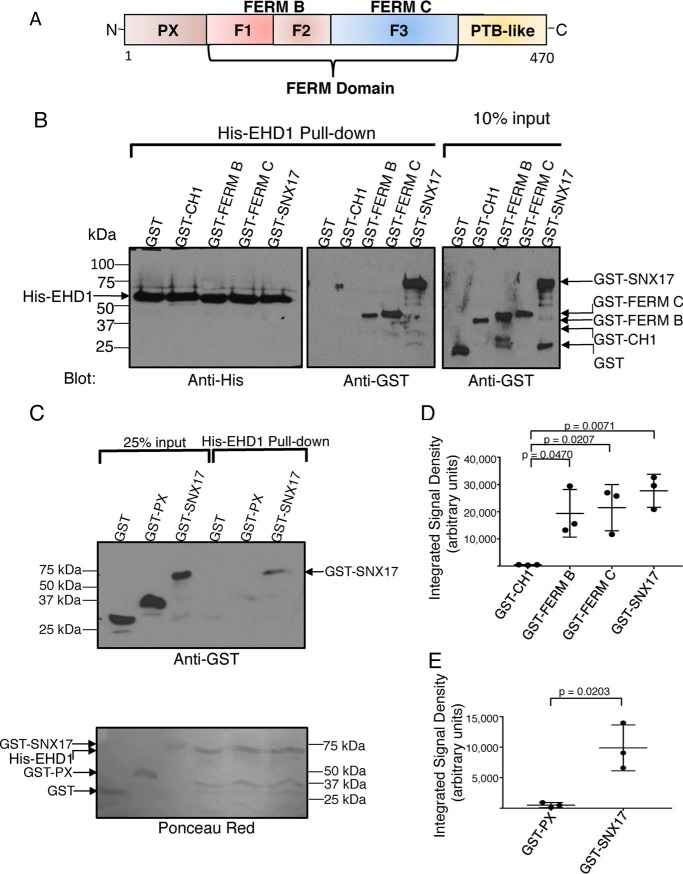
Given that SNX17 and EHD1 interact directly (at least in vitro), we aimed to delineate the mode of this interaction and define the specific domains or protein regions involved. Accordingly, in addition to full-length SNX17, we generated constructs comprised of the SNX17 FERM B or SNX17 FERM C regions (Fig. 2A), which are capable of interaction with multiple proteins containing NPXY/NXXY motifs (28). Using equal concentrations of His-EHD1 as bait (Fig. 2B, left panel), we observed that, similar to full-length GST-SNX17, both GST-FERM B and GST-FERM C domains were precipitated, whereas controls (GST alone or GST fused to the CH1 domain of MICAL-L1) were not precipitated (Fig. 2B, center panel). Moreover, densitometric analysis from three experiments demonstrated that full-length SNX17 as well as each of the individual FERM domains showed significant binding to His-EHD1 compared with the control MICAL-L1 CH1 domain (Fig. 2D). However, although SNX17 W321 is essential for the interaction with NPXY/NXXY motifs in receptor tails (28), mutation of SNX17 to W321A had no effect on its ability to bind EHD1 (Fig. S1B), likely indicating a different mechanism for SNX17 interactions with NPXY/NXXY motifs and EHD1 and also raising the possibility that SNX17 might be able to simultaneously couple receptors and the fission machinery. In addition, we also assessed the ability of the SNX17 PX domain, a region implicated in interactions with phosphoinositides (29, 30), to interact with EHD1. As expected, unlike the full-length SNX17 and its FERM domains, no interaction with EHD1 was observed with this lipid-binding domain (Fig. 2C, quantified in Fig. 2E). These experiments suggest that the SNX17 atypical FERM domain directly binds to EHD1.
Figure 2.
Delineation of the SNX17 and EHD1 domains required for their interaction. A, schematic illustrating the domain architecture of SNX17. B and C, purified His-EHD1 was bound to Ni2+-NTA beads for 2 h at 4 °C, as described under “Experimental procedures.” The His-EHD1 and purified GST-fusion target proteins (GST alone, GST-CH1, GST-FERM B, GST-FERM C, GST-PX, and GST-SNX17) were treated with micrococcal nuclease at 30 °C for 10 min. His-EHD1 was then incubated with GST fusion proteins for 2 h at 4 °C. Samples were washed, eluted, and separated by SDS-PAGE. B, left panel, immunoblotting was done with anti-His-HRP antibody, showing equivalent amounts of His-EHD1 used to incubate with GST-fusion proteins. B, center and right panels, immunoblotting was done with anti-GST antibody, as in the top panel in C. C, bottom panel, levels of the purified proteins as stained by Ponceau Red. Input refers to the amounts of purified GST, GST-CH1, GST-FERM B, GST-FERM C, GST-PX, and GST-SNX17 used for incubation with His-EHD1 bound to beads. D and E, densitometric quantification of purified GST-CH1, GST-PX, GST-FERM B, GST-FERM C, and GST-SNX17 protein levels precipitated by purified His-EHD1. Error bars denote standard deviation. The p values were determined by Student's two-tailed t test. Data shown are representative of three independent experiments. PTB, phosphotyrosine binding.
We next asked which EHD1 region is required for binding to SNX17. Most interaction partners of EHD1 interact with the C-terminal EH domain (Fig. S2A), usually via an NPF motif (which is lacking in SNX17) (22, 31, 32), so we used His-EHD1 (full-length), a construct containing only the EHD1 EH domain (His–EH-1), or His-EHD1, lacking the EH domain (His-EHD1 ΔEH), to pull down GST-SNX17 or GST (control) (Fig. S2B and quantified in Fig. S2C). As demonstrated, His-EHD1 ΔEH pulled down similar levels of GST-SNX17 as those observed upon full-length His-EHD1 pulldown. Moreover, when used as bait, His–EH-1 showed decreased SNX17 pulldown. These results support the notion that the EHD1 EH domain is not required for interaction with SNX17.
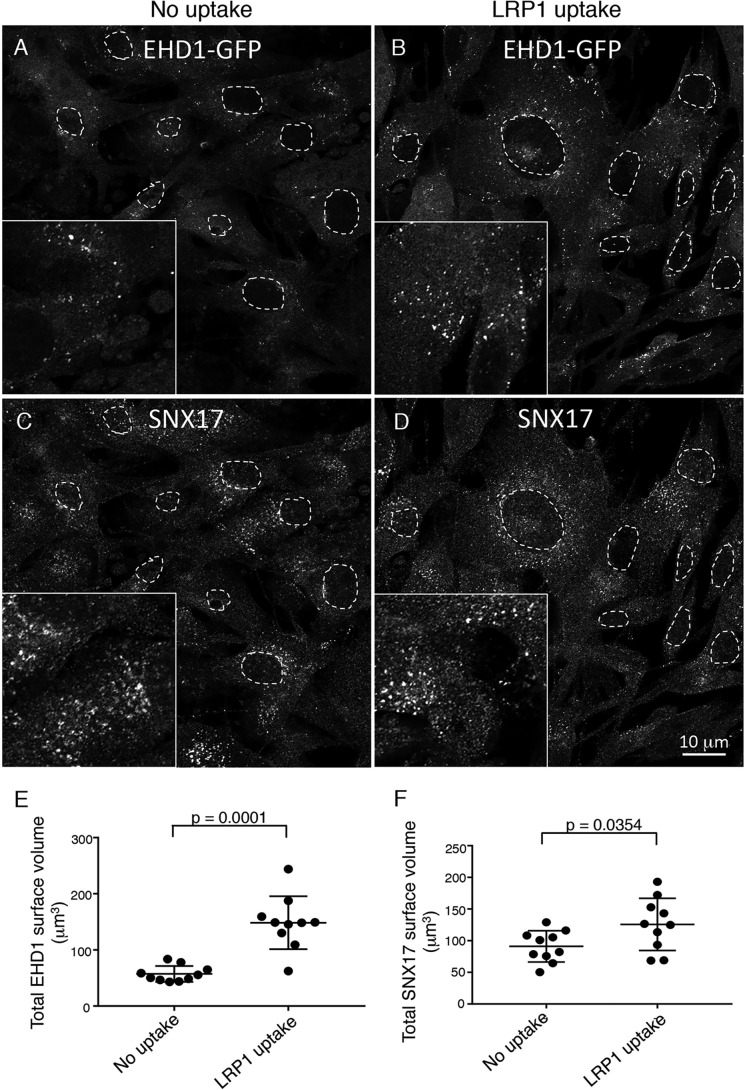
EHD1 is an ATPase that dimerizes and cycles between the cytoplasm and endosomes (16, 33–36). To date, however, the potential triggers for EHD1 recruitment to endosomes have not been examined. Accordingly, we rationalized that, because EHD1 appears to be recruited to SE to facilitate fission of cargo in conjunction with SNX17-based cargo sorting, internalization of SNX17 cargo molecules might serve to induce EHD1 recruitment to SE. Accordingly, we used our NIH3T3 CRISPR/Cas9 gene-edited EHD1-GFP cells (37, 38) to address this question. At steady state, in the absence of select uptake, we visualized EHD1 in the cytoplasm and on a smattering of puncta and small tubules, consistent with SE (Fig. 3A, see inset for more detail). When incubated with an antibody to the SNX17-sorted cargo receptor low-density lipoprotein receptor–related protein 1 (LRP1) (9, 39, 40) to induce its internalization, we observed a dramatic increase in the number and size of EHD1-containing puncta and small tubules (Fig. 3B, see inset for more detail). Typically, we observed that the LRP1 antibody/LRP1 complex is internalized within 15–30 min and observed in endosomes (Fig. S3, A and B, quantified in G). Moreover, we demonstrated that the receptor–antibody complex recycles and reappears on the plasma membrane within an hour of internalization and peaks on the cell surface at 2 h before undergoing additional uptake and removal from the plasma membrane (Fig. S3, C–F, and quantified in G). Using 3D surface rendering of z-stacks with IMARIS software, we quantified the total surface volume of EHD1-containing endosomes in cells that internalized LRP1 compared with untreated cells, and we observed a mean ∼300% increase per cell (Fig. 3E). However, SNX17, which interacts with phosphoinositides directly through its PX domain and is mostly found on endosomal membranes (41), showed only a modest mean ∼20% recruitment upon induction of LRP1 internalization (Fig. 3, C and D, and quantified in F). Although additional experimentation will be needed to test a wide range of receptors and determine whether their internalization induces EHD1 recruitment to endosomes, addition of a control antibody to the V5 epitope had no significant impact on EHD1 recruitment compared with LRP1 uptake (Fig. S4, A–C, and quantified in D). These data suggest that internalization of an SNX17-sorted receptor can lead to EHD1 recruitment to SE, potentially to facilitate fission events and recycling.
Figure 3.
EHD1 is recruited to endosomes upon LRP1 uptake. A–D, CRISPR/Cas9 gene-edited NIH3T3 cells expressing EHD1-GFP were either mock-treated (A and C, no uptake) or incubated with anti-LRP1 antibody (B and D, 30 min on ice and 30 min at 37 °C) prior to fixation and immunostaining with anti-SNX17 antibody and imaging by confocal microscopy. Representative images consisting of a field of cells are displayed. Regions of interest are shown in the insets, and dashed ovals outline the nuclei of the cells. E, 3D surface rendering was carried out from z-sections to capture and quantify the total surface volume of EHD1 (E) or SNX17 (F) (see “Experimental procedures” for details). Error bars denote standard deviation. Two-tailed t tests were performed to derive p values. Data shown are representative of three independent experiments, each using 10 images with seven z-sections each.
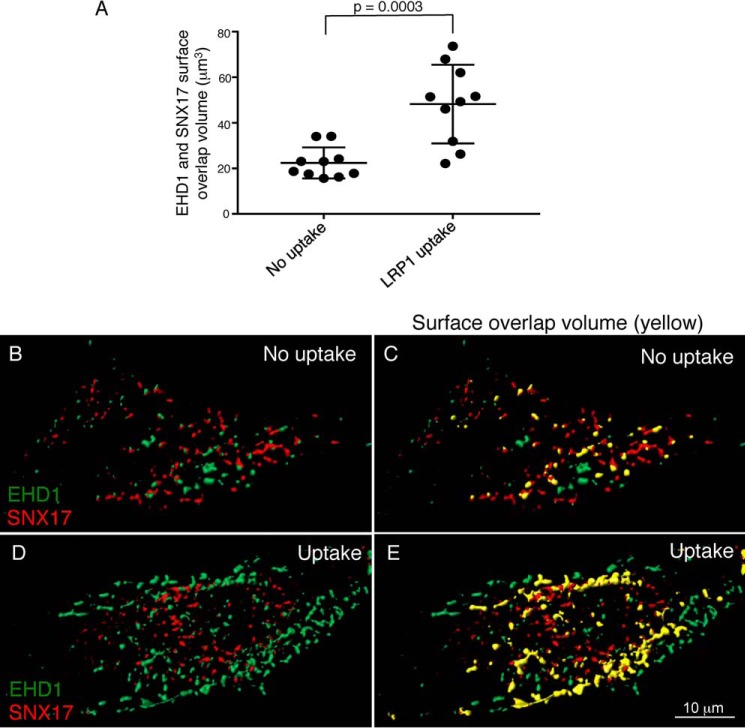
Given that EHD1 and SNX17 directly interact and coimmunoprecipitate, we next determined whether the two proteins displayed a degree of colocalization in cells. Accordingly, we used untreated cells (no uptake) and cells that were incubated with antibodies against LRP1 to induce receptor uptake and EHD1 recruitment to endosomes (LRP1 uptake). As demonstrated in Fig. S5, A–F, insets), induced recruitment of EHD1 to SNX17-containing endosomes led to increased colocalization. To quantify the overlap between SNX17 and EHD1, we used IMARIS software to measure 3D surface overlap volume of SNX17 with EHD1 (Fig. 4A). Following extensive calibrations and comparisons with both 2D and 3D measurements of Pearson's correlation and Manders overlap coefficient, we found that use of multiple z-sections and measurement of voxel overlap in this system provides the most accurate assessment of colocalization within the cell. Essentially, automated analysis renders 3D surfaces from both channels, removing “haze” from the cytoplasm, and measures the degree of overlap between the two sets of 3D surfaces. Representative micrographs display surface-rendered EHD1 (green) and SNX17 (red) in the absence of uptake (Fig. 4, B and C, and Video S1, with surface overlap volume shown in yellow) and following LRP1 uptake (Fig. 4, D and E, and Video S2, with surface overlap volume shown in yellow). As indicated, the percentage of SNX17 surface volume overlap with EHD1 increases significantly by 2- to 3-fold upon LRP1 uptake. Overall, these data support the notion that ∼20% of SNX17 overlaps with EHD1 at steady state and that inducing LRP1 internalization and EHD1 recruitment leads to significantly more SNX17 overlap with EHD1.
Figure 4.
EHD1 and SNX17 surface overlap volume increases upon LRP1 uptake. CRISPR/Cas9 gene-edited NIH3T3 cells expressing EHD1-GFP were mock-treated (no uptake) or incubated with anti-LRP1 antibody as described in Fig. 3. The cells were fixed and stained with anti-SNX17 antibody and imaged by confocal microscopy. Z-stacks were acquired and processed by IMARIS. 3D surface reconstruction was performed simultaneously for both channels to capture EHD1 and SNX17 voxels. Surface–surface overlap volume was assessed using the IMARIS XT bundle Kiss and Run by integrating it with MATLAB Compiler Runtime and launching on IMARIS. A, EHD1 surfaces were selected as target surfaces, and SNX17 surfaces were tracked for any overlapping voxels with those of EHD1. The total surface overlap volume between EHD1 and SNX17 surfaces was quantified and plotted for the no uptake and LRP1 uptake conditions. Two-tailed t tests were performed. B–E, representative images showing 3D surface reconstruction for EHD1 and SNX17 surfaces without (B and C) and with LRP1 uptake (D and E). The overlap is indicated in yellow (C and E). Data shown are representative of three independent experiments.
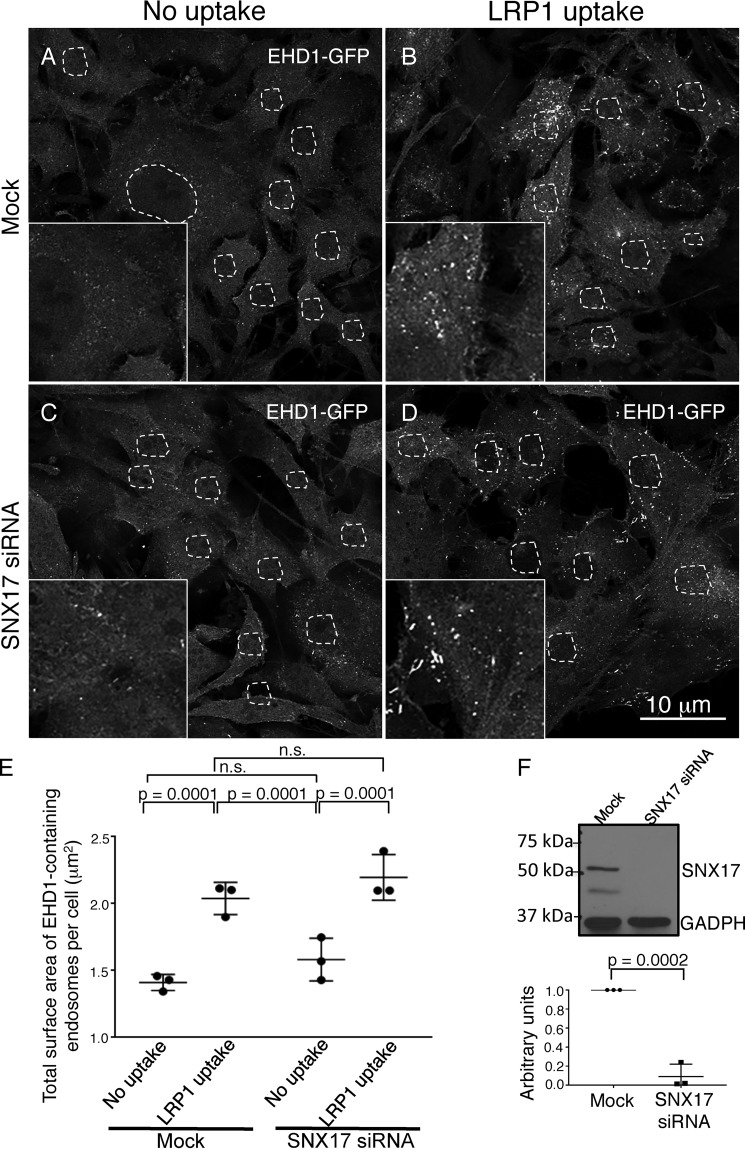
Although we rationalize that coupling between SNX17 and EHD1 is required to connect the sorting and fission machineries at SE, EHD1 could initially be recruited to SE through its interactions with SNX17 or independent of SNX17. To test whether SNX17 is required for EHD1 recruitment to SE, we used mock (untreated) NIH3T3 CRISPR/Cas9 gene-edited EHD1-GFP cells and the same cells after SNX17 siRNA knockdown (decreased SNX17 expression validated and quantified in Fig. 5F). As we saw previously, mock-treated cells contained cytoplasmic EHD1 as well as some puncta and small tubules representing endosomes (Fig. 5A, total surface area of EHD1-containing endosomes quantified in E) and internalization of LRP1 significantly increased recruitment of EHD1 to endosomal membranes (Fig. 5B and quantified in E). For SNX17 siRNA-treated cells without internalization of LRP1, the distribution of EHD1 remained similar to that observed for mock-treated cells, with a mostly cytoplasmic distribution pattern and a smattering of vesicles and tubules (Fig. 5C and quantified in E). However, when SNX17 knockout cells were subjected to LRP1 internalization, EHD1 was again recruited to endosomes in a manner similar to that seen in mock-treated cells (Fig. 5D and quantified in E). Overall, these data suggest that EHD1 recruitment to SE is independent of SNX17 and may rely on other interaction partners that are localized to SE.
Figure 5.
EHD1 is recruited to endosomes in the absence of SNX17. A–D, CRISPR/Cas9 gene-edited NIH3T3 cells expressing EHD1-GFP were mock-treated (A and B) or subjected to SNX17 siRNA (C and D) and incubated with LRP1 antibodies (B and D) or left untreated as a control (A and C). E, z-sections obtained from confocal microscopy were processed with IMARIS software to construct 3D surfaces for EHD1, as discussed under “Experimental procedures,” and the total EHD1 surface area per cell was calculated. The graph depicts the total surface area of EHD1-containing endosomes per cell in mock and SNX17 knockdown cells with or without LRP1 uptake. Two-tailed t tests were performed to derive p values. n.s., not significant. F, immunoblot showing reduced SNX17 expression in CRISPR/Cas9 gene-edited NIH3T3 EHD1-GFP cells and densitometric quantification of SNX17 protein levels in cells subjected to SNX17 siRNA treatment compared with untreated cells (mock) plotted. Error bars denote standard deviation, and p values were determined by Student's two-tailed t test. Data shown are representative of three independent experiments.
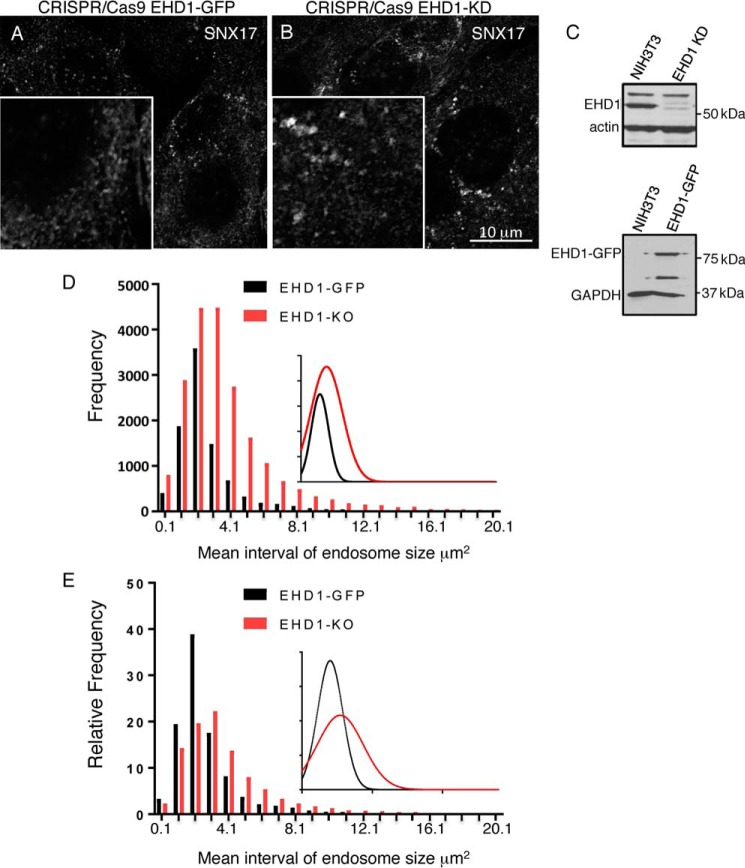
We postulate that SNX17 selects and sorts cargo into budding structures on SE and that EHD1 is recruited to the SE membrane to catalyze fission and release transport vesicles to be targeted for recycling. Accordingly, we rationalized that, if EHD1 is depleted from cells, then SNX17-containing SE would undergo a decreasing amount of fission and display an enlarged endosomal size distribution. To test this idea, we again used CRISPR/Cas9 gene-edited NIH3T3 cells and initially compared the size of SNX17-containing endosomes in EHD1-GFP and EHD1 knockout cells (Fig. 6, A–C). As shown in the representative micrographs (Fig. 6, A and B), EHD1 knockout cells indeed appeared to have larger SNX17-containing endosomes. To measure the precise distribution of SNX17-containing SE in the presence and absence of EHD1, we applied IMARIS software to obtain reconstructed 3D surfaces and calculate the frequency and size of thousands of SE from tens of micrographs and plotted the frequency over the mean interval of SNX17-containing SE endosome size (Fig. 6D, inset). The distribution plot is shifted to the right upon EHD1 knockout and shows that, as the endosomes increase in size (particularly above a threshold of 3 μm2), the relative distribution of endosomes is increased in EHD1 knockout cells. Calculation of the relative frequency of endosome size (where the size distribution of endosomes for each cell type is plotted from a total of 100%) further highlights the notion that, in the absence of EHD1, the ratio of larger to smaller endosomes is higher compared with EHD1-GFP–containing cells (Fig. 6E, see shift to the right for the knockout in the inset). Overall, these data support the idea that EHD1 is recruited to SE, where it interacts with SNX17 and facilitates fission of transport vesicles.
Figure 6.
SNX17 endosome size increases in the absence of EHD1. A, CRISPR/Cas9 gene-edited NIH3T3 cells expressing EHD1-GFP were immunostained with anti-SNX17 antibody and imaged. B, CRISPR/Cas9 gene-edited NIH3T3 cells lacking EHD1 (EHD1-KO) were fixed, immunostained with anti-SNX17 antibody, and imaged. Insets are included to highlight the endosomal size difference. C, immunoblot demonstrating loss of EHD1 in NIH3T3 EHD1 knockout cells (top panel) and expression of EHD1-GFP (molecular weight, ∼87 kDa) in NIH3T3 EHD1-GFP cells (bottom panel). D, 3D surface rendering was carried out to encompass SNX17 voxels, and their total surface area was quantified. A grouped frequency distribution bar graph and curve (inset) for SNX17 endosome size (square micrometers) are plotted to compare endosome size distribution in EHD1-GFP (black) and EHD1-KO (red) cells. E, the relative frequency of SNX17 endosome size (of 100%) was quantified and plotted to compare the relative frequency of endosome size distribution in EHD1-GFP (black) and EHD1-KO (red) cells. Data shown are representative of six independent experiments.
Discussion
Significant advances in our understanding of receptor sorting at the endosome have been made in recent years (reviewed in Refs. 1, 2, 42–44). Not only have studies demonstrated that receptor recycling from endosomes is an active process (45), but key sorting proteins of the sorting nexin family, including SNX17, SNX27, and others, have been identified as hubs that can bind to multiple receptors and channel them to select pathways (5–7, 9–12, 46). However, a central remaining question is the mode by which sorted receptors are coupled with the endosomal fission machinery leading to transport vesicle formation and recycling.
Here we demonstrated for the first time that a key sorting nexin, SNX17, can couple internalized receptors with EHD1, a protein implicated in membrane fission. Binding of SNX17 to EHD1 occurs through the SNX17 atypical FERM domain, which can bind an array of receptor cargos and other molecules (28). Although it remains unclear whether SNX17 can simultaneously bind both receptors and EHD1, these proteins likely localize to common endosomal membrane complexes slated for budding and fission.
EHD1 binding to SNX17 is atypical because most of the EHD1 interaction partners identified to date interact via NPF motifs (22, 23, 47). However, proteins that interact with EHD1 without an NPF motif have been identified (48).
To better address the mode by which SNX17 and EHD1 coordinately function in the cell, we sought to better visualize their localization and potential overlap at endosomal membranes. Initial attempts to quantify “colocalization” by imaging in two or three dimensions followed by application of Pearson's or Manders' coefficients (49) did not accurately reflect the degree of overlap observed, likely because of the significant degree of cytoplasmic EHD1 haze that interferes with the analysis, especially in cells imaged without LRP1 internalization. Accordingly, we developed a method using IMARIS surface rendering and Kiss and Run software in which 3D images of cells with EHD1-GFP and immunostained endogenous SNX17 were converted to “3D surfaces,” allowing us to effectively filter out most of the cytoplasmic protein haze. We were then able to selectively measure EHD1 and SNX17 surface overlap volume in a reliable and automated manner. Our data clearly show that ∼20% of SNX17 and EHD1 display overlap in 3D at steady state. Remarkably, however, we observed a 2- to 3-fold increase in the surface overlap volume following 30-min internalization of LRP1. Significantly, because total SNX17 surface volume is only modestly increased upon LRP1 uptake, the increase in overlap with EHD1 comes primarily from a massive 3-fold recruitment of EHD1 to membrane surfaces upon LRP1 internalization. Indeed, to our knowledge, this is the first evidence that EHD1 and endosomal fission machinery can be recruited to endosomal membranes by a specific physiologic signal.
EHD1 is a highly dynamic protein that cycles on and off of membranes, similar to many ATPases and GTPases (16). Although the mechanism of recruitment of EHD1 from the cytoplasm to membranes is not understood, it has been posited that posttranslational events such as ATP binding, dimerization, and potentially phosphorylation may be required (23). Given the direct interaction of EHD1 with SNX17, one possibility was that SNX17 is needed for recruitment/anchoring of EHD1 to endosomes. However, because SNX17 knockdown had little impact on the recruitment of EHD1 to endosomes, this suggests the following scenario (Fig. 7). Upon LRP1 internalization, EHD1 is “activated” in the cytoplasm in a manner that might include ATP binding, dimerization, and/or phosphorylation and potentially other posttranslational modifications. The EHD1 dimers might then interact with an endosomal EHD1 interaction partner (EEIP) at the endosomal membrane. Examples of EEIPs include Rabenosyn-5 (50), Rabankyrin-5 (51), MICAL-L1 (20, 52–55), Rab11-FIP2 (34), SNAP29 (56, 57), and Syndapin2 (53, 58). Recruitment of EHD1 to endosomes by one or more of these EEIPs would explain why SNX17 knockdown did not affect recruitment. After recruitment, EHD1 might be able to diffuse along the endosomal membrane, still bound to the EEIP via EH domain NPF motifs, until it comes into contact with and binds to SNX17 (in an EH domain–independent mechanism), which is itself bound to a receptor cargo. Ultimately, EHD1 might play a significant role in fission of vesicles slated for recycling.
Figure 7.
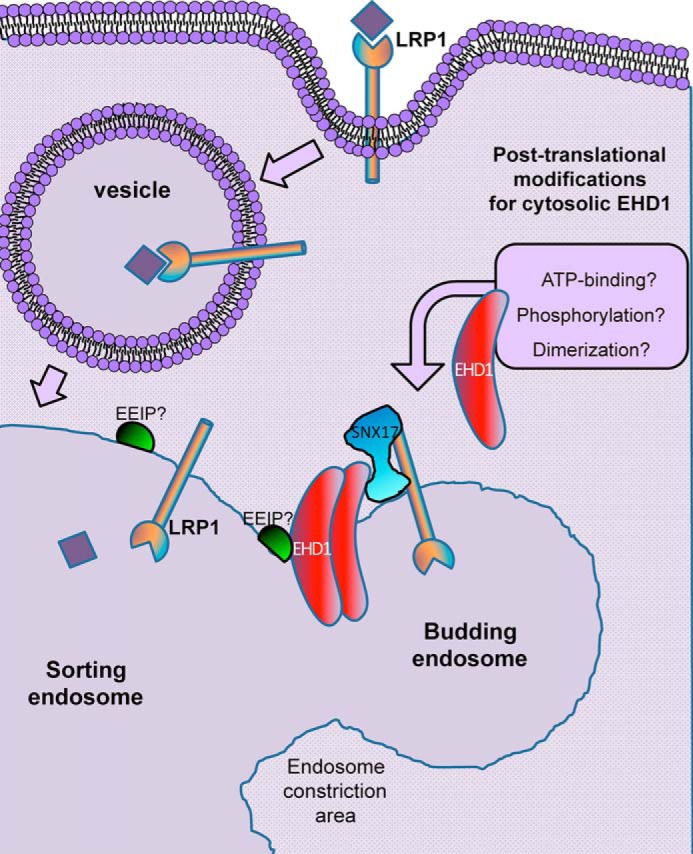
Model depicting potential mechanisms for EHD1 endosomal recruitment and coordination of membrane fission with SNX17.
The process of endosomal fission mediated by EHD1 is highly complex (17, 18) and may include a variety of additional proteins involved in membrane curvature, such as cPLA2α (14) and the BAR (Bin, Amphiphysin, Rvs) domain–containing protein GRAF1 (15). However, EHD1 is only one of several key mechanisms that regulate endosomal fission; in particular, the Wiskott-Aldrich syndrome protein and SCAR homolog (WASH) complex has been implicated in activation of the Arp2/3 actin polymerization nucleator at endosomes, a process thought to precede dynamin-controlled endosomal fission (59). How EHD1-mediated endosomal fission can be integrated into the latter processes remains to be determined.
Overall, we identified a novel direct interaction between the sorting nexin SNX17 and the endosomal fission protein EHD1. Our study demonstrates that receptor-mediated endocytosis of the LRP1 receptor induces massive recruitment of EHD1 to endosomal membranes. Although recruitment occurs in a SNX17-independent manner, our data demonstrate that, in the absence of EHD1, SNX17-containing endosomes increase in size, likely as a result of impaired fission. This study provides new insight into coupling of the endosomal sorting and fission machineries, highlighting a significant role of EHD1 in the endosomal fission process.
Experimental procedures
Cell lines
The HeLa cervical cancer cell line was obtained from the ATCC and grown in DMEM (high-glucose) containing 10% FBS, 1× penicillin–streptomycin (Invitrogen), and 2 mm glutamine. CRISPR/Cas9 was applied to generate the NIH3T3 cell line expressing endogenous levels of EHD1 with GFP attached to its C terminus as well as EHD1 knockout cells, as described previously (37). WT and gene-edited NIH3T3 cells were cultured at 37 °C in 5% CO2 in DMEM containing 10% FBS with 2 mm l-glutamine and 100 units/ml penicillin/streptomycin. For treatments, cells were plated on fibronectin-coated coverslips. All cell lines were routinely tested for Mycoplasma infection.
Antibodies
The following antibodies were used: anti-EHD1 (109311, Abcam), anti-SNX17 (NBP1-92417, Novus, for immunoblotting; HPA043867, Atlas, for immunofluorescence), anti-EB3 (126953, Abcam), anti-V5 (R960-25, Invitrogen), anti-SNX27 (77799, Abcam), anti-HA (600-401384S, Rockland), anti-GST-HRP (A01380, GenScript), anti-His-HRP (66005, Proteintech), anti-GAPDH-HRP (HRP-60004, Proteintech), anti-LRP1 (NB100-64808, Novus), anti-GFP (11814460001, Roche), anti-caveolin (3238, Cell Signaling), donkey anti-mouse-HRP (715-035-151, Jackson ImmunoResearch Laboratories), mouse anti-rabbit IgG light chain–HRP (211-032-171, Jackson), Alexa Fluor 568–conjugated goat anti-rabbit (A11036, Molecular Probes), and Alexa Fluor 568–conjugated goat anti-mouse (A11031, Molecular Probes).
DNA constructs, cloning, and site-directed mutagenesis
pGEX-4T-1-SNX17 (bp 1–1413) was obtained from GenScript (clone S80141), and pET28a-EHD1 was generated from the original EHD1 constructs designed (16). Primers were designed using the New England Biolabs primer design tool for PCR amplification of the ORF encoding human SNX17 FERM B (bp 328–780), FERM C (bp 781–1200), EHD1ΔEH (bp 1–1329), and the EH domain of EHD1 (bp 1330–1603). The amplified products, SNX17 FERM B and FERM C, were cloned in pGEX-4T-1, whereas EHD1ΔEH and the EH domain of EHD1 were cloned in pET-28a. pGEX-4T-1-SNX17 was mutated from glutamine to a STOP at amino acid position 110 (Q110STOP), which eventually coded for the GST-PX domain of SNX17, and pGEX-4T-1-FERM C was mutated from tryptophan to alanine at amino acid 321 (W321A) using the QuikChange site-directed mutagenesis kit (Stratagene, catalog no. 200518) following the manufacturer's protocol.
Coimmunoprecipitation
HeLa cells were grown in 100-mm dishes until confluent. Cells were lysed with lysis buffer containing 50 mm Tris (pH 7.4), 100 mm NaCl, 0.5% Triton X-100, and 1× protease inhibitor mixture (Millipore) on ice for 30 min. Lysates were incubated with a specific antibody at 4 °C overnight. Protein G beads for antibodies raised against rabbit antigens (GE Healthcare) or protein L beads for antibodies raised against mouse antigens (I91R-844, ABT) were added to the lysate–antibody mixture at 4 °C for 4 h. Samples were then washed three times with the same lysis buffer. Proteins were eluted from the beads by boiling in the presence of 4× loading buffer (250 mm Tris (pH 6.8), 8% SDS, 40% glycerol, 5% β-mercaptoethanol, and 0.2% bromphenol blue) for 10 min. Eluted proteins were then detected by immunoblotting.
Recombinant gene expression and protein purification
The recombinant DNA constructs were expressed in the Escherichia coli Rosetta (R2) strain and purified by affinity chromatography in separate experiments. Briefly, a freshly transformed colony of E. coli was inoculated in 50 ml of Luria-Bertani broth (with 50 μg/ml kanamycin for recombinant pET-28a expression plasmids and 100 μg/ml ampicillin for recombinant pGEX-4T-1 expression plasmids) and cultured overnight at 37 °C with continuous shaking (primary culture). Next, the primary culture was inoculated in 1000 ml of fresh Luria-Bertani broth at 1:100 dilution and incubated at 37 °C with continuous shaking until readings of 0.4–0.6 at A600 nm. The culture was then induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside overnight either at 18 °C for recombinant pGEX-4T-1 expression plasmids or 25 °C for recombinant pET-28a expression plasmids. The cells were then centrifuged at 2100 × g for 15 min at 4 °C. The bacterial pellet obtained was resuspended in ice-cold lysis buffer containing 1 tablet/10 ml of protease inhibitor mixture (Roche). The composition of lysis buffer for recombinant pET-28a expression plasmids was 50 mm Tris, 200 mm NaCl, and 50 mm imidazole (pH 8.0), whereas for recombinant pGEX-4T-1 expression plasmids, the composition of lysis buffer was 1× PBS (pH 7.4). Sample lysis was performed by six cycles of sonication on ice (2-min bursts/2 min cooling/200–200 watts in a Branson sonicator). The lysate was centrifuged at 18,000 × g for 30 min at 4 °C, which allowed separation of clear supernatant and cellular debris (inclusion bodies were pelleted by centrifugation). The supernatant was then mixed and allowed to bind with either Ni2+-NTA (pET-28a plasmids) or GSH-Sepharose resin (pGEX-4T-1 plasmids) for 4 h at 4 °C. To ensure the removal of any nonspecifically bound proteins, the beads were then washed extensively with 10 bed volumes of wash buffer (for pGEX-4T-1 plasmids, three times with 2× PBS followed by a final wash with 1× PBS; for pET-28a plasmids, three times with buffer containing 100 mm imidazole, 50 mm Tris, and 200 mm NaCl (pH 8.0)) by centrifuging at 3000 × g for 3 min at 4 °C. Finally, the bound histidine-tagged and GST-tagged proteins were subjected to elution for 4 h at 4 °C in elution buffer containing 300 mm imidazole, 50 mm Tris, and 200 mm NaCl (pH 8.0) and 30 mm GSH (reduced) in 50 mm Tris-HCl (pH 8.0), respectively, followed by centrifugation at 2100 × g for 5 min at 4 °C. The purified proteins were then dialyzed against dialysis buffer (50 mm Tris (pH 8.0), 200 mm NaCl, and 0.1 mm PMSF) overnight at 4 °C.
Direct interaction assay
For the protein–protein interaction assays, 20 μl of a slurry of Ni2+-NTA beads was washed four times with 10 bed volumes of beads with TGEM buffer (20 mm Tris-HCl (pH 7.9), 20% glycerol, 1 mm EDTA, 5 mm MgCl2, 0.1% NP-40, 1 mm DTT, 0.2 mm PMSF, and 0.1 m NaCl) by centrifugation at 13,000 rpm for 30 s, followed by addition of three bed volumes of TGEM to packed beads to which 0.5 μg of the purified histidine-tagged proteins was incubated for 2 h at 4 °C in a tube rotator. The immobilized His-tagged bait slurry was then centrifuged at 13,000 rpm for 30 s, followed by two washes with 10 bed volumes of beads with TGEM buffer (20 mm Tris-HCl (pH 7.9), 20% glycerol, 1 mm EDTA, 5 mm MgCl2, 0.1% NP-40, 1 mm DTT, 0.2 mm PMSF, and 1 m NaCl) and then twice with TGMC buffer (20 mm Tris-HCl (pH 7.9), 20% glycerol, 5 mm CaCl2, 0.1% NP-40, 1 mm DTT, 0.2 mm PMSF, and 0.1 m NaCl). After the last wash, two bed volumes of TGMC were added to the bait along with 1 unit of micrococcal nuclease and incubated at 30 °C for 10 min, as described by Nguyen and Goodrich (60). 0.5 μg of target GST fusion proteins was diluted in TGMC to obtain a 40-μl sample (target) volume per reaction and treated with 1 unit of micrococcal nuclease at 30 °C for 10 min. The nuclease-treated GST-target proteins were then incubated with bait for 2 h at 4 °C in a tube rotator. The samples were then washed four times with 10 bed volumes of TGEM and subjected to SDS-PAGE.
siRNA treatment
CRISPR/Cas9 gene-edited NIH3T3 cells expressing EHD1 were plated on fibronectin-coated coverslips and grown for 24 h in DMEM containing 10% FBS with 2 mm l-glutamine and 100 units/ml penicillin/streptomycin. The cells were then treated with mouse SNX17 siRNA oligonucleotides (Dharmacon, On-TARGETplus SMARTpool, catalog no. L-054627-02-0005) for 48 h at 37 °C using Lipofectamine RNAiMax transfection reagent (Thermo Fisher Scientific), following the manufacturer's protocol.
Immunofluorescence and LRP1 uptake
CRISPR/Cas9 gene-edited NIH3T3 cells expressing EHD1 or lacking EHD1 were treated as indicated. Briefly, LRP1 uptake was performed in NIH3T3 cells expressing EHD1-GFP by diluting LRP1 antibody (1:70) in DMEM containing 10% FBS with 2 mm l-glutamine and 100 units/ml penicillin–streptomycin on ice for 30 min, followed by three washes with 1× PBS. Coverslips were switched to 37 °C for another 30 min and then washed again three times with PBS. Following treatment, cells were fixed in 4% paraformaldehyde in PBS for 10 min at room temperature. After fixation, cells were rinsed three times in PBS then incubated with primary antibody in staining buffer (PBS containing 0.5% BSA and 0.2% saponin) for 1 h at room temperature. Cells were then washed three times in PBS, followed by incubation with the appropriate fluorochrome-conjugated secondary antibodies diluted in staining buffer for 30 min. Cells were washed three times in PBS and mounted in Fluoromount. Z-stack confocal imaging was performed using a Zeiss LSM 800 confocal microscope with a ×63/1.4 numerical aperture oil objective, and more than 30 cells from three independent experiments were assessed using IMARIS.
Image processing, 3D reconstruction, and Kiss and Run analysis
Z-sections of images (six slices) acquired from the confocal microscope were imported into IMARIS x64 9.1.2 software (Bitplane AG, Zurich, Switzerland) coupled with custom MATLAB (2009 and 2014) programming for 3D surface rendering and quantitative analysis, as indicated. Briefly, the image display was adjusted for both of the channels (EHD1, green; SNX17, red), and the rendering quality was set to 100%. Surfaces were created by selecting source channel and smooth surface detail set at 0.198 μm. Background subtraction was set to 0.743 μm, and the threshold was reduced for surfaces to fully cover all voxels. The surface area and volume of the surfaces generated were quantified by IMARIS for both of the channels, and the values were exported into Excel for graphical and statistical analysis using GraphPad Prism. To quantify surface overlap volume between two surfaces (EHD1 and SNX17), the IMARIS XT bundle Kiss and Run was first integrated with MATLAB and launched in IMARIS. 3D surface-reconstructed images were then processed for Kiss and Run analysis using the surface–surface overlap module, which uses a surface mask for the target and tracks the surface and determines overlap for each surface independently. This particular Xtension program analyzes contact events between surfaces that are defined by having at least one overlapping voxel. Volume of overlap for each surface was then quantified and exported to Excel for further analysis.
Graphical and statistical analysis
Total surface overlap volume was calculated using the IMARIS XT bundle Kiss and Run surface–surface colocalization module with EHD1 set as target surfaces and SNX17 as tracked surfaces. A bar graph was plotted, and a two-tailed t test was performed with significance, as indicated in the figures. A frequency distribution (interleaved) graph was plotted for SNX17 endosome size in cells expressing EHD1-GFP or lacking EHD1, with the first bin starting at 0.1 μm2 up to 20.1 μm2 and bin width set at 0.00001 μm2. Gaussian curves were also plotted. National Institutes of Health ImageJ was used to quantify signal integrated density. All graphical and statistical tests were done using GraphPad Prism 7.
Author contributions
K. D. data curation; K. D., N. N., and S. C. formal analysis; K. D. investigation; K. D. and S. C. visualization; K. D. methodology; K. D. and S. C. writing-original draft; K. D., N. N., and S. C. writing-review and editing; N. N. and S. C. conceptualization; N. N. and S. C. supervision; N. N. and S. C. funding acquisition; S. C. validation; S. C. project administration.
Supplementary Material
Acknowledgments
We thank the University of Nebraska Medical Center Advanced Microscopy Core Facility, which receives partial support from NIGMS, National Institutes of Health INBRE P20 GM103427 and COBRE P30 GM106397 grants as well as NCI, National Institutes of Health for The Fred and Pamela Buffett Cancer Center Support Grant P30 CA036727 and the Nebraska Research Initiative. We also thank Jason Sughroue for technical assistance.
This work was supported by National Institutes of Health Grants 1R01GM123557 and P20 GM103427 (to S. C.) and Nebraska Department of Health and Human Services Grant 2019-41 (to N. N.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S5 and Videos S1 and S2.
- SE
- sorting endosome(s)
- PX
- Phox homology
- IP
- immunoprecipitation
- EEIP
- endosomal EHD1 interaction partner
- WASH
- Wiskott-Aldrich syndrome protein and SCAR homolog
- Ni2+-NTA
- nickel-nitrilotriacetic acid
- EH
- Eps15 homology
- FERM
- 4.1 protein, Ezrin, Radixin, Moesin.
References
- 1. Naslavsky N., and Caplan S. (2018) The enigmatic endosome: sorting the ins and outs of endocytic trafficking. J. Cell Sci. 10.1242/jcs.216499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang J., Fedoseienko A., Chen B., Burstein E., Jia D., and Billadeau D. D. (2018) Endosomal receptor trafficking: retromer and beyond. Traffic 19, 578–590 10.1111/tra.12574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Seaman M. N., McCaffery J. M., and Emr S. D. (1998) A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 142, 665–681 10.1083/jcb.142.3.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arighi C. N., Hartnell L. M., Aguilar R. C., Haft C. R., and Bonifacino J. S. (2004) Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J. Cell Biol. 165, 123–133 10.1083/jcb.200312055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clairfeuille T., Mas C., Chan A. S., Yang Z., Tello-Lafoz M., Chandra M., Widagdo J., Kerr M. C., Paul B., Mérida I., Teasdale R. D., Pavlos N. J., Anggono V., and Collins B. M. (2016) A molecular code for endosomal recycling of phosphorylated cargos by the SNX27-retromer complex. Nat. Struct. Mol. Biol. 23, 921–932 10.1038/nsmb.3290 [DOI] [PubMed] [Google Scholar]
- 6. Gallon M., Clairfeuille T., Steinberg F., Mas C., Ghai R., Sessions R. B., Teasdale R. D., Collins B. M., and Cullen P. J. (2014) A unique PDZ domain and arrestin-like fold interaction reveals mechanistic details of endocytic recycling by SNX27-retromer. Proc. Natl. Acad. Sci. U.S.A. 111, E3604–E3613 10.1073/pnas.1410552111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lauffer B. E., Melero C., Temkin P., Lei C., Hong W., Kortemme T., and von Zastrow M. (2010) SNX27 mediates PDZ-directed sorting from endosomes to the plasma membrane. J. Cell Biol. 190, 565–574 10.1083/jcb.201004060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Temkin P., Lauffer B., Jäger S., Cimermancic P., Krogan N. J., and von Zastrow M. (2011) SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat. Cell Biol. 13, 715–721 10.1038/ncb2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Donoso M., Cancino J., Lee J., van Kerkhof P., Retamal C., Bu G., Gonzalez A., Cáceres A., and Marzolo M. P. (2009) Polarized traffic of LRP1 involves AP1B and SNX17 operating on Y-dependent sorting motifs in different pathways. Mol. Biol. Cell 20, 481–497 10.1091/mbc.e08-08-0805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Osborne D. G., Piotrowski J. T., Dick C. J., Zhang J. S., and Billadeau D. D. (2015) SNX17 affects T cell activation by regulating TCR and integrin recycling. J. Immunol. 194, 4555–4566 10.4049/jimmunol.1402734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Steinberg F., Heesom K. J., Bass M. D., and Cullen P. J. (2012) SNX17 protects integrins from degradation by sorting between lysosomal and recycling pathways. J. Cell Biol. 197, 219–230 10.1083/jcb.201111121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stockinger W., Sailler B., Strasser V., Recheis B., Fasching D., Kahr L., Schneider W. J., and Nimpf J. (2002) The PX-domain protein SNX17 interacts with members of the LDL receptor family and modulates endocytosis of the LDL receptor. EMBO J. 21, 4259–4267 10.1093/emboj/cdf435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McNally K. E., Faulkner R., Steinberg F., Gallon M., Ghai R., Pim D., Langton P., Pearson N., Danson C. M., Nägele H., Morris L. L., Singla A., Overlee B. L., Heesom K. J., Sessions R., et al. (2017) Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling. Nat. Cell Biol. 19, 1214–1225 10.1038/ncb3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cai B., Caplan S., and Naslavsky N. (2012) cPLA2α and EHD1 interact and regulate the vesiculation of cholesterol-rich, GPI-anchored, protein-containing endosomes. Mol. Biol. Cell 23, 1874–1888 10.1091/mbc.e11-10-0881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cai B., Xie S., Caplan S., and Naslavsky N. (2014) GRAF1 forms a complex with MICAL-L1 and EHD1 to cooperate in tubular recycling endosome vesiculation. Front. Cell Dev. Biol. 2, 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Caplan S., Naslavsky N., Hartnell L. M., Lodge R., Polishchuk R. S., Donaldson J. G., and Bonifacino J. S. (2002) A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 21, 2557–2567 10.1093/emboj/21.11.2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deo R., Kushwah M. S., Kamerkar S. C., Kadam N. Y., Dar S., Babu K., Srivastava A., and Pucadyil T. J. (2018) ATP-dependent membrane remodeling links EHD1 functions to endocytic recycling. Nat. Commun. 9, 5187 10.1038/s41467-018-07586-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kamerkar S. C., Roy K., Bhattacharyya S., and Pucadyil T. J. (2019) A screen for membrane fission catalysts identifies the ATPase EHD1. Biochemistry 58, 65–71 10.1021/acs.biochem.8b00925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rapaport D., Auerbach W., Naslavsky N., Pasmanik-Chor M., Galperin E., Fein A., Caplan S., Joyner A. L., and Horowitz M. (2006) Recycling to the plasma membrane is delayed in EHD1 knockout mice. Traffic 7, 52–60 10.1111/j.1600-0854.2005.00359.x [DOI] [PubMed] [Google Scholar]
- 20. Sharma M., Giridharan S. S., Rahajeng J., Naslavsky N., and Caplan S. (2009) MICAL-L1 links EHD1 to tubular recycling endosomes and regulates receptor recycling. Mol. Biol. Cell 20, 5181–5194 10.1091/mbc.e09-06-0535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jakobsson J., Ackermann F., Andersson F., Larhammar D., Löw P., and Brodin L. (2011) Regulation of synaptic vesicle budding and dynamin function by an EHD ATPase. J. Neurosci. 31, 13972–13980 10.1523/JNEUROSCI.1289-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Naslavsky N., and Caplan S. (2005) C-terminal EH-domain-containing proteins: consensus for a role in endocytic trafficking, EH? J. Cell Sci. 118, 4093–4101 10.1242/jcs.02595 [DOI] [PubMed] [Google Scholar]
- 23. Naslavsky N., and Caplan S. (2011) EHD proteins: key conductors of endocytic transport. Trends Cell Biol. 21, 122–131 10.1016/j.tcb.2010.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grant B., Zhang Y., Paupard M. C., Lin S. X., Hall D. H., and Hirsh D. (2001) Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat. Cell Biol. 3, 573–579 10.1038/35078549 [DOI] [PubMed] [Google Scholar]
- 25. Grant B. D., and Caplan S. (2008) Mechanisms of EHD/RME-1 protein function in endocytic transport. Traffic 9, 2043–2052 10.1111/j.1600-0854.2008.00834.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin S. X., Grant B., Hirsh D., and Maxfield F. R. (2001) Rme-1 regulates the distribution and function of the endocytic recycling compartment in mammalian cells. Nat. Cell Biol. 3, 567–572 10.1038/35078543 [DOI] [PubMed] [Google Scholar]
- 27. Lee S., Chang J., and Blackstone C. (2016) FAM21 directs SNX27-retromer cargoes to the plasma membrane by preventing transport to the Golgi apparatus. Nat. Commun. 7, 10939 10.1038/ncomms10939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ghai R., Bugarcic A., Liu H., Norwood S. J., Skeldal S., Coulson E. J., Li S. S., Teasdale R. D., and Collins B. M. (2013) Structural basis for endosomal trafficking of diverse transmembrane cargos by PX-FERM proteins. Proc. Natl. Acad. Sci. U.S.A. 110, E643–E652 10.1073/pnas.1216229110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jia Z., Ghai R., Collins B. M., and Mark A. E. (2014) The recognition of membrane-bound PtdIns3P by PX domains. Proteins 82, 2332–2342 10.1002/prot.24593 [DOI] [PubMed] [Google Scholar]
- 30. Ghai R., and Collins B. M. (2011) PX-FERM proteins: a link between endosomal trafficking and signaling? Small GTPases 2, 259–263 10.4161/sgtp.2.5.17276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kieken F., Jović M., Naslavsky N., Caplan S., and Sorgen P. L. (2007) EH domain of EHD1. J. Biomol. NMR 39, 323–329 10.1007/s10858-007-9196-0 [DOI] [PubMed] [Google Scholar]
- 32. Kieken F., Jović M., Tonelli M., Naslavsky N., Caplan S., and Sorgen P. L. (2009) Structural insight into the interaction of proteins containing NPF, DPF, and GPF motifs with the C-terminal EH-domain of EHD1. Protein Sci. 18, 2471–2479 10.1002/pro.258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee D. W., Zhao X., Scarselletta S., Schweinsberg P. J., Eisenberg E., Grant B. D., and Greene L. E. (2005) ATP binding regulates oligomerization and endosome association of RME-1 family proteins. J. Biol. Chem. 280, 17213–17220 10.1074/jbc.M412751200 [DOI] [PubMed] [Google Scholar]
- 34. Naslavsky N., Rahajeng J., Sharma M., Jovic M., and Caplan S. (2006) Interactions between EHD proteins and Rab11-FIP2: a role for EHD3 in early endosomal transport. Mol. Biol. Cell 17, 163–177 10.1091/mbc.e05-05-0466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Daumke O., Lundmark R., Vallis Y., Martens S., Butler P. J., and McMahon H. T. (2007) Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature 449, 923–927 10.1038/nature06173 [DOI] [PubMed] [Google Scholar]
- 36. Melo A. A., Hegde B. G., Shah C., Larsson E., Isas J. M., Kunz S., Lundmark R., Langen R., and Daumke O. (2017) Structural insights into the activation mechanism of dynamin-like EHD ATPases. Proc. Natl. Acad. Sci. U.S.A. 114, 5629–5634 10.1073/pnas.1614075114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yeow I., Howard G., Chadwick J., Mendoza-Topaz C., Hansen C. G., Nichols B. J., and Shvets E. (2017) EHD proteins cooperate to generate caveolar clusters and to maintain caveolae during repeated mechanical stress. Curr. Biol. 27, 2951–2962.e5 10.1016/j.cub.2017.07.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xie S., Reinecke J. B., Farmer T., Bahl K., Yeow I., Nichols B. J., McLamarrah T. A., Naslavsky N., Rogers G. C., and Caplan S. (2018) Vesicular trafficking plays a role in centriole disengagement and duplication. Mol. Biol. Cell 29, 2622–2631 10.1091/mbc.E18-04-0241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van Kerkhof P., Lee J., McCormick L., Tetrault E., Lu W., Schoenfish M., Oorschot V., Strous G. J., Klumperman J., and Bu G. (2005) Sorting nexin 17 facilitates LRP recycling in the early endosome. EMBO J. 24, 2851–2861 10.1038/sj.emboj.7600756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Farfán P., Lee J., Larios J., Sotelo P., Bu G., and Marzolo M. P. (2013) A sorting nexin 17-binding domain within the LRP1 cytoplasmic tail mediates receptor recycling through the basolateral sorting endosome. Traffic 14, 823–838 10.1111/tra.12076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Knauth P., Schlüter T., Czubayko M., Kirsch C., Florian V., Schreckenberger S., Hahn H., and Bohnensack R. (2005) Functions of sorting nexin 17 domains and recognition motif for P-selectin trafficking. J. Mol. Biol. 347, 813–825 10.1016/j.jmb.2005.02.004 [DOI] [PubMed] [Google Scholar]
- 42. Burd C., and Cullen P. J. (2014) Retromer: a master conductor of endosome sorting. Cold Spring Harb. Perspect. Biol. 6, pii: a016774 10.1101/cshperspect.a016774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cullen P. J. (2008) Endosomal sorting and signalling: an emerging role for sorting nexins. Nat. Rev. Mol. Cell Biol. 9, 574–582 10.1038/nrm2427 [DOI] [PubMed] [Google Scholar]
- 44. Chen K. E., Healy M. D., and Collins B. M. (2019) Towards a molecular understanding of endosomal trafficking by Retromer and Retriever. Traffic 20, 465–478 10.1111/tra.12649 [DOI] [PubMed] [Google Scholar]
- 45. Hsu V. W., Bai M., and Li J. (2012) Getting active: protein sorting in endocytic recycling. Nat. Rev. Mol. Cell Biol. 13, 323–328 10.1038/nrm3332 [DOI] [PubMed] [Google Scholar]
- 46. Steinberg F., Gallon M., Winfield M., Thomas E. C., Bell A. J., Heesom K. J., Tavaré J. M., and Cullen P. J. (2013) A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat. Cell Biol. 15, 461–471 10.1038/ncb2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kieken F., Sharma M., Jovic M., Giridharan S. S., Naslavsky N., Caplan S., and Sorgen P. L. (2010) Mechanism for the selective interaction of C-terminal Eps15 homology domain proteins with specific Asn-Pro-Phe-containing partners. J. Biol. Chem. 285, 8687–8694 10.1074/jbc.M109.045666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shi A., Pant S., Balklava Z., Chen C. C., Figueroa V., and Grant B. D. (2007) A novel requirement for C. elegans Alix/ALX-1 in RME-1-mediated membrane transport. Curr. Biol. 17, 1913–1924 10.1016/j.cub.2007.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McDonald J. H., and Dunn K. W. (2013) Statistical tests for measures of colocalization in biological microscopy. J. Microsc. 252, 295–302 10.1111/jmi.12093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Naslavsky N., Boehm M., Backlund P. S. Jr, and Caplan S. (2004) Rabenosyn-5 and EHD1 interact and sequentially regulate protein recycling to the plasma membrane. Mol. Biol. Cell 15, 2410–2422 10.1091/mbc.e03-10-0733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang J., Reiling C., Reinecke J. B., Prislan I., Marky L. A., Sorgen P. L., Naslavsky N., and Caplan S. (2012) Rabankyrin-5 interacts with EHD1 and Vps26 to regulate endocytic trafficking and retromer function. Traffic 13, 745–757 10.1111/j.1600-0854.2012.01334.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Giridharan S. S., Cai B., Naslavsky N., and Caplan S. (2012) Trafficking cascades mediated by Rab35 and its membrane hub effector, MICAL-L1. Commun. Integr. Biol. 5, 384–387 10.4161/cib.20064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Giridharan S. S., Cai B., Vitale N., Naslavsky N., and Caplan S. (2013) Cooperation of MICAL-L1, syndapin2, and phosphatidic acid in tubular recycling endosome biogenesis. Mol. Biol. Cell 24, 1776–1790, S1–S15 10.1091/mbc.e13-01-0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rahajeng J., Giridharan S. S., Cai B., Naslavsky N., and Caplan S. (2012) MICAL-L1 is a tubular endosomal membrane hub that connects Rab35 and Arf6 with Rab8a. Traffic 13, 82–93 10.1111/j.1600-0854.2011.01294.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sharma M., Giridharan S. S., Rahajeng J., Caplan S., and Naslavsky N. (2010) MICAL-L1: An unusual Rab effector that links EHD1 to tubular recycling endosomes. Commun. Integr. Biol. 3, 181–183 10.4161/cib.3.2.10845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rotem-Yehudar R., Galperin E., and Horowitz M. (2001) Association of insulin-like growth factor 1 receptor with EHD1 and SNAP29. J. Biol. Chem. 276, 33054–33060 10.1074/jbc.M009913200 [DOI] [PubMed] [Google Scholar]
- 57. Xu Y., Shi H., Wei S., Wong S. H., and Hong W. (2004) Mutually exclusive interactions of EHD1 with GS32 and syndapin II. Mol. Membr. Biol. 21, 269–277 10.1080/09687680410001716871 [DOI] [PubMed] [Google Scholar]
- 58. Braun A., Pinyol R., Dahlhaus R., Koch D., Fonarev P., Grant B. D., Kessels M. M., and Qualmann B. (2005) EHD proteins associate with syndapin I and II and such interactions play a crucial role in endosomal recycling. Mol. Biol. Cell 16, 3642–3658 10.1091/mbc.e05-01-0076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Derivery E., Sousa C., Gautier J. J., Lombard B., Loew D., and Gautreau A. (2009) The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev. Cell 17, 712–723 10.1016/j.devcel.2009.09.010 [DOI] [PubMed] [Google Scholar]
- 60. Nguyen T. N., and Goodrich J. A. (2006) Protein-protein interaction assays: eliminating false positive interactions. Nat. Methods 3, 135–139 10.1038/nmeth0206-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gomez T. S. and Billadeau D. D. (2009) A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev Cell 17, 699–711 10.1016/j.devcel.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Harbour M. E., Breusegem S. Y., and Seaman M. N. (2012) Recruitment of the endosomal WASH complex is mediated by the extended 'tail' of Fam21 binding to the retromer protein Vps35. Biochem J 442, 209–220 10.1042/BJ20111761 [DOI] [PubMed] [Google Scholar]
- 63. Jia D., Gomez T. S., Metlagel Z., Umetani J., Otwinowski Z., Rosen M. K., and Billadeau D. D. (2010) WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proc Natl Acad Sci U S A 107, 10442–10447 10.1073/pnas.0913293107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jia D., Gomez T. S., Billadeau D. D., and Rosen M. K. (2012) Multiple repeat elements within the FAM21 tail link the WASH actin regulatory complex to the retromer. Mol Biol Cell 23, 2352–2361 10.1091/mbc.E11-12-1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.