Abstract
Human milk oligosaccharides (HMOs) promote the development of the neonatal intestinal, immune, and nervous systems and has recently received considerable attention. Here we investigated how the maternal diet affects HMO biosynthesis and how any diet-induced HMO alterations influence the infant gut microbiome and immunity. Using capillary electrophoresis and MS-based analyses, we extracted and measured HMOs from breast milk samples and then correlated their levels with results from validated 24-h diet recall surveys and breast milk fatty acids. We found that fruit intake and unsaturated fatty acids in breast milk were positively correlated with an increased absolute abundance of numerous HMOs, including 16 sulfonated HMOs we identified here in humans for the first time. The diet-derived monosaccharide 5-N-glycolyl-neuraminic acid (Neu5Gc) was unambiguously detected in all samples. To gain insights into the potential impact of Neu5Gc on the infant microbiome, we used a constrained ordination approach and identified correlations between Neu5Gc levels and Bacteroides spp. in infant stool. However, Neu5Gc was not associated with marked changes in infant immune markers, in contrast with sulfonated HMOs, whose expression correlated with suppression of two major Th2 cytokines, IL-10 and IL-13. The findings of our work highlight the importance of maternal diet for HMO biosynthesis and provide as yet unexplored targets for future studies investigating interactions between HMOs and the intestinal microbiome and immunity in infants.
Keywords: glycobiology, oligosaccharide, microbiome, sialic acid, cytokine, diet, MS, capillary electrophoresis, prebiotic
Introduction
The most abundant solid component of human milk is lactose, followed by lipids, free human milk oligosaccharides (HMOs),6 and glycoproteins. To date, over 200 HMOs have been identified in human milk (1), and a typical infant will consume several grams per day. HMOs, in contrast to lactose, are indigestible by infants (2–4), but they are often readily digested by bacteria present in the large intestine and colon, provided they express the requisite glycolytic enzymes (5, 6). By providing these microbes with a nutrient source, HMOs actively promote colonization of the neonatal gastrointestinal (GI) tract (7–11). Because HMOs are often chemically similar to the protective mucus layer lining the GI tract, they may also serve as decoys for pathogenic bacteria, mimicking the glycoepitopes to which pathogens prefer to bind (12). Additionally, some HMOs directly impact immune signaling events (13, 14), which, in light of the fact that they have been shown to enter the bloodstream (15), is suggestive of systemic immune functions (16). Identifying the principal bioactive HMOs and defining their health-promoting effects is imperative, given the clear links between these compounds and proper establishment of the infant microbiome, alterations of which are associated with numerous immunological disorders and infectious diseases (17, 18).
HMOs vary significantly among women (19), with genetic factors affecting the expression of the glycosyltransferases required for HMO biosynthesis being an important source of this variance. HMOs are biosynthesized by a nontemplate-directed process in which the numerous glycosyltransferases expressed within the cells of the mammary gland elongate lactose with other monosaccharides, including galactose (Gal), fucose (Fuc), GlcNAc, and 5-N-acetylneuraminic acid (Neu5Ac). The best-known genetic source of HMO variance is a woman's Lewis blood group and secretor (Se) status, which are dictated by polymorphisms α1,4 and α1,2-fucosyltransferase genes, respectively (19–21). This variation is correlated with pediatric infectious disease susceptibility (22) and gut microbiome diversity (23).
A recent international study observed that HMOs from ethnically similar mothers varied geographically, suggesting an environmental and/or dietary influence on HMO biosynthesis (24). Obesity (21), malnourishment (7), and hyperglycemia (21) impact both HMO concentrations and structures; it is possible that these conditions affect the levels of nucleotide-activated GlcNAc and Neu5Ac available for HMO biosynthesis, as obesity and hyperglycemia are both linked to altered metabolic flux through the hexosamine biosynthetic pathway (25). The levels of Gal and Fuc in the maternal diet may likewise influence the levels of these monosaccharides incorporated into HMOs, as there is evidence that both Gal (26) and Fuc (27) may be directly recycled by specific monosaccharide salvage pathways in mammalian cells. Incorporation of diet-derived Neu5Ac into new glycoconjugates has been well-established. Dietary Neu5Ac is primarily observed glycosidically bound to other biomolecules, but it must be released into its free form by neuraminidases expressed by the GI mucosa or microbes before incorporation into infant tissue (28). Among the clearest evidence of salvaging of dietary Neu5Ac in vivo comes from studies of the structurally related analog 5-N-glycolyl-neuraminic acid (Neu5Gc). Humans are unable to biosynthesize Neu5Gc, instead obtaining it from the diet, with red meat and dairy products being particularly rich sources; however, human metabolic enzymes do not differentiate between Neu5Gc or Neu5Ac in vitro (29) or in vivo (30). Experiments with mice unable to biosynthesize Neu5Gc have demonstrated that diet-derived Neu5Gc that is glycosidically bound is more bioavailable than free Neu5Gc (31).
To date, Neu5Gc incorporation into HMOs has not been demonstrated, although it has been detected in a single sample of whole, pooled human milk (32). Establishing the capacity of diet-derived Neu5Gc to be incorporated into newly biosynthesized HMOs is relevant to infant health because the human immune system is able to distinguish between Neu5Ac- and Neu5Gc-containing glycans, specifically recognizing the latter as foreign (33). Experiments in rodents have led to the proposal that the anti-Neu5Gc antibodies, appearing in human neonates around 6 months after birth (34), are linked to cancer (35), atherosclerosis (36), and autoimmune disease (37).
Recently, we tested the potential influence of diet on milk oligosaccharide biosynthesis in dairy cattle (38). Cow milk shares some oligosaccharides in common with human milk, although the concentration and variety of those observed in cow milk are lower than that of human milk. This research led to identification of nine previously undescribed sulfonated milk oligosaccharides in bovine milk; these sulfonated glycoepitopes chemically resemble the O-glycans lining the infant GI tract (39) and the ligands for cell adhesion molecules borne by immune cells (40). Accordingly, the research presented here sought to address three questions. Does the human diet impact the biosynthesis of HMOs? Are sulfonated HMOs present in human milk? If so, do these have an observable prebiotic effect in exclusively breast-fed infants? Our observations indicate that all three questions are answered in the affirmative.
Results
Nonhuman monosaccharides are present in HMOs but not associated specifically with dairy intake
Red meat, (bovine) milk, and dairy products represent the most prevalent dietary sources of the nonhuman sialic acid Neu5Gc in the human diet (30, 41). It has been hypothesized that in women, milk and dairy products would be the major dietary source of Neu5Gc and that this monosaccharide would consequently be observed at elevated levels in the HMOs biosynthesized by women consuming cow's milk rather than alternative, almond-based beverages. Accordingly, breast milk samples from two groups of women (n = 8 for each), classified as milk or almond beverage consumers, as self-assessed on prestudy questionnaires, were selected from a larger cohort of samples obtained from donors who had completed a detailed diet recall survey for the 24-h period preceding milk collection. Close analysis of participants' diets after group selection revealed that, although not all the women in the almond group refrained entirely from dairy products, these groups were different in their mean intake of milk (0.2 ± 0.06 versus 1.2 ± 0.45 food group equivalents (FTE), p = 0.028, one-tailed Student's t test) and total dairy products (0.8 ± 0.32 versus 2.3 ± 0.39 FTE, p = 0.005). Based on the diet recalls, estimated levels of ingested Neu5Ac and Neu5Gc were predicted for each participant (Fig. 1 and Table S1) by multiplying the FTE (i.e. mass) of each Neu5Ac or Neu5Gc source by its putative abundance in common sources, as evaluated previously (30, 41). Overall, the estimated amounts of ingested Neu5Ac and Neu5Gc were similar in the milk- and almond-consuming groups; however, milk consumers obtained a greater fraction of Neu5Gc from dairy products than almond beverage consumers (Fig. 1a). Across both groups, the ratio of ingested Neu5Gc to Neu5Ac remained essentially constant. The actual levels of Neu5Ac and Neu5Gc glycosidically bound to HMOs were quantitated (42) in all samples (Fig. 1a). Intriguingly, low amounts of Neu5Gc were detected in all (n = 16) samples tested, representing, to our knowledge, the first conclusive report of the presence of this diet-derived monosaccharide on HMOs. However, despite estimated amounts of Neu5Gc being higher in dairy consumers, their levels in HMOs were unchanged relative to the almond group and, thus, not specifically associated with dairy intake.
Figure 1.
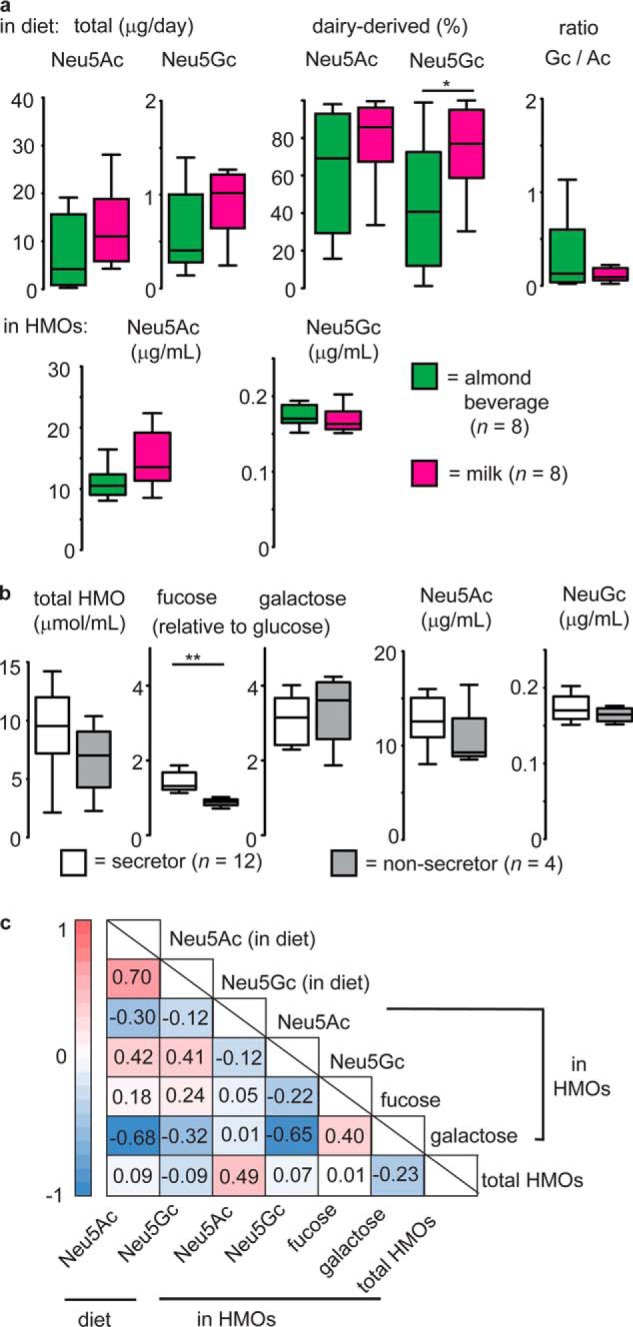
Diet-derived Neu5Gc was present as a constituent of HMOs in all (n = 16) samples tested. a, milk (i.e. bovine milk) was hypothesized to be the most significant source dietary Neu5Gc. It was determined that milk-consuming women did not ingest significantly more Neu5Gc than those consuming an almond milk beverage (top panels), nor did these groups significantly differ in median levels of HMO-borne Neu5Ac or Neu5Gc (bottom panels). b, maternal secretor status did not appear to affect levels of Neu5Gc in HMOs. *, p < 0.05; **, p < 0.01; Mann-Whitney U test. For all box-and-whisker plots, center lines depict medians, and box limits indicate 25th and 75th percentiles. Whiskers extend to 1.5 times the interquartile range. c, Spearman rank correlation heatmap depicting correlations between selected HMO monosaccharides and calculated Neu5Ac and Neu5Gc dietary intake levels over the 24-h period prior to sample collection. Color indicates directionality (red, positive; blue, negative); values for ρcrit are provided for each association and are indicated in bold font when exceeding the critical value (p < 0.01, n = 16) for a two-tailed test.
Differences in the levels of HMO-bound Neu5Gc or Neu5Ac between women in the milk- and almond beverage–consuming groups may have been obscured by genetic variability with respect to the Se status of individuals. Accordingly, monosaccharide levels, including Neu5Ac and Neu5Gc, were compared (Fig. 1b) between Se+ or Se− individuals, groups that were defined on the basis of relative HMO levels as described below. However, Neu5Gc and Neu5Ac levels in HMOs did not vary significantly with secretor type. When considering a subset of Se+ samples only, it was observed that HMOs from women in the milk-consuming group (n = 5) contained essentially equivalent amounts of Neu5Gc as observed for almond beverage consumers (n = 3), although the former did contain higher average levels of Neu5Ac (p = 0.055, two-tailed Student's t test). Because of the human inability to biosynthesize Neu5Gc, its presence in human milk is clear evidence of a direct dietary influence on HMO biosynthesis (Fig. S1). Spearman rank correlations were therefore determined to uncover potential associations between estimated levels of ingested Neu5Gc (from all possible sources) and the concentration of HMO-bound Neu5Gc in human milk (Fig. 1c). A positive but not significant association was observed between ingested and observed (on HMO) Neu5Gc levels. Interestingly, the opposite trend emerged for Neu5Ac, where a negative correlation was observed between the estimated levels of ingested Neu5Ac and the abundance of Gal in HMOs. Overall, these results reveal that, although Neu5Gc in HMOs depends on dietary intake, a clear correlation between estimated dietary intake within the previous 24 h and HMO prevalence was not apparent.
Elucidation of secretor status and discovery of sulfonated HMOs by CE
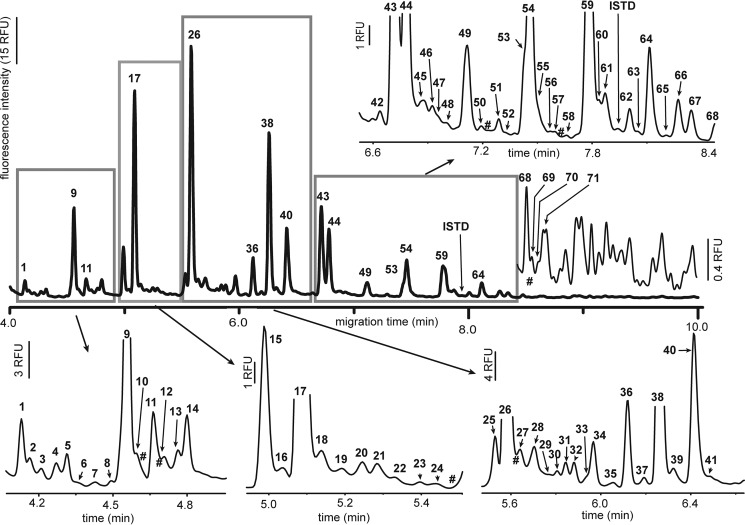
We (38) and others (43) have previously used capillary electrophoresis (CE) with laser-induced fluorescence (LIF) detection to rapidly profile milk oligosaccharides. CE-LIF analysis of HMOs extracted from our 16 samples permitted up to 93 (median = 87) different oligosaccharides to be distinguished within 10 min (Fig. 2), 21 of which could be identified on the basis of their co-migration with HMO standards (Fig. S2). The absolute and relative peak areas for 70 HMOs present in the majority of samples were recorded (Table S2). Traces of lactose (Fig. 2, HMO17) remained in all samples, accounting for, on average, 13% ± 6% of the total integrated peak area; this peak was excluded in determination of relative HMO peak areas.
Figure 2.
Representative CE electropherogram of fluorescently labeled HMOs. Peaks attributable to HMOs were sequentially numbered (bold font); # indicates the migration times of HMOs observed in only one to three (of 16) samples. An original unscaled electropherogram is depicted with the bold line, and the boxed regions and the region from 8.4 to 10 min are individually scaled to more clearly illustrate the HMO peaks. Peak areas were recorded for all numbered HMOs; those above 71 could not be reliably aligned between different samples and were recorded collectively as the sum of all HMO peaks in this region. RFU, relative fluorescence unit.
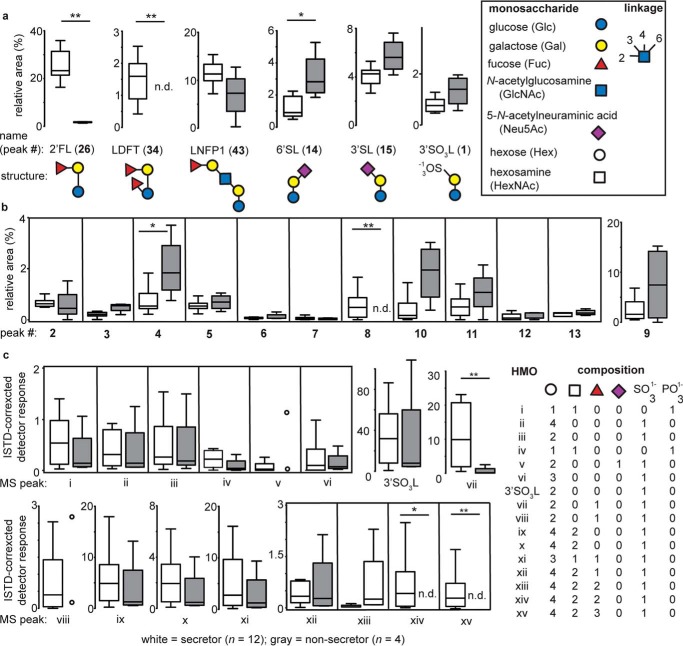
The relative abundance of several HMOs, including 2′-fucosyllactose (2′FL), lactodifucotetraose (LDFT), and lacto-N-fucopentaose 1 (LNFP1), has been demonstrated to accurately establish an individual's Se status (44). Determination, by CE, of the median levels of 2′FL and LDFT in the 16 samples in this study permitted them to be clearly differentiated into Se+ (n = 12) and Se− (n = 4) groups (Fig. 3a). Semiquantitative analysis of the same HMOs by HPLC-MS also classified samples into the same secretor groups as predicted by CE (Fig. S3). By CE, 2′FL and 3FL could not be clearly resolved; however, HPLC-MS could distinguish between these HMOs, demonstrating that, among samples identified as Se+ by CE, the median levels of 2′FL were nearly 300-fold higher than those of 3FL. In contrast, Se− samples contained median levels of 3FL over four times above those observed for 2′FL. Mean absolute concentrations for 11 selected HMOs were determined by CE-LIF, yielding values that were of comparable magnitude and precision as those reported previously by other groups (Table S3).
Figure 3.
Relative quantitation of sulfonated and phosphorylated HMOs by CE and HPLC-MS. a, CE analysis of HMOs permitted identification of maternal Se status based on significant differences in the relative levels of 2′FL, LDFT, and 6′SL (44). Numbers in bold font refer to CE peaks as numbered in Fig. 2. 3′SL and 3′SO3L, a sulfonated HMO previously identified in dairy milk samples, did not significantly vary with maternal secretor status. For all box-and-whisker plots, center lines depict medians, and box limits indicate 25th and 75th percentiles. Whiskers extend to 1.5 times the interquartile range. Open circles are used rather than box-and-whisker plots when fewer than four samples contained a specific HMO. b, with a single exception, all human milk samples contained a class of sulfate- or phosphate-bearing HMOs (HMO1–HMO6 and HMO10) with identical electrophoretic mobilities as those identified previously for the first time in bovine milk samples (38). The occurrence of several other peaks in this region (e.g. 7–9 and 11–13) suggests the presence of additional sulfonated HMOs. c, HPLC-MS analysis indicates the presence of HMOs with accurate masses (m/z) consistent with phosphorylated and sulfonated HMOs. HMOs i–vi and 3′SO3L have been identified previously in bovine milk; HMOs with m/z consistent with xiii–xv have been documented previously in pooled human milk (46). In all cases, the reducing-end residue was a hexose (i.e. glucose). Statistical differences between groups were assessed using a Mann–Whitney U test. *, p < 0.05; **, p < 0.01; n.d., not detected.
We previously documented the presence of nine sulfonated and two phosphorylated milk oligosaccharides in bovine milk, the availability of a 3′-sulfolactose (3′SO3L) standard permitting level-one identification (45) of this oligosaccharide by both CE-LIF and HPLC-MS. CE-LIF also permitted detection and relative quantitation of 3′SO3L (Fig. 2, HMO1) in every human milk sample tested (Table S2). Relative levels of 3′SO3L did not significantly differ between Se+ and Se− samples, although they tended to be higher in the latter (Fig. 3a). As observed previously with bovine milk samples, CE analysis of human milk revealed the presence of 13 HMOs of higher electrophoretic mobility than the well-known anionic HMOs 3′- or 6′-sialyllactose (3′/6′SL; Fig. 2, HMO15 and HMO14, respectively). These HMOs have been putatively identified as sulfonated or phosphorylated milk oligosaccharides on the basis of their sensitivity to chemical or enzyme-catalyzed hydrolysis; seven of these HMOs (HMO1–HMO6 and HMO10) possessed electrophoretic mobilities identical to oligosaccharides observed previously in bovine milk, whereas one (HMO9) appeared to be unique to human milk (Fig. S4). Among these anionic HMOs, the median levels of only two differed significantly between the Se+ and Se− groups, as assessed by CE (Fig. 3b); HMO4 was elevated in Se− samples, whereas HMO8 was nondetectable. HMO3 and HMO9–HMO11 all tended to have higher median levels in Se− individuals; indeed, when considered as a group (i.e. HMO1–HMO13), these anionic HMOs comprised a significantly greater fraction of the total HMO pool than in Se+ individuals (median levels, 15.7% and 7.1%, respectively). None of these HMOs (1–13) differed significantly between the almond- or milk-consuming groups.
HMOs were also analyzed by HPLC-MS to further confirm the presence of a class of previously uncharacterized sulfonated and/or phosphorylated HMOs. Monoisotopic masses of 16 unique HMOs were detected in our set of human milk samples (Figs. S5 and S6); for each secretor type, median levels were determined relative to a common internal standard (ISTD; Fig. 3c). To our knowledge, there has been only one previous report of the existence of sulfonated HMOs in which structures consistent with monoisotopic m/z values for xiii–xv (Fig. 3c) were identified in a sample extracted from 20 liters of pooled human milk (46). We observed oligosaccharides equivalent to HMOs i–vi and 3′SO3L in bovine milk (Table S4), i.e. structures matching both HPLC retention times and monoisotopic mass (38).
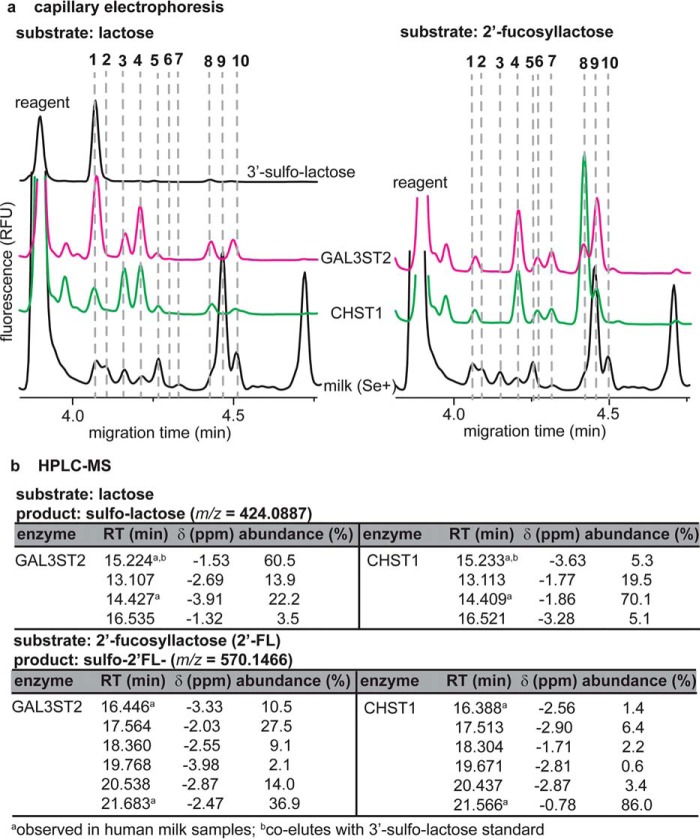
To further establish the identities of putative sulfonated HMOs in the absence of authentic standards, several neutral HMOs were tested as substrates for recombinant human galactose-3-O-sulfotransferase 2 (GAL3ST2) and carbohydrate sulfotransferase 1 (CHST1). GAL3ST2 and CHST1 have been demonstrated previously to transfer sulfate residues to lactose (47) and siallyllactosamine-containing oligosaccharides (48), respectively. CE and HPLC-MS analyses revealed that these enzymes were capable of producing a range of sulfonated HMO analogs from lactose and 2′FL (Fig. 4). More specifically, both enzymes produced at least four products from lactose, with CE mobilities identical to HMO1, HMO3, HMO4, and HMO5; similarly, 2′FL yielded products with mobilites identical to HMO8–HMO10. HPLC-MS analyses of these sulfotransferase mixtures similarly indicated that lactose and 2′FL were converted into four and six distinct sulfonated products, respectively; for each substrate, only two products were detectible in human milk samples by HPLC-MS (Fig. 3c). Tandem MS analyses (Fig. S7) demonstrated that, in vitro, sulfotransferases were able to transfer a sulfonate moiety to both the reducing and nonreducing ends of lactose and 2′FL. These data support the existence of a large, previously unrecognized class of at least 16 (Fig. 3c) sulfonated HMOs in human milk, at least 13 of which (Fig. 2, HMO1–HMO13) were quantifiable by CE.
Figure 4.
Human sulfotransferases accept lactose and 2′-fucosyllactose as substrates, creating multiple sulfonated HMOs. a, CE analysis of sulfotransferase reaction mixtures reveals at least four products (HMO1, HMO3, HMO4, and HMO5) that are also observed in human milk samples; five products (HMO6–HMO10) are detectible by CE when 2′FL is used as a substrate. Additional products are attributed to traces of lactose in the 2′FL. b, consistent with CE, HPLC-MS analysis demonstrates that human sulfotransferases convert lactose and 2′FL into four and six unique sulfonated products, respectively.
Correlative analysis between HMO levels, maternal diet, and breast milk fatty acids
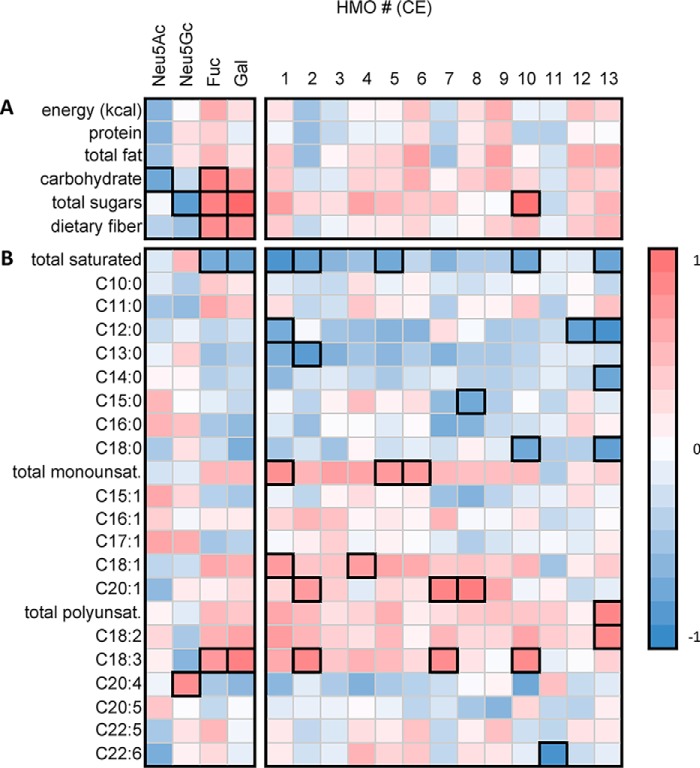
Spearman's rank correlation coefficients were determined to assess correlations between the maternal diet and the absolute CE detector responses of HMOs, of particular interest being anionic, putative sulfonated HMOs (1–13) with electrophoretic mobilities exceeding that of 6′SL (Fig. 5 and Fig. S8). Only Se+ individuals were assessed to eliminate the major source of genetic HMO variability. Several correlations were observed between this group of HMOs and breast milk fatty acids, which are known to directly reflect their levels in the maternal diet (49). First, significant negative correlations were observed between HMO1, HMO2, HMO5, HMO10, and HMO13 and the total saturated fats quantitated in breast milk. HMO1 and HMO12 negatively correlated with breast milk dodecanoic acid (C12:0), whereas HMO2 negatively correlated with tridecylic acid (C13:0). HMO8 negatively correlated with pentadecylic acid (C15:0), HMO10 negatively correlated with stearic acid (C18:0), and HMO13 negatively correlated with dodecanoic acid, myristic acid (C14:0), and stearic acid. No notable correlations were observed between HMO1–HMO13 and capric (C10:0), undecylic (C11:0), and palmitic (C16:0) acid. Interestingly, unsaturated fats showed opposite correlations. Total monounsaturated fats in breast milk positively correlated with HMO1, HMO6, and HMO7. Oleic acid (C18:1) was positively associated with HMO1 and HMO4, whereas gondoic acid (C20:1) correlated with HMO2, HMO7, and HMO8. Total polyunsaturated fatty acids and linoleic acid (C18:2n-6) positively correlated with HMO13, whereas α linolenic acid (C18:3n-3) correlated with Fuc, Gal, HMO2, HMO7, and HMO11. Arachidonic acid positively correlated with Neu5Gc. Docosahexanoic acid (C22:6n-3) was the only unsaturated fat that negatively correlated with HMO11. Dietary cholesterol levels, as assessed from 24-h recalls, appeared to negatively influence the levels of all 13 sulfonated/phosphorylated HMOs, and, when the 70 quantitated HMOs are considered, significant inverse correlations between cholesterol and 18 different HMOs were observed (Fig. S8).
Figure 5.
The total dietary intake of sugars and saturated fats significantly impacted the absolute levels of HMOs and monosaccharide HMO constituents observed in human milk. a and b, Spearman rank correlation heatmap for only Se+ samples (n = 12), depicting the associations between estimated dietary intake of selected macronutrients (a) and GC quantified breast milk fatty acids (b) and indicating that the relative levels of Fuc and Gal in HMOs were positively correlated with both total sugars (p < 0.01) and total dietary fiber (p < 0.05) ingested within the 24-h period prior to milk collection. Similarly, several of the putative sulfonated/phosphorylated HMOs were observed to be positively correlated with breast milk monounsaturated fats and polyunsaturated fats and negatively correlated with levels of saturated fats. A complete heatmap depicting the associations between all HMOs and an extensive list of nutrients is shown in the Fig. S8. Note that the intensity of each color indicates the directionality of the associations; all associations exceeding ρcrit (p = < 0.05) are highlighted in black. Numbers in bold font refer to CE peaks as labeled in Fig. 2.
Ingested carbohydrates, including both simple sugars and dietary fiber, were observed to significantly correlate positively with relative levels of both Gal and Fuc present in HMOs to the decrement of Neu5Ac (total carbohydrates) and Neu5Gc (total sugars, Fig. 5). Similar correlations were observed for the total amount of ingested fruit, a major dietary source of both sugars and fiber, the latter of which is a known source of Fuc (Fig. S8); specifically, significant positive correlations were observed for Gal and Fuc, whereas Neu5Ac levels were significantly lower in HMOs biosynthesized by women consuming high amounts of fruit. Total fruit intake was positively correlated with the absolute levels of 15 different HMOs (including putative sulfonated HMO1, HMO4, HMO5, and HMO10). Similar correlations were not observed for any other food group, with the exception of cured meats, which were negatively correlated with the abundance of 22 HMOs. Finally, it was hypothesized that Neu5Gc levels would be highest in milk samples from women who had consumed the largest amounts of red meat and dairy products. In contrast, the data indicate that only cheese intake positively correlated with Neu5Gc levels. Overall, these results suggest that, in Se+ women, unsaturated fat- and fruit-derived dietary fiber are correlated with increases in multiple HMOs, whereas the opposite trend was observed for saturated fat and cholesterol. With the exception of cheese intake, no positive associations were observed between known Neu5Gc sources and its levels in HMOs, suggesting that class of biomolecule (i.e. glycolipid versus glycoprotein) might influence Neu5Gc bioavailability.
Secretor status and HMOs explain overall bacterial community composition in infants
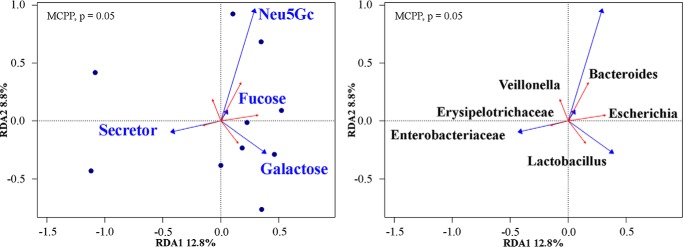
Redundancy analysis (RDA) ordination with a Monte Carlo permutation test indicated that Se status, as defined by levels of 2′FL, LDFT, and LNFP1 (44) (Fig. 3a), and levels of Fuc, Gal, and Neu5Gc were marginally (Monte Carlo permutation procedure, p = 0.05) related to infant gut microbial composition (Fig. 6). Stringent adjustments for multiple testing revealed Escherichia to be the only significant genus associated with maternal HMOs among the 43 genera included. Without adjusting for multiple testing, several taxa exhibited a positive association. While acknowledging the increased potential for type-1 errors, these taxa included: Bacteroides, Escherichia, Lactobacillus, Enterobacteriaceae, Erysipelotrichaceae, and Veillonella spp. Overall, ∼20% of the total variation in the microbial community could be explained by the selected HMOs.
Figure 6.
RDA ordination of microbial community constrained by selected explanatory variables. Arrow length indicates the importance of each infant microbial community and its relationship with HMOs. Only the predictor variables that significantly explained variability in microbial community structures are displayed. The HMOs (left, blue arrows) and bacterial genera (right, red arrows) are presented separately for clarity; however, they are derived from the same RDA model. In total, RDA1 and RDA2 explain over 20% of the total variation in β diversity. The global model's p value was significant (p = 0.05), as calculated by a Monte Carlo permutation procedure (MCPP) (n = 10).
Our results indicate that the abundance of Enterobacteriaceae was associated with maternal Se status, whereas Bacteroides spp. correlated with HMOs bearing more Neu5Gc and Fuc. Likewise, Fuc positively correlated with Escherichia abundance, whereas Gal positively correlated with Lactobacillus spp. along RDA2. Other taxa that aligned with RDA2 but were not correlated with the selected predictors included Veillonella and Erysipelotrichaceae. These results suggest that HMO type in breast milk can predict the type of microbes in the infant gut.
HMOs correlate with distinct immune markers
The absolute levels of HMOs correlated with distinct immune markers expressed in breast milk and infant stool. The Spearman rank correlation heatmap of breast milk (Fig. 7) depicts a positive association (p < 0.05) between HMO9 with the pro-inflammatory cytokine tumor necrosis factor α and the chemokine IL-8. 6′SL associated with pro-inflammatory cytokine IL-6, whereas HMO2 was negatively associated with IL-13, a major anti-inflammatory cytokine. In contrast, Neu5Ac negatively correlated with the pleotropic cytokine IL-6 and the chemokines myocyte chemoattractant protein-1 (MCP-1) and IL-8. Neu5Gc negatively correlated with the granulocyte-macrophage colony-stimulating factor (GM-CSF). Finally, Gal levels exhibited a negative association with the pro-inflammatory cytokines IL-2 and IL-12.
Figure 7.
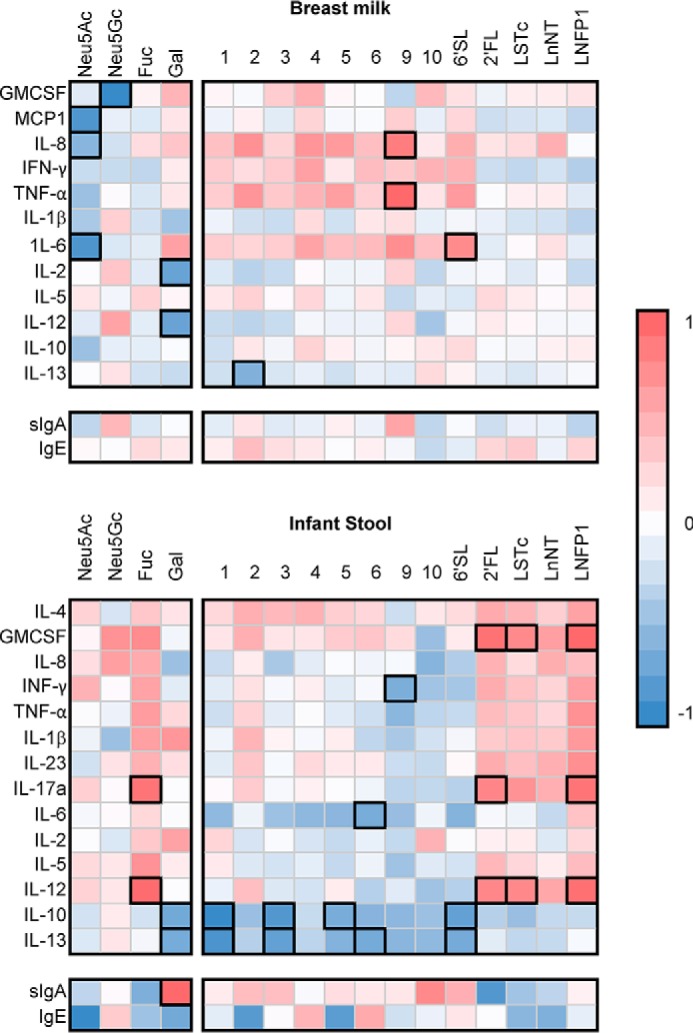
The Spearman rank correlation heatmap for both Se+ and Se− samples (n = 16) depicts the associations between the levels of HMOs and oligosaccharides in human milk with immune markers in breast milk (top) and infant stool (bottom). Note that the intensity of each color indicates the directionality of the associations; all associations with p = < 0.05 are highlighted in black. HMOs are numbed as in Fig. 2.
In infant stool, there was a negative association between absolute abundance of HMO1, HMO3, HMO5, 6′SL, and galactose with expression of the anti-inflammatory cytokine IL-10. Similarly, HMO1, HMO3, HMO6, 6′SL, and galactose were all negatively associated with anti-inflammatory IL-13 in infant stool. Uniquely, there was a positive association between galactose and stool secretory IgA. Following a similar trend as the other sulfonated/phosphorylated HMOs, HMO6 negatively correlated with IL-6, whereas HMO9 negatively correlated with the pro-inflammatory cytokine IFN-γ. In stark contrast, 2′FL, siallyllacto-N-tretraose c (LSTc), and LNFP1 were associated with higher expression of pro-inflammatory cytokines in infant stool. 2′FL, LSTc, and LNFP1 positively correlated with GM-CSF expression. Similarly, 2′FL, LSTc, LNFP1, and fucose positively associated with pro-inflammatory IL-12. Likewise, 2′FL, LNFP1, and fucose were associated with IL-17a expression. These results suggest that HMO type in breast milk is associated with the overall immune status of the infant.
Discussion
The neonatal period is a critical time in mammalian life, particularly with respect to nutrition and development of the intestinal microbiome. The intestinal microbiome is a diverse microbial community composed of 500–1,000 different species that lie at the interface of the intestinal and the external environment, forming a relationship between host epithelial cells and dietary antigens. Collectively, these microbes provide a multitude of benefits to their mammalian host, including nutrient absorption, immune system maturation, and pathogen defense. However recent evidence suggests that any disturbance in the host–microbiota relationship, termed dysbiosis, is associated with diseases such as inflammatory bowel disease, metabolic syndrome, obesity, and diabetes. Consequently, there has been an increase in research aimed at manipulating the intestinal microbiota in an attempt to promote health and prevent dysbiosis-associated diseases. Of all of the exogenous factors implicated in structuring the phylogenetic makeup of the gut microbiota, diet is the most extensively studied. However, this area of research remains to be fully elucidated.
Breast milk is considered the “gold standard” of infant nutrition and is known to provide bioactive components that help infants develop their immune system and microbiome. Of these bioactive components, HMOs are thought to play a vital role in nourishing the developing infant gut microbiome. Despite the high abundance and diversity of HMOs, the structure and function of individual HMOs as well as factors that influence their biosynthesis remain unclear.
Here, CE-LIF analysis was used to detect a median of 87 distinct HMOs (n = 16 samples) within 10 min (Fig. 2). The presence of 13 HMOs of electrophoretic mobility in excess of that of 6′SL (HMO14) suggested that human milk, like cow milk (38), contains small, highly charged HMOs consistent with the presence of either phosphate- or sulfate-bearing oligosaccharides. Comigration of a CE-LIF peak with a 3′-sulfolactose standard in all 16 samples and HPLC-MS evidence of 16 unique oligosaccharides bearing sulfate and/or phosphate groups (Figs. 3c and 4b) provide further evidence of a large and, with a sole exception (46), previously undescribed class of sulfate-bearing HMOs. The abundance of sulfonated HMOs in human milk appears to be due to the ability of human sulfotransferases to produce multiple products from lactose and/or other neutral HMOs (Fig. 4). Sulfonation of mucus-linked oligosaccharides in the colon has been shown to be correlated with a decrease in susceptibility to parasitic infections (50) and inflammation associated with human inflammatory bowel disease or colon cancer (51).
Because the sulfonated HMOs identified here in many instances (Fig. S7) bear the sulfate moiety on a nonreducing hexose (i.e. galactose) residue, the predominant site of oligosaccharide sulfonation in the human colon (52), we hypothesize that these unique HMOs fill a similar protective role in maintaining colon homeostasis. Nonsulfonated HMOs such as 2′FL, LSTc, and LNP1 associated with higher Th1 and Th17 immune mediators in infant stool, whereas sulfonated HMOs correlated with suppression of two major Th2 cytokines, IL-10 and IL-13. Considering this in addition to correlation of HMOs to Enterobacteriaceae, microbes that drive Th1 immune responses in the gut, this study highly suggests that both sulfonated and nonsulfonated HMOs stimulate Th1 immunity in the infant gut through unique immune mediators. Curiously, this was in the absence of strong immune stimulation in the breast milk, suggesting that this is a mild or specific effect in the naive infant gut. Given that the infant immune system is under development during the postnatal period, we propose that HMOs are important modulators in training of the infant immune system from moving away from Th2 dominance through Th1 stimulation. HMO ingestion by the infant thus has far-reaching consequences for protection against infectious disease susceptibility as well as chronic immune conditions, including allergies. As discussed by Xu and Knight (53), a challenge in understanding the dietary effects on the gut microbiota is the confounding variations in host genotypes. Thus, here we used the relative abundance of 2′FL, LDFT, and LNFP1 to determine and account for maternal Se status. In this study, maternal genetics (as assessed by HMO analysis) had modest effects on the infant fecal microbiome. For example, Se status was associated with Enterobacteriaceae in infants, suggesting a defining role of genetics in establishment of early colonizers in infants. Although this work focuses on HMOs, it should be noted that other components in breast milk may also directly or indirectly influence the microbiome. For instance, Hill et al. (54) have shown that low-molecular-weight hyaluronan reported in human milk is able to alter expression of β-defensins in the gastrointestinal tract, which could conceivably alter gut microbial communities. Thus, future studies should seek to isolate the HMOs identified here for mechanistic studies.
In addition to genetic factors, maternal dietary intake during lactation appears to influence the community composition of the infant microbiome. Long-term dietary intake of animal protein and saturated fats is associated with high abundance of Bacteroides taxa (55). Red meat, such as beef, pork, and lamb, contains considerable amounts of Neu5Gc, which has recently been shown to incorporate into tissues. We show for the first time that diet-derived Neu5Gc is also incorporated into HMOs. To facilitate insights into the potential impact of Neu5Gc on the infant microbiome, an RDA was used to understand assemblage composition differences in relation to predictors of interest. The RDA indicated that Neu5Gc correlates with Bacteroides spp. abundance in infant stool. However, Bacteroides spp. are known for their ability to metabolize a variety of HMOs (56) and, therefore, this observation should be tested in a controlled setting. Alongside Neu5Gc, our study shows that fucose levels in breast milk are associated with Bacteroides and Escherichia spp. in infant stool. These findings align with previous literature showing that some bacteria of the Bacteroides genus, such as Bacteroides thetaiotaomicron, produce multiple fucosidases that cleave fucose from host glycans, resulting in free fucose available in the intestinal lumen (57). Certain species of Escherichia carry a fucose-sensing signal transduction system that can sense the monosaccharide and use it to their competitive advantage (58), suggesting a rationale for the observed association between Bacteroides and Escherichia spp. and dietary fucose. Finally, our results show that Lactobacillus spp. in infant stool correlated with total galactose concentration in human milk. This is unexpected, given that most Lactobacillus spp. do not grow well on HMOs and do not typically utilize them as a carbon source (59). Rather, Lactobacillus spp. tend to metabolize the glucose moiety of lactose and release galactose and lactic acid into their environment.
Given that the gut microbiome and immune responses are tightly interrelated, this study also sought to generate hypotheses with respect to HMO–immune associations. We observed a distinct separation between the sulfonated/phosphorylated HMOs from 2′FL, LSTc, LnNT, and LNFP1 in infant stool which, similar to the bacterial associations, should be investigated further.
Overall, the findings from this exploratory study suggest that maternal diet affects the biosynthesis of HMOs and, in turn, may influence their infants' bacteriome and immune development. However, interactions between host genetics, diet, and the gut microbiota are convoluted and multifaceted (53), and the effect size in this analysis is small compared with the huge variability intrinsic in human data. We acknowledge that large-scale studies with greater statistical power are required to reveal definite contributions of maternal diet to the infant gut microbiome. Future studies may benefit from using the HMO–microbe–immune associations presented here as a possible a priori interest.
Experimental procedures
Study design
This study was conducted according to the Declaration of Helsinki guidelines, and all procedures were approved by the University of British Columbia Clinical Research Ethics Board and British Columbia Interior Health Ethics Board. Informed signed consent was obtained from each participant at enrollment.
A prospective cohort clinical study was conducted in the Okanagan Valley, Canada. The full study design has been published previously (60). Prior to enrollment, participants were screened to include healthy, exclusively breastfeeding mother–infant pairs (self-reported). Participants were excluded if the infant had any disease or was born prematurely. In total, 109 women–infant pairs were recruited. Of these, women who exclusively drank dairy milk were selected for this study (n = 8) and matched with women who did not drink dairy (n = 8) to encompass a range of maternal dietary patterns and limiting other covariates, such as maternal age, education, and socioeconomic status.
Stool and breast milk sample collection
The method for infant stool collection, storage, and processing has been described previously (61). For this study, only samples collected at 5 months of age were utilized. To collect breast milk samples, mothers were instructed to clean the nipple and surrounding area with warm water and soap and manually express a few drops of milk before collecting 10 ml of foremilk on the morning of infant stool collection. Participants were given a sterile collection kit and instructed to store the breast milk and stool samples in their home freezer for a maximum of 3 days. Samples were transported to the research facility on dry ice and stored at −80 °C until further analysis.
HMO extraction and analyses
HMOs were extracted from milk samples essentially as described previously (38). In brief, milk samples were thawed at 4 °C, and a 500-μl aliquot was fortified with 40 μg of maltoheptaose (Sigma) as an ISTD. HMOs were subsequently removed from the proteins and lipids by liquid–liquid extraction and dried before salts and the majority of lactose were removed by solid-phase extraction using graphitized carbon cartridges (ENVI-Carb, Sigma). HMO-containing fractions were freeze-dried, redissolved in 18 mΩcm−1 water, and divided into five equal portions for quantitation of total reducing sugars (62), quantitation of Neu5Ac and Neu5Ac (42), neutral monosaccharide analysis (38), HMO profiling by CE-LIF, and targeted HMO analysis by HPLC-MS (38). All monosaccharide standards were purchased from Carbosynth, and HMO standards were purchased from Carbosynth or Dextra Laboratories, Inc. Where possible, HMO identification was made based on comigration (CE-LIF) or coelution and m/z (HPLC-MS) to these commercially available standards.
CE-LIF data were acquired under normal (monosaccharides) and reverse (HMOs) polarity, exactly as described previously, using a ProteomeLab PA800 (Beckman Coulter) (38). Peak areas were determined by manual integration using 32 Karat software (version 7.0); in instances where peaks could not be detected, the relevant region of baseline was integrated, and the background signal was recorded. All CE peaks areas were divided by their migration times and subsequently normalized to the area of the ISTD (maltoheptaose) to permit comparison of absolute HMO levels; similarly, the extracted ion chromatogram peak areas for HMOs detected by HPLC-MS were normalized to the area of the ISTD present in each sample. Relative HMO levels, as assessed by CE-LIF, were calculated by expressing each peak area as a fraction of the total detectible area of all HMOs, with the exception of residual lactose. HPLC-MS analysis of HMOs (38) and Neu5Ac/Neu5Gc (42) was performed as described previously. HPLC was conducted on an Agilent 1290 Infinity system (Agilent Technologies) with a 1290 Infinity binary pump, 1290 Infinity autosampler, and 1290 Infinity column compartment. HMOs were separated on a Thermo Scientific Hypercarb 100 × 2.1 mm column (3-μm particle size) at a temperature of 50 °C. Samples were analyzed using an injection volume of 5 μl at a flow rate of 0.250 ml/min. Mobile phases A and B were H2O and acetonitrile, respectively, each with 0.1% formic acid. Gradient elution was programmed with a total run time of 30 min as follows: 0–5 min, 0–5% B; 5–15 min, 5–20% B; 15–20 min, 20–40% B; 20–25 min, 40–80% B; 25–27 min, 80% B; 27–27.1 min, 80–5% B; 27.1–29 min, 5% B; and 29–30 min, 5–0% B. Mass spectrometry was conducted in negative ion mode on an Agilent 6530 QToF mass spectrometer with an Agilent Jet Stream electrospray ionization source. Source parameters were as follows: drying gas (N2) temperature 300 °C with a flow rate of 10 liters/min, sheath gas (N2) temperature 350 °C with a flow rate of 10 liters/min, nebulizer pressure 35 pounds per square inch, capillary voltage 3,500 V, nozzle voltage 1,000 V, and fragmenter voltage 175 V. A reference ion solution containing 10 μm purine and 2.0 μm HP-0921 (m/z 121.0509 and 922.0098, respectively) in 95:5 ACN:H2O was added post-column at 6 μl/min by an Agilent 1260 Infinity II isocratic pump.
The QToF was tuned and calibrated in 2-GHz extended dynamic range mode for the 100–3200 m/z range immediately prior to sample analysis. MS/MS spectra were acquired using data-dependent Auto MS/MS mode with a preferred precursor ion list containing calculated m/z for each HMO and sulfonated HMO. Precursor ions were selected for MS/MS product ion scans with absolute and relative thresholds set to 1,000 counts and 0.01%, respectively. Active exclusion was disabled for HMO standards and enabled for human samples, with precursors excluded after 10 spectra and released after 0.75 min. Abundance-dependent accumulation was enabled for all samples; scan speed was varied based on precursor abundance with a target of 30,000 counts per spectrum for HMO standards and 20,000 counts per spectrum for human samples. Full-scan MS spectra were collected at a rate of 2 Hz with a mass range of 100–1100 m/z for HMO standards and 100–3200 m/z for human samples. All MS/MS spectra were collected at a rate of 2 Hz with the isolation width set to ∼1.3 m/z. The mass range for MS/MS spectra was adjusted depending on the sample to maximize the number of transients per spectrum, with the minimum set to 100 m/z for all samples and the maximum set to that of the highest-m/z sulfonated HMO precursor ion, rounded up in increments of 50 m/z units.
HPLC-MS data acquisition and analysis were performed using MassHunter Work Station software (Agilent Technologies): Data Acquisition Work Station (v. B.06.01, SP1) and Qualitative Analysis (v. B.07.00, SP2). The Find-by-Formula algorithm in Qualitative Analysis was used to generate extracted ion chromatograms for analyte m/z values with a ± 15.00 ppm mass accuracy limit and ± 0.300 min retention time window, as established by analyzing HMO standards and enzyme-treated HMO standards without retention time filtering. Peak areas and retention times were processed further in Microsoft Excel. In silico fragmentation of HMOs and sulfonated HMOs was performed in GlycoWorkbench (v. 2.1, build 146). MS/MS data were processed, and spectra were generated and annotated in R (v. 3.6.1, R Core Team) with major B/Y and C/Z fragments from GlycoWorkbench.
Dietary intake and breast milk analysis
To understand the relationship between dietary intake and HMOs, a validated 24-h dietary recall was given to all participants to track food consumption. Participants were asked to fill out the recall on sample collection days; all dietary data were assessed and entered into the Self-Administered 24 h (ASA24) Dietary Assessment Tool, a web-based tool for dietary analysis (63). The data output from ASA24 includes a summary of dietary nutrients consumed by the participant. Because dietary recalls are not always good indicators of breast milk fatty acids, the medium- and long-chain fatty acids in breast milk were quantitated using GC exactly as described previously (64).
Synthesis of sulfonated HMOs
3.8 mg of each sulfotransferase (CHST1 and GAL3ST2, R&D Systems) were mixed with 10 nmol of either lactose or 2′FL, 49.25 nmol adenosine 3′-phosphate,5′-phosphosulfate (Sigma) and reacted at 37 °C. Reactions were conducted in 50 mm Tris, 500 mm NaCl, and 15 mm MgCl2 (pH 7.5) (CHST1) or 25 mm MES and 15 mm MgCl2 (pH 5.6) (GAL3ST2) in total reaction volumes of 50 μl. Following overnight incubation, samples were applied directly to conditioned ENVI-Carb solid-phase extraction cartridges and subsequently prepared for CE-LIF and HPLC-MS/MS analysis exactly as described above.
Immune function analysis
The expression of secretory IgA, IgE, and cytokines (GM-CSF, IFN-γ, IL-1β, IL-2, IL-6, IL-8, IL-10, IL-12 (p70), MCP-1, tumor necrosis factor α, IL-13, and IL-5) was measured in infant stool and breast milk by Eve Technologies (Calgary, AB, Canada).
High-throughput sequencing
Sequencing methods
The QIAamp DNA Stool Mini Kit (Qiagen, catalog no. 51504) was used to extract total DNA fecal samples following the manufacturer's specifications as described previously (65). Briefly, 5 ng of extracted DNA was used in a PCR reaction to amplify the V3-V4 region of the 16S ribosomal DNA using 341F and 805R primers (66) attached to the Illumina adapter overhang. A Nextera XT Dual Index Kit was used to attach unique identifiers to 5′ and 3′ ends using a second PCR reaction. PCR products were cleaned using Agencourt Ampure XP beads (Beckman Coulter), and pooled amplicons were checked for quality and quantity on the Experion Automated Electrophoresis System (Bio-Rad). The resulting 2 × 300-bp paired-end reads were sequenced on a MiSeq system at the Applied Genomics Core (Edmonton, AB, Canada).
Bioinformatics and microbial analysis
All bioinformatics processing was performed within QIIME2 (67). Forward sequences underwent quality filtering, dereplication, chimera removal, denoising, and merging using the Deblur plugin with default settings. Using the most recent version of Greengenes available (13_8), a naïve Bayes classifier was trained on a specific region targeted by our primer sets. MAFFT (Multiple Alignment using Fast Fourier Transform)-aligned sequences (68) were used to produce a phylogeny tree using FastTree2 (69) with default settings for all microbiome analyses requiring phylogenetic information. This resulted in 10 samples being included for microbial comparisons. All used software packages, versions, and parameters are available under the “provenance” section of the QIIME2 feature-table artifact available at the Open Science Framework with the project ID 2xvgh. The file can be viewed by dragging and dropping onto the QIIME 2 View (RRID: SCR_018074).
Statistics
Median HMO levels between different secretor groups were deemed statistically significant if the two-tailed Mann–Whitney U values were lower than Ucrit at p < 0.05 or 0.01. Associations between absolute HMO levels (assessed by CE-LIF) and estimated dietary metrics were determined by calculating the two-tailed Spearman rank correlations for each comparison. Following a previous approach (65), an RDA, performed using R version 3.4.1, was used to simultaneously evaluate the relationship between dietary components, HMOs, and the infant microbiome. This constrained ordination looks at composition differences in the infant microbiome in relation to potential predictors. Given our low sample size (n = 10), we limited our predictor variables (n = 70) to include maternal secretor status (44), Neu5Ac, Neu5Gc, fucose, and galactose. Microbial data were first Hellinger-transformed (70) to accommodate the high occurrences of zero values in the count data using the “Vegan” package. The “ordistep” function was used for variable selection using default settings. To identify genera that contributed to the variance in microbial data, a Spearman correlation was used between genus abundance and the first two RDA axes. Any significant correlation evaluated at an α level of 0.05 was displayed on the RDA plot with type II scaling. For comparisons between immune markers and HMOs, a Spearman rank correlation was conducted using the “rcorr” function in the “Hmisc” package in R and presented as a heatmap. Holm's method was applied to correct for multiple inference.
Data Availability
Microbiome, HPLC-MS, and tandem MS data have been deposited to the publicly accessible repository Open Science Framework with the project ID 2xvgh.
Author contributions
C. Q., S. D. V., D. L. G., and W. F. Z. formal analysis; C. Q. and W. F. Z. validation; C. Q., S. D. V., and W. F. Z. investigation; C. Q., D. M. V., and D. L. G. ran clinical trials; C. Q. and D. L. G. writing-original draft; C. Q., D. L. G., and W. F. Z. writing-review and editing; S. D. V., N. A. M., A .M .G., D. M. V., D. L. G., and W. F. Z. data curation; S. D. V. and W. F. Z. methodology; D. L. G. and W. F. Z. resources; D. L. G. and W. F. Z. software; D. L. G. and W. F. Z. supervision; D. L. G. and W. F. Z. funding acquisition; D. L. G. and W. F. Z. project administration; W. F. Z. conceptualization; W. F. Z. visualization.
Supplementary Material
Acknowledgments
We thank Benjamin Noyovitz for assistance with MS/MS. We thank Interior Health, D. Erland (RN), and Dr. M. Docherty (MD) for recruitment assistance and the Okanagan community for promoting and participating in the study.
The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S8 and Tables S1–S4.
- HMO
- human milk oligosaccharide
- GI
- gastrointestinal
- Gal
- galactose
- FTE
- food group equivalent
- Fuc
- fucose
- Neu5Gc
- 5-N-glycolyl-d-neuraminic acid
- CE
- capillary electrophoresis
- LIF
- laser-induced fluorescence
- 2′FL
- 2′-fucosyllactose
- 3FL
- 3-fucosyllactose
- LDFT
- lactodifucotetraose
- 3′SO3L
- 3′-sulfolactose
- SL
- siallyllactose
- ISTD
- internal standard
- RDA
- redundancy analysis
- GM-CSF
- granulocyte-macrophage colony-stimulating factor
- LSTc
- siallyllacto-N-tretraose c
- Neu5Ac
- 5-N-acetyl-d-neuraminic acid
- Se
- secretor.
References
- 1. Urashima T., Hirabayashi J., Sato S., and Kobata A. (2018) Human milk oligosaccharides as essential tools for basic and application studies on galectins. Trends Glycosci. Glycotechnol. 30, SE51–SE65 10.4052/tigg.1734.1SE [DOI] [Google Scholar]
- 2. Engfer M. B., Stahl B., Finke B., Sawatzki G., and Daniel H. (2000) Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am. J. Clin. Nutr. 71, 1589–1596 10.1093/ajcn/71.6.1589 [DOI] [PubMed] [Google Scholar]
- 3. Gnoth M. J., Kunz C., Kinne-Saffran E., and Rudloff S. (2000) Human milk oligosaccharides are minimally digested in vitro. J. Nutr. 130, 3014–3020 10.1093/jn/130.12.3014 [DOI] [PubMed] [Google Scholar]
- 4. Albrecht S., Schols H. A., van den Heuvel E. G., Voragen A. G., and Gruppen H. (2011) Occurrence of oligosaccharides in feces of breast-fed babies in their first six months of life and the corresponding breast milk. Carbohydr. Res. 346, 2540–2550 10.1016/j.carres.2011.08.009 [DOI] [PubMed] [Google Scholar]
- 5. Karav S., Le Parc A., Leite Nobrega de Moura Bell J. M., Frese S. A., Kirmiz N., Block D. E., Barile D., and Mills D. A. (2016) Oligosaccharides released from milk glycoproteins are selective growth substrates for infant-associated bifidobacteria. Appl. Environ. Microbiol. 82, 3622–3630 10.1128/AEM.00547-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Asakuma S., Hatakeyama E., Urashima T., Yoshida E., Katayama T., Yamamoto K., Kumagai H., Ashida H., Hirose J., and Kitaoka M. (2011) Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J. Biol. Chem. 286, 34583–34592 10.1074/jbc.M111.248138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Charbonneau M. R., O'Donnell D., Blanton L. V., Totten S. M., Davis J. C., Barratt M. J., Cheng J., Guruge J., Talcott M., Bain J. R., Muehlbauer M. J., Ilkayeva O., Wu C., Struckmeyer T., Barile D., et al. (2016) Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell 164, 859–871 10.1016/j.cell.2016.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davis J. C., Lewis Z. T., Krishnan S., Bernstein R. M., Moore S. E., Prentice A. M., Mills D. A., Lebrilla C. B., and Zivkovic A. M. (2017) Growth and morbidity of Gambian infants are influenced by maternal milk oligosaccharides and infant gut microbiota. Sci. Rep. 7, 40466 10.1038/srep40466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davis J. C., Totten S. M., Huang J. O., Nagshbandi S., Kirmiz N., Garrido D. A., Lewis Z. T., Wu L. D., Smilowitz J. T., German J. B., Mills D. A., and Lebrilla C. B. (2016) Identification of oligosaccharides in feces of breast-fed infants and their correlation with the gut microbial community. Mol. Cell. Proteomics 15, 2987–3002 10.1074/mcp.M116.060665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Matsuki T., Yahagi K., Mori H., Matsumoto H., Hara T., Tajima S., Ogawa E., Kodama H., Yamamoto K., Yamada T., Matsumoto S., and Kurokawa K. (2016) A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat. Commun. 7, 11939 10.1038/ncomms11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lewis Z. T., Totten S. M., Smilowitz J. T., Popovic M., Parker E., Lemay D. G., Van Tassell M. L., Miller M. J., Jin Y. S., German J. B., Lebrilla C. B., and Mills D. A. (2015) Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome 3, 13 10.1186/s40168-015-0071-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu Y., Mishra S., Song X., Lasanajak Y., Bradley K. C., Tappert M. M., Air G. M., Steinhauer D. A., Halder S., Cotmore S., Tattersall P., Agbandje-McKenna M., Cummings R. D., and Smith D. F. (2012) Functional glycomic analysis of human milk glycans reveals the presence of virus receptors and embryonic stem cell biomarkers. J. Biol. Chem. 287, 44784–44799 10.1074/jbc.M112.425819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Noll A. J., Gourdine J. P., Yu Y., Lasanajak Y., Smith D. F., and Cummings R. D. (2016) Galectins are human milk glycan receptors. Glycobiology 26, 655–669 10.1093/glycob/cww002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He Y., Liu S., Leone S., and Newburg D. S. (2014) Human colostrum oligosaccharides modulate major immunologic pathways of immature human intestine. Mucosal Immunol. 7, 1326–1339 10.1038/mi.2014.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rudolff S., Pohlentz G., Borsch C., Lentze M. J., and Kunz C. (2012) Urinary excretion of in vivo 13C-labelled milk oligosaccharides in breastfed infants. Br. J. Nutr. 107, 957–963 10.1017/S0007114511004016 [DOI] [PubMed] [Google Scholar]
- 16. Oliveros E., Ramirez M., Vazquez E., Barranco A., Gruart A., Delgado-Garcia J. M., Buck R., Rueda R., and Martin M. (2016) Oral supplementation of 2′-fucosyllactose during lactation improves memory and learning in rats. J. Nutr. Biochem. 31, 20–27 10.1016/j.jnutbio.2015.12.014 [DOI] [PubMed] [Google Scholar]
- 17. Cho I., and Blaser M. J. (2012) The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270 10.1038/nrg3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Victora C. G., Bahl R., Barros A. J., França G. V., Horton S., Krasevec J., Murch S., Sankar M. J., Walker N., Rollins N. C., and Lancet Breastfeeding Series Group (2016) Breastfeeding in the 21st century: Epidemiology, mechanisms, and lifelong effect. Lancet 387, 475–490 10.1016/S0140-6736(15)01024-7 [DOI] [PubMed] [Google Scholar]
- 19. Thurl S., Munzert M., Henker J., Boehm G., Müller-Werner B., Jelinek J., and Stahl B. (2010) Variation of human milk oligosaccharides in relation to milk groups and lactational periods. Br. J. Nutr. 104, 1261–1271 10.1017/S0007114510002072 [DOI] [PubMed] [Google Scholar]
- 20. Blank D., Dotz V., Geyer R., and Kunz C. (2012) Human milk oligosaccharides and Lewis blood group: individual high-throughput sample profiling to enhance conclusions from functional studies. Adv. Nutr. 3, 440S–449S 10.3945/an.111.001446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smilowitz J. T., Lebrilla C. B., Mills D. A., German J. B., and Freeman S. L. (2014) Breast milk oligosaccharides: structure-function relationships in the neonate. Annu. Rev. Nutr. 34, 143–169 10.1146/annurev-nutr-071813-105721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang X., Huang P., Zhong W., Tan M., Farkas T., Morrow A. L., Newburg D. S., Ruiz-Palacios G. M., and Pickering L. K. (2004) Human milk contains elements that block binding of noroviruses to human histo-blood group antigens in saliva. J. Infect. Dis. 190, 1850–1859 10.1086/425159 [DOI] [PubMed] [Google Scholar]
- 23. Wacklin P., Mäkivuokko H., Alakulppi N., Nikkilä J., Tenkanen H., Räbinä J., Partanen J., Aranko K., and Mättö J. (2011) Secretor genotype (FUT2 gene) is strongly associated with the composition of bifidobacteria in the human intestine. PLoS ONE 6, e20113 10.1371/journal.pone.0020113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McGuire M. K., Meehan C. L., McGuire M. A., Williams J. E., Foster J., Sellen D. W., Kamau-Mbuthia E. W., Kamundia E. W., Mbugua S., Moore S. E., Prentice A. M., Kvist L. J., Otoo G. E., Brooker S. L., Price W. J., et al. (2017) What's normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am. J. Clin. Nutr. 105, 1086–1100 10.3945/ajcn.116.139980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hebert L. F. Jr., Daniels M. C., Zhou J., Crook E. D., Turner R. L., Simmons S. T., Neidigh J. L., Zhu J. S., Baron A. D., and McClain D. A. (1996) Overexpression of glutamine:fructose-6-phosphate amidotransferase in transgenic mice leads to insulin resistance. J. Clin. Invest. 98, 930–936 10.1172/JCI118876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bosch A. M. (2011) Classic galactosemia: dietary dilemmas. J. Inherit. Metab. Dis. 34, 257–260 10.1007/s10545-010-9157-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marquardt T., Lühn K., Srikrishna G., Freeze H. H., Harms E., and Vestweber D. (1999) Correction of leukocyte adhesion deficiency type II with oral fucose. Blood 94, 3976–3985 10.1182/blood.V94.12.3976.424k06_3976_3985 [DOI] [PubMed] [Google Scholar]
- 28. Röhrig C. H., Choi S. S., and Baldwin N. (2017) The nutritional role of free sialic acid, a human milk monosaccharide, and its application as a functional food ingredient. Crit. Rev. Food Sci. Nutr. 57, 1017–1038 10.1080/10408398.2015.1040113 [DOI] [PubMed] [Google Scholar]
- 29. Bardor M., Nguyen D. H., Diaz S., and Varki A. (2005) Mechanism of uptake and incorporation of the non-human sialic acid N-glycolylneuraminic acid into human cells. J. Biol. Chem. 280, 4228–4237 10.1074/jbc.M412040200 [DOI] [PubMed] [Google Scholar]
- 30. Tangvoranuntakul P., Gagneux P., Diaz S., Bardor M., Varki N., Varki A., and Muchmore E. (2003) Human uptake and incorporation of an immunogenic nonhuman dietary sialic acid. Proc. Natl. Acad. Sci. U.S.A. 100, 12045–12050 10.1073/pnas.2131556100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Banda K., Gregg C. J., Chow R., Varki N. M., and Varki A. (2012) Metabolism of vertebrate amino sugars with N-glycolyl groups: mechanisms underlying gastrointestinal incorporation of the non-human sialic acid xeno-autoantigen N-glycolylneuraminic acid. J. Biol. Chem. 287, 28852–28864 10.1074/jbc.M112.364182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lacomba R., Salcedo J., Alegría A., Barberá R., Hueso P., Matencio E., and Lagarda M. J. (2011) Sialic acid (N-acetyl and N-glycolylneuraminic acid) and ganglioside in whey protein concentrates and infant formulae. Int. Dairy J. 21, 887–895 10.1016/j.idairyj.2011.05.008 [DOI] [Google Scholar]
- 33. Padler-Karavani V., Yu H., Cao H., Chokhawala H., Karp F., Varki N., Chen X., and Varki A. (2008) Diversity in specificity, abundance, and composition of anti-Neu5Gc antibodies in normal humans: Potential implications for disease. Glycobiology 18, 818–830 10.1093/glycob/cwn072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Taylor R. E., Gregg C. J., Padler-Karavani V., Ghaderi D., Yu H., Huang S., Sorensen R. U., Chen X., Inostroza J., Nizet V., and Varki A. (2010) Novel mechanism for the generation of human xeno-autoantibodies against the nonhuman sialic acid N-glycolylneuraminic acid. J. Exp. Med. 207, 1637–1646 10.1084/jem.20100575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Samraj A. N., Pearce O. M., Läubli H., Crittenden A. N., Bergfeld A. K., Banda K., Gregg C. J., Bingman A. E., Secrest P., Diaz S. L., Varki N. M., and Varki A. (2015) A red meat-derived glycan promotes inflammation and cancer progression. Proc. Natl. Acad. Sci. U.S.A. 112, 542–547 10.1073/pnas.1417508112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pham T., Gregg C. J., Karp F., Chow R., Padler-Karavani V., Cao H., Chen X., Witztum J. L., Varki N. M., and Varki A. (2009) Evidence for a novel human-specific xeno-auto-antibody response against vascular endothelium. Blood 114, 5225–5235 10.1182/blood-2009-05-220400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mahajan V. S., and Pillai S. (2016) Sialic acids and autoimmune disease. Immunol. Rev. 269, 145–161 10.1111/imr.12344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vicaretti S. D., Mohtarudin N. A., Garner A. M., and Zandberg W. F. (2018) Capillary electrophoresis analysis of bovine milk oligosaccharides permits an assessment of the influence of diet and the discovery of nine abundant sulfated analogues. J. Agric. Food Chem. 66, 8574–8583 10.1021/acs.jafc.8b01041 [DOI] [PubMed] [Google Scholar]
- 39. Bum-Erdene K., Leffler H., Nilsson U. J., and Blanchard H. (2016) Structural characterisation of human galectin-4 N-terminal carbohydrate recognition domain in complex with glycerol, lactose, 3′-sulfo-lactose, and 2′-fucosyllactose. Sci. Rep. 6, 20289 10.1038/srep20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bruehl R. E., Bertozzi C. R., and Rosen S. D. (2000) Minimal sulfated carbohydrates for recognition by L-selectin and the MECA-79 antibody. J. Biol. Chem. 275, 32642–32648 10.1074/jbc.M001703200 [DOI] [PubMed] [Google Scholar]
- 41. Chen Y., Pan L., Liu N., Troy F. A., and Wang B. (2014) LC-MS/MS quantification of N-acetylneuraminic acid, N-glycolylneuraminic acid and ketodeoxynonulosonic acid levels in the urine and potential relationship with dietary sialic acid intake and disease in 3-to 5-year-old children. Br. J. Nutr. 111, 332–341 10.1017/S0007114513002468 [DOI] [PubMed] [Google Scholar]
- 42. Wylie A. D., and Zandberg W. F. (2018) Quantitation of sialic acids in infant formulas by liquid chromatography-mass spectrometry: an assessment of different protein sources and discovery of new analogues. J. Agric. Food Chem. 66, 8114–8123 10.1021/acs.jafc.8b01042 [DOI] [PubMed] [Google Scholar]
- 43. Albrecht S., Schols H. A., Van Den Heuvel E. G. H. M., Voragen A. G., and Gruppen H. (2010) CE-LIF-MSn profiling of oligosaccharides in human milk and feces of breast-fed babies. Electrophoresis 31, 1264–1273 10.1002/elps.200900646 [DOI] [PubMed] [Google Scholar]
- 44. Totten S. M., Zivkovic A. M., Wu S., Ngyuen U., Freeman S. L., Ruhaak L. R., Darboe M. K., German J. B., Prentice A. M., and Lebrilla C. B. (2012) Comprehensive profiles of human milk oligosaccharides yield highly sensitive and specific markers for determining secretor status in lactating mothers. J. Proteome Res. 11, 6124–6133 10.1021/pr300769g [DOI] [PubMed] [Google Scholar]
- 45. Schymanski E. L., Jeon J., Gulde R., Fenner K., Ruff M., Singer H. P., and Hollender J. (2014) Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environ. Sci. Technol. 48, 2097–2098 10.1021/es5002105 [DOI] [PubMed] [Google Scholar]
- 46. Guérardel Y., Morelle W., Plancke Y., Lemoine J., and Strecker G. (1999) Structural analysis of three sulfated oligosaccharides isolated from human milk. Carbohydr. Res. 320, 230–238 10.1016/S0008-6215(99)00153-6 [DOI] [PubMed] [Google Scholar]
- 47. Suzuki A., Hiraoka N., Suzuki M., Angata K., Misra A. K., McAuliffe J., Hindsgaul O., and Fukuda M. (2001) Molecular cloning and expression of a novel human β-Gal-3-O-sulfotransferase that acts preferentially on N-acetyllactosamine in N- and O-glycans. J. Biol. Chem. 276, 24388–24395 10.1074/jbc.M103135200 [DOI] [PubMed] [Google Scholar]
- 48. Torii T., Fukuta M., and Habuchi O. (2000) Sulfation of sialyl N-acetyllactosamine oligosaccharides and fetuin oligosaccharides by keratan sulfate Gal-6-sulfotransferase. Glycobiology 10, 203–211 10.1093/glycob/10.2.203 [DOI] [PubMed] [Google Scholar]
- 49. Kim H., Kang S., Jung B. M., Yi H., Jung J. A., and Chang N. (2017) Breast milk fatty acid composition and fatty acid intake of lactating mothers in South Korea. Br. J. Nutr. 117, 556–561 10.1017/S0007114517000253 [DOI] [PubMed] [Google Scholar]
- 50. Hasnain S. Z., Dawson P. A., Lourie R., Hutson P., Tong H., Grencis R. K., McGuckin M. A., and Thornton D. J. (2017) Immune-driven alterations in mucin sulphation is an important mediator of Trichuris muris helminth expulsion. PLoS Pathog. 13, e1006218 10.1371/journal.ppat.1006218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Corfield A. P., Myerscough N., Bradfield N., Corfield Cdo A., Gough M., Clamp J. R., Durdey P., Warren B. F., Bartolo D. C., King K. R., and Williams J. M. (1996) Colonic mucins in ulcerative colitis: evidence for loss of sulfation. Glycoconj. J. 13, 809–822 10.1007/BF00702345 [DOI] [PubMed] [Google Scholar]
- 52. Robbe C., Capon C., Maes E., Rousset M., Zweibaum A., Zanetta J. P., and Michalski J. C. (2003) Evidence of regio-specific glycosylation in human intestinal mucins: presence of an acidic gradient along the intestinal tract. J. Biol. Chem. 278, 46337–46348 10.1074/jbc.M302529200 [DOI] [PubMed] [Google Scholar]
- 53. Xu Z., and Knight R. (2015) Dietary effects on human gut microbiome diversity. Br. J. Nutr. 113, S1–S5 10.1017/S0007114514004127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hill D. R., Kessler S. P., Rho H. K., Cowman M. K., and de la Motte C. A. (2012) Specific-sized hyaluronan fragments promote expression of human β-defensin 2 in intestinal epithelium. J. Biol. Chem. 287, 30610–30624 10.1074/jbc.M112.356238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu G. D., Chen J., Hoffmann C., Bittinger K., Chen Y. Y., Keilbaugh S. A., Bewtra M., Knights D., Walters W. A., Knight R., Sinha R., Gilroy E., Gupta K., Baldassano R., Nessel L., et al. (2011) Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Marcobal A., Barboza M., Froehlich J. W., Block D. E., German J. B., Lebrilla C. B., and Mills D. A. (2010) Consumption of human milk oligosaccharides by gut-related microbes. J. Agric. Food Chem. 58, 5334–5340 10.1021/jf9044205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xu J., Bjursell M. K., Himrod J., Deng S., Carmichael L. K., Chiang H. C., Hooper L. V., and Gordon J. I. (2003) A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076 10.1126/science.1080029 [DOI] [PubMed] [Google Scholar]
- 58. Pacheco A. R., Curtis M. M., Ritchie J. M., Munera D., Waldor M. K., Moreira C. G., and Sperandio V. (2012) Fucose sensing regulates bacterial intestinal colonization. Nature 492, 113–117 10.1038/nature11623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schwab C., and Gänzle M. (2011) Lactic acid bacteria fermentation of human milk oligosaccharide components, human milk oligosaccharides and galactooligosaccharides. FEMS Microbiol. Lett. 315, 414–418 10.1111/j.1574-6968.2010.02185.x [DOI] [PubMed] [Google Scholar]
- 60. Quin C., Estaki M., Vollman D. M., Barnett J. A., Gill S. K., and Gibson D. L. (2018) Probiotic supplementation and associated infant gut microbiome and health: a cautionary retrospective clinical comparison. Sci. Rep. 8, 8283 10.1038/s41598-018-26423-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gorzelak M. A., Gill S. K., Tasnim N., Ahmadi-Vand Z., Jay M., and Gibson D. L. (2015) Methods for improving human gut microbiome data by reducing variability through sample processing and storage of stool. PLoS ONE 10, e0134802 10.1371/journal.pone.0134802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mopper K., and Gindler E. M. (1973) A new noncorrosive dye reagent for automatic sugar chromatography. Anal. Biochem. 56, 440–442 10.1016/0003-2697(73)90210-8 [DOI] [PubMed] [Google Scholar]
- 63. Subar A. F., Kirkpatrick S. I., Mittl B., Zimmerman T. P., Thompson F. E., Bingley C., Willis G., Islam N. G., Baranowski T., McNutt S., and Potischman N. (2012) The automated self-administered 24-Hour Dietary Recall (ASA24): a resource for researchers, clinicians, and educators from the National Cancer Institute. J. Acad. Nutr. Diet. 112, 1134–1137 10.1016/j.jand.2012.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kang J. X., and Wang J. (2005) A simplified method for analysis of polyunsaturated fatty acids. BMC Biochem. 6, 5 10.1186/1471-2091-6-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Estaki M., Pither J., Baumeister P., Little J. P., Gill S. K., Ghosh S., Ahmadi-Vand Z., Marsden K. R., and Gibson D. L. (2016) Cardiorespiratory fitness as a predictor of intestinal microbial diversity and distinct metagenomic functions. Microbiome 4, 42 10.1186/s40168-016-0189-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., and Glöckner F. O. (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1 10.1093/nar/gks808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Peña A. G., Goodrich J. K., Gordon J. I., Huttley G. A., Kelley S. T., Knights D., Koenig J. E., Ley R. E., et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Katoh K., and Standley D. M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Price M. N., Dehal P. S., and Arkin A. P. (2010) FastTree 2: approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Legendre P., and Gallagher E. D. (2001) Ecologically meaningful transformations for ordination of species data. Oecologia 129, 271–280 10.1007/s004420100716 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microbiome, HPLC-MS, and tandem MS data have been deposited to the publicly accessible repository Open Science Framework with the project ID 2xvgh.