Abstract
Native American genetic variation remains underrepresented in most catalogs of human genome sequencing data. Previous genotyping efforts have revealed that Mexico’s Indigenous population is highly differentiated and substructured, thus potentially harboring higher proportions of private genetic variants of functional and biomedical relevance. Here we have targeted the coding fraction of the genome and characterized its full site frequency spectrum by sequencing 76 exomes from five Indigenous populations across Mexico. Using diffusion approximations, we modeled the demographic history of Indigenous populations from Mexico with northern and southern ethnic groups splitting 7.2 KYA and subsequently diverging locally 6.5 and 5.7 KYA, respectively. Selection scans for positive selection revealed BCL2L13 and KBTBD8 genes as potential candidates for adaptive evolution in Rarámuris and Triquis, respectively. BCL2L13 is highly expressed in skeletal muscle and could be related to physical endurance, a well-known phenotype of the northern Mexico Rarámuri. The KBTBD8 gene has been associated with idiopathic short stature and we found it to be highly differentiated in Triqui, a southern Indigenous group from Oaxaca whose height is extremely low compared to other Native populations.
Keywords: Native Americans, exome sequencing, demographic inference, adaptive evolution
Introduction
Comprehensive genome sequencing projects of human populations have demonstrated that a vast majority of human genetic variation has arisen in the past 10,000 years and is, therefore, specific to the continental and subcontinental regions in which they arose. As a result, the majority of rare variation contributing to disease burden is expected to be population specific and influenced by the local demographic history and evolutionary processes of each population (Gravel et al. 2011; Martin et al. 2017). Furthermore, it is recognized that there is a strong bias toward the inclusion of individuals of European descent in biomedical research, which is problematic for medical, scientific, and ethical reasons and should be counter balanced by including underrepresented populations in large genomic surveys of genetic variation (Bustamante et al. 2011; Popejoy and Fullerton 2016).
Despite recent large-scale sequencing projects like the Exome Aggregation Consortium (ExAC) (Lek et al. 2016), which considerably expanded the knowledge on the patterns of protein-coding variation worldwide, little is known about the distribution of population-specific genetic variants that may underlie important evolutionary and biomedical traits of understudied groups. In particular, populations in the Americas of Indigenous ancestry are expected to show exacerbated genetic divergence due to extreme isolation and serial founder effects during the continental peopling, leading to an increased fraction of population-specific variation that remains to be characterized (The 1000 Genomes Project Consortium 2015; Martin et al. 2017; Bergström et al. 2019). Present-day Mexico represents one of the largest reservoirs of Native American variation. However, studies targeting such variation have used either genotyping arrays with markers ascertained mainly in non-native American populations (Silva-Zolezzi et al. 2009; Moreno-Estrada et al. 2014) or with whole-genome sequencing on a small number (n = 2) of individuals per indigenous population (Romero-Hidalgo et al. 2017). The former approach prevents the discovery of new variants, while the latter does not allow reliable estimation of allele frequencies; consequently, there is a need to harness high coverage sequencing with population-level sampling. This will shed light on the consequences of functional variation in protein-coding genes as well as the adaptive and demographic processes that have shaped Native Mexican (NM) genomes.
To fulfill this need, we sequenced the exomes of 78 individuals from five different indigenous groups from Northern (Rarámuri [TAR] or Tarahumara, and Huichol [HUI]), Central (Nahua [NAH]), South (Triqui [TRQ]), and Southeast (Maya [MYA]) Mexico. We characterized the protein-coding genetic variation from these populations to infer the broad demographic history of pre-Hispanic Mexico and to search for signatures of adaptive evolution.
Results
Genetic Variation in 76 Native Mexican Exomes
According to previous genetic characterizations of indigenous Mexican groups, Mexico’s Native American ancestry is substructured into three major geographical components: Northern, Central and Southern (Gorostiza et al. 2012; Moreno-Estrada et al. 2014). In order to capture such substructure, we obtained protein-coding genetic variation from the sequences of 78 exomes from five NM populations representing all three major genetic regions of Mexico: HUI (n = 14), MYA (n = 13), NAH (n = 17), TAR (n = 19), and TRQ (n = 15). Most exomes were sequenced at >70× (average depth 90.3×), except for six individuals with depths between 31× and 35× (supplementary fig. S1, Supplementary Material online). We found no correlation between average depth and number of called heterozygous sites (p = 0.13, P-value = 0.274), ruling out a potential bias of the lower-depth sequences in downstream analyses (supplementary fig. S2, Supplementary Material online). We used the Genome Analysis Tool Kit (GATK) (McKenna et al. 2010) to call variants jointly with an exome data set including 103 Han Chinese (CHB) individuals from 1000 Genomes Project (TGP) (The 1000 Genomes Project Consortium 2015). We jointly called with CHB exome data as we used the variants in this population for downstream analysis involving tests for selection in the NM groups. We identified 120,735 single-nucleotide variants (SNV) and computed the genotype concordance between these and previously generated data from Affymetrix 6.0 (Moreno-Estrada et al. 2014) and Axiom World IV (Galanter et al. 2014) SNP arrays available for the NM individuals. Concordance was above 93% for all individuals except for one TRQ and one HUI individual, which were excluded from all downstream analyses (supplementary fig. S3 and table S1, Supplementary Material online). A predominance of Native American genetic ancestry in the remaining 76 NM individuals was corroborated with ADMIXTURE (Alexander et al. 2009) and principal components analysis (PCA) (Patterson et al. 2006) (supplementary table S2 and fig. S4, Supplementary Material online). Fifty-nine individuals displayed some non-Native ancestry ranging from 0.1% to 13%, and therefore we masked this fraction in the admixed exomes (see Material and Methods) for downstream analyses (supplementary tables S2 and S3, Supplementary Material online).
After masking, a total of 58,918 biallelic SNV were retained in the 76 NM exomes with a transition/transversion (Ti/Tv) ratio of 3.025, in agreement with the value observed in human-exome sequencing data (Bainbridge et al. 2011). Of all sites, 62% fell within exonic regions and 31% in intronic, while the remaining 6% consisted of UTR, ncRNA, intergenic, splicing, upstream and downstream annotations (supplementary table S4, Supplementary Material online). A subset of 4,181 SNVs was absent from public data sets (ExAC, TGP, and dbSNP v.142). The number of novel variant sites per exome ranged between 29 and 118 (median 84). Most of these novel SNVs are nonsynonymous (67.5%) and found at low frequencies: approximately 80% are singletons, while the rest are found at less than 5% frequency in the NM exomes (supplementary figs. S5 and S6, Supplementary Material online). The number of singletons per population was 5,262 for the HUI (average per individual 405), 6,093 for the MYA (average per individual 469), 8,108 for the NAH (average per individual 476), 5,454 for the TAR (average per individual 287), and 5,166 for the TRQ (average per individual 369).
Population History of Native Mexicans
We used the site frequency spectrum (SFS) to infer the demographic history of four NM populations: TAR, HUI, TRQ, and MYA. The NAH population was excluded from this analysis due to genetic substructure found within this linguistic group, which introduces noise in this type of analysis (see Discussion). A total of 20,991 “neutral sites” (see Materials and Methods for a description of filters used) were used as input for demographic inference. We utilized a diffusion approximation approach implemented in the software δαδι (Gutenkunst et al. 2009) to infer the best-fit topology and demographic parameters of the four populations (see Materials and Methods). The best-fitting topology joins Northern populations together (HUI and TAR), as well as Southern populations (TRQ and MYA) stemming from a shared branch (fig. 1) (supplementary table S5, Supplementary Material online). The same topology is recovered when inferring split patterns with the program TreeMix (supplementary fig. S7, Supplementary Material online) (Pickrell and Pritchard 2012).
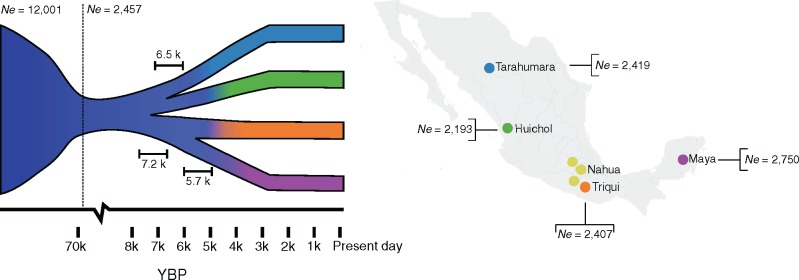
FIg. 1.
Sampling locations and inferred demographic model for NM populations. Inferred split times are shown on the demographic model and effective population sizes (Ne) are shown on the map. Each branch represents one of the populations used in the demographic inference; colors correspond to those shown in the map displaying the sampling locations of the participant NM. The Nahua were not included in the demographic inference (see Discussion).
For all models, we fixed a population bottleneck around 70 KYA, representing the Out of Africa bottleneck (Gutenkunst et al. 2009). Our best-fit model inferred an ancestral effective population (Ne) of 12,000 individuals for all NM (95% CI: 11,438–12,570), which is reduced to a Ne of 2,457 individuals (95% CI: 2,340–2,589) after the bottleneck. We inferred that the split between northern (TAR and HUI) and southern (TRQ and MYA) NM populations occurred 7,219 years ago (95% CI: 6,454–7,857) (fig. 1, supplementary table S5, Supplementary Material online). We find the two subsequent splits occurring within 1,500 years of each other: TAR and HUI diverged from each other 6,523 years ago (95% CI: 5,669–7,249), followed by the TRQ and MYA split 5,715 years ago (95% CI: 4,829–6,501) (fig. 1, supplementary table S5, Supplementary Material online). We estimated all four populations to have similarly small Ne. The MYA has the largest Ne (2,750, 95% CI: 2,310–3,256), followed by the TAR (2,419, 95% CI: 2,117–2,729), TRQ (2,407, 95% CI: 2,050–2,759), and HUI (2,193, 95% CI: 1,865–2,493) (fig. 1, supplementary table S5, Supplementary Material online). A total of 95% confidence intervals for these parameters were determined with 1000 bootstrapped replicates (supplementary table S6, Supplementary Material online). We note that this model assumes a constant population size since the last split. We were not able to estimate population growth rates due to the small sample size.
Adaptive Evolution in Native Mexicans
For inferring adaptive evolution, we calculated the Population Branch Statistic (PBS) for each gene as in Yi et al. (2010). This Fst-based statistic allows the identification of genes with strong differentiation between a test population and a closely related one since their divergence; using a third more distantly related population to detect changes affecting the test population. To this end, we considered the variant sites in the CHB and NM exomes that remained after masking non-Native American ancestry in NM, and oriented these to the ancestral/derived state, which reduced the number of sites to 117,644 (i.e., it was not possible to infer the ancestral state for 3,091 sites). We refer to this analysis as “Ancestry-Specific PBS.” We estimated a per-gene PBS in NM considering the CHB data used for joint variant calling (see Materials and Methods) as well exome data available as part of the TGP for individuals of European ancestry (CEU), as the second and third populations, respectively. This allowed the detection of genes likely under selection in all NM since divergence from the CHB.
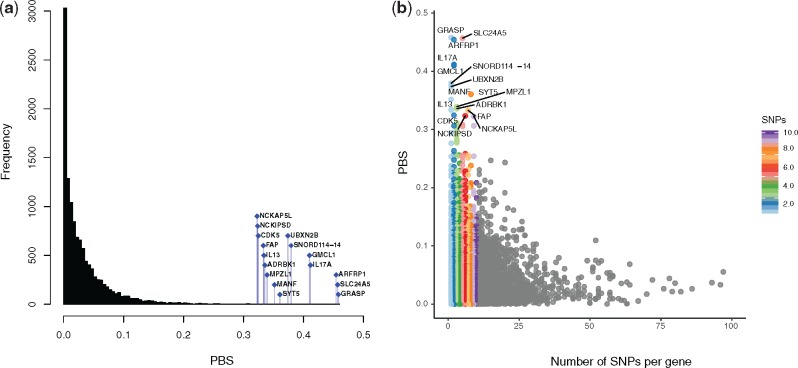
We defined the genes in the 99.9th percentile of the empirical distribution of the PBS values as being candidates of adaptive evolution (fig. 2 and supplementary table S7, Supplementary Material online). Because PBS distribution is different for different gene sizes (i.e., for different numbers of SNPs per gene), we selected this stringent filter and demonstrate that with it, only the genes at the top of their size distribution are retained (fig. 2b). Interestingly, some of these genes had previously been identified as targets of selection in other populations. These genes include SLC24A5, involved in skin pigmentation, and FAP, which was previously suggested to be under adaptive archaic introgression in Peruvians (Racimo et al. 2016) and Melanesians (Vernot et al. 2016). Of interest, three genes were involved in immune response. These include SYT5, implicated in innate immune response, and interleukins IL17A and IL13. The remaining candidate genes were involved in signal transduction (MPZL1), protein localization and transport (GRASP and ARFRP1), cell differentiation and spermatogenesis (GMCL), Golgi apparatus organization (UBXN2B), neuron differentiation (MANF), signaling and cardiac muscle contraction (ADRBK1), cell cycle (CDK5), microtubule organization and stabilization (NCKAP5L), and stress fiber formation (NCKIPSD); supplementary table S7, Supplementary Material online).
FIg. 2.
Genes likely under adaptive evolution in NM. (a) Distribution of gene-based PBS values. Genes with extreme PBS values (99.9th percentile) are highlighted in blue. The x-axis shows the value of the PBS and the y-axis represents the frequency at which that value was observed in all NM. (b) Distribution of gene-based PBS values as a function of the number of SNPs per gene. Colors represent different SNP/gene bins, from one to ten SNPs/gene. Same genes as in (a) are shown, illustrating that they have PBS values at are at the top of their respective bin category.
Population-Specific Adaptive Evolution within Mexico
To investigate genes under selection specific to each of the four NM populations for which we had a demographic model (HUI, MYA, TAR, and TRQ), we calculated PBS using the Han Chinese (CHB) as the third population in the form of NM1, NM2, and CHB for all 12 combinations. To evaluate the significance of these PBS values, we compared the observed data to PBS values obtained from simulations under the inferred demographic model (see Materials and Methods). We were thus able to assign P values to each gene and rank them by significance.
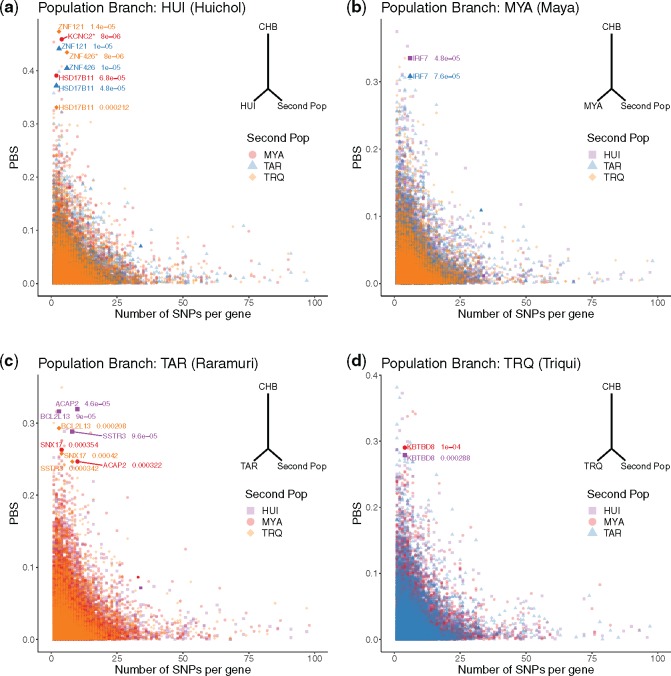
Only two genes with extreme PBS values passed a significance threshold of P < 10−5 (fig. 3a–c). These were identified in HUI and were involved in cellular proliferation and differentiation (KCNC2) and transcriptional repression of herpesvirus promoters (ZNF426). To identify additional genes in the remaining populations, we looked for genes in the top 1% of the PBS distribution and in the lowest 1% of the P-value distribution that were shared by at least two of the three possible pairwise comparisons between populations. This yielded eight additional candidate genes for adaptive evolution specific to different NM populations (supplementary table S8, Supplementary Material online).
FIg. 3.
PBS as a function of the number of SNPs in each gene for (a) HUI, (b) MYA, (c) TAR, and (d) TRQ. Colors and symbols represent the population used as the second population for the computation of the PBS. Genes under likely adaptive evolution are shown in bold with their corresponding P-value (the few genes with low PBS scores that seem to appear as bold are not candidate genes, but rather multiple data points overlapping on the same position of the plot). A tree in each panel illustrates the topology used for the PBS calculation. CHB stands for Han Chinese; this population was used as the third, distantly related, population in all comparisons.
Among these new candidate genes for adaptive evolution, we noticed gene BCL2L13 (BCL2 like 13), in the TAR, who are known for their cultural practice of high-endurance long-distance running (Balke and Snow 1965). BCL2L13 encodes for a pro-apoptotic protein that localizes in the mitochondria, is highly expressed in skeletal muscle (GTex Version 7, supplementary fig. S8, Supplementary Material online), and found in a locus previously associated to osteoarthritis (OA) risk in Mexican Americans (Coan et al. 2013). In addition, in TRQ we identified KBTBD8 (Kelch Repeat and BTB Domain Containing 8), a gene involved in ubiquitination and found in a locus previously associated to idiopathic short stature in Koreans (Kim et al. 2010). This is relevant because the TRQ (from the southern state of Oaxaca) display a particularly short stature (Faulhaber 2014) and the SNV driving the selection signal (rs13096789) causes a nonsynonymous change classified as “possibly damaging” by PolyPhen. Lastly, we found gene HSD17B11 (Hydroxysteroid 17-beta dehydrogenase 11) as an additional candidate of adaptive evolution in HUI. HSD17B11is a short-chain alcohol dehydrogenase that metabolizes secondary alcohols and ketones, and it has been suggested to participate in androgen metabolism during steroidogenesis.
To evaluate if genes showing extreme PBS values were enriched in any functional category or metabolic pathway, we took the intersection of the genes with a P-value < 0.05 in all three pairwise comparisons for each NM population (see Materials and Methods). This way we compiled a list of 203 genes for HUI, 211 for MYA, 165 for TAR, and 183 for TRQ (supplementary tables S9–S12, Supplementary Material online). We evaluated functional enrichment for the three Gene Ontology (GO) project categories: biological processes, cellular components, and molecular function (Ashburner et al. 2000; The Gene Ontology Consortium 2017), as well as pathway over-representation using the IMPaLa tool (Kamburov et al. 2011). We observed a functional enrichment with a significance of P < 0.05 in HUI involving lipid intestinal absorption (GO: 1904729, GO: 0030300, GO: 1904478) (supplementary fig. S9 and table S13, Supplementary Material online). Consistently, the IMPaLa pathway over-representation analysis revealed an enrichment of genes involved in lipid metabolism and transport, specifically the Statin pathway, for the same population (pathway source: Wikipathways, P-value [8.32e-06], Q-value 0.0382).
Discussion
We carried out the most comprehensive characterization of potentially adaptive functional variation in Indigenous peoples from the Americas to date. We identified in these populations over 4,000 new variants, most of them singletons, with neutral, regulatory, as well as protein-truncating and missense annotations. The average number of singletons per individual was higher in NAH and MYA, which is expected given these two Indigenous groups embody the descendants of the largest civilizations in Mesoamerica, and that today NAH and MYA languages are the most spoken Indigenous languages in Mexico (INEGI 2015). Furthermore, the generated data also allowed us to propose a demographic model inferred from genomic data in Native Mexicans and to identify possible events of adaptive evolution in pre-Columbian Mexico.
Demography
We propose, to our knowledge, the first demographic model that uses genetic data to estimate split times between ancestral populations within Mexico. By using the SFS of putatively neutral SNVs using a diffusion approximation approach, we inferred a split between northern and southern NM at approximately 6.5–7.9 KYA, followed by regional differentiation in the north at 5.7–7.2 KYA, and 4.8–6.5 KYA in the south of Mexico (95% bootstrap CI, supplementary table S5, Supplementary Material online). We note that the confidence intervals for the TAR/HUI split and the TRQ/MYA split are overlapping, and the second best fit-model infers these splits to occur at the same time (supplementary table S6, Supplementary Material online). This northern/southern split and a northwest to southeast cline is consistent with previous reports based on whole-genome and microarray genotype data from NM (Moreno-Estrada et al. 2014; Romero-Hidalgo et al. 2017). Furthermore, these split times are also coherent with previous estimates of ancestral Native Americans diverging ∼17.5–14.6 KYA into Southern Native Americans or “Ancestral A” (AncA, comprising Central and Southern Native Americans) and Northern Native Americans or “Ancestral B” (AncB) (Reich et al. 2012; Rasmussen et al. 2014; Raghavan et al. 2015; Moreno-Mayar et al. 2018; Scheib et al. 2018), and with an initial settlement of Mexico occurring at least 12,000 years ago, as suggested by the earliest skeletal remains dated to approximately this age found in Central Mexico (Gonzalez et al. 2003) and the Yucatán peninsula (Chatters et al. 2014). Studies on genome-wide data from ancient remains from Central and South America reveal genetic continuity between ancient and modern populations in some parts of the Americas over the last 8,500 years (Raghavan et al. 2015; Posth et al. 2018), though two ancient genomes from Belize (dated to 7,740 BP and 9,300 BP, respectively) do not show specific allele sharing with present-day populations from that geographic area, instead they display similar affinities to different present-day populations from Central and South American populations, respectively (Posth et al. 2018). This suggests that, by that time, the ancestral population of MYA was not yet genetically differentiated from others, so our estimates of northern/southern split at 7.2 KYA and MYA/TRQ divergence at 5.7 KYA fit with this scenario.
Our SFS-based demographic insights are further supported with D-statistics (Patterson et al. 2012), which in the forms D (Anzick1, Athabascan, NM, YRI), reveal that all NM are indeed closer to the AncA branch, represented by the ∼12.8k years old Anzick1 individual from Montana (Rasmussen et al. 2014), than to two present-day Athabascans from British Columbia, who are representatives of branch AncB (Raghavan et al. 2014) (supplementary fig. S10, Supplementary Material online). Although all comparisons reveal this trend, it is important to acknowledge that the tests were not significant, most likely due to the limited number of sites, derived from only considering exomic regions, available to calculate these D-statistics. Furthermore, tests in the forms D (NM1, NM2, Anzick1, YRI) revealed that all NM populations are equally related to Anzick1. Interestingly, tests in the form D (NM1, NM2, Athabascan, YRI) reveal that MYA, TRQ, and NAH are closer to Athabascans when using HUI and TAR as the second NM population; however the test is only significant when using HUI (supplementary fig. S10, Supplementary Material online). This pattern could be explained by gene flow from an unsampled Native American population diverging before the AncA and AncB split into the Northern Mexican populations after they split from the Southern Mexican ones. A similar model has been proposed by Moreno-Mayar and colleagues, who observe the contribution of an unsampled population “Unsampled population A” (UPopA) to the NM Mixe (Moreno-Mayar et al. 2018). Another possibility is the contribution of an Athabascan-like population into the ancestors of the Central/Southern Mexican populations, which could bring MYA, TRQ, and NAH closer to Athabascans. Despite these interesting observations, we caution that exome data are not ideal for demographic inference at the level of resolution required to test these or other more complex scenarios. The availability of whole-genome data from additional ancient and present-day populations is necessary to untangle their deep population history.
Altogether these observations based on archeological and paleogenomic data are consistent with our time estimates of population splits within Mexico, which involve a divergence of the Northern and Southern NM occurring at least two thousand years after the settlement, and a divergence time within these branches occurring approximately between 600 and 2,000 years later, respectively.
Regarding effective population sizes (Ne), we inferred an ancestral Ne of 2,457 for all NM, which is in line with a recent Ne estimate of 2,000 based on Markovian coalescent analyses of whole-genome data from 12 NM (Romero-Hidalgo et al. 2017) as well as from Native ancestry segments in admixed Mexicans (Schiffels and Durbin 2014). Both studies show a low Ne around 2,000 sustained for the last 20 ka, in agreement with genomic and archeological evidence pointing to a population bottleneck ca. 20,000 years ago experienced by the Native American ancestors when crossing the Bering Strait into the Americas (Goebel et al. 2008; Moreno-Estrada et al. 2014; Raghavan et al. 2015).
In addition, our model inferred low Ne for present-day NMs ranging between ∼2,200 and ∼2,800. The NM with the largest Ne was the MYA (95% CI: 2,310–3,256) followed by TAR (95% CI: 2,117–2,729), TRQ (95% CI: 2,050–2,759), and HUI (95% CI: 1,865–2,493) (supplementary table S6, Supplementary Material online). Using runs of homozygosity, Moreno-Estrada et al. (2014) inferred slightly higher variation in Ne among different Indigenous groups, but with overlapping confidence intervals. On the other hand, using whole-genome data, Raghavan et al. (2015) inferred a HUI Ne to a similar 2,500. Of notice, the census size of these populations is also the largest for the MYA and TAR (INEGI 2015). These observations are noteworthy since low Ne combined with founder effects can exacerbate the disproportionate accumulation of deleterious and clinically relevant variants in the population (Belbin et al. 2018).
One caveat of our demographic inference is that we did not include the NAH in the model. Initial tests including this population resulted in extremely high Ne and ambiguous location in the tree. Furthermore, ADMIXTURE analyses showed that NAH are constituted by components from multiple populations within each individual (supplementary fig. S4a, Supplementary Material online), an observation made also in a recent study by Romero-Hidalgo et al. (2017). This likely reflects NAH being genetically heterogeneous as a consequence of their past history involving continuous colonization and extended domination of multiple distinct groups by the NAH-speaking Aztec empire right before European colonization (Brumfiel 1983; Romero-Hidalgo et al. 2017).
Overall, our inferences describe, to our knowledge, the most detailed demographic history model based on genetic data for NM to date. We caution, however, that as with any inferred demographic model, the assumptions have certain caveats that could lead to errors. Specifically, our model assumes constant population sizes since the most recent split. Certainly, the availability of genome-wide data from present day, as well as from ancient populations from throughout Mexico spanning these time frames, will contribute to draw a more refined picture of past population history and genetic structure.
Targets of Adaptive Evolution in NM
We implemented an ancestry-specific approach of the widely used FST-based PBS to identify genes with strong differentiation in all NM since their divergence from CHB, as well as genes differentiated in each NM population branch. The first approach revealed selection signals previously found in Native Americans and other populations, as well as genes not previously identified to be under selection.
We identified a strong differentiation on FAP (Fibroblast Activator Protein Alpha). The locus harboring this gene, together with IFIH1 (interferon induced with helicase C domain 1) is suggested to be under adaptive archaic introgression in Peruvians from the TGP (PEL), a population with a high proportion of Native American genetic ancestry (Racimo et al. 2016) and Melanesians (Vernot et al. 2016). This locus has been associated with type 1 diabetes (Liu et al. 2008) and susceptibility to diverse viral infections (Fumagalli et al. 2010). The fact that this locus also has an adaptive signal in NM is consistent with a previous study that suggested that the loci harboring IFIH1 suffered recent positive selection in South Americans (Fumagalli et al. 2010). We confirmed that this haplotype is present in NM by comparing the Neandertal haplotype (Marnetto and Huerta-Sánchez 2017) with whole-genome sequence data available for other Native Americans from an independent study (Romero-Hidalgo et al. 2017). The archaic haplotype was found in 7 out of 24 chromosomes in TAR and MYA individuals as well as in some Mexican individuals from LA (MXL) and other NM (Tepehuano, Totonaco, and Zapotec) (supplementary fig. S11, Supplementary Material online).
SLC24A5 (solute Carrier family 24 Member 5) has also been previously identified as target of selection. This gene is involved in melanogenesis (Lamason et al. 2005) and has been vastly studied in European populations, where it displays one of the strongest signals of selection in humans (Mallick et al. 2013). Specifically, the derived allele of SNP rs1426654 within this gene leads to a decrease in skin pigmentation. However, the ancestral allele, which might be advantageous in latitudes with higher exposure UV radiation, is nearly fixed in NM populations, driving the extreme PBS signal and suggesting either a relaxation of selection or an adaptive event favoring the ancestral state (the derived allele is fixed in CEU, and found at 0.03 and 0.007 frequencies in CHB and NM, respectively).
In addition, we found three genes with extreme PBS values involved in immunity and defense against pathogens (SYT5, IL13, and IL17A). Selection on these genes could be explained by the strong selective pressure posed by pathogens brought by Europeans on the Native population during colonization; it is estimated that up to 90% of the Native population died as a consequence of infections during this period (Rodolfo Acuna-Soto et al. 2002).
Regarding population-specific signals of adaptive evolution, we identified few genes with significant (P < 10−5) in HUI only. These genes were involved in repression of herpesvirus transcription (ZNF426), and cellular proliferation and differentiation (KNC2), When we expanded our search to consider genes above the significance threshold, but with consistent extreme PBS and low P values in two or more population comparisons, we identified some interesting instances in TRQ, HIU, and TAR, which we speculate could be related to some characteristic traits in these populations.
Previous anthropometric studies have revealed that the TRQ (together with other Indigenous groups in Oaxaca and neighboring states of Veracruz and Chiapas) exhibit the lowest average stature values in Mexico (females mean = 142.5 cm, males mean = 155.1) (Faulhaber 2014). This observation becomes relevant as one of the genes (KBTBD8) identified here as likely being under adaptive evolution in this population, lies within a locus previously associated to idiopathic short stature in Koreans (Kim et al. 2010).
Regarding HUI, we found a gene (HSD17B11) involved in the metabolism of steroids and retinoids, reported to have high expression in tissues related to steroidogenesis (adrenal gland and testis) and detoxification (liver, lung, kidney, and small intestine) (Lundová et al. 2016). Interestingly, the ceremonial intake of peyote cactus (Lophophora williamsii) is a cultural tradition that traces back to centuries in the Mexican region settled by the HUI population. The psychoactive compound in peyote, the alkaloid mescaline, is metabolized by the liver enzymes and can cause severe toxicity when consumed in high amounts. Furthermore, the GO and pathway enrichment analyses in this population returned genes APOA1, APOA2, APOA4, APOA5, and ABCG5 (supplementary table S14, Supplementary Material online) involved in regulation of intestinal cholesterol absorption. Of notice, genes APOA1, APOA4, and APOA5 form a cluster on chromosome 11, so it is possible that the SNPs driving the signal are in LD, resulting in correlated PBS values. However, genes APOA2 and ABCG5 are in independent chromosomes (chromosome 1 and 2, respectively), suggesting that the enriched pathway might in fact be under adaptive evolution. Variants in some of these genes have been associated to high levels of LDL and total cholesterol (Aulchenko et al. 2009; Bandarian et al. 2013) as well as high triglyceride levels (Pennacchio et al. 2002; Ouatou et al. 2014; Zhu et al. 2014), both factors leading to cardiovascular disease. The derived allele of rs3135506 in APOA-5 has the highest frequency in HUI compared to the other populations from the study and the TGP (supplementary table S14, Supplementary Material online). This missense mutation (S19W) has been associated with increased triglyceride levels and elevated risk of developing coronary artery disease in several populations including one labeled as “Hispanic” (Pennacchio et al. 2002; Ouatou et al. 2014; Zhu et al. 2014). Moreover, HUI has the highest frequency of the missense mutation rs6756629 in ABCG5, which has been associated to increased total cholesterol and LDL a risk factor for coronary heart disease (Aulchenko et al. 2009). Together, these observations point some possible adaptation to a low cholesterol-lipid diet (such as a reduced meat consumption) or a manner to regulate the intake of lipids from animal source foods.
Lastly, among the most remarkable results, we found a candidate gene under selection in TAR (BCL2L13) with annotations related to joint and bone physiology, namely OA, which could be related to the outstanding physical endurance in the Rarámuri. Interestingly, a recent study (Romero-Hidalgo et al. 2017) detected an enrichment of genes harboring novel promoter and missense variants with pathway and GO annotations related to musculoskeletal function in the same population. In agreement with this, we recapitulated similar observations when looking for GO enrichment in novel nonsynonymous variants in our 19 TAR exomes (supplementary table S15, Supplementary Material online). These two approaches represent independent evidence from both highly diverged and novel functional variation that converge in musculoskeletal traits as the potential underlying mechanism for Raramuri’s endurance. Taken together, these observations could imply that this cultural trait has imposed a selective pressure on this population. However, additional in-depth studies in the TAR incorporating genomic and detailed phenotype data are needed to disentangle the genetic architecture and the molecular pathways behind this complex trait.
In conclusion, we generated a rich catalog of Native American genetic variation from Mexican populations, the analysis of which has yielded novel estimates for ancestral population splits as well as candidate genes likely under adaptive evolution in both the general NM population and in specific NM groups. Our demographic inference is consistent with previous archeological and genetic knowledge on the peopling of the Americas, while adding temporal resolution to the population dynamics occurring thousands of years ago in the Mexican mainland. This demographic model also allowed us to compare the estimated PBS values of genes against a simulated null distribution under such model, and to identify the instances with significant extreme values. Genes with extreme values in specific populations have annotations that hint a likely role in characteristic phenotype or cultural practices in the NM included in this study. However, it remains to be tested, if these high values indeed derive from adaptive events and if these adaptations are in fact involved with the observed traits in these NM populations.
Materials and Methods
Samples
Most of the samples sequenced in this study were previously collected and sampling procedures are described in Moreno-Estrada et al. (2014). Specifically, a subset of samples from four of the studied populations were selected for having the highest proportions of Native American ancestry according to Affymetrix 6.0 SNP array data generated therein. After filtering for DNA quality control, a total of 19 Tarahumara (Chihuahua), 13 HUI (Jalisco), 15 TRQ (Oaxaca), and 12 MYA (Quintana Roo) individuals were included in this study. Additionally, 18 NAH samples from three sampling locations in Central Mexico previously genotyped with Affymetrix Axiom World Array IV (Galanter et al. 2014) were selected for exome sequencing based on their proportions of Native American ancestry and passing DNA quality control. In both sampling schemes, Institution Review Board (IRB) approval was obtained from Stanford University, and individuals were consented according to the approved protocol. All individuals gave written consent. DNA was extracted from blood and ethnographic information including family, ancestry, and place of birth were collected for all individuals. In agreement with the informed consents obtained from participants of the Indigenous communities and to respect their privacy for the transfer of genetic data, individual-level exome vcf files for the samples sequenced in this study are available through a data access agreement by contacting A.M.-E. and M.C.A.-A.
Exome Sequencing
Exome regions were captured using the Agilent SureSelect 44Mb human all-exon array v2.0 for the 76 individuals. Genotype data from previous studies (Galanter et al. 2014; Moreno-Estrada et al. 2014) were available for these individuals (Affymetrix 6.0 SNP array data for HUI, MYA, Tarahumara, and TRQ, and Axiom World Array IV data for the NAH).
Each individual was sequenced in a 5-plex library on an Illumina HiSeq 2000 producing 101-bp paired end reads. Reads were processed according to a standard pipeline informed by the best practices described by the TGP (The 1000 Genomes Project Consortium 2015). Briefly, reads were mapped to the human reference genome (hg19) using bwa (version 0.6.2). Duplicate read pairs were identified using Picard (http://broadinstitute.github.io/picard/; last accessed December 8, 2019). Base qualities were empirically recalibrated and indel realignment was performed jointly across all samples using the GATK (version 1.6) (McKenna et al. 2010). Variants were filtered to the exome capture region. We corroborated the concordance of the variants with the previously generated array genotype data and identified two samples with low concordance (HUI75 and TRQ41) that, due to mislabeling, were actually replicates of other individuals. We therefore excluded these two samples from further downstream analyses. Furthermore, we carried out a Pearson correlation test in the software R (cor.test) (R Core Team 2014), to investigate if the difference in depths could be affecting our ability to retrieve heterozygous sites. We found no such correlation.
Masking of Non-Native Genomic Segments
To corroborate the Native American ancestry of the individuals, we combined the genotype data available for these individuals (Galanter et al. 2014; Moreno-Estrada et al. 2014) with genotype data from European (CEU) and African (YRI) individuals generated as part of the 1000 Genomes consortium (TGP) (The 1000 Genomes Project Consortium 2015) and performed PCA using the tool smartpca from the EIGENSOFT package (Patterson et al. 2006; Price et al. 2006). Additionally, to identify the extent of non-Native genetic ancestry in each individual, we used the maximum-likelihood–based clustering algorithm ADMIXTURE to infer three (K = 3) ancestral components in each individual (Alexander et al. 2009). For PCA and ADMIXTURE analyses, the genotype data were pruned with plink (Purcell et al. 2007) using the commands –geno 0.1 and –indep 50 5 2.
To avoid the inclusion of markers with European and African genetic ancestry in downstream analyses, we carried out a masking approach of the non-Native genomic segments in admixed individuals. We did this by calling local ancestry tracts in the available genotype data for the individuals in the study and used these calls to mask their respective exome data. To this end, first we merged the genotyping array data available for the admixed HUI, MYA, TAR, and TRQ (Moreno-Estrada et al. 2014) with a reference panel consisting of 30 individuals with 100% Indigenous ancestry, from the same study, as well as with 30 CEU and 30 YRI genotyped on the same array (Affymetrix 6.0) as part of the TGP (supplementary table S2, Supplementary Material online). For admixed NAH, we combined their genotype data from Galanter et al. (2014) with a reference panel consisting of genotype data of 20 individuals with 100% Indigenous ancestry genotyped with the same array (Affymetrix Axiom World Array IV, also known as LAT array for its informativeness in Latino populations), as well as with 20 HapMap CEU and 20 HapMap YRI genotyped in Affymetrix Axiom arrays (supplementary table S3, Supplementary Material online). The number of individuals in each reference population was down sampled to the data available for individuals with 100% Indigenous ancestry for each array (i.e., there were 30 individuals with 100% Indigenous ancestry in the Affymetrix 6.0 array, and 20 in the Axiom LAT) to avoid a potential bias of having more individuals in a reference population than other (Maples et al. 2013).
Genotype data for each set was then phased using SHAPEIT (version 2) (O’Connell et al. 2014) with default parameters. Local ancestry was estimated for the resulting haplotypes using the ‘‘PopPhased’’ routine of RFMix version 1.0.2 (Maples et al. 2013) with parameters –correct-phase (for phase correction) and –G15 (to assume 15 generations since admixture) and no EM iterations. Local ancestry calls in the Viterbi files were then used to mask (make missing) the sites in the exome vcf files that were not part of homozygous Native American-ancestry blocks. We note that, even though it is recommended to use EM iterations in RFMix (Maples et al. 2013), we were confident that the masking of the called European and African segments yielded reliable Native American segments. We corroborated this by a posteriori calling local ancestry with RFMix using two EM iterations on the Affymetrix 6.0 data set and comparing the amount of Native American ancestry detected. We observed that using two EM iterations called more non-Native American segments than when using zero (supplementary table S2, Supplementary Material online). We further inspected the few regions with inconsistent calls between the two runs (with and without EM iterations). For most cases, the European or African segments identified in the run with zero EM iterations were called as Native American in the run with two, suggesting that we would be mainly missing Native American variants and not including European and African ones in the analysis, which would still leave our results free of a bias introduced by including missed European and African segments. Furthermore, we manually inspected the regions in the exome data where the inconsistency was reversed, i.e., regions called as Native with the zero EM iterations and as European or African with two iterations and found that such did not affect the genes identified as likely being under adaptive evolution. Therefore, we conclude that this lack of EM iterations in RFMix is not affecting the inferences made on the masked data set.
Variant Analysis and Annotation
The vcf file was annotated using the tool ANNOVAR (version 2015Jun17) (Wang et al. 2010) with the following reference data sets: refGene, esp6500siv2_all, 1000g2015aug_all, exac03, avsnp142, and ljb26_all.
The script table_annovar.pl was used with the following parameters:
table_annovar.pl -- vcfinput $pop.vcf annovar_humandb/–buildver hg19 -- out $pop - –remove - –protocol refGene, esp6500siv2_all, 1000g2015aug_all, exac03, snp142, clinvar_20160302, dbscsnv11, dbnsfp30a – - operation g, f, f, f, f, f, f, f - –nastring.
New variants were defined as those not present in ExAC (Lek et al. 2016) NHLBI Exome Sequencing Project (ESP) (https://esp.gs.washington.edu; last accessed December 8, 2019), TGP (The 1000 Genomes Project Consortium 2015), and NCBI’s dbSNP142 data sets. These were discovered in the entire data set of 76 exomes and also per population. The annotation of these new variants was retrieved and classified according to their potential effect on transcripts using RStudio (RStudio Team 2015).
Treemix
We used the program Treemix version 1.12 (Pickrell and Pritchard 2012) to infer a tree topology for the relationships among the Indigenous populations in this study. First, we masked the Affymetrix 6.0 genotype data for HUI, MYA, TAR, and TRQ to retain only segments with homozygous Native American ancestry, and merged these with data from CHB individuals genotyped on the same array as part of the TGP (The 1000 Genomes Project Consortium 2015). We used plink to set a missingness filter of 1% (–geno .01), and to calculate allele frequencies (–freq). We used the script plink2treemix.py (provided with treemix) to generate the input file. We then ran 100 bootstrap replicates of treemix setting the root to CHB, selecting the -global and -bootstrap options, requesting 1000 blocks, and defining a randomly generated seed. We then obtained a consensus of the 100 trees using the Geneious Prime 2 2019.2.1 (https://www.geneious.com; last accessed December 8, 2019), and averaged the values for the covariance, covariance errors, and covariance according to the model matrices to plot the residuals of each inferred topology. To infer migrations, we ran treemix with one and two migrations and used the consensus from the 100 bootstrap replicates as the previously generated tree (-g option) (supplementary fig. S7, Supplementary Material online).
Demographic Inference
For demographic inference we used the exome sequencing data from the four NM populations HUI, MYA, TAR, and TRQ, as well as data from Han Chinese (CHB) from TGP. Non-native ancestry was masked in each NM sample as described above. To include only putatively “neutral sites” in the analysis, we limited these to 4-fold (synonymous) and intronic sites determined via SNPEff (Cingolani et al. 2012). While background selection has been reported to have an effect in CDS regions (Naidoo et al. 2018) we don’t expect a biased impact in our demographic inferences given the joint analysis with CHB. Inference was made on the unfolded SFS. We used the panTro4 reference sequence as an outgroup and implemented a context-dependent correction for ancestral misidentification (Hernandez et al. 2007). We estimate the chimpanzee reference genome to have a 0.012 divergence from the human reference (hg19) in our target regions. After removing triallelic sites and sites with a missing outgroup allele, the callable sequence length was 8,889,201 bp and the number of sites used was 20,991.
Dadi
The demographic model was inferred via an approximation to the forward diffusion equation implemented in δαδι (Gutenkunst et al. 2009). This approach infers the best-fitting parameters given a specific demographic model and calculates the log-likelihood of the model fit based on a comparison of the expected to observed SFS. δαδι can handle a maximum of three populations and has difficulty optimizing with more than two populations. Due to this, we optimized over the composite likelihood of six pairwise two-population allele frequency distributions extending the approach from Gravel et al. (2011).We used grids of 40, 50, and 60 grid points per population, and we projected population allele frequencies to the following number of haplotypes: TAR 26; MYA 14; TRQ 16; HUI, 16.
We fixed a population bottleneck in the ancestral population at around 70 KYA (δαδι parameter: t = 0.09), and inferred all other parameters. The timing of the bottleneck was fixed as it has been estimated in previous studies (Li and Durbin 2011) and because δαδι often has difficulties inferring the time and size of a bottleneck. As the expected SFS of multiple bottlenecks looks nearly identical to the SFS of one bottleneck (with a different magnitude), we expect this bottleneck to encompass the loss of diversity in the out-of-Africa expansion and the crossing of the Bering strait (similar to Raghavan et al. 2015). We utilized the topology inferred via Treemix (supplementary fig. S7, Supplementary Material online, Pickrell and Pritchard 2012), and confirmed this as the best-fitting topology in δαδι (supplementary table S6, Supplementary Material online). To convert best-fit parameters to interpretable values, we assumed a generation time of 29 years (Fenner 2005) and a mutation rate of 1.25e-8 mutations per base pair per generation.
Confidence intervals were determined via 1,000 bootstrapped replicates. For each replicate, we divided the genome into 500 kb blocks and removed the blocks that contained no target regions. Then we randomly sampled blocks with replacement and inferred δαδι parameters.
D-Statistics
We computed genotype-based D-statistics using ADMIXTOOLS (Patterson et al. 2012) to test hypothesis of tree-like relationships between populations. We evaluated relationships to the “Ancestral A” (AncA) and “Ancestral B” (AncB) branches as defined in Scheib et al. (2018). We used genomic data from individual Anzick1 from Montana (∼12.8k years old) (Rasmussen et al. 2014) to represent the AncA branch, and genomic data from two present-day Athabascans (Raghavan et al. 2014) to represent the AncB branch. In addition, we included 108 Yoruba individuals (YRI) from the TGP to be used as the outgroup population. We merged these data with the variant data from the exomes of NM and CHB and only considered sites with a minor allele frequency (maf) of 0.10 in YRI. After merging and applying the maf filter, we estimated D-statistics in the forms D (NM1, NM2; AncA, YRI), D (NM1, NM2; AncB, YRI), D (NM1, NM2; CHB, YRI), and D (AncB, AncA; NM, YRI).
Population Branch Statistic
Variants were annotated as ancestral/derived using the orthologous regions in a great ape and rhesus macaque phylogeny as reported by Ensembl Compara and used by the TGP (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase1/analysis_results/supporting/ancestral_alignments/). To estimate PBS for genes in all NM, we used the exome data available as vcf files in the TGP for CHB (used as the second population) and CEU (used as the third population). We used VCFtools (Danecek et al. 2011) to calculate allele frequencies of the derived alleles for each population and then computed pairwise FST values for each gene using Reynold’s FST formula (Reynolds et al. 1983). Only sites with depth above 10× and at least 10 chromosomes per population were included in the calculation. Also, only sites that were polymorphic in at least one of the three populations were considered.
PBS values for each gene in NM since divergence from the CHB were computed using the formula:
where
We used R (R Core Team 2014) to select genes at the top 1% of the overall PBS distribution and to confirm these were also at the top of their gene size (SNPs/gene) bin (fig. 2b).
To compute PBS values for genes in a specific NM population (NM1) since their divergence from a second NM (NM2) population, we implemented the same filters as above (only sites with depth ≥10× and at least ten chromosomes per population) and used CHB as the third population and the formula:
Since we considered four NM (HUI, MYA, TAR, and TRQ), there was a total of 12 possible NM1-NM2 comparisons. To evaluate the significance of the PBS values in specific NM populations, we compared them to a null distribution of simulated neutral sequences and followed the method of Yi et al. (2010). These simulated sequences were generated in ms (Hudson 2002) with our inferred 4-population NM demography and used CHB as an outgroup. We estimated the CHB-NM split time by averaging the inferred split time and bottleneck Ne between the CHB and each NM population.
Allele frequencies were calculated from sequences comprising 700k simulated genes with 1–80 SNPs per gene. PBS values on the simulated data were then calculated using the same filters and formula as for the observed data. Because of filters (at least ten chromosomes per population and only polymorphic sites), some of the simulated sites were disregarded causing a change in the number of available simulations for each SNPs/gene bin. Because of this we randomly subsampled 500k simulations for each SNPs/gene bin category, which covered all bins between 1 and 71 SNPs/gene.
Observed PBS values were then compared to the simulated values in their corresponding SNPs/gene bin category. A P-value was calculated by observing the fraction of simulated PBS values larger than the observed PBS. For example, if there was only one simulated PBS value larger than the observed one, the P-value corresponded to a P-value of 0.000002 (1/500,000). For genes with 1–71 SNPs/gene, observed values were compared to their respective simulated bins. Observed PBS values for genes with >71 SNPs were compared to the simulated PBS distribution of 71 SNPs/gene.
Functional Enrichment Analysis
To perform the enrichment analysis, we first defined an intersecting subset of genes for each population considering only the genes in all pairwise comparison showing extreme PBS values (P-value < 0.05) (e.g., the intersection of the subsets TAR-HUI, TAR-MYA, and TAR-TRQ generates list of intersecting genes for the Tarahumara).
For GO enrichment, we used the online tool in http://www.geneontology.org/page/go-enrichment-analysis; last accessed December 8, 2019. We analyzed each of the four gene lists with the three GO categories (biological processes, cellular components, and molecular function) using FRD correction (ran on August 25, 2019).
To test for enrichment in GO categories among genes with novel missense SNV, we used WebGestalt online tool (Wang et al. 2013) as reported in Romero-Hidalgo et al. (2017).
We ran a pathway over-representation analysis on the same gene sets using the IMPaLA online tool (Kamburov et al. 2011) available at: http://impala.molgen.mpg.de (ran on August 25, 2019) and considered only pathways with a Q-value less than 0.05. Most results had a Q-value of 1, representing an enriched category with a 100% of probability of being a false positive, even when they exhibited low P-value scores. We selected the threshold of a 0.05 score as it implies 5% of the results with that corresponding P values are false positives.
Supplementary Material
Acknowledgments
We thank the participants and volunteers who donated DNA samples and the fieldwork teams led by Hector Rangel, Victor Acuña, and Leonor Buentello on the various sampling expeditions. We thank M.C. Yee, M.L. Carpenter, Alejandra Castillo Carbajal, and Carina Uribe Díaz for technical support, and E. Huerta-Sánchez, Diego Ortega del Vecchyo, and Federico Sánchez-Quinto for input on early versions of the manuscript. We deeply thank the generous support from the Beijing Genomics Institute (BGI) for contributing with sequencing capacity, and the Stanford Center for Computational, Evolutionary and Human Genomics (CEHG) for supporting the initial stages of this project. We thank IT support from Luis Aguilar, Alejandro de León, Carlos S. Flores, and Jair García of the Laboratorio Nacional de Visualización Científica Avanzada at UNAM. This work was supported by Mexico’s CONACYT Basic Research Program (grant number CB-2015-01-251380 awarded to A.M.-E.) and the International Center for Genetic Engineering and Biotechnology (ICGEB) (grant number CRP/MEX15-04_EC awarded to A.M.-E). M.C.A.-A. was partially supported with a fellowship from the 2012 George Rosenkranz Prize for Health Care Research in Developing Countries awarded to A.M.-E. M.C.A.-A.’s laboratory was supported by Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica, Universidad Nacional Autónoma de México (grant number IA206817) and CONACYT Infrastructure (grant number 26944).
References
- Acuna-Soto R, Stahle DW, Cleaveland MK, Therrell MD.. 2002. Megadrought and megadeath in 16th century Mexico. Emerg Infect Dis. 8(4):360.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19(9):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene Ontology: tool for the unification of biology. Nat Genet. 25(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, Penninx BWJH, Janssens ACJW, Wilson JF, Spector T, et al. ; the ENGAGE Consortium. 2009. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 41(1):47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge MN, Wang M, Wu Y, Newsham I, Muzny DM, Jefferies JL, Albert TJ, Burgess DL, Gibbs RA.. 2011. Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities. Genome Biol. 12(7):R68.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balke B, Snow C.. 1965. Anthropological and physiological observations on Tarahumara endurance runners. Am J Phys Anthropol. 23(3):293–301. [DOI] [PubMed] [Google Scholar]
- Bandarian F, Hedayati M, Daneshpour MS, Naseri M, Azizi F.. 2013. Genetic polymorphisms in the APOA1 gene and their relationship with serum HDL cholesterol levels. Lipids 48(12):1207–1216. [DOI] [PubMed] [Google Scholar]
- Belbin GM, Nieves-Colón MA, Kenny EE, Moreno-Estrada A, Gignoux CR.. 2018. Genetic diversity in populations across Latin America: implications for population and medical genetic studies. Curr Opin Genet Dev. 53:98–104. [DOI] [PubMed] [Google Scholar]
- Bergström A, McCarthy SA, Hui R, Almarri MA, Ayub Q, Danecek P, Chen Y, Felkel S, Hallast P, Kamm J, et al. 2019. Insights into human genetic variation and population history from 929 diverse genomes. bioRxiv: 674986. [DOI] [PMC free article] [PubMed]
- Brumfiel EM. 1983. Aztec state making: ecology, structure, and the origin of the state. Am Anthropol. 85(2):261–284. [Google Scholar]
- Bustamante CD, De La Vega FM, Burchard EG.. 2011. Genomics for the world. Nature 475(7355):163–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatters JC, Kennett DJ, Asmerom Y, Kemp BM, Polyak V, Blank AN, Beddows PA, Reinhardt E, Arroyo-Cabrales J, Bolnick DA, et al. 2014. Late Pleistocene human skeleton and mtDNA link paleoamericans and modern Native Americans. Science 344(6185):750–754. [DOI] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM.. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: sNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coan HB, Curran JE, Dyer TD, Kent JW, Choudary A, Nicolella DP, Carless MA, Kumar S, Almeida MA, Duggirala R, et al. 2013. Variation in osteoarthritis biomarker serum comp levels in Mexican Americans is associated with SNPs in a region of chromosome 22q encompassing MICAL3, BCL2L13, and BID. Osteoarthritis Cartilage 21:S172. [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulhaber J. 2014. Anthopometry of Living Indians In: Handbook of Middle American Indians. Vol. 9. Austin (TX: ): University of Texas Press; p. 82–104. [Google Scholar]
- Fenner JN. 2005. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am J Phys Anthropol. 128(2):415–423. [DOI] [PubMed] [Google Scholar]
- Fumagalli M, Cagliani R, Riva S, Pozzoli U, Biasin M, Piacentini L, Comi GP, Bresolin N, Clerici M, Sironi M.. 2010. Population genetics of IFIH1: ancient population structure, local selection, and implications for susceptibility to type 1 diabetes. Mol Biol Evol. 27(11):2555–2566. [DOI] [PubMed] [Google Scholar]
- Galanter JM, Gignoux CR, Torgerson DG, Roth LA, Eng C, Oh SS, Nguyen EA, Drake KA, Huntsman S, Hu D, et al. 2014. GWAS and admixture mapping identify different asthma-associated loci in Latinos: the GALA II Study. J Allergy Clin Immunol. 134(2):295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel T, Waters MR, O'Rourke DH.. 2008. The Late Pleistocene dispersal of modern humans in the Americas. Science 319(5869):1497–1502. [DOI] [PubMed] [Google Scholar]
- Gonzalez S, Jiménez-López JC, Hedges R, Huddart D, Ohman JC, Turner A, Pompa y Padilla JA.. 2003. Earliest humans in the Americas: new evidence from México. J Hum Evol. 44(3):379–387. [DOI] [PubMed] [Google Scholar]
- Gorostiza A, Acunha-Alonzo V, Regalado-Liu L, Tirado S, Granados J, Sámano D, Rangel-Villalobos H, González-Martín A.. 2012. Reconstructing the history of mesoamerican populations through the study of the mitochondrial DNA control region. PLoS One 7(9):e44666.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Henn BM, Gutenkunst RN, Indap AR, Marth GT, Clark AG, Yu F, Gibbs RA, Bustamante CD, Altshuler DL, et al. 2011. Demographic history and rare allele sharing among human populations. Proc Natl Acad Sci U S A. 108(29):11983–11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD.. 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5(10):e1000695.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez RD, Williamson SH, Bustamante CD.. 2007. Context dependence, ancestral misidentification, and spurious signatures of natural selection. Mol Biol Evol. 24(8):1792–1800. [DOI] [PubMed] [Google Scholar]
- Hudson RR. 2002. Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics 18(2):337–338. [DOI] [PubMed] [Google Scholar]
- INEGI. 2015. Encuesta intercensal 2015. Available from: https://www.inegi.org.mx/programas/intercensal/2015/, last accessed December 8, 2019.
- Kamburov A, Cavill R, Ebbels TMD, Herwig R, Keun HC.. 2011. Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics 27(20):2917–2918. [DOI] [PubMed] [Google Scholar]
- Kim J-J, Lee H-I, Park T, Kim K, Lee J-E, Cho NH, Shin C, Cho YS, Lee J-Y, Han B-G, et al. 2010. Identification of 15 loci influencing height in a Korean population. J Hum Genet. 55(1):27–31. [DOI] [PubMed] [Google Scholar]
- Lamason RL, Mohideen M-A, Mest JR, Wong AC, Norton HL, Aros MC, Jurynec MJ, Mao X, Humphreville VR, Humbert JE, et al. 2005. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310(5755):1782–1786. [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536(7616):285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2011. Inference of human population history from individual whole-genome sequences. Nature 475(7357):493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Wang H, Jin Y, Podolsky R, Reddy MPL, Pedersen J, Bode B, Reed J, Steed D, Anderson S, et al. 2008. IFIH1 polymorphisms are significantly associated with type 1 diabetes and IFIH1 gene expression in peripheral blood mononuclear cells. Hum Mol Genet. 18(2):358–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundová T, Štambergová H, Zemanová L, Svobodová M, Havránková J, Šafr M, Wsól V.. 2016. Human dehydrogenase/reductase (SDR family) member 8 (DHRS8): a description and evaluation of its biochemical properties. Mol Cell Biochem. 411(1–2):35–42. [DOI] [PubMed] [Google Scholar]
- Mallick CB, Iliescu FM, Möls M, Hill S, Tamang R, Chaubey G, Goto R, Ho SYW, Romero IG, Crivellaro F, et al. 2013. The light skin allele of SLC24A5 in South Asians and Europeans shares identity by descent. PLoS Genet. 9:e1003912.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maples BK, Gravel S, Kenny EE, Bustamante CD.. 2013. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet. 93(2):278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnetto D, Huerta-Sánchez E.. 2017. Haplostrips: revealing population structure through haplotype visualization. Methods Ecol Evol. 8(10):1389–1392. [Google Scholar]
- Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, Daly MJ, Bustamante CD, Kenny EE.. 2017. Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet. 100(4):635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Estrada A, Gignoux CR, Fernández-López JC, Zakharia F, Sikora M, Contreras AV, Acuña-Alonzo V, Sandoval K, Eng C, Romero-Hidalgo S, et al. 2014. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science 344(6189):1280–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Mayar JV, Vinner L, Peter de Barros D, Fuente C, de la Chan J, Spence JP, Allentoft ME, Vimala T, Racimo F, Pinotti T, et al. 2018. Early human dispersals within the Americas. Science 362(6419):eaav2621. [DOI] [PubMed] [Google Scholar]
- Naidoo T, Sjödin P, Schlebusch C, Jakobsson M.. 2018. Patterns of variation in cis-regulatory regions: examining evidence of purifying selection. BMC Genomics 19(1):95.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell J, Gurdasani D, Delaneau O, Pirastu N, Ulivi S, Cocca M, Traglia M, Huang J, Huffman JE, Rudan I, et al. 2014. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 10:e1004234.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouatou S, Ajjemami M, Charoute H, Sefri H, Ghalim N, Rhaissi H, Benrahma H, Barakat A, Rouba H.. 2014. Association of APOA5 rs662799 and rs3135506 polymorphisms with arterial hypertension in Moroccan patients. Lipids Health Dis. 13(1):60.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D.. 2006. Population structure and eigenanalysis. PLoS Genet. 2(12):e190.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson NJ, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D.. 2012. Ancient admixture in human history. Genetics 112:145037.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Olivier M, Hubacek JA, Krauss RM, Rubin EM, Cohen JC.. 2002. Two independent apolipoprotein A5 haplotypes influence human plasma triglyceride levels. Hum Mol Genet. 11(24):3031–3038. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK.. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8:e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popejoy AB, Fullerton SM.. 2016. Genomics is failing on diversity. Nat News 538(7624):161.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posth C, Nakatsuka N, Lazaridis I, Skoglund P, Mallick S, Lamnidis TC, Rohland N, Nägele K, Adamski N, Bertolini E, et al. 2018. Reconstructing the deep population history of Central and South America. Cell. 175(5):1185–1197.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D.. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 38(8):904–909. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team 2014. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Available from: http://www.R-project.org/; last accessed December 8, 2019.
- Racimo F, Marnetto D, Huerta-Sánchez E.. 2016. Signatures of archaic adaptive introgression in present-day human populations. Mol Biol Evol. 34:296–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan M, DeGiorgio M, Albrechtsen A, Moltke I, Skoglund P, Korneliussen TS, Grønnow B, Appelt M, Gulløv HC, Friesen TM, et al. 2014. The genetic prehistory of the New World Arctic. Science 345(6200):1255832. [DOI] [PubMed] [Google Scholar]
- Raghavan M, Steinrücken M, Harris K, Schiffels S, Rasmussen S, DeGiorgio M, Albrechtsen A, Valdiosera C, Ávila-Arcos MC, Malaspinas A-S, et al. 2015. Genomic evidence for the Pleistocene and recent population history of Native Americans. Science 349(6250):aab3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M, Anzick SL, Waters MR, Skoglund P, DeGiorgio M, Stafford TW Jr,, Rasmussen S, Moltke I, Albrechtsen A, Doyle SM, et al. 2014. The genome of a Late Pleistocene human from a Clovis burial site in western Montana. Nature 506(7487):225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Patterson N, Campbell D, Tandon A, Mazieres S, Ray N, Parra MV, Rojas W, Duque C, Mesa N, et al. 2012. Reconstructing Native American population history. Nature 488(7411):370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC.. 1983. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics 105(3):767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Hidalgo S, Ochoa-Leyva A, Garcíarrubio A, Acuña-Alonzo V, Antúnez-Argüelles E, Balcazar-Quintero M, Barquera-Lozano R, Carnevale A, Cornejo-Granados F, Fernández-López JC, et al. 2017. Demographic history and biologically relevant genetic variation of Native Mexicans inferred from whole-genome sequencing. Nat Commun. 8(1):1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Team. 2015. RStudio: integrated development environment for R. Boston (MA): RStudio, Inc. Available from: http://www.rstudio.com/; last accessed December 8, 2019.
- Scheib CL, Li H, Desai T, Link V, Kendall C, Dewar G, Griffith PW, Mörseburg A, Johnson JR, Potter A, et al. 2018. Ancient human parallel lineages within North America contributed to a coastal expansion. Science 360(6392):1024–1027. [DOI] [PubMed] [Google Scholar]
- Schiffels S, Durbin R.. 2014. Inferring human population size and separation history from multiple genome sequences. Nat Genet. 46(8):919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Zolezzi I, Hidalgo-Miranda A, Estrada-Gil J, Fernandez-Lopez JC, Uribe-Figueroa L, Contreras A, Balam-Ortiz E, Bosque-Plata L, del Velazquez-Fernandez D, Lara C, et al. 2009. Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico. Proc Natl Acad Sci U S A. 106(21):8611–8616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. 2017. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 45:D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernot B, Tucci S, Kelso J, Schraiber JG, Wolf AB, Gittelman RM, Dannemann M, Grote S, McCoy RC, Norton H, et al. 2016. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science 352(6282):235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Duncan D, Shi Z, Zhang B.. 2013. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 41(W1):W77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H.. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZXP, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, et al. 2010. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329:75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Wang C, Liang L, Shen Z, Fu J, Liu P, Lv L, Zhu Y.. 2014. Triglyceride-raising APOA5 genetic variants are associated with obesity and non-HDL-C in Chinese children and adolescents. Lipids Health Dis. 13(1):93.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.