Abstract
Deregulation of microRNAs (miRs) has been demonstrated to contribute to the development and malignant progression of nasopharyngeal carcinoma (NPC). Recently, miR-93 was reported to be significantly upregulated in NPC tissues and cell lines, and promote the proliferation, migration and invasion of NPC cells in vitro, as well as tumor growth in vivo. However, whether there is any clinical value of serum miR-93 expression in NPC still remains unclear. Therefore, the present study aimed to explore the clinical significance of serum miR-93 expression in NPC. A total of 85 serum samples from NPC patients and 30 from healthy controls were collected. Reverse transcription-quantitative polymerase chain reaction data demonstrated that the serum expression of miR-93 was significantly increased in NPC patients, when compared with those in healthy controls. Following receiving chemo-radiotherapy, the serum miR-93 levels were significantly decreased in NPC patients. Furthermore, the increased serum levels of miR-93 were significantly associated with advanced grade, clinical stage, lymph node metastasis, as well as worse 5-year overall survival of NPC patients. Furthermore, the serum miR-93 expression was demonstrated to be an independent factor for predicating the prognosis of NPC. In vitro experiments demonstrated that knockdown of miR-93 caused a decrease in NPC cell proliferation, whereas overexpression of miR-93 promoted NPC cell proliferation. PDCD4 was then identified as a direct target of miR-93 in NPC cells. Overexpression of PDCD4 significantly eliminated the promoting effects of miR-93 overexpression on NPC cell proliferation. Taken together, these findings suggest that the serum miR-93 expression could be used as a predicator for the clinical outcome of NPC patients, and suggest that miR-93 may also become a potential therapeutic target for NPC treatment.
Keywords: nasopharyngeal carcinoma, microRNA-93, serum, prognosis, proliferation, PDCD4
Introduction
Nasopharyngeal carcinoma (NPC) is rare in America and Europe, but a common head and neck cancer in Southeast Asia, particularly in Southern China, with an incidence rate <0.03% (1,2). Genetic susceptibility and Epstein-Barr virus (EBV) infection may contribute to the development of NPC (2-4). In early stages of NPC, there is no obvious clinical manifestation, and the location of the tumor is hidden, thus it is difficult to detect (5,6). Although detection of serum EBV infection is the main clinical screening method at present, the sensitivity and specificity of diagnosis is not high (7). Therefore, exploring novel diagnostic and prognostic biomarkers for NPC is urgently needed.
microRNAs (miRNAs or miRs), are 22-25 nucleotide-long small non-coding RNAs, and are important regulators for gene expression via epigenetic mechanisms (8). They can inhibit gene expression at the post-transcriptional level, via directly binding to the 3'-untranslated region (UTR) of their target mRNAs, and thus lead to translation inhibition or mRNA degradation (8). Through mediation of various genes expression, miRs participate in a variety of biological processes, such as cell survival, proliferation, differentiation, apoptosis, migration and invasion (9). Furthermore, as many oncogenes or tumor suppressors are also their targets, miRs have been demonstrated to contribute to the development and progression of many types of human cancers, including NPC (10-12). For instance, the oncogenic miR-17-92 cluster and miR-155 are significantly upregulated in NPC, whereas the tumor suppressive miRs, including miR-34 family, miR-143 and miR-145, are significantly downregulated in NPC (10). In addition, miR-218 has been demonstrated to suppress NPC progression through inhibition of survivin and the slit guidance ligand 2-roundabout guidance receptor 1 pathway (11). miR-378 is able to promote cell proliferation, colony formation, migration and invasion of NPC cells in vitro, and tumor growth in vivo (13).
miR-93 has been demonstrated to be frequently deregulated and serve key roles in certain common human cancers, such as lung cancer, gastric cancer, glioma, esophageal cancer and osteosarcoma (14-18). Qu et al (14) recently demonstrated that miR-93 promoted TGF-β-induced epithelial-to-mesenchymal transition (EMT) in lung cancer cells through targeting neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase. It can also promote the proliferation of osteosarcoma cells via targeting of phosphatase and tensin homolog (17). Recently, miR-93 was demonstrated to be significantly upregulated in NPC tissues and cell lines, and to promote the proliferation, migration and invasion of NPC cells in vitro, as well as tumor growth in vivo (19,20). However, little is currently known about the clinical significance of the serum miR-93 in NPC, and the molecular targets of miR-93 in NPC cells should also be revealed.
Therefore, the present study aimed to explore the clinical significance of serum miR-93 levels in NPC, as well as its value for predicating the prognosis of NPC patients. In addition, the regulatory mechanism of miR-93 in NPC cell proliferation was also evaluated.
Materials and methods
Ethics approval and clinical samples collection
The present study was approved by the Ethics Committee of First Affiliated Hospital of Xinjiang Medical University (Urumqi, China). Serum samples from 85 NPC patients (age range, 32-66 years; male, 48; female, 37) and 30 healthy controls (age range, 29-61 years; male, 18; female, 12) were collected at the Department of Otolaryngology, First Affiliated Hospital of Xinjiang Medical University.
Differentiation grade, lymph node metastasis, T stage, advanced clinical stage and Epstein-Barr virus (EBV) infection were recorded (21). All patients were treated by radiotherapy and serum samples were collected at 1 day prior to and 1 day following radiotherapy. Blood samples were collected (10 ml) and serum was obtained by centrifugation (1,500 x g; 5 min; 4˚C). All NPC patients and healthy controls recruited for the present study did not receive any therapy prior to blood collection. The serum samples were stored at -70˚C prior to use. The clinical characteristics of NPC patients are presented in Table I. Written informed consent was obtained from all patients. Patient follow-ups were conducted and no patients were lost during the 60 months follow-up period.
Table I.
Association between serum miR-93 levels and clinicopathological characteristics of patients with nasopharyngeal carcinoma.
| Variable | Total | Low miR-93 | High miR-93 | P-value |
|---|---|---|---|---|
| Age, years | ||||
| <55 | 39 | 23 | 15 | 0.511 |
| ≥55 | 46 | 24 | 22 | |
| Gender | ||||
| Male | 48 | 28 | 20 | 0.660 |
| Female | 37 | 19 | 18 | |
| Grade | ||||
| G1-2 | 48 | 33 | 15 | 0.008 |
| G3 | 37 | 14 | 23 | |
| T stage | ||||
| T1-2 | 45 | 28 | 17 | 0.196 |
| T3-4 | 40 | 19 | 21 | |
| Lymph node metastasis | ||||
| No | 50 | 35 | 15 | 0.002 |
| Yes | 35 | 12 | 23 | |
| Clinical stage | ||||
| I-II | 45 | 33 | 12 | 0.001 |
| III-IV | 40 | 14 | 26 | |
| EBV infection | ||||
| No | 14 | 9 | 5 | 0.563 |
| Yes | 71 | 38 | 33 |
Data are presented as the number of patients (n). High miR-93, ≥3.88 and low miR-93 <3.88. EBV, Epstein-Barr virus.
Cell culture
NPC cell line C666-1 and a normal nasopharyngeal epithelial cell line NP69 were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco's modified Eagle's medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) in a 37˚C humidified atmosphere of 5% CO2.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissue and cell samples using a QIAamp RNA Blood kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's protocol. RNA was then reverse transcribed into cDNA using a miRNA Reverse Transcription Kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. qPCR was then performed to detect the miR-93 expression on an ABI 7300 Plus Thermocycler (Applied Biosystems; Thermo Fisher Scientific, Inc.) using a miRNA Q-PCR Detection kit (GeneCopoeia, Inc., Rockville, MD, USA), according to the manufacturer's protocol. U6 gene was used as an internal reference. The PCR conditions were 95˚C for 5 min, followed by 45 cycles of 95˚C for 30 sec and 60˚C for 30 sec. Each sample was examined in triplicate. The relative expression was analyzed by the 2-ΔΔCq method (22). Primers for miR-93 and U6 were purchased from Guangzhou Fulengen Co., Ltd. (Guangzhou, China) with the following sequences: miR-93, forward, 5'-AGGCCCAAAGTGCTGTTCGT-3' and reverse, 5'-GTGCAGGGTCCGAGGT-3'; and U6, forward, 5'-CTCGCTTCGGCAGCACA-3' and reverse, 5'-AACGCTTCACGAATTTGCGT-3'.
Cell transfection
For transfection, C666-1 cells were cultured as described to 70% confluence and transfected with 100 nM miR-93 inhibitor (cat. no., HmiR-AN0836-SN-10), negative control (NC) inhibitor (cat. no., CmiR-AN0001-SN), miR-93 mimic (cat. no., HmiR0110-MR04) or scramble miR mimic (miR-NC; cat. no., CmiR0001-MR04; all Guangzhou FulenGen Co., Ltd., Guangzhou, China), or co-transfected with miR-93 mimic and 200 ng pcDNA3.1-programmed cell death 4 (PDCD4) open reading frame plasmid (www.amspring.com; Amspring, Changsha, China), using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Following experiments were conducted following 48 h.
MTT assay
Cells (5x104) were seeded in 96-well plates and 100 µl of fresh serum-free medium containing 0.5 g/l MTT (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) were added to each well. Following incubation at 37˚C for 0, 24, 48 and 72 h, medium was removed, 50 µl of dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA) was added and samples were incubated at 37˚C for 10 min. The absorbance at 570 nm was recorded.
Bioinformatics analysis and Dual Luciferase reporter assay
Bioinformatics analysis software, including TargetScanHuman 7.2 (www.targetscan.org), miRanda 1.0 (www.microrna.org) and PicTar 1.0 (pictar.mdc-berlin.de) were used to predict the putative target genes of miR-93 according to the manufacturers' protocols. Amspring provided the wild type (WT)-PDCD4 and mutant type (MT)-PDCD4 reporter plasmid. The WT or MT of PDCD4 3'-UTR were constructed by PCR and a Quick-Change Site-Directed Mutagenesis kit (Agilent Technologies, Inc., Santa Clara, CA, USA), which was then inserted into the multiple cloning sites in the psiCHECK™2 vector (Promega Corporation, Madison, WI, USA). C666-1 cells were cultured as described to 70% confluence in a 24-well plate, and co-transfected with 100 ng WT-PDCD4 or MT-PDCD4 vector, and 50 nM miR-93 mimics or scramble miR (NC) using Lipofectamine 2000. The xDual-Luciferase® Reporter Assay system (Promega Corporation) was used to determine the luciferase activities at 48 h following transfection. Renilla luciferase activity was normalized to firefly luciferase activity.
Western blotting
Cells were solubilized in cold radioimmunoprecipitation assay lysis buffer (Thermo Fisher Scientific, Inc.) to extract protein. The protein concentration was determined using a bicinchoninic acid Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Protein (50 µg) was separated by 10% SDS-PAGE (Pierce; Thermo Fisher Scientific, Inc.), and transferred onto a polyvinylidene difluoride membrane (Thermo Fisher Scientific, Inc.). Following incubation with PBS containing 5% non-fat milk at 4˚C overnight, the membrane was incubated with rabbit anti-human PDCD4 primary antibody (1:50; ab80590; Abcam, Cambridge, MA, USA) or rabbit anti-human GAPDH primary antibody (1:100; ab9485; Abcam), respectively, at room temperature for 3 h. Following washing with PBS-Tween-20 for 15 min, the membrane was incubated with the horseradish peroxidase-conjugated mouse anti-rabbit secondary antibody (1:10,000; ab99697; Abcam) at room temperature for 1 h. Chemiluminescent detection was conducted using an Novex™ ECL Chemiluminescent Substrate Reagent kit (Pierce; Thermo Fisher Scientific, Inc). The protein expression was analyzed using Image-Pro Plus software 6.0 (Media Cybernetics, Inc., Rockville, MD, USA), represented as the density ratio vs. GAPDH.
Statistical analyses
Data in the present study are expressed as the mean ± standard deviation. The differential expression of serum miR-93 between NPC patients and healthy controls was analyzed using an independent sample t-test, whereas the differential expression of serum miR-93 between pre-treated and post-treated NPC patients was analyzed using a paired Student's t-test. The clinical association between serum miR-93 expression and clinical variables was analyzed using a χ² test. Kaplan-Meier method was used to analyze the association between serum miR-93 expression and survival time. Univariate and multivariate analysis were used to identify the independent predicators for the prognosis of NPC patients. The statistical association of data between groups was analyzed by Student's t-test or one-way analysis of variance. SPSS 19.0 software (IBM Corp., Armonk, NY, USA) was used to perform the statistical analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Upregulation of serum miR-93 expression in NPC patients
miR-93 has been reported to be significantly upregulated in NPC tissues and cell lines (19,20); however, whether the serum expression of miR-93 is also upregulated in NPC still remains unclear. To explore the clinical significance of serum miR-93 expression in NPC, RT-qPCR was used to detect the serum miR-93 expression in 85 NPC patients and 30 healthy controls. As indicated in Fig. 1, the serum miR-93 expression was significantly increased in NPC patients compared with healthy controls.
Figure 1.
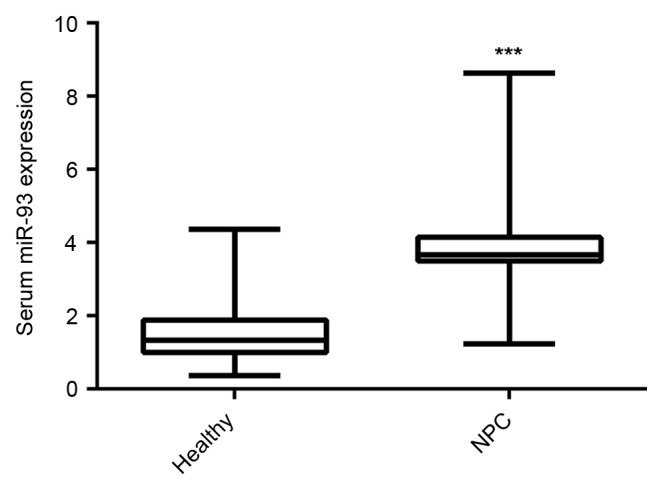
Reverse transcription-quantitative polymerase chain reaction was used to detect the serum miR-93 expression in NPC patients and healthy controls. ***P<0.001 vs. healthy. miR, microRNA; NPC, nasopharyngeal carcinoma.
Chemo-radiotherapy reduces serum miR-93 expression in NPC patients
Radiotherapy is a conventional treatment strategy for NPC, and all of the patients recruited for the present study received radiotherapy. Serum samples were collected from the NPC patients following treatment, and the serum miR-93 levels were compared between the NPC patients prior to and following treatment. The data demonstrated that following receiving the standard chemo-radiotherapy, the serum miR-93 expression was significantly decreased (Fig. 2).
Figure 2.
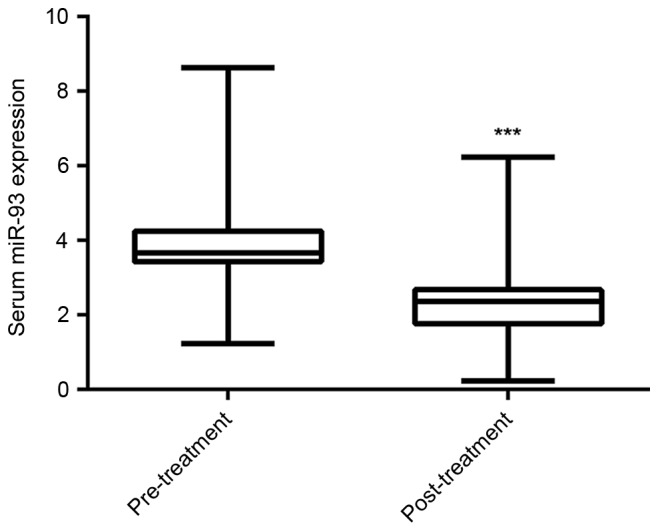
Reverse transcription-quantitative polymerase chain reaction was used to detect the serum miR-93 expression in nasopharyngeal carcinoma patients prior to and following receiving chemo-radiotherapy. ***P<0.001 vs. pre-treatment. miR, microRNA.
Increased serum miR-93 expression is associated with the malignant progression of NPC
Subsequently, the clinical association between serum miR-93 expression and clinical characteristics in NPC patients was analyzed. The mean expression (3.88±1.75) of serum miR-93 was defined as a cut-off point and NPC patients were divided into two groups, high serum miR-93 group (n=48) and low serum miR-93 group (n=47). A χ² test demonstrated that high serum miR-93 expression was significantly associated with a poor differentiation grade, lymph node metastasis and advanced clinical stage, but not associated with age, gender, T stage, and EBV infection (Table I). These findings suggest that increased serum levels of miR-93 are significantly associated with the malignant progression of NPC.
Increased serum miR-93 expression is associated with poor survival time of NPC patients
The clinical association between serum miR-93 expression and survival time of NPC patients was further evaluated using the Kaplan-Meier method. The data indicated that those NPC patients with higher serum miR-93 levels exhibited a shorter overall survival time, when compared with those with lower serum miR-93 expression (Fig. 3). Accordingly, the increased serum miR-93 expression was associated with worse prognosis of NPC patients.
Figure 3.
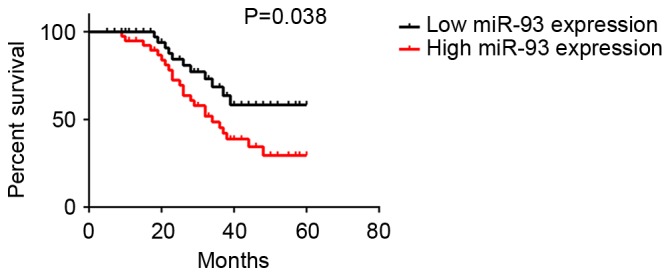
The association between serum miR-93 expression and 5-year overall survival rate of nasopharyngeal carcinoma patients. miR, microRNA.
Serum miR-93 expression is an independent factor for predicating the prognosis of NPC patients
The potential factors that may predicate the prognosis were further analyzed using univariate and multivariate analyses. As indicated in Table II, several clinical factors including grade, lymph node metastasis, clinical stage, and serum miR-93 expression were significantly associated with the survival time. Furthermore, multivariate analysis indicated that serum miR-93 expression level, grade, lymph node metastasis, and clinical stage were independent factors for predicating the prognosis of NPC patients (Table III).
Table II.
Univariate analysis of prognostic factors of nasopharyngeal carcinoma.
| Variable | Hazard ratio | P-value |
|---|---|---|
| Age, years (≥55/<55) | 1.15 | 0.802 |
| Gender (male/female) | 1.11 | 0.844 |
| Grade (G3/G1-2) | 2.84 | 0.017 |
| T stage (T3-4/T1-2) | 1.56 | 0.186 |
| Lymph node metastasis (yes/no) | 3.12 | 0.008 |
| Clinical stage (III-IV/I-II) | 4.32 | 0.001 |
| EBV infection (yes/no) | 1.36 | 0.543 |
| Serum miR-93 levels (high/low) | 2.95 | 0.011 |
EBV, Epstein-Barr virus; miR, microRNA.
Table III.
Multivariate analysis of independent prognostic factors of nasopharyngeal carcinoma.
| Variable | Hazard ratio | P-value |
|---|---|---|
| Grade | 2.03 | 0.043 |
| Lymph node metastasis | 2.57 | 0.028 |
| Clinical stage | 3.93 | 0.001 |
| Serum miR-93 levels | 2.78 | 0.019 |
miR, microRNA.
miR-93 promotes NPC cell proliferation
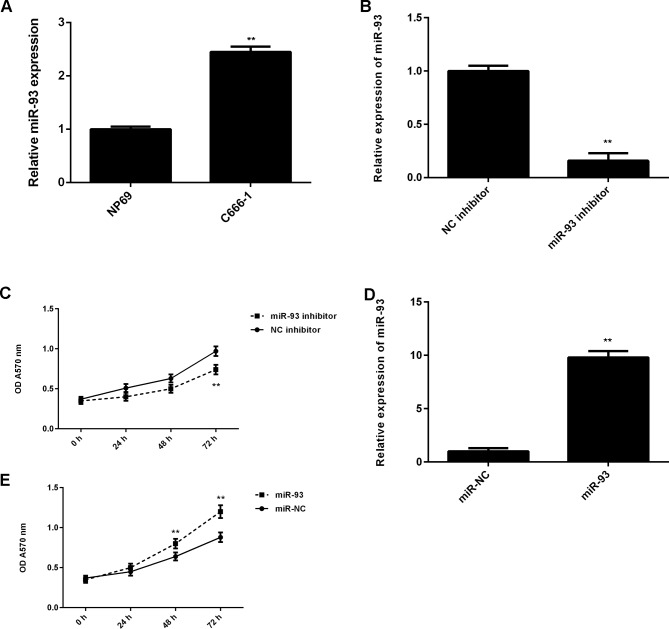
These findings suggest that miR-93 may serve a promoting role in NPC progression. Subsequently, the expression levels of miR-93 in NPC cell line C666-1 were evaluated, and the normal nasopharyngeal epithelial cell line NP69 was used as a control. As presented in Fig. 4A, qPCR data indicated that miR-93 was also significantly upregulated in C666-1 cells compared with NP69 cells. C666-1 cells were then transfected with miR-93 inhibitor or NC inhibitor as a control. As indicated in Fig. 4B, transfection with miR-93 inhibitor caused a significant reduction in miR-93 levels, when compared with the NC inhibitor group. MTT assay data further demonstrated that knockdown of miR-93 significantly decreased the proliferation of C666-1 cells, when compared with the NC inhibitor group (Fig. 4C). To further support these data, C666-1 cells were transfected with miR-93 mimic or miR-NC as the control group. Following transfection, the miR-93 levels were significantly increased in the miR-93 group compared with the miR-NC group (Fig. 4D). Furthermore, overexpression of miR-93 significantly promoted C666-1 cell proliferation (Fig. 4E). These findings suggest that miR-93 serves a promoting role in NPC growth.
Figure 4.
(A) RT-qPCR was used to detect the miR-93 levels in NPC cell line C666-1 compared with the normal nasopharyngeal epithelial cell line NP69. **P<0.01 vs. NP69. (B) RT-qPCR was used to detect the miR-93 levels in C666-1 cells transfected with negative control (NC) inhibitor or miR-93 inhibitor, and (C) MTT assay was used to determine the cell proliferation. **P<0.01 vs. NC inhibitor. (D) RT-qPCR was used to detect the miR-93 levels in C666-1 cells transfected with miR-NC or miR-93 mimic, and (E) MTT assay was used to determine the cell proliferation. **P<0.01 vs. miR-NC. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; miR, microRNA; NPC, nasopharyngeal carcinoma; NC, negative control; miR-NC, scramble miR; OD, optical density.
PDCD4 is a target gene of miR-93 in NPC cells
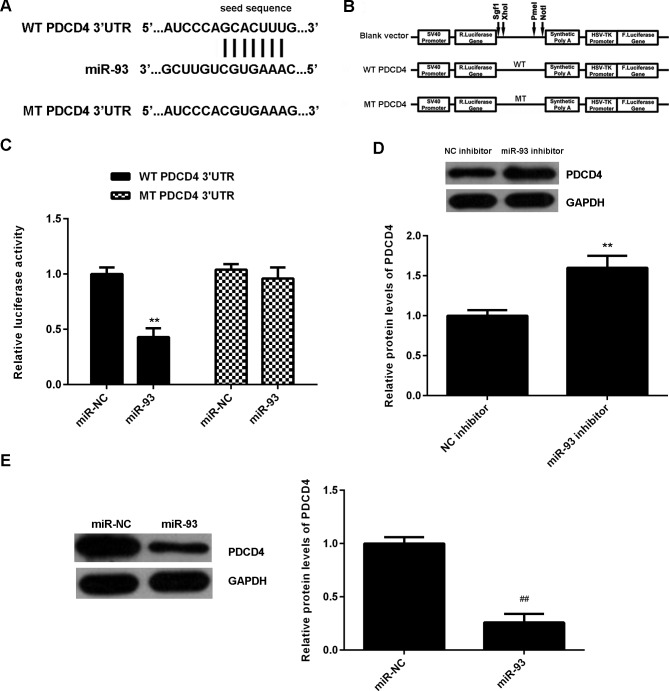
The potential target of miR-93 in NPC was then evaluated. Bioinformatics analysis indicated that PDCD4 was a target gene of miR-93 (Fig. 5A). To confirm this targeting association, luciferase vectors containing WT or MT PDCD4 3'-UTR were generated (Fig. 5A and B). Luciferase reporter assay data indicated that the luciferase activity was significantly reduced in C666-1 cells co-transfected with the WT-PDCD4 luciferase reporter vector and miR-93 mimics, when compared with the control group (Fig. 5C). However, these effects were eliminated by transfection with MT-PDCD4 luciferase reporter vector. Therefore, miR-93 was demonstrated to directly bind to the 3'UTR of PDCD4 mRNA. The effects of miR-93 on PDCD4 protein expression were further determined. Western blotting data demonstrated that knockdown of miR-93 significantly increased the protein level of PDCD4 in C666-1 cells (Fig. 5D). Conversely, overexpression of miR-93 significantly reduced the PDCD4 protein expression in C666-1 cells (Fig. 5E). Accordingly, miR-93 negatively regulates the protein expression of PDCD4 via directly binding to the 3'UTR of its mRNA in C666-1 cells.
Figure 5.
(A and B) The luciferase reporter vectors containing WT or MT of the PDCD4 3'-UTR were generated. (C) The luciferase activity was significantly reduced in C666-1 cells co-transfected with the WT-PDCD4-3'UTR luciferase reporter vector and miR-93 mimics, when compared with the control group. However, these effects were eliminated by transfection with MT-PDCD4-3'UTR luciferase reporter vector. **P<0.01 vs. miR-NC. (D and E) Western blotting was used to examine the protein levels of PDCD4 in C666-1 cells transfected with NC inhibitor, miR-93 inhibitor, miR-NC or miR-93 mimic, respectively. **P<0.01 vs. NC inhibitor; ##P<0.01 vs. miR-NC. WT, wild type; MT, mutant type; UTR, untranslated region; miR, microRNA; NC, negative control; miR-NC, scramble miR.
PDCD4 acts as a downstream effector in miR-93-mediated NPC cell proliferation
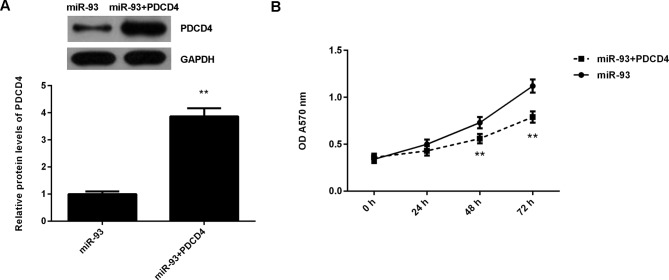
It was then speculated that the promoting effects of miR-93 on NPC cell proliferation may be through negatively regulating PDCD4 expression. To clarify this speculation, C666-1 cells were co-transfected with miR-93 mimic and pcDNA3.1-PDCD4 expression plasmid. Transfection with miR-93 mimic was used as the control group. Following transfection, the protein expression levels of PDCD4 were examined. As demonstrated in Fig. 6A, the protein levels of PDCD4 were significantly higher in the miR-93+PDCD4 group when compared with those in the miR-93 group. MTT assay data further indicated that the proliferation of C666-1 cells was significantly reduced in the miR-93+PDCD4 group when compared with the miR-93 group (Fig. 6B). These findings suggest that miR-93 promotes NPC cell proliferation, at least partly, through directly targeting PDCD4.
Figure 6.
C666-1 cells were transfected with miR-93 mimic, or co-transfected with miR-93 mimic and pcDNA3.1-PDCD4 expression plasmid, respectively. (A) Western blotting was conducted to examine the protein levels of PDCD4. (B) MTT assay was used to determine cell proliferation. **P<0.01 vs. miR-93. miR, microRNA.
Discussion
Exploring novel biomarkers in body fluids shows promise for detecting and predicating the prognosis in NPC. For instance, serum cathepsin B and cathepsin D concentrations have been demonstrated to have a diagnostic value in NPC (23). In the present study, it was demonstrated that the serum expression of miR-93 was significantly increased in NPC patients compared with healthy controls, and was significantly decreased following receiving chemo-radiotherapy in NPC patients. Furthermore, the increased serum levels of miR-93 were significantly associated with the advanced grade, clinical stage, lymph node metastasis, and worse overall survival of NPC patients. Furthermore, the serum miR-93 expression was demonstrated to be an independent factor for predicating the prognosis of NPC. Furthermore, it was demonstrated that miR-93 served a promoting role in NPC cell proliferation through directly targeting PDCD4.
Accumulating evidence has demonstrated that multiple types of miRNA are deregulated in NPC and contribute to the malignant progression, and some of them may become promising biomarkers for the diagnosis and prognosis in NPC. For example, miR-205 promotes proliferation, migration and invasion of NPC cells by activation of AKT signaling, and decreased radiation-induced apoptosis (24). miR-1 suppresses tumor growth and metastasis in NPC by targeting K-ras (25). In the present study, it was demonstrated that the serum miR-93 levels were significantly higher in NPC patients compared with healthy controls. Consistent with the present findings, miR-93 has also been demonstrated to be significantly upregulated in NPC tissues and cell lines (19). In addition, miR-93 was also reported to promote cell growth and metastasis in NPC by targeting DAB2 and transforming growth factor β receptor 2 (TGFβR2) (19,20). Furthermore, it was demonstrated that the serum miR-93 expression in NPC patients was significantly decreased following receiving chemo-radiotherapy. Similar findings were also reported in miR-774(26), suggesting that both the serum miR-93 and miR-774 expression may become useful biomarkers for evaluating the treatment response.
miR-93 has been demonstrated to serve an oncogenic role in different cancer types. Increased miR-93 expression is associated with poor prognosis in head and neck squamous cell carcinoma (27). Du et al (28) previously reported that miR-93, together with miR-98 and miR-197, could directly target FUS1, an important tumor suppressor. miR-93 also promotes tumor growth, metastasis, and angiogenesis by inhibition of integrin-β8 and large tumor suppressor kinase 2 (29,30). Knockdown of miR-93 effectively decreased the proliferation and clone formation capacity of hepatocellular carcinoma cells (31). Conversely, a number of previous studies have demonstrated that miR-93 acted as a tumor suppressor in colon cancer. miR-93 was downregulated in colon cancer tissues compared with non-tumor colon tissues, and its decreased expression was significantly associated with unfavorable clinicopathological features and short overall survival in patients (32). Yu et al (33) previously demonstrated that miR-93 suppressed the proliferation and colony formation of human colon cancer stem cells. Tang et al (34) previously reported that miR-93 suppressed colorectal cancer development via inhibition of the Wnt/β-catenin pathway. Yang et al (35) previously demonstrated that miR-93 inhibits tumor growth and early relapse of human colorectal cancer by inhibiting the protein expression of cyclin B1, leading to cell cycle arrest in the G2 phase. Accordingly, the function of miR-93 is cancer-specific, likely as it regulates different target genes in different tumor microenvironments.
Further investigation demonstrated that the serum miR-93 expression was significantly associated with grade, lymph node metastasis, and clinical stage in NPC. Furthermore, those NPC patients with higher serum miR-93 expression demonstrated worse survival time compared with those with lower serum miR-93 expression. These findings suggest that the increased serum expression of miR-93 may become a potential biomarker for not only evaluating the malignant progression of NPC, but also predicating the prognosis of NPC patients. Indeed, the present multivariate analysis demonstrated that the serum miR-93 expression, as well as lymph node metastasis and clinical stage, were independent predicators for the prognosis in NPC.
Several previous studies have demonstrated that miR-93 had promoting effects on the malignant phenotypes of NPC cells in vitro. For instance, Xu et al (19) reported that miR-93 promotes NPC cell growth and invasion by targeting disabled homolog-2. Lyu et al (20) also demonstrated that miR-93, directly targets TGFβR2, thus promoting cell proliferation, invasion and metastasis in vitro and in vivo. In the present study, it was also demonstrated that miR-93 was significantly upregulated in NPC cells, and promoted NPC cell proliferation, consistent with the previous findings (19,20). Furthermore, PDCD4 was identified as a novel target of miR-93 in NPC cells. PDCD4, an important tumor suppressor, can bind to the eukaryotic translation initiation factor 4A1 and inhibit its function by preventing RNA binding (36,37). Recently, PDCD4 was demonstrated to serve a suppressive role in NPC. Zhen et al (36) previously demonstrated that PDCD4 modulated the miR-184-mediated C-MYC and B cell lymphoma 2 expression, decreasing NPC cell growth and survival. In addition, PDCD4 was also demonstrated to suppress cell growth, metastasis, and sensitizes NPC to cisplatin, through mediating the miR-374a-CCND1-pPI3K/AKT-c-JUN feedback loop (37). To the best of our knowledge, the present study demonstrated for the first time that miR-93 promoted NPC cell proliferation, partly at least, through directly targeting PDCD4, thus elucidating the role of miRNAs/PDCD4 signaling in NPC.
In conclusion, to the best of our knowledge, the present study demonstrated for the first time that increased serum levels of miR-93 are significantly associated with the advanced progression of NPC, as well as worse survival time of NPC patients. These findings suggest that the serum miR-93 expression may be used as a predicator for the clinical outcome of NPC patients, and it may also become a promising therapeutic target for NPC treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
JS collected clinical tissues and wrote the manuscript. HZ designed the study and revised the manuscript. JS and JY performed the experiments. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of First Affiliated Hospital of Xinjiang Medical University (Urumqi, China). Informed, written consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Wei WI, Sham JS. Nasopharyngeal carcinoma. Lancet. 2005;365:2041–2054. doi: 10.1016/S0140-6736(05)66698-6. [DOI] [PubMed] [Google Scholar]
- 3.Yip TT, Ngan RK, Fong AH, Law SC. Application of circulating plasma/serum EBV DNA in the clinical management of nasopharyngeal carcinoma. Oral Oncol. 2014;50:527–538. doi: 10.1016/j.oraloncology.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 4.Sze H, Blanchard P, Ng WT, Pignon JP, Lee AW. Chemotherapy for nasopharyngeal carcinoma-current recommendation and controversies. Hematol Oncol Clin North Am. 2015;29:1107–1122. doi: 10.1016/j.hoc.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Xiao L, Xiao T, Wang ZM, Cho WC, Xiao ZQ. Biomarker discovery of nasopharyngeal carcinoma by proteomics. Expert Rev Proteomics. 2014;11:215–225. doi: 10.1586/14789450.2014.897613. [DOI] [PubMed] [Google Scholar]
- 6.Lee AW, Lin JC, Ng WT. Current management of nasopharyngeal cancer. Semin Radiat Oncol. 2012;22:233–244. doi: 10.1016/j.semradonc.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Zeng X, Xiang J, Wu M, Xiong W, Tang H, Deng M, Li X, Liao Q, Su B, Luo Z, et al. Circulating miR-17, miR-20a, miR-29c, and miR-223 combined as non-invasive biomarkers in nasopharyngeal carcinoma. PLoS One. 2012;7(e46367) doi: 10.1371/journal.pone.0046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Chen HC, Chen GH, Chen YH, Liao WL, Liu CY, Chang KP, Chang YS, Chen SJ. MicroRNA deregulation and pathway alterations in nasopharyngeal carcinoma. Br J Cancer. 2009;100:1002–1011. doi: 10.1038/sj.bjc.6604948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alajez NM, Lenarduzzi M, Ito E, Hui AB, Shi W, Bruce J, Yue S, Huang SH, Xu W, Waldron J, et al. MiR-218 suppresses nasopharyngeal cancer progression through downregulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res. 2011;71:2381–2391. doi: 10.1158/0008-5472.CAN-10-2754. [DOI] [PubMed] [Google Scholar]
- 12.Ye JJ, Cao J. MicroRNAs in colorectal cancer as markers and targets: Recent advances. World J Gastroenterol. 2014;20:4288–4299. doi: 10.3748/wjg.v20.i15.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu BL, Peng XH, Zhao FP, Liu X, Lu J, Wang L, Li G, Chen HH, Li XP. MicroRNA-378 functions as an onco-miR in nasopharyngeal carcinoma by repressing TOB2 expression. Int J Oncol. 2014;44:1215–1222. doi: 10.3892/ijo.2014.2283. [DOI] [PubMed] [Google Scholar]
- 14.Qu MH, Han C, Srivastava AK, Cui T, Zou N, Gao ZQ, Wang QE. miR-93 promotes TGF-β-induced epithelial-to-mesenchymal transition through downregulation of NEDD4L in lung cancer cells. Tumour Biol. 2016;37:5645–5651. doi: 10.1007/s13277-015-4328-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabbri E, Brognara E, Montagner G, Ghimenton C, Eccher A, Cantù C, Khalil S, Bezzerri V, Provezza L, Bianchi N, et al. Regulation of IL-8 gene expression in gliomas by microRNA miR-93. BMC Cancer. 2015;15(661) doi: 10.1186/s12885-015-1659-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ansari MH, Irani S, Edalat H, Amin R, Mohammadi Roushandeh A. Deregulation of miR-93 and miR-143 in human esophageal cancer. Tumour Biol. 2016;37:3097–3103. doi: 10.1007/s13277-015-3987-9. [DOI] [PubMed] [Google Scholar]
- 17.Kawano M, Tanaka K, Itonaga I, Ikeda S, Iwasaki T, Tsumura H. microRNA-93 promotes cell proliferation via targeting of PTEN in Osteosarcoma cells. J Exp Clin Cancer Res. 2015;34(76) doi: 10.1186/s13046-015-0192-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Jiang M, Yuan W, Tang H. Prognostic value of miR-93 overexpression in resectable gastric adenocarcinomas. Acta Gastroenterol Belg. 2012;75:22–27. [PubMed] [Google Scholar]
- 19.Xu YF, Mao YP, Li YQ, Ren XY, He QM, Tang XR, Sun Y, Liu N, Ma J. MicroRNA-93 promotes cell growth and invasion in nasopharyngeal carcinoma by targeting disabled homolog-2. Cancer Lett. 2015;363:146–155. doi: 10.1016/j.canlet.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 20.Lyu X, Fang W, Cai L, Zheng H, Ye Y, Zhang L, Li J, Peng H, Cho WC, Wang E, et al. TGFβR2 is a major target of miR-93 in nasopharyngeal carcinoma aggressiveness. Mol Cancer. 2014;13(51) doi: 10.1186/1476-4598-13-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Ou X, Hu C. Validation and suggestion of eighth T classifications of the UICC/AJCC staging system for nasopharyngeal carcinoma patients: A retrospective analysis. Jpn J Clin Oncol. 2018;48:927–933. doi: 10.1093/jjco/hyy109. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Tan G, Liu Q, Tang X, Kang T, Li Y, Lu J, Zhao X, Tang F. Diagnostic values of serum cathepsin B and D in patients with nasopharyngeal carcinoma. BMC Cancer. 2016;16(241) doi: 10.1186/s12885-016-2283-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao Y, Wu S, Zhao R, Deng Q. MiR-205 promotes proliferation, migration and invasion of nasopharyngeal carcinoma cells by activation of AKT signalling. J Int Med Res. 2016;44:231–240. doi: 10.1177/0300060515576556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, Shi J, Zhong J, Huang Z, Luo X, Huang Y, Feng S, Shao J, Liu D. miR-1, regulated by LMP1, suppresses tumour growth and metastasis by targeting K-ras in nasopharyngeal carcinoma. Int J Exp Pathol. 2015;96:427–432. doi: 10.1111/iep.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu Q, Zhang F, Du Z, Xiang Y. Up-regulation of serum miR-744 predicts poor prognosis in patients with nasopharyngeal carcinoma. Int J Clin Exp Med. 2015;8:13296–13302. [PMC free article] [PubMed] [Google Scholar]
- 27.Li G, Ren S, Su Z, Liu C, Deng T, Huang D, Tian Y, Qiu Y, Liu Y. Increased expression of miR-93 is associated with poor prognosis in head and neck squamous cell carcinoma. Tumour Biol. 2015;36:3949–3956. doi: 10.1007/s13277-015-3038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du L, Schageman JJ, Subauste MC, Saber B, Hammond SM, Prudkin L, Wistuba II, Ji L, Roth JA, Minna JD, Pertsemlidis A. miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol Cancer Res. 2009;7:1234–1243. doi: 10.1158/1541-7786.MCR-08-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang L, Deng Z, Shatseva T, Yang J, Peng C, Du WW, Yee AJ, Ang LC, He C, Shan SW, Yang BB. MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-beta8. Oncogene. 2011;30:806–821. doi: 10.1038/onc.2010.465. [DOI] [PubMed] [Google Scholar]
- 30.Fang L, Du WW, Yang W, Rutnam ZJ, Peng C, Li H, O'Malley YQ, Askeland RW, Sugg S, Liu M, et al. MiR-93 enhances angiogenesis and metastasis by targeting LATS2. Cell Cycle. 2012;11:4352–4365. doi: 10.4161/cc.22670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu D, He XX, Chang Y, Sun SZ, Xu CR, Lin JS. Downregulation of MiR-93 expression reduces cell proliferation and clonogenicity of HepG2 cells. Hepatogastroenterology. 2012;59:2367–2373. doi: 10.5754/hge12458. [DOI] [PubMed] [Google Scholar]
- 32.Xiao ZG, Deng ZS, Zhang YD, Zhang Y, Huang ZC. Clinical significance of microRNA-93 downregulation in human colon cancer. Eur J Gastroenterol Hepatol. 2013;25:296–301. doi: 10.1097/MEG.0b013e32835c077a. [DOI] [PubMed] [Google Scholar]
- 33.Yu XF, Zou J, Bao ZJ, Dong J. miR-93 suppresses proliferation and colony formation of human colon cancer stem cells. World J Gastroenterol. 2011;17:4711–4717. doi: 10.3748/wjg.v17.i42.4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang Q, Zou Z, Zou C, Zhang Q, Huang R, Guan X, Li Q, Han Z, Wang D, Wei H, et al. MicroRNA-93 suppress colorectal cancer development via Wnt/β-catenin pathway downregulating. Tumour Biol. 2015;36:1701–1710. doi: 10.1007/s13277-014-2771-6. [DOI] [PubMed] [Google Scholar]
- 35.Yang IP, Tsai HL, Hou MF, Chen KC, Tsai PC, Huang SW, Chou WW, Wang JY, Juo SH. MicroRNA-93 inhibits tumor growth and early relapse of human colorectal cancer by affecting genes involved in the cell cycle. Carcinogenesis. 2012;33:1522–1530. doi: 10.1093/carcin/bgs166. [DOI] [PubMed] [Google Scholar]
- 36.Zhen Y, Liu Z, Yang H, Yu X, Wu Q, Hua S, Long X, Jiang Q, Song Y, Cheng C, et al. Tumor suppressor PDCD4 modulates miR-184-mediated direct suppression of C-MYC and BCL2 blocking cell growth and survival in nasopharyngeal carcinoma. Cell Death Dis. 2013;4(e872) doi: 10.1038/cddis.2013.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhen Y, Fang W, Zhao M, Luo R, Liu Y, Fu Q, Chen Y, Cheng C, Zhang Y, Liu Z. miR-374a-CCND1-pPI3K/AKT-c-JUN feedback loop modulated by PDCD4 suppresses cell growth, metastasis, and sensitizes nasopharyngeal carcinoma to cisplatin. Oncogene. 2017;36:275–285. doi: 10.1038/onc.2016.201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.