Abstract
The Molecular Evolutionary Genetics Analysis (MEGA) software enables comparative analysis of molecular sequences in phylogenetics and evolutionary medicine. Here, we introduce the macOS version of the MEGA software. This new version eliminates the need for virtualization and emulation programs previously required to use MEGA on Apple computers. MEGA for macOS utilizes memory and computing resources efficiently for conducting evolutionary analyses on macOS. It has a native Cocoa graphical user interface that is programmed to provide a consistent user experience across macOS, Windows, and Linux. MEGA for macOS is available from www.megasoftware.net free of charge.
Keywords: software, phylogenetics, cross-platform
Introduction
The use of Apple’s macOS is widespread in research and education in the biological sciences, including evolution, genomics, and bioinformatics. We previously supported researchers and students using Apple computers by bundling the Molecular Evolutionary Genetics Analysis (MEGA) software for Microsoft Windows with the Wineskin tool built on top of the WINE compatibility layer, which is capable of running Windows applications on other operating systems (Kumar et al. 2016). Many researchers and students showed interest in the MEGA+Wineskin package for macOS, as evidenced by >270,000-lifetime downloads.
However, MEGA+Wineskin packaging suffers from many limitations. First, it introduces an additional execution layer that degrades computational performance and stability. Second, MEGA+Wineskin packaging supported only 32-bit applications, so the 64-bit versions of MEGA for handling memory-intensive analyses of large contemporary data sets were not available to macOS users (Kumar et al. 2016, 2018). Third, the graphical user interface of the MEGA+Wineskin package is for Windows, resulting in a suboptimal experience for users accustomed to the graphical interface and keyboard commands on macOS. Also, continued production of MEGA+Wineskin is no longer possible due to the decreasing stability of Wineskin implementations over the years and the recent announcement that Apple will not support 32-bit applications as of macOS version 10.15 (Catalina).
We have now developed a version of MEGA that runs natively on macOS, eliminating the need for Wineskin virtualization and emulation. MEGA for macOS has a 64-bit computing architecture, and its user interface is programmed with Apple’s Cocoa widget set. Beta test version of MEGA for macOS was downloaded at a fast pace: 4,000 times/month (MS Windows version is being downloaded ∼19,000 times/month). Therefore, we expect this new addition to the MEGA software to benefit many existing users and enable new ones to consider utilizing MEGA in their research and teaching endeavors. In the following, we present a brief description of MEGA for macOS.
Results
64-Bit Architecture of MEGA on macOS
MEGA for macOS is now a 64-bit application, which eliminates the 32-bit limitation of the previous versions that could only use a maximum of 4 GB RAM. This advance makes MEGA suitable for analyzing sizable contemporary data sets with intensive memory requirements on macOS. For example, MEGA for macOS successfully produced a phylogeny of 43 arthropod species (15,447 amino acids) by using maximum likelihood method and the Jones–Taylor–Thornton (JTT) substitution model with gamma-distributed evolutionary rates (4 categories). This analysis required 5 h and 38 min to run and used >10 GB of memory. MEGA+Wineskin version could not analyze this data on the same computer, because its 32-bit architecture could not use >4 GB memory.
By using MEGA for macOS, users can now benefit from built-in parallelization of computationally intensive operations (e.g., Maximum Likelihood analysis) that effectively employ all the CPU cores and threads available, resulting in substantial time savings. Furthermore, the speed of this new version of MEGA for macOS is significantly faster than the previous MEGA+Wineskin version. For instance, we compared the time taken to evaluate 56 amino acid substitution models for alcohol dehydrogenase (ADH) genes for 11 Drosophila species and found that the MEGA+Wineskin version took ∼280 s to complete, whereas the new MEGA for macOS, running on the same machine, required ∼200 s.
Therefore, macOS users of MEGA now have the same facilities as their Windows and Linux counterparts to analyze large-scale genomic data using many sophisticated methods in the MEGA software.
Native macOS User Interface
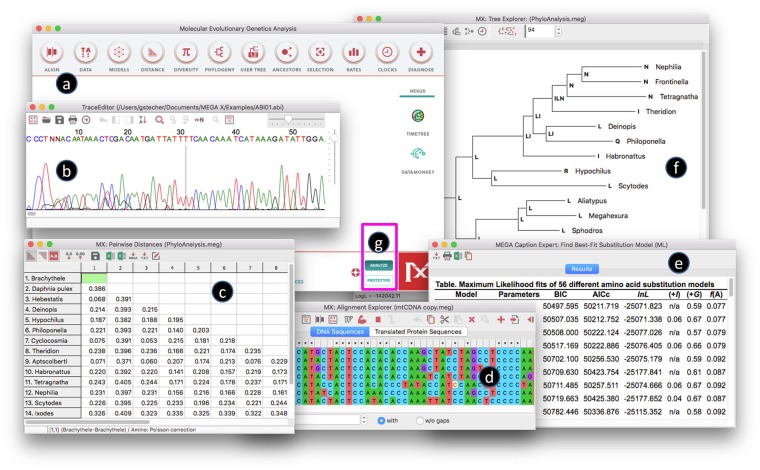
The user interface of MEGA for macOS is programmed with the 64-bit Cocoa API and widget set, which makes the graphical controls intuitive for macOS users. This upgrade eliminates confusion and frustration caused by the MEGA+Wineskin package due to its Windows interface. For instance, the tab control in the Alignment Explorer window for toggling between DNA and translated sequences is centered and short with rounded corners instead of shifted to the left and tall with square corners (fig. 1). Menus are now located at the top of the screen, where macOS users naturally expect them to be, instead of under the title bar for a given window. The close, minimize, and maximize buttons are located on the left side instead of the right side. Also, the keyboard shortcuts for MEGA on macOS have now been updated with standard macOS keyboard combinations. For instance, the keyboard combination to copy selected data to the clipboard is now Command + C instead of Ctrl + C.
FIg. 1.
MEGA for macOS graphical user interface, developed with 64-bit Cocoa API and widgets. The close, minimize, and maximize buttons are located on the left side of the title bar, and native toolbars, scroll bars, spin edits, and other visual controls are used. (a) The main form has the same look in macOS, Windows, and Linux. (b) The Trace Editor form uses an entirely custom-drawn GUI control for data display. (c) The Distance Matrix Explorer uses a native Cocoa view for displaying the pairwise distance matrix table. (d) The Alignment Editor uses the native Cocoa tab control for switching between the DNA Sequences and Translated Protein Sequences grids. (e) The Caption Expert form uses an HTML5 compliant web browser control to display results and detailed analysis information. (f) The Tree Explorer also uses a custom-drawn graphical control for rendering trees while the display is uniform across operating systems. (g) On the main form, the Analysis and Prototype buttons are used to toggle between GUI and CLI (Command Line Interface) execution modes (see Kumar et al. 2018).
Challenges Experienced in Porting MEGA to macOS
Porting of MEGA to macOS was a part of our plan to advance the MEGA software to be fully cross-platform. In this effort, we previously released a version of MEGA that worked on both Windows and Linux (Kumar et al. 2018), which represented a significant milestone for cross-platform compatibility. Despite this preparation, the porting of MEGA to macOS required more than two years of refactoring, reprogramming, debugging, and testing. This was because the 64-bit Cocoa API for macOS is relatively new, and its support in software development has been evolving. Also, MEGA has many sophisticated input data and output results explorer windows (e.g., Alignment Editor, Tree Explorer, and Genes/Domains dialog box) that are rich with visual controls, some of which were custom-developed for MEGA for Windows. Their porting to macOS was complicated and time-consuming, notably because the timing and ordering of mouse and keyboard actions and the drawing of visual controls differ between macOS, Windows, and Linux. To make the visual controls work correctly on macOS, we needed to access the Cocoa API directly to develop fixes and workarounds through fine-grained access to the Objective-C runtime.
Also, a significant refactoring and customization effort of the MEGA web browser code was required to utilize the Chromium Embedded Framework’s (Greenblatt 2019) shared libraries effectively in macOS, including integration with the target platform message loop, management of subprocess initialization and lifetime, and application layout. Their usage in macOS differs significantly from that on Windows and Linux platforms. Additionally, we needed to develop custom scripts to codesign, notarize, and package MEGA and associated resources for distribution on the macOS platform. Apple’s development tools are not cross-platform and could not be used for MEGA. A score of other system-level updates was needed to port MEGA to macOS successfully.
Distributions of MEGA for macOS
The MEGA macOS has been tested extensively on 10.13 (High Sierra) and newer releases. For users that wish to run MEGA through a command-line interface (MEGA-CC), MEGA for macOS generates a configuration file specifying analysis options for user-desired analysis by using the computing core of MEGA that is useful for high throughput and iterative analysis (Kumar et al. 2012).
Conclusion
With the porting of MEGA to the macOS operating system, MEGA GUI now runs natively on Windows, Linux, and macOS and provides a consistent user-experience across platforms. These developments address the need for cross-platform solutions because of technology heterogeneity that exists in contemporary research laboratories and classroom environments.
Acknowledgments
We thank Christina Grigoryev, Louise Huuki, and many beta testers for providing feedback and locating bugs. This work was supported in part by research grants from National Science Foundation (ABI 1661218), National Institutes of Health (R01GM126567-03), and Japan Society for the Promotion of Science (JSPS) grants-in-aid for scientific research (DB5) to K.T.
References
- Greenblatt M. 2019. Chromium embedded framework. Available from: https://bitbucket.org/chromiumembedded/cef (last accessed on December 27, 2019).
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Peterson D, Tamura K.. 2012. MEGA-CC: computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics 28(20):2685–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]