Abstract
Diamond-Blackfan anemia (DBA) is a rare congenital bone marrow disorder with mutations in ribosomal protein genes. Several animal models have been developed to study the pathological mechanism of DBA. Previously, we reported that the complete knock-out of both Rpl5 and Rps24 alleles were lethal, while heterozygous Rpl5+/- and Rps24+/- mice showed normal phenotype. To establish a more efficient mouse model for mimicking DBA symptoms, we have taken advantage of RNAi technology to generate an inducible mouse model utilizing tetracycline-induced down-regulation of Rpl5. After two weeks of treatment with doxycycline in drinking water, a subset of treated shRNA Rpl5+/- adult mice developed mild anemia while control mice had normal complete blood counts. Similarly, treated shRNA Rpl5+/- mice developed reticulocytopenia and bone marrow erythroblastopenia. Detection of DBA symptoms in these mice make them a valuable DBA model for studying the pathological mechanism underlying DBA and for further assessment of the disease and drug testing for novel therapies.
Keywords: Diamond-Blackfan anemia, Ribosomal Protein L5, Rpl5-Inducible Mouse Model.
Diamond-Blackfan anemia (DBA) is a rare congenital bone marrow disorder inherited in an autosomal dominant pattern and resulting from haploinsufficiency of ribosomal proteins. It is characterized by macrocytic anemia, reticulocytopenia, and bone marrow erythroblastopenia as well as congenital malformations in about 50% of patients1.
The first major breakthrough in the molecular pathogenesis of DBA came from the discovery of the ribosomal protein S19 gene2, followed by identification of mutations in 24 additional ribosomal protein (RP) genes including RPL53. Additionally, mutations in two non-ribosomal protein genes, GATA1, encoding a critical transcription factor for red blood cell maturation, and TSR2, encoding a pre-RNA processing protein, have been reported in a subset of patients4-6. Furthermore, we reported a significant decrease in the expression level of GATA1 protein but not mRNA in primary hematopoietic cells from patients with mutations in RP genes6.
To address pathological mechanisms underlying DBA, several animal models have been generated. Homozygous Rps19-/- mice are embryonic lethal while heterozygotes had similar levels of RPS19 protein and mRNA as wild-type litermates7. We have reported that knocking-out Rpl5 and Rps24 alleles also leads to embryonic lethality, while heterozygous Rpl5+/- and Rps24+/- mice showed normal phenotypes at birth and throughout their development with no detectable differences between the expression levels of RPL5 and RPS24 mRNA and protein compared to those of wild-type mice8. Interestingly, a small number of these mice developed soft tissue sarcomas, also seen in some of patients with DBA8. Due to the severity of symptoms associated with RPL5 mutations in patients, we decided to focus our studies on the molecular mechanism of RPL5 deficiency induced postnataly3. Here, we report the generation of a conditional Rpl5 mouse line using an RNA interference (RNAi) approach. All animal studies were approved by Boston Children's Hospital's Institutional Animal Care and Use Committee.
A validated short-hairpin RNA (shRNA) construct targeting the mouse Rpl5 mRNA transcript was cloned into a TRE3G-based Col1A1-targeting vector (pColA1-TRE3G-GFP-miR30) and co-electroporated into pre-engineered KH2 embryonic stem cells, which contain a reverse tet-transactivator (rtTA) cassette integrated into the Rosa26 locus9. In parallel, we obtained an shRNA-luciferase (shRNA Renilla,shRNA Ren) mouse line to use as a non-specific shRNA control line. Both mouse lines were generated by Mirimus Laboratories (Mirimus Inc., Brooklyn, NY) on the C57BL/6 background.
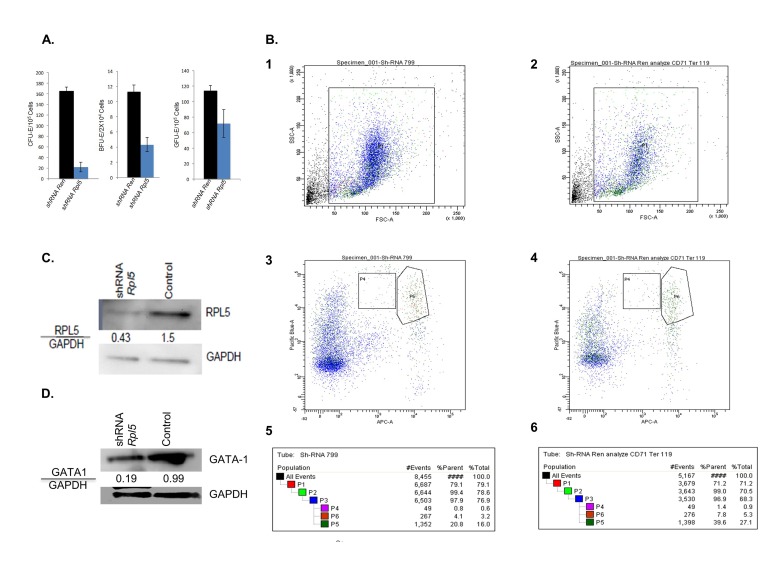
To assess the effects of Rpl5 down-regulation, mice either heterozygous or homozygous for the shRNA at the ColA1 and Rosa26 loci were generated. To increase the tetracycline effect for a stronger mRNA knockdown, we used mice that were heterozygous for ColA1 and homozygous for Rosa26, which would increase the production of reverse tetracycline transactivator and therefore, the effect of tetracycline treatment (as described above). Henceforth, we will refer to shRNA Rpl5+/-mice as shRNA Rpl5 and shRNA Ren+/- as control mice. Five to eight-week old female and male shRNA Rpl5mice were treated with 2mg/mL of doxycycline in drinking water10. After 2 weeks of treatment, a mild anemia was detected in about 20% of the treated shRNA Rpl5 mice while controlmice were normal (Table 1). Further hematological studies revealed marked reticulocytopenia in all treated shRNA Rpl5 mice (n=3) (reticulocytes 1.77%, 0.1x106/ul) versus controlmice (n=3) (reticulocytes 3.8%, 0.24x106/ul) and bone marrow erythroblastopenia (myeloid to erythroid linage 4.9 in all shRNA Rpl5 mice and 3.4 in control mice). To further investigate erythropoiesis in shRNA Rpl5 and control mice, methylcellulose colony assays11 were performed on bone marrow cells isolated from these mice to quantify the number of burst-forming unit- erythroid BFU-E and colony forming unit-erythroid CFU-E colonies as well as colony forming-unit granulocyte macrophage (CFU-GM). Our results showed a decrease in the number of BFU-E and CFU-E colonies in shRNA Rpl5 mice compared to control, which correlates to a decrease in the proliferation level of erythroid progenitor cells and a slight decrease of CFU-GM colonies (Figure 1A). We next performed flow cytometry on freshly isolated bone marrow cells from shRNA Rpl5and controlmice to compare the percentage of differentiated erythroid cells in each cell population. In our experiment, we examined two erythroid populations: less mature CD71highTer119med and more mature CD71highTer119high12. shRNA Rpl5 mice had lower numbers of CD71highTer119med (0.6% vs. 0.9%) and CD71highTer119high (3.2% vs. 5.3%) cells as compared to control mice (Figure 1B). GATA-1 is a necessary factor for the survival and terminal differentiation of erythroid progenitors13.
Table 1. Complete blood count in shRNA Rpl5 and shRNA Ren mice treated with doxycycline.
RBC: red blood cells (P=0.13); Hb: hemoglobin (P=0.0008); HTC: Hematocrit (P=0.117); MCV: corpuscular volume (P=0.3); WBC: white blood cell (P=0.46); NE: neutrophils (P=0.34); LY: lymphocytes (P=0.27); MO: monocytes (P=0.15); EO: eosinophils (P=0.45); BA: basophils (P=0.15); PLT: platelet count (P=0.92). Graphpad T test calculator website has been used to determine the P values based on standard division.
| Parameter (Units) | shRNA Rpl5 (Mean Result, n=3) | shRNA Ren (Mean Results, n=3) | Normal Range |
|---|---|---|---|
| RBC (M/µL) | 5.7 ±0.4 | 8.3±1.3 | 6.36-9.42 |
| Hb (g/dL) | 5.9±0.3 | 11.3±0.5 | 11.0-15.1 |
| HTC (%) | 26.8±1.9 | 41.8±7.3 | 35.1-45.4 |
| MCV (fl) | 47.3±0.9 | 50.3±2.3 | 45.4-60.3 |
| WBC (K/µL) | 6.0±2.7 | 12.6±7.6 | 1.8-10.7 |
| NE (K/µL) | 1.0±0.2 | 4.2±3.0 | 0.1-2.4 |
| LY (K/µL) | 2.9±0.8 | 7.4±3.4 | 0.9-9.3 |
| MO (K/µL) | 0.07±0.005 | 0.54±0.66 | 0.0-0.4 |
| EO (K/µL) | 0.02±0.01 | 0.38±0.43 | 0.0-0.2 |
| BA (K/µL) | 0.01±0.0 | 0.17±0.09 | 0.0-0.2 |
| PLT (K/µL) | 674.3±292.3 | 626.3±349.7 | 592-2972 |
Figure 1. A. Methylcellulose colony assay on bone marrow cells from doxycycline treated shRNA Rpl5 and control mice (n=3), (CFU-E, P=0.0003; BFU-E, P=1; GFU-GM, P=0.3); B. FACS analysis of bone marrow cells from shRNA Rpl5 and control mice. Total bone marrow cells from shRNA Rpl5 (B.1) and control (B.2) mice; viable cells were gated for analysis and sorting. We used the cell-surface markers CD71 (Y-axis) and Ter119 (X-axis) to separate mature erythroids and erythroid precursors. The most mature erythroblasts are CD71lowTer119high, while the erythroid precursors are CD71highTer119intermediate cells. In our FACS analysis the gated population comprises CD71highTer119med cells (P4) and CD71highTer119high cells (P5) from shRNA Rpl5 (B.3 and B.5) and control (B4 and B6) mice; the results here are collected from one mouse per group and are representative of 6 independent experiments; in each experiment, cells were collected from 1-3 mice for each group and sorted independently; C. Western Blot analysis of RPL5 protein expressions in CD71highTer119high cells. There was a significant decrease in the expression level of RPL5 in the treated shRNA Rpl5 compared to the control mice; D. Western Blot analysis of GATA1 protein expressions in CD71highTer119high cells. There was a significant decrease in the expression of GATA1 in the shRNA Rpl5 group compared to the control mice. In these experiments, GAPDH was used as a loading control, and they are representative of 3 independent experiments.
To assess expression levels of RPL5 and GATA1 proteins in shRNA Rpl5mice, western blots were performed on CD71highTer119high cell population total cell lysates using anti-RPL5 (Novus Biologic, NBP1-31413) or anti-GATA-1 antibodies (Sc-1234)8. These results demonstrated a significant decrease in expression of both RPL5 (Figure 1C) and GATA1 (Figure 1D) in cells from the treated shRNA Rpl5mice compared to control mice. On the other hand, quantitative PCR on two sorted erythroid populations showed similar levels of Gata1 mRNA expression in shRNA Rpl5and control mice (data not shown), as was shown in human cells6. The reduction of GATA1 protein is likely due to the reduction of RPL5, impaired ribosomal erythropoiesis, lower numbers of ribosomes, and altered GATA1 translation, as reported in human cells6,14.
In summary, we have generated and characterized a novel DBA mouse model, which allows an inducible and graded down-regulation of RpL5 gene expression. These mice recapitulate the major features of DBA including anemia, reticulocytopenia, and bone marrow erythroblastopenia1. Therefore, these shRNA Rpl5+/-mice may provide an effective approach for studying DBA and testing novel therapies.
Acknowledgments
These studies were funded in part by generous support from the Mauch Family, a Manton Center for Orphan Disease Research Junior Investigator Award, and by NIH R01HL107558 and NIH K02HL111156 to HTG. SK and HTG performed the experiments, analyzed the data, and wrote the manuscript. DY performed the experiments and edited the manuscript. MSA designed the technical aspects of the mouse line and edited the manuscript. AHB and HTG edited the manuscript and advised with experiments. We also thank Vasilis Toxavidis and John Tigges of the Beth Israel Deaconess Medical Center Flow Cytometry Core Facility for their help in sorting bone marrow cells.
Footnotes
Conflict of interests: The authors declare no competing financial interests.
Diamond-Blackfan Anemia (DBA); Colonyforming Unit-Erythroid (CFU-E); Colony-forming Unit-Granulocyte and Monocyte (CFU-GM); Burstforming Unit-Erythroid (BFU-E); Ribosomal Protein (RP).
DISCOVERIES is a peer-reviewed, open access, online, multidisciplinary and integrative journal, publishing high impact and innovative manuscripts from all areas related to MEDICINE, BIOLOGY and CHEMISTRY
References
- 1.Critical Issues in Diamond-Blackfan Anemia and Prospects for Novel Treatment. Li Hojun, Lodish Harvey F., Sieff Colin A. Hematology/Oncology Clinics of North America. 2018;32(4):701-712. doi: 10.1016/j.hoc.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Draptchinskaia Natalia, Gustavsson Peter, Andersson Björn, Pettersson Monica, Willig Thiébaut-Noël, Dianzani Irma, Ball Sarah, Tchernia Gil, Klar Joakim, Matsson Hans, Tentler Dimitri, Mohandas Narla, Carlsson Birgit, Dahl Niklas. Nature Genetics. 1999;21(2):169-175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 3.The Genetic Landscape of Diamond-Blackfan Anemia. Ulirsch JC, Verboon JM, Kazerounian S, Guo MH, Yuan D, Ludwig LS. Am J Hum Genet. 2018;103(6):930–947. doi: 10.1016/j.ajhg.2018.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. Sankaran Vijay G., Ghazvinian Roxanne, Do Ron, Thiru Prathapan, Vergilio Jo-Anne, Beggs Alan H., Sieff Colin A., Orkin Stuart H., Nathan David G., Lander Eric S., Gazda Hanna T. Journal of Clinical Investigation. 2012;122(7):2439-2443. doi: 10.1172/JCI63597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diamond-Blackfan anemia with mandibulofacial dystostosis is heterogeneous, including the novel DBA genesTSR2andRPS28. Gripp Karen W., Curry Cynthia, Olney Ann Haskins, Sandoval Claudio, Fisher Jamie, Chong Jessica Xiao-Ling, Pilchman Lisa, Sahraoui Rebecca, Stabley Deborah L., Sol-Church Katia. American Journal of Medical Genetics Part A. 2014;164(9):2240-2249. doi: 10.1002/ajmg.a.36633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altered translation of GATA1 in Diamond-Blackfan anemia. Ludwig Leif S, Gazda Hanna T, Eng Jennifer C, Eichhorn Stephen W, Thiru Prathapan, Ghazvinian Roxanne, George Tracy I, Gotlib Jason R, Beggs Alan H, Sieff Colin A, Lodish Harvey F, Lander Eric S, Sankaran Vijay G. Nature Medicine. 2014;20(7):748-753. doi: 10.1038/nm.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Targeted Disruption of the Ribosomal Protein S19 Gene Is Lethal Prior to Implantation. Matsson H., Davey E. J., Draptchinskaia N., Hamaguchi I., Ooka A., Leveen P., Forsberg E., Karlsson S., Dahl N. Molecular and Cellular Biology. 2004;24(9):4032-4037. doi: 10.1128/MCB.24.9.4032-4037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Development of Soft Tissue Sarcomas in Ribosomal Proteins L5 and S24 Heterozygous Mice. Kazerounian Shideh, Ciarlini Pedro D.S.C., Yuan Daniel, Ghazvinian Roxanne, Alberich-Jorda Meritxell, Joshi Mugdha, Zhang Hong, Beggs Alan H., Gazda Hanna T. Journal of Cancer. 2016;7(1):32-36. doi: 10.7150/jca.13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.A Rapid and Scalable System for Studying Gene Function in Mice Using Conditional RNA Interference. Premsrirut Prem K., Dow Lukas E., Kim Sang Yong, Camiolo Matthew, Malone Colin D., Miething Cornelius, Scuoppo Claudio, Zuber Johannes, Dickins Ross A., Kogan Scott C., Shroyer Kenneth R., Sordella Raffaella, Hannon Gregory J., Lowe Scott W. Cell. 2011;145(1):145-158. doi: 10.1016/j.cell.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mice with ribosomal protein S19 deficiency develop bone marrow failure and symptoms like patients with Diamond-Blackfan anemia. Jaako Pekka, Flygare Johan, Olsson Karin, Quere Ronan, Ehinger Mats, Henson Adrianna, Ellis Steven, Schambach Axel, Baum Christopher, Richter Johan, Larsson Jonas, Bryder David, Karlsson Stefan. Blood. 2011;118(23):6087-6096. doi: 10.1182/blood-2011-08-371963. [DOI] [PubMed] [Google Scholar]
- 11.Defective Ribosomal Protein Gene Expression Alters Transcription, Translation, Apoptosis, and Oncogenic Pathways in Diamond-Blackfan Anemia. Gazda Hanna T., Kho Alvin T., Sanoudou Despina, Zaucha Jan M., Kohane Isaac S., Sieff Colin A., Beggs Alan H. Stem Cells. 2006;24(9):2034-2044. doi: 10.1634/stemcells.2005-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Identification and Analysis of Mouse Erythroid Progenitors using the CD71/TER119 Flow-cytometric Assay. Koulnis Miroslav, Pop Ramona, Porpiglia Ermelinda, Shearstone Jeffrey R., Hidalgo Daniel, Socolovsky Merav. Journal of Visualized Experiments. 2011;(54) doi: 10.3791/2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Pevny Larysa, Simon M. Celeste, Robertson Elizabeth, Klein William H., Tsai Shih-Feng, D'Agati Vivette, Orkin Stuart H., Costantini Frank. Nature. 1991;349(6306):257-260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 14.Ribosome Levels Selectively Regulate Translation and Lineage Commitment in Human Hematopoiesis. Khajuria Rajiv K., Munschauer Mathias, Ulirsch Jacob C., Fiorini Claudia, Ludwig Leif S., McFarland Sean K., Abdulhay Nour J., Specht Harrison, Keshishian Hasmik, Mani D.R., Jovanovic Marko, Ellis Steven R., Fulco Charles P., Engreitz Jesse M., Schütz Sabina, Lian John, Gripp Karen W., Weinberg Olga K., Pinkus Geraldine S., Gehrke Lee, Regev Aviv, Lander Eric S., Gazda Hanna T., Lee Winston Y., Panse Vikram G., Carr Steven A., Sankaran Vijay G. Cell. 2018;173(1):90-103.e19. doi: 10.1016/j.cell.2018.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]