Abstract
Regulatory networks control the spatiotemporal gene expression patterns that give rise to and define the individual cell types of multicellular organisms. In eumetazoa, distal regulatory elements called enhancers play a key role in determining the structure of such networks, particularly the wiring diagram of “who regulates whom.” Mutations that affect enhancer activity can therefore rewire regulatory networks, potentially causing adaptive changes in gene expression. Here, we use whole-tissue and single-cell transcriptomic and chromatin accessibility data from mouse to show that enhancers play an additional role in the evolution of regulatory networks: They facilitate network growth by creating transcriptionally active regions of open chromatin that are conducive to de novo gene evolution. Specifically, our comparative transcriptomic analysis with three other mammalian species shows that young, mouse-specific intergenic open reading frames are preferentially located near enhancers, whereas older open reading frames are not. Mouse-specific intergenic open reading frames that are proximal to enhancers are more highly and stably transcribed than those that are not proximal to enhancers or promoters, and they are transcribed in a limited diversity of cellular contexts. Furthermore, we report several instances of mouse-specific intergenic open reading frames proximal to promoters showing evidence of being repurposed enhancers. We also show that open reading frames gradually acquire interactions with enhancers over macroevolutionary timescales, helping integrate genes—those that have arisen de novo or by other means—into existing regulatory networks. Taken together, our results highlight a dual role of enhancers in expanding and rewiring gene regulatory networks.
Keywords: enhancer, de novo genes, regulatory networks, gene regulation, transcription
Introduction
Enhancers are a defining characteristic of eumetazoan gene regulatory networks. They recruit transcription factors and cofactors that “loop out” DNA to bind core promoters and increase the expression of target genes (Catarino and Stark 2018; Haberle and Stark 2018), thus mediating interactions between genes. Such interactions are highly dynamic throughout development, facilitating the differential deployment of distinct regulatory subnetworks in different cells, which helps define cell-type–specific spatiotemporal gene expression patterns (Davidson and Levine 2008; Spitz and Furlong 2012).
Enhancer activity is dynamic not only throughout development but also throughout evolution (Villar et al. 2015). The reason is that mutations in enhancer sequences can create or ablate interactions with regulatory proteins, thus enabling modifications in gene use without affecting gene product (Prud’homme et al. 2007; Carroll 2008). Such changes alter a regulatory network’s wiring diagram of “who regulates whom,” which can cause changes in gene expression patterns that embody or lead to evolutionary adaptations or innovations (Peter and Davidson 2011). Examples include the archetypical pentadactyl limb anatomy of extant tetrapods (Kherdjemil et al. 2016), ocular regression in subterranean rodents (Partha et al. 2017; Roscito et al. 2018), limb loss in snakes (Kvon et al. 2016; Roscito et al. 2018), convergent pigmentation patterns in East African cichlids (Kratochwil et al. 2018), the diversity of butterfly wing patterns (Barton et al. 2016), the mammalian neocortex (Emera et al. 2016), and cell-type diversity in eumetazoans (Sebé-Pedrós, Chomsky, et al. 2018; Sebé-Pedrós, Saudemont, et al. 2018).
Regulatory networks evolve not only via rewiring but also via the addition of new genes (Teichmann and Babu 2004). Gene duplication, fusion, retrotransposition, the domestication of genomic parasites, and horizontal gene transfer are all means by which new genes can arise from preexisting genes (Kaessmann 2010), and thus expand gene regulatory networks. In addition, it is becoming increasingly appreciated that new genes can arise de novo from noncoding regions of the genome (Carvunis et al. 2012; Betran et al. 2013; Li et al. 2014; Kim et al. 2015; McLysaght and Hurst 2016; Van Oss and Carvunis 2019; Willemsen et al. 2019). For protein-coding genes, the essential prerequisites of this process are the formation of an open reading frame (ORF), together with the transcription and translation of that ORF. Because much of the genome is transcribed (Kapranov et al. 2007; Neme and Tautz 2016) and many lineage-specific transcripts containing ORFs show evidence of translation (Wilson and Masel 2011; Ingolia et al. 2014; Ruiz-Orera et al. 2014, 2018; Prabh and Rödelsperger 2016; Schmitz et al. 2018; Ruiz-Orera and Alba 2019; Zhang et al. 2019), the de novo evolution of new protein-coding genes is also a likely contributor to the growth of gene regulatory networks.
An important question concerning new genes—those that have arisen de novo or by other means—is how they integrate into existing regulatory networks, and what role enhancers may play in this process. It has been hypothesized that enhancer acquisition allows new genes to expand their breadth of expression, providing opportunities to acquire new functions in different cellular contexts (Tautz and Domazet-Loso 2011). Enhancers may therefore help new genes integrate into existing regulatory networks via edge formation and rewiring. Less appreciated is the role enhancers may play in the origin of de novo genes (Wu and Sharp 2013), and thus in the growth of gene regulatory networks. The physical proximity between active enhancers and their target genes (Levine et al. 2014)—facilitated by DNA looping—creates a transcriptionally permissive environment that is engaged with RNA polymerase II, which may lead to the transcription of DNA near the enhancer, or to the transcription of the enhancer itself, producing so-called enhancer RNA (De Santa et al. 2010; Kim et al. 2010; Li et al. 2016; Haberle and Stark 2018). If the transcript contains an ORF, then such increased transcription will increase the likelihood of interaction with ribosomes, and because enhancers are typically active in a small number of cell types (He et al. 2014), interactions with ribosomes will occur in a limited diversity of cellular contexts. This may help purge toxic peptides and enrich for benign peptides, a process that has been hypothesized to increase the likelihood of de novo gene birth (Wilson and Masel 2011). Moreover, similarities in the architectures of enhancers and promoters may facilitate the regulatory repurposing of the former into the latter (Carelli et al. 2018), reinforcing the transcription of new ORFs that emerge near enhancers. Thus, enhancers may play a dual role in the evolution of de novo genes, and consequently in the evolution of gene regulatory networks. By creating a transcriptionally permissive environment, enhancers may facilitate the origin of de novo genes; by physically interacting with gene promoters, enhancers may facilitate the integration of new genes—those emerging de novo or by other means—into existing regulatory networks.
The first evidence that enhancers can facilitate de novo gene birth was recently provided using whole-animal transcriptomic and epigenetic data from the nematode Pristionchus pacificus (Werner et al. 2018). Specifically, the transcription start sites of expressed genes that were in open chromatin and private to P. pacificus were found to be in closer proximity to histone modifications indicative of enhancers than the transcription start sites of expressed genes that were in open chromatin and shared with other nematode species. Although this evidence is compelling, additional systematic analyses are required to draw firm conclusions and to address remaining open questions. For example, we do not yet know about the generality of this mechanism, specifically whether it applies to other clades of eumetazoa. Furthermore, information on the stability of the transcribed ORFs or their potential for translation is still lacking. We also do not know about the cell-type specificity of the enhancers that facilitate de novo gene birth (because the data used to study P. pacificus were derived from the whole animal) or how the facilitating role of enhancers in de novo gene birth differs from that of other means of pervasive transcription (Neme and Tautz 2016). Finally, we do not know how enhancers integrate new genes into existing cellular networks, especially over macroevolutionary timescales.
Here, we take an integrative approach to address these open questions and to study the potential dual role of enhancers in the evolution of gene regulatory networks. We leverage whole-tissue and single-cell transcriptomic and functional genomics data from mouse that describe gene expression levels, chromatin accessibility, and chemical modifications to histones, as well as phylostratigraphic estimates of the ages of transcribed ORFs. We find young ORFs are preferentially located near enhancers, whereas older ORFs are not. Some of these young ORFs likely are enhancers, as evidenced by their balanced bidirectional transcription—a hallmark of enhancer activity. Mouse-specific intergenic ORFs that are proximal to enhancers are more highly and stably transcribed than mouse-specific intergenic ORFs that are not proximal to enhancers or promoters, and they are transcribed in more cellular contexts, thus highlighting fundamental differences between the facilitating role of enhancers versus other forms of pervasive transcription in de novo gene birth. We find the transcripts of enhancer-proximal ORFs often associate with ribosomes, and we uncover several instances of mouse-specific intergenic ORFs that are proximal to promoters that are likely repurposed enhancers. Finally, we show the number of enhancer interactions per ORF increases with ORF age, which correlates with an increase in expression breadth, even across macroevolutionary timescales. In sum, our findings support a dual role for enhancers in the origin of de novo genes and the integration of genes into regulatory networks.
Results
Mouse-Specific Intergenic ORFs Are Often Proximal to Enhancers
We considered a set of 56,262 ORFs from transcripts expressed in the liver, brain, and testis of mouse. Previous work assigned phylogenetic ages to these ORFs (Schmitz et al. 2018), based on the presence of homologous sequences in the transcriptomes of other mammalian species, including rat, human, and opossum (fig. 1A). We further classified the mouse-specific ORFs as genic or intergenic, based on whether or not they are proximal to older, annotated genes (Materials and Methods). We use the term proximal to mean within 500 bp (in the supplementary material, Supplementary Material online, we show our findings are qualitatively insensitive to changing this definition to 250 and 1,000 bp, see supplementary figs. S5–S9, Supplementary Material online), and we use an ORF’s first exon to calculate its distance from other genomic features. To characterize the regulatory background of an ORF, we considered data describing histone modifications that are indicative of promoters and enhancers (Heintzman et al. 2007). Specifically, we merged chromatin immunoprecipitation followed by DNA sequencing (ChIP-seq) data for H3K27ac, H3K4me1, and H3K4me3 obtained from 23 mouse tissues and cell types (ENCODE 2012). We considered enhancers to be those genomic regions where H3K27ac and/or H3K4me1 peaks do not overlap H3K4me3 peaks in any tissue, and promoters to be those genomic regions with H3K4me3 peaks (Creyghton et al. 2010; Berthelot et al. 2018) (Materials and Methods).
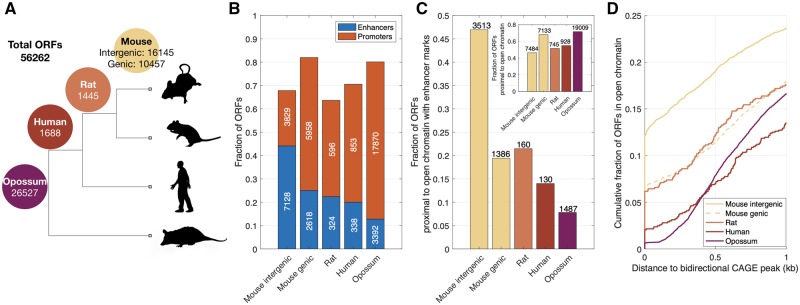
FIg. 1.
Mouse-specific intergenic ORFs are often proximal to enhancers. (A) Phylogeny showing the four age classes of the 56,262 ORFs. The numbers on the branches indicate the number of ORFs that are either mouse-specific or shared with rat, human, and opossum. Mouse-specific ORFs are further classified as intergenic or genic. (B) Fraction of ORFs that are proximal to ChIP-seq peaks indicative of enhancers (H3K27ac and/or H3K4me1 without overlapping H3K4me3) or promoters (H3K4me3), shown in relation to ORF class. (C) Fraction of ORFs that are proximal to regions of open chromatin that contain enhancers, but not promoters, shown in relation to ORF class. The inset shows the fraction of ORFs that are proximal to regions of open chromatin, regardless of whether those regions contain promoters or enhancers. (D) Cumulative fraction of ORFs that are proximal to regions of open chromatin, shown in relation to their distance to the closest bidirectional CAGE peak. The raw data underlying these and all subsequent visualizations are provided in supplementary data files 1–5, Supplementary Material online, along with the Matlab scripts used to generate the visualizations.
The majority of ORFs in each age class are proximal to a promoter or an enhancer (fig. 1B). Remarkably, mouse-specific intergenic ORFs are the only class of ORFs that are more likely to be proximal to enhancers than to promoters. Although the first exons of nearly 45% (7,128) of mouse-specific intergenic ORFs are proximal to enhancers, fewer than 25% of rat, human, and opossum-shared ORFs are proximal to enhancers. Similar trends are observed when we restrict our attention to ORFs that are within, or proximal to, genomic regions of open chromatin in at least one of 13 mouse tissues (fig. 1C; Materials and Methods). Specifically, ∼47% (3,513 out of 7,484) of mouse-specific intergenic ORFs are proximal to regions of open chromatin that harbor histone modifications indicative of enhancers, but not promoters, whereas fewer than 21% of rat, human, and opossum-shared ORFs are proximal to such regions. Similar trends are also observed when we consider histone modification data from individual tissues, as opposed to merging data across cell and tissue types. Specifically, 25% (281 ORFs), ∼36% (897 ORFs), and ∼20% (537 ORFs) of intergenic mouse-specific ORFs that are in open chromatin and expressed in liver, brain, and testis, respectively, are proximal to an enhancer in that tissue, as compared with <10% of genic and older ORFs, which are instead preferentially proximal to promoters (supplementary fig. S2D–F, Supplementary Material online). Finally, mouse-specific intergenic ORFs are more likely to show evidence of balanced bidirectional transcription—a hallmark of enhancer activity (Andersson et al. 2014)—than any other class of ORFs (fig. 1D), with 12% of the ORFs overlapping a bidirectional capped analysis of gene expression (CAGE) peak and nearly 20% (1,429) of the ORFs proximal to a bidirectional CAGE peak (supplementary fig. S1, Supplementary Material online, shows that these trends are not driven by exon length). Taken together, these results support a model in which enhancers facilitate the expression of young ORFs (Wu and Sharp 2013; Werner et al. 2018).
Intergenic ORFs That Are Proximal to Enhancers Are Highly and Stably Transcribed, Relative to Intergenic ORFs That Are Not Proximal to Enhancers or Promoters
We next asked what differentiates the facilitating role of enhancers in de novo gene birth from other forms of pervasive transcription taking place away from promoters and enhancers. We hypothesized that because enhancers are regularly engaged with the transcriptional machinery, they may confer higher levels of expression and greater expression stability. To test this hypothesis, we compared the expression levels and stabilities of intergenic mouse-specific ORFs that are proximal to enhancers with those of intergenic mouse-specific ORFs that are not proximal to enhancers or promoters, using transcriptomic, histone modification, and chromatin accessibility data from liver, brain, and testis (Materials and Methods).
In all three tissues, we observed that mouse-specific intergenic ORFs that are proximal to enhancers have a higher median expression level than mouse-specific intergenic ORFs that are not proximal to enhancers or promoters (fig. 2A; Wilcoxon signed-rank test, P < 0.001 in liver, P = 0.003 in brain, and P = 0.02 in testis). To measure expression stability, we calculated the entropy of expression across biological replicates (Materials and Methods). When this measure equals its minimum of 0, the ORF is expressed in only one of the replicates; when it equals its maximum of 1, the ORF is expressed at equal levels across replicates. In all three tissues, we observed that expression stability is higher for mouse-specific intergenic ORFs that are proximal to enhancers than for mouse-specific intergenic ORFs that are not proximal to enhancers or promoters (fig. 2B; Wilcoxon’s signed-rank test, P < 0.001). These observations support the hypothesis that enhancers confer higher expression levels and greater expression stability to proximal ORFs than do other forms of pervasive transcription away from promoters and enhancers.
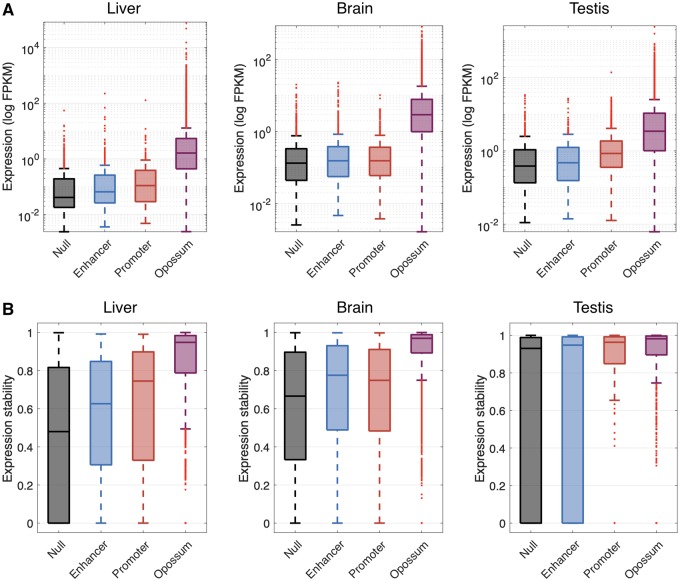
FIg. 2.
Mouse-specific intergenic ORFs that are proximal to enhancers are more highly expressed and have greater expression stability than mouse-specific intergenic ORFs that are not proximal to enhancers or promoters. (A) Expression level of mouse-specific intergenic ORFs proximal to enhancers, promoters, or neither (“Null”) in liver, brain, and testis. (B) Expression stability of mouse-specific intergenic ORFs proximal to enhancers, promoters, or neither (“Null”) in liver (eight replicates), brain (eight replicates), and testis (two replicates). The expression levels and stabilities of opossum-shared ORFs are shown as a point of comparison.
In liver and testis, we observed that mouse-specific intergenic ORFs that are proximal to enhancers have lower median expression levels and stabilities than mouse-specific intergenic ORFs that are proximal to promoters (fig. 2A and B; Wilcoxon signed-rank test, P = 0.03 and P < 0.001 for expression level, and P = 0.02 and P < 0.001 for expression stability, in liver and testis, respectively). This observation is consistent with previous analyses of transcription emerging from enhancers and promoters, which showed that enhancers drive lower and less stable expression than promoters, despite the architectural similarities of these regulatory elements (Core et al. 2014). The increased expression levels and stabilities of promoter-associated transcription may derive from the sequence features of the corresponding transcripts, including the presence or absence of early polyadenylation sites and splicing signals, which are conducive to transcriptional elongation and may contribute to a positive feedback loop wherein elongation promotes subsequent rounds of initiation (Core et al. 2014).
Intergenic ORFs That Are Proximal to Enhancers Are More Likely to Associate with Ribosomes than Intergenic ORFs That Are Not Proximal to Enhancers or Promoters
Many noncoding transcripts associate with ribosomes (Wilson and Masel 2011; Ingolia et al. 2014; Ruiz-Orera et al. 2014; Zhang et al. 2015). It has been suggested that this may enrich the pool of transcribed ORFs for benign peptides, thus increasing the likelihood of de novo gene birth (Wilson and Masel 2011). We hypothesized that because of their increased expression levels and stabilities, mouse-specific intergenic ORFs that are proximal to enhancers will be more likely to associate with ribosomes than mouse-specific intergenic ORFs that are not proximal to enhancers or promoters. To test this hypothesis, we considered liver, brain, and testis data from a ribosomal profiling assay called ribo-seq, which describes the transcriptome-wide binding patterns of ribosomes to RNA molecules (Ruiz-Orera et al. 2018; Ruiz-Orera and Alba 2019) (Materials and Methods).
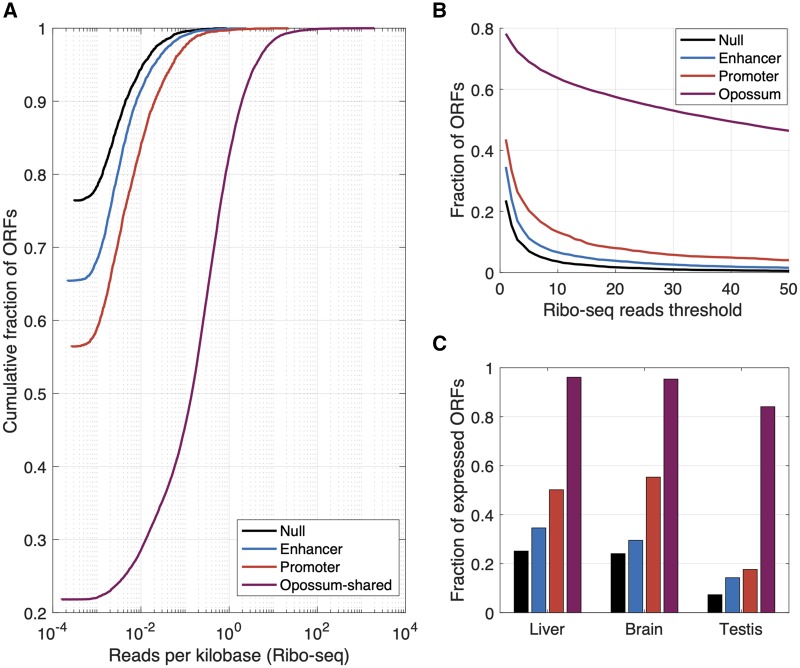
Following Schmitz et al. (2018), we first consider a permissive definition of ribosomal association: at least one read mapping to the first exon of an ORF. We found that mouse-specific intergenic ORFs that are proximal to enhancers are ∼10% more likely to associate with ribosomes than mouse-specific intergenic ORFs that are not proximal to enhancers or promoters, and ∼10% less likely to associate with ribosomes than mouse-specific intergenic ORFs that are proximal to promoters (fig. 3A). When we apply more conservative thresholds for ribosomal association, mouse-specific intergenic ORFs that are proximal to enhancers remain more likely to associate with ribosomes than mouse-specific intergenic ORFs that are not proximal to enhancers or promoters, and less likely than mouse-specific intergenic ORFs that are proximal to promoters, although the differences in ribosomal association between these classes decreases as the threshold for ribosomal association increases, both when evaluating reads per kilobase mapped to the first exon (fig. 3A), or simply total number of reads mapped to the first exon (fig. 3B). These trends remain when considering tissue-specific transcriptomic, histone modification, and ribosomal association data for liver, brain, and testis (fig. 3C).
FIg. 3.
Mouse-specific intergenic ORFs that are proximal to enhancers are more likely to associate with ribosomes than mouse-specific intergenic ORFs that are not proximal to enhancers or promoters. (A) Cumulative fractions of mouse-specific intergenic ORFs that are proximal to an enhancer, a promoter, or neither (“Null”), or to opossum-shared ORFs, shown in relation to the number of ribo-seq reads mapped per kilobase to the first exon of each ORF. (B) Fraction of ORFs with ribosomal association, shown in relation to the minimum threshold for the number of reads mapped. (C) Fraction of ORFs expressed in liver, brain, and testis for which at least one tissue-specific ribo-seq read could be mapped to their first exon. The color scheme is the same as in (A).
Intergenic ORFs That Are Proximal to Enhancers Are Expressed in More Cellular Contexts than Intergenic ORFs That Are Not Proximal to Enhancers or Promoters
In the model of enhancer-facilitated de novo gene birth studied here, ORFs emerging near enhancers are likely to have their expression restricted to cells where those enhancers are active. Enhancers are often specific to a small number of cell types (He et al. 2014), which may reduce the potential for enhancer-proximal ORFs to have deleterious pleiotropic effects, while simultaneously exposing the ORFs to a range of cellular contexts in which they may confer a selective advantage. To study the breadth of expression of ORFs, we considered two sources of data: whole tissue measurements of total RNA across 10 tissues and single-cell measurements of open chromatin across 38 cell types (Materials and Methods).
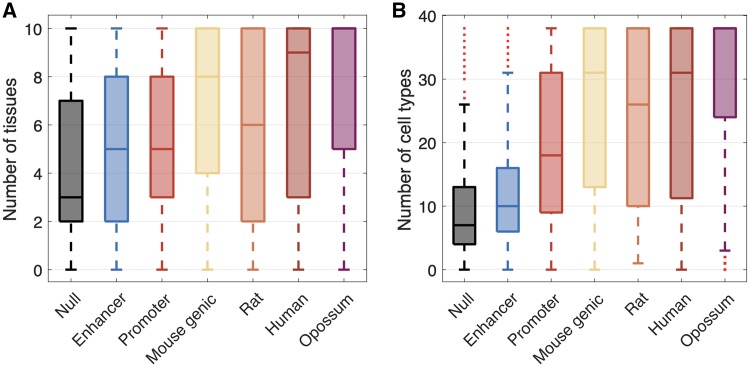
We found that mouse-specific intergenic ORFs that are proximal to enhancers are expressed in more tissues (Wilcoxon’s signed-rank test, P < 0.001; fig. 4A) and are in open chromatin in more cell types (Wilcoxon’s signed-rank test, P < 0.001; fig. 4B) than mouse-specific intergenic ORFs that are not proximal to enhancers or promoters. However, these ORFs are expressed in fewer tissues (Wilcoxon’s signed-rank test, P < 0.001; fig. 4A) and are in open chromatin in fewer cell types (Wilcoxon’s signed-rank test, P < 0.001; fig. 4B) than mouse-specific intergenic ORFs that are proximal to promoters. This result is expected, because enhancers tend to be active in fewer tissues than promoters (Colbran et al. 2019). ORFs emerging near enhancers are therefore transcribed in more cellular contexts than ORFs emerging away from promoters and enhancers, but in fewer cellular contexts than ORFs associated with promoters. This may help balance the reward of sampling a diversity of cellular environments with the risk of the pleiotropic effects of broad expression.
FIg. 4.
Mouse-specific intergenic ORFs that are proximal to enhancers are expressed in a limited diversity of cellular contexts. (A) Number of tissues in which ORFs have an average FPKM > 0 across replicates. (B) Number of cell types in which ORFs are in regions of open chromatin. In both panels, the “Null,” “Enhancer,” and “Promoter” categories correspond to mouse-specific intergenic ORFs.
Some Intergenic ORFs Are Proximal to Promoters That Show Evidence of Being Repurposed Enhancers
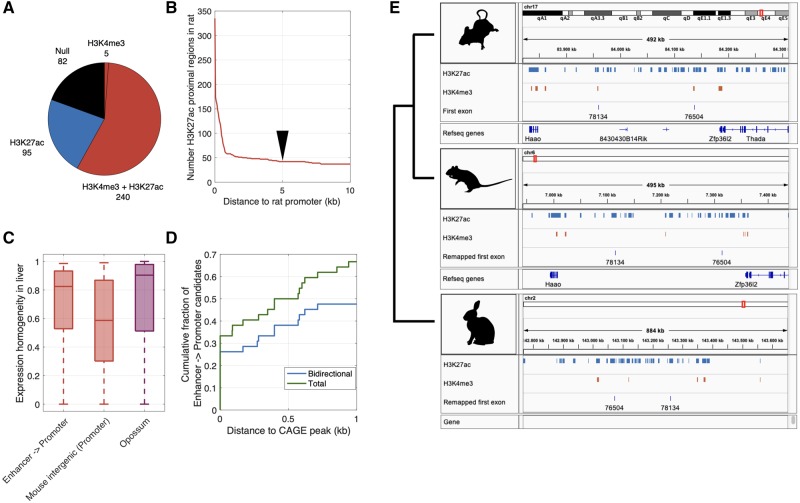
Similarities in the architectures of enhancers and promoters can facilitate the regulatory repurposing of enhancers into promoters (Wu and Sharp 2013; Carelli et al. 2018), which could reinforce the transcription of ORFs emerging near enhancers. We next assessed whether the mouse-specific intergenic ORFs that are proximal to promoters are cases of ORFs transcribed from enhancers that were repurposed into promoters. To do so, we considered 422 mouse-specific intergenic ORFs that are expressed and proximal to an active promoter in mouse liver (Materials and Methods). Subsequently, we assessed the chromatin modification status in the rat liver of those mapped genomic regions, using ChIP-seq data for H3K27ac and H3K4me3, marking enhancers and promoters, respectively.
Of the regions mapped to the rat genome, 335 are proximal to H3K27ac peaks and 245 are proximal to H3K4me3 peaks identified from rat liver samples. The majority (∼72%) of the regions that are proximal to H3K27ac peaks are also proximal to H3K4me3 peaks (fig. 5A and B), implying they act as promoters in the liver of both mouse and rat. However, some mapped genomic regions are at such distances from H3K4me3 peaks that they could well be enhancers in rat and may therefore have been repurposed into promoters on the lineage to mouse (fig. 5B). Considering those mapped genomic regions with H3K27ac peaks that are separated from an H3K4me3 peak by a conservative threshold of at least 5 kb, we found 42 candidates for the repurposing of rat enhancers to mouse promoters (10% of the 422 ORFs; fig. 5B). The ORFs corresponding to these mapped genomic regions show evidence of stable transcription, both in terms of expression stability across biological replicates (fig. 5C) and in terms of their proximity to CAGE peaks (fig. 5D), which provides evidence that the transcripts are 5′-capped. Of note, many of these CAGE peaks are bidirectional, despite their proximity to H3K4me3 (promoter) peaks, which further supports the hypothesis of an enhancer origin (fig. 5D). Finally, ∼81% of the 42 ORFs show evidence of association with ribosomes in mouse (using our most permissive criterion), which is more than we would expect by randomly sampling mouse-specific intergenic ORFs that are proximal to promoters and expressed in liver (fig. 3C; binomial test P < 0.001). Together, these observations give further support to a model in which enhancers provide fertile ground for de novo gene birth.
FIg. 5.
Some mouse-specific intergenic ORFs that are proximal to promoters show evidence of being repurposed enhancers. (A) Distribution of histone modification marks among genomic regions in rat. These regions are orthologous to genomic regions in mouse that harbor ORFs that are expressed in liver and are proximal to promoters. (B) Number of genomic regions in rat liver with H3K27ac peaks, shown in relation to their distance to the closest H3K4me3 peak. We use a conservative threshold of 5 kb (black arrow) between a promoter and an enhancer mark to determine that an enhancer is not a promoter. This results in 42 candidate promoters that were potentially repurposed from enhancers. (C) Expression homogeneity in liver of these 42 ORFs (“Enhancer -> Promoter”), mouse-specific intergenic ORFs that are expressed and proximal to promoters in liver, and opossum-shared ORFs. (D) Cumulative fraction of the 42 ORFs shown in relation to their distance to the nearest CAGE peak (“Total”) or the nearest bidirectional CAGE peak (“Bidirectional”). (E) Example repurposed enhancers. Orthologous genomic regions in mouse, rat, and rabbit that in mouse include the first exon of an intergenic mouse-specific ORF. The blue tracks represent H3K27ac peaks (enhancers) and the red tracks represent H3K4me3 peaks (promoters), both measured in liver samples from each organism. Annotated refseq genes are also indicated.
An alternative interpretation of these data is that promoters were repurposed as enhancers on the rat lineage, rather than enhancers being repurposed as promoters on the mouse lineage. To study the directionality of the repurposing, we considered ChIP-seq data for H3K27ac and H3K4me3 from the liver of rabbit, which served as an outgroup (Villar et al. 2015). Of the 42 candidate genomic regions, 11 could be mapped to the rabbit genome using liftOver and were proximal to an H3K27ac peak in rabbit liver. Of these, ten were proximal to an H3K27ac peak that was separated from an H3K4me3 peak by at least 5 kb (see e.g., fig. 5E). This provides further support for the hypothesis of an ancestral enhancer state, at least for these ten ORFs. Of note, all of the ORFs corresponding to these mapped genomic regions are surrounded by other enhancer marks in the mouse liver (supplementary fig. S3, Supplementary Material online), which hints that enhancer redundancy may help prevent conflicts that arise in the repurposing of enhancers into promoters, a possibility we revisit in the discussion.
Enhancer Interactions Are Gradually Acquired over Macroevolutionary Timescales
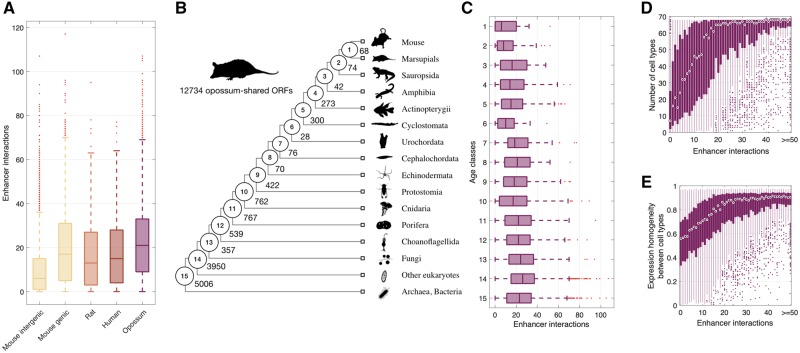
We next explored a distinct and complementary role of enhancers in the life cycle of genes, namely how enhancers help to integrate genes into regulatory networks. To do so, we considered an enhancer–promoter interaction map derived from single-cell chromatin accessibility data in 13 murine tissues (Cusanovich et al. 2018) (Materials and Methods). We uncovered a positive correlation between the age of an ORF and its number of enhancer interactions (Spearman’s correlation coefficient ρ = 0.23, P < 0.001; fig. 6A), with the number of enhancer interactions gradually increasing from a median of 10 for mouse-specific ORFs to a median of 13, 15, and 21 for ORFs that are shared with rat, human, and opossum, respectively. Among mouse-specific ORFs, intergenic ORFs had a median of 6 enhancer interactions, whereas genic ORFs had a median of 17, which makes them more similar to nonmouse-specific ORFs in their number of enhancer interactions. This suggests that many of the mouse-specific ORFs of genic origin may be coopting the regulatory interactions of their host gene, or of nearby genes.
FIg. 6.
Enhancers facilitate the integration of genes into regulatory networks. (A) Number of enhancer interactions per ORF. (B) Phylogeny adapted from (Neme and Tautz 2013). The numbered circles indicate lineages representative of the age classes to which we assigned 12,734 opossum-shared ORFs, each of which corresponds to an annotated gene. The numbers on each branch represent the total number of annotated genes assigned to each age class. (C) Number of enhancer interactions per gene, shown in relation to the age classes depicted in (B). (D) Expression breadth and (E) homogeneity of opossum-shared annotated genes as a function of the number of enhancer interactions.
Our observation that enhancers are gradually acquired across ∼160 My of mammalian evolution is consistent with the hypothesis that enhancers help integrate de novo genes into regulatory networks. Ideally, we would have high-coverage transcriptomic data across a shallower phylogeny of mouse taxa, which would provide more convincing support for this hypothesis by facilitating the estimation of ORF age for the mouse-specific intergenic ORFs. To our knowledge, no such data exist. However, low-coverage transcriptomic data from brain are available for mouse taxa spanning ∼10 My of evolution (Neme and Tautz 2016) (supplementary fig. S4A, Supplementary Material online). These data facilitate the estimation of ORF age based on expression breadth across the phylogeny, although their limited depth precludes the assurance that every mapped transcript contains the ORF of interest and makes it difficult to unequivocally age the onset of transcription (Materials and Methods). Keeping these caveats in mind, we uncovered that mouse-specific intergenic ORFs whose expression can be detected in more modern branches of the recent mouse phylogeny have fewer enhancer interactions than ORFs whose expression can be detected at more basal branches, considering both permissive (supplementary fig. S4B, Supplementary Material online) and more stringent (supplementary fig. S4C, Supplementary Material online) thresholds for the number of reads assigned to each ORF.
To explore the pace at which enhancer interactions are acquired over macroevolutionary timescales, we shifted our focus to opossum-shared ORFs: We considered 12,734 opossum-shared ORFs corresponding to annotated genes with different first exons and separated them into 15 new age classes dating back to the origin of cellular life (Neme and Tautz 2013) (fig. 6B). We again found a significant correlation between the age of a gene and its number of enhancer interactions (Spearman’s correlation coefficient ρ = 0.09, P < 0.001; fig. 6C). We have thus uncovered a positive correlation between the age of an ORF and its number of enhancer interactions across three evolutionary timescales, a shallow phylogeny spanning ∼10 My of murine evolution, a phylogeny spanning ∼160 My of mammalian evolution, and a deep phylogeny dating back to the origin of cellular life. We interpret these observations as support for the hypothesis that enhancers help to integrate new genes—those evolved de novo or by other means—into regulatory networks, and that this integration process continues throughout the evolutionary lifetimes of genes.
We next explored the consequences of enhancer acquisition. First, we studied the expression breadth of opossum-shared annotated genes using single-cell transcriptomic data from 68 cell types of 10 murine tissues (Tabula Muris Consortium 2018), for which single-cell chromatin accessibility data were also available (Materials and Methods). We found that expression breadth increases with the number of enhancer interactions (Spearman’s correlation coefficient ρ = 0.49, P < 0.001; fig. 6D) and with gene age (Spearman’s correlation coefficient ρ = 0.12, P < 0.001). The latter observation corroborates previous findings based on transcriptomic data from whole tissues (Kryuchkova-Mostacci and Robinson-Rechavi 2015). We next quantified the expression homogeneity of each gene across all cell types where expression was measurable (Materials and Methods), uncovering a positive correlation between expression homogeneity and the number of enhancer interactions (Spearman’s correlation coefficient ρ = 0.45, P < 0.001; fig. 6E), as well as gene age (Spearman’s correlation coefficient ρ = 0.10, P < 0.001).
Taken together, these results show that genes acquire enhancer interactions gradually over macroevolutionary timescales, a process that correlates with expression breadth and homogeneity across cell types. Enhancers thus facilitate the integration of genes into regulatory networks.
Discussion
Our study provides empirical support for the hypothesis that enhancers facilitate de novo gene evolution, which to our knowledge was first proposed upon the discovery of enhancer RNA (Kim et al. 2010) and later expanded upon in a perspective piece by Wu and Sharp (2013). Our findings complement recent work on the regulatory architecture of the nematode P. pacificus, which showed that young genes—those private to P. pacificus—are in closer proximity to enhancers than genes with one-to-one orthologs in other nematode species (Werner et al. 2018). The observation that many young ORFs are proximal to enhancers in both nematodes and mammals suggests that this mode of gene evolution dates back to at least the common ancestor of Bilateria, and possibly even earlier, since cnidarians, ctenophores, and sponges also employ distal regulatory elements (Schwaiger et al. 2014; Gaiti et al. 2017; Sebé-Pedrós, Chomsky, et al. 2018; Sebé-Pedrós, Saudemont, et al. 2018). As the complexity of gene regulation increased during the evolution of some lineages, such as the lineage to vertebrates (Marletaz et al. 2018), we speculate that enhancer-facilitated de novo gene birth may have played an increasingly prominent role in the expansion of gene regulatory networks.
The facilitating role of enhancers in de novo gene birth is conceptually similar to the facilitating role of the permissive chromatin state of meiotic spermatocytes and postmeiotic round spermatids that underlies the “out-of-testis hypothesis,” which proposes the testis as a primary tissue for the origination of new genes (Kaessmann 2010; Witt et al. 2019). Both scenarios envision regions of open chromatin that are exposed to the transcriptional machinery, and thus produce a transcriptionally active environment that is conducive to the evolution of new genes. The two scenarios differ, however, in at least three ways. First, genes that emerge from or near enhancers may rapidly acquire their own promoters, due to the similar architectural and functional features of enhancers and promoters, a similarity that facilitates the repurposing of the former to the latter (Wu and Sharp 2013). Indeed, we report several mouse-specific intergenic ORFs that are proximal to promoters that show evidence of being repurposed enhancers, complementing recent analyses of enhancer repurposing in primates and rodents (Carelli et al. 2018). Second, enhancers are often deployed in multiple cell types or developmental stages (Kvon et al. 2014), exposing enhancer-proximal young ORFs to selection in a limited diversity of cellular contexts. This may help to purge toxic peptides (Wilson and Masel 2011) and balance the benefit of expression in distinct cellular environments with the cost of pleiotropic effects. Third, because enhancers are often active in somatic cell types, de novo genes emerging near enhancers are more likely to be involved in physiological or morphological traits than de novo genes emerging from testis, which are more likely to be involved in reproductive traits. We emphasize that the enhancer-facilitated and out-of-testis scenarios are not mutually exclusive; in fact, they may be complementary or even interactive. Indeed, we found many young transcribed ORFs that associate with ribosomes in testis that are also proximal to enhancers (supplementary figs. S2F and S3C, Supplementary Material online).
The three points that differentiate enhancer-facilitated de novo gene birth from the out-of-testis scenario also differentiate enhancer-facilitated de novo gene birth from pervasive transcription (Clark et al. 2011) taking place away from promoters or enhancers. An additional difference is the relatively high and stable expression levels of enhancers, which increases the chances of ORF-bearing transcripts that stem from or near enhancers to associate with ribosomes. This is indeed what we observe when comparing ribosomal association among mouse-specific intergenic ORFs that are proximal to enhancers with mouse-specific intergenic ORFs that are not proximal to enhancers or promoters. However, we note that this observation may be a technological artifact. If the likelihood of the ribo-seq assay to detect ribosomal association increases with the level or stability of expression, then we would expect to see increased ribosomal association for mouse-specific intergenic ORFs that are proximal to enhancers, relative to mouse-specific intergenic ORFs that are not proximal to enhancers or promoters, even if these two classes of ORFs tend to associate with ribosomes to the same extent. Thus, the reason why enhancer proximity increases the likelihood of ribosomal association is the same reason why we cannot rule out the possibility that we observe this association due to a technological artifact.
An additional facet to enhancer-facilitated de novo gene birth is conflict between the enhancer and the emerging gene. If the enhancer is repurposed as a promoter to enforce directional transcription, then the ancestral function of the enhancer may be compromised. There are at least two ways to resolve this conflict. One is to maintain enhancer function; indeed, many promoters also act as enhancers (Medina-Rivera et al. 2018). Another is enhancer redundancy. Genes are often targeted by multiple enhancers, and in many of these cases, only a subset of the enhancers are necessary to drive correct expression under normal growth conditions (Osterwalder et al. 2018). Thus, we hypothesize that redundant enhancers are less likely to face conflict in facilitating de novo gene birth. Although our observation that repurposed enhancers tend to be surrounded by other enhancers provides anecdotal support for this hypothesis (supplementary fig. S3, Supplementary Material online), more systematic analyses are warranted.
The hypothesis that enhancers help de novo genes integrate into existing regulatory networks was previously proposed in the context of the out-of-testis hypothesis, as a means to expand a new gene’s breadth of expression (Tautz and Domazet-Loso 2011). Using single-cell chromatin accessibility and transcriptomic data, our study provides empirical support for the hypothesis that genes—those emerging de novo or via other means—gradually acquire enhancer interactions over time, and that this acquisition increases expression breadth and homogeneity. These findings complement related studies of gene integration into cellular networks, such as networks of protein–protein interactions (Capra et al. 2010; Abrusán 2013). Our observation that genes continue to acquire enhancer interactions over macroevolutionary timescales mirrors similar increases in other aspects of gene regulation, such as in the number of proximal transcription factor binding sites, alternative transcript isoforms, and miRNA targets (Warnefors and Eyre-Walker 2011).
Regulatory networks drive the spatiotemporal gene expression patterns that give rise to and define the numerous and distinct cellular identities characteristic of Metazoan life. Enhancers play an integral role in this process, mediating cell-type–specific gene–gene interactions, thus facilitating the combinatorial deployment of different genes and network modules in different contexts. Genetic changes that affect such interactions are responsible for myriad evolutionary adaptations and innovations (Carroll 2001, 2008; Prud’homme et al. 2007; Peter and Davidson 2011). Our results suggest that the power of enhancers in creating such evolutionary novelties lies not only in their ability to rewire gene regulatory networks but also in their ability to expand them, by providing fertile ground for de novo gene birth.
Materials and Methods
ORF Age and Classification
Schmitz et al. (2018) identified a set of 58,864 ORFs from the transcriptomes of three murine tissues: liver, brain, and testis. Blasting against the transcriptomes of four other mammalian species (rat, human, kangaroo rat, and opossum), they estimated the age of each ORF by phylostratigraphic methods (Domazet-Lošo et al. 2007; Schmitz et al. 2018). Because of the small number of ORFs shared with the kangaroo rat (49 ORFs), we merged these ORFs together with those from the rat age class. We used the genomic coordinates of the first exon of each ORF in the mm10 mouse genome reference to study the regulatory properties of ORFs of different ages, for example, to study their distance to the nearest enhancer. We only considered ORFs that were transcribed from nuclear chromosomes and whose first exon was longer than 30 base pairs. If first exons were shared between more than one ORF, we only retained the oldest of the ORFs. Our filtered data set contained 56,262 ORFs.
Schmitz et al. (2018) annotated each ORF as belonging to one of eight different categories: “intergenic,” “close to promoter same strand,” “close to promoter opposite strand,” “overlapping same strand,” “overlapping opposite strand,” “overlapping coding sequence same strand,” “overlapping coding sequence opposite strand,” and “overlapping annotated gene in frame.” We considered all categories except “intergenic” to be “genic” in order to separate ORFs that were born within or near existing genes from those that were not. This resulted in five classes: mouse-specific intergenic ORFs, mouse-specific genic ORFs, rat-shared ORFs, human-shared ORFs, and opossum-shared ORFs.
Proximity to Enhancers and Promoters
We obtained ChIP-seq data for H3K27ac, H3K4me1, and H3K4me3 modifications from 23 different tissues and cell types from the ENCODE project (bone marrow, cerebellum, cortex, heart, kidney, liver, lung, olfactory bulb, placenta, spleen, small intestine, testis, thymus, embryonic whole brain, embryonic liver, embryonic limb, brown adipose tissue, macrophages, MEL, MEF, mESC, CH12 cell line, and E14 embryonic mouse) (ENCODE 2012). We used liftOver (Kent et al. 2002) to convert the genomic coordinates of the peaks from mm9 to mm10. We used the “merge” function of bedtools (Quinlan and Hall 2010) with default parameters to collate the peaks for all tissues and cell types, considering any overlapping H3K27ac and H3K4me1 peak as part of the same enhancer. We used the “intersect” function of bedtools with default parameters to separate H3K27ac and H3K4me1 peaks that overlapped any length of H3K4me3 peaks from those that did not. This resulted in 172,930 H3K27ac and 277,187 H3K4me1 peaks that did not overlap H3K4me3 peaks. We considered genomic regions with H3K4me3 peaks to be promoters, and those exclusively with H3K27ac and/or H3K4me1 peaks to be enhancers (Berthelot et al. 2018). We measured the distance in base pairs between the first exon of an ORF to an enhancer or promoter using the “closest” function of bedtools with the “-t first” option activated. We considered an ORF to be proximal to an enhancer if the distance to the first exon was shorter than 500 bp and there was no promoter within that distance. When controlling for the length of the first exon, we considered the distance to windows of 750-bp up- and down-stream of the central nucleotide of the first exon, rather than to the first exon itself.
We followed the same procedures when measuring the distance of ORFs to enhancers and promoters in liver, brain, and testis tissues separately. For brain tissue, we merged ChIP-seq data from embryonic whole brain and cortex. The ORFs we considered as expressed in each tissue (supplementary fig. S2A–C, Supplementary Material online) were those with a mean fragments per kilobase per million mapped reads (FPKM) > 0 across replicates of total RNA transcriptomic data (eight replicates for liver and brain and two replicates for testis) (Li et al. 2017).
Chromatin Accessibility
We used single-cell ATAC-seq data from 13 different mouse tissues (bone marrow, cerebellum, large intestine, heart, small intestine, kidney, liver, lung, cortex, spleen, testes, thymus, and whole brain). We obtained the data from the Mouse ATAC atlas (Cusanovich et al. 2018), which composed 436,206 peaks of open chromatin. We used liftOver to convert the genome coordinates from mm9 to mm10. A total of 29 peaks could not be converted. Using the “closest” function of bedtools with the “-t first” option activated, we calculated the distance between ORFs and regions of open chromatin. We annotated regions of open chromatin as enhancers if they overlapped H3K27ac and/or H3K4me1 peaks but not H3K4me3 peaks, or as promoters if they overlapped H3K4me3 peaks. To do so, we used the bedtools intersect function with the -u option activated.
Cusanovich et al. (2018) used these single-cell ATAC-seq data to identify clusters of cells with similar patterns of chromatin accessibility. They assigned the clusters to 38 distinct cell types based on the chromatin accessibility of marker genes indicative of each cell type. We used these data to identify tissues and cell types where ORFs are in accessible chromatin. We considered an ORF-containing region of the genome to be in open chromatin in a certain cell type if it was accessible in at least 1% of the cells that made up at least one of the clusters of that cell type (Cusanovich et al. 2018).
5′-Capping
We used CAGE data from the FANTOM5 consortium from 1,016 mouse samples including cell lines, primary cells, and tissues (Lizio et al. 2015; Noguchi et al. 2017). This method is based on the capture of 5′-capped ends of mRNA, which allows the mapping of regions of transcription initiation genome wide (Shiraki et al. 2003). Using the “closest” function from bedtools with the “-t first” option activated (Quinlan and Hall 2010), we measured the distance between an ORF’s first exon and its closest CAGE peak. In the same manner, we also considered a subset of CAGE peaks which were annotated as bidirectional and transcribed from enhancers (Andersson et al. 2014; Dalby et al. 2018).
Expression Level and Stability
We measured the expression levels and stabilities of ORFs. To do so, we aligned paired reads produced by RNAseq from total RNA from ten tissues (liver, testis, brain, muscle, bone, small intestine, thymus, heart, lung and spleen) (Li et al. 2017) using STAR 2.5.3a (Dobin et al. 2013) to the mm10 build of the mouse genome. We chose these tissues because ChIP-seq data for histone modifications were also available. For each ORF, we calculated FPKM as the number of reads mapped to the first exon divided by a millionth of the number of reads sequenced in each sample and then by the length of the exon in kilobases. We considered an ORF to be expressed if it had an average FPKM > 0 across replicates.
We calculated the expression stability of ORF k as
where n is the number of replicates for a given tissue (8 for liver and brain and 2 for testis). We refer to this measure as expression homogeneity when calculated across tissues or cell types, rather than across replicates for the same tissue or cell type.
Ribosome Association
We used ribosome profiling (ribo-seq) data from mouse liver, brain, and testis (Ingolia 2014). We obtained the coordinates of mRNA segments detected by ribo-seq from GWIPS-viz (Michel et al. 2014), a database that includes such data from different studies. From this source, we considered samples from liver (three samples from three studies), brain (five samples from two studies), and testis (one sample) (details provided in supplementary information 1, Supplementary Material online). We combined the data sets for each tissue and merged the provided genomic coordinates using the bedtools merge function; we did so with the options “-c” and “-o absmax” activated. Following Ruiz-Orera et al. (2018), we removed all merged coordinates shorter than 26 bp, because these could be anomalous reads. We subsequently mapped these merged coordinates on the first exon of our set of ORFs using the bedtools “map” function and we summed the number of reads from each of the mapped merged coordinates. In this way, we were able to assign a number of ribo-seq reads to each ORF, which allowed us to estimate ribosomal association and thus potential for translation.
Enhancer Repurposing
We considered the set of 544 mouse-specific intergenic ORFs that were transcribed (average FPKM > 0) and proximal to an H3K4me3 peak in mouse liver. We filtered this set to the 456 ORFs that were proximal to what Villar et al. (2015) considered to be replicated H3K4me3 peaks in mouse, in order to facilitate comparison with the histone methylation data from rat and rabbit that were generated for the same study. We used liftOver to map the genomic coordinates of these ORFs to the rat and rabbit genomes (builds r5 and oryCun2), requiring a minimum fraction of remapped bases of 0.6 and 0.4, respectively (Carelli et al. 2018). This resulted in 422 and 152 presumably orthologous genomic regions in rat and rabbit, respectively. Considering H3K27ac and H3K4me3 ChIP-seq peaks in the livers of mouse, rat, and rabbit (Villar et al. 2015), we then calculated the distance between these mapped regions and H3K4me3 and H3K27ac peaks using the bedtools “closest” function. We considered the promoter of an ORF in mouse to show evidence of being a repurposed enhancer if its mapped genomic region in rat or rabbit was proximal to an H3K27ac peak, yet more than 5 kb from an H3K4me3 peak, in rat or rabbit liver.
Enhancer Interactions
Cusanovich et al. (2018) used single-cell ATAC-seq data to predict physical interactions between regions of open chromatin (Pliner et al. 2018), thus creating an atlas of enhancer interactions in single murine cells. We downloaded these data from the Mouse ATAC atlas (Cusanovich et al. 2018), which includes the cell clusters where the interactions occur, as well as the coaccessibility scores of pairs of regions of open chromatin—a measure of interaction strength. We disregarded cell clusters classified as “unknown” or “collisions,” as well as interactions with a coaccessibility score <0.25, following Pliner et al. (2018). We also filtered out interactions with regions of open chromatin that overlapped ChIP-seq peaks for H3K4me3 marks or no enhancer marks, in order to focus solely on interactions with enhancers. An interaction was assigned to an ORF if the ORF’s first exon was included in the interaction.
Expression within the Mouse Lineage
We considered the transcriptomes of brain from ten different mouse taxa that diverged after the mouse-rat split (three populations of Mus musculus domesticus, two populations of M. m. musculus, and one from M. m. castaneus, M. spicilegus, M. spretus, M. mattheyi, and Apodemus uralensis) (Neme and Tautz 2016). The data consisted of read counts from the transcriptomes of each taxon mapped to 200-bp windows of the mm10 mouse reference genome. We assigned each ORF to one of the 200-bp windows if the middle point of the ORF’s first exon mapped to that window. For this analysis, we only considered mouse-specific intergenic ORFs that overlapped regions of open chromatin. We considered two different thresholds to evidence transcription of an ORF. Using the first, more permissive threshold, we only considered ORFs that had at least one read mapping to its 200-bp window in at least one of the three samples of M. m. domesticus. This resulted in 4,104 ORFs. Using the second, more stringent threshold, we only considered ORFs that had at least 20 reads from across the three samples of M. m. domesticus mapping to the ORF’s 200-bp window. This resulted in 2,864 ORFs. We separated these ORFs into four age categories (supplementary fig. S4A, Supplementary Material online), depending on whether expression could be detected using a highly conservative threshold of just a single read in 1) at least one of the three M. m. domesticus samples, 2) in M. m. domesticus, and also in at least one of the other Mus musculus subspecies, 3) in M. m. domesticus, at least one other subspecies of M. musculus, and at least one other Mus species, but not in the A. uralensis sample, or 4) in M. m. domesticus and in A. uralensis. We assigned a total of 3,980 ORFs to each of these categories when considering ORFs with at least 1 read detectable in the M. m. domesticus clade (∼97%), and 2,855 ORFs when considering ORFs with at least 20 reads detectable in the M. m. domesticus clade (∼99.7%).
Because of the low coverage of the transcriptomic data (1× sequencing depth), there is increased uncertainty in our estimation of ORF ages relative to the other phylogenies considered in this study. This is especially true of ORFs that are expressed at low levels, which are less likely to be detected across the phylogeny and are therefore more susceptible to the underestimation of their ages. We were therefore concerned by the observed positive correlation between the expression level of an ORF and its estimated age (Spearman’s correlation coefficient ρ = 0.20, P < 0.001). To ameliorate this concern, we determined the probability of underestimating the age of an ORF in the M. m. domesticus clade under our most stringent detection limit of 20 reads per 200-bp window. Specifically, for each ORF assigned to the M. m. domesticus clade, we used the binomial formula to calculate the probability of underestimating the ORF’s age due to lack of detection in the other seven clades, under the assumption that the ORF actually emerged at the base of the phylogeny and is expressed at the same low level across the phylogeny. This probability is low: given 27.76 × 108 trials (the total number of reads from the seven samples not in the M. m. domesticus clade) and a probability of success of 20/10.75 × 108 (the minimum fraction of reads from the three samples in the M. m. domesticus clade mapping to the ORF’s 200-bp window), the probability of observing zero reads mapping to the ORF’s 200-bp window in all of the seven samples from the outgroup is 1.4 × 10−13, after Bonferroni correction for 14 tests (the number of ORFs assigned to M. m. domesticus).
Age of Annotated Genes
To study how genes acquire enhancer interactions over macroevolutionary timescales, we considered the subset of ORFs that belong to the opossum age class in Schmitz et al. (2018) and that are annotated as genes in the latest version of Ensembl (release 95) (Cunningham et al. 2019). We matched these genes to age estimates reported by Neme and Tautz (2013), based on a phylostratigraphic analysis of 20 lineages spanning 4 Gy from the last universal common ancestor to the common ancestor of mouse and rat. We further filtered the data set to only include ORFs that emerged in the first 15 of the 20 phylostrata, in order to focus on ORFs that are considered to have emerged before the split between the common ancestor of placental mammals and marsupials by both Schmitz et al. (2018) and Neme and Tautz (2013). This left us with ∼16,000 ORFs corresponding to 12,734 unique annotated genes that emerged prior to the origin of placental mammals.
Expression Breadth and Homogeneity of Annotated Genes
To study the transcription of annotated genes, we used the expression data reported by the Tabula Muris Consortium (2018) for the single-cell RNA sequencing performed with FACS-based cell capture in plates, for 20 different mouse tissues. The data include the log-normalization of 1 + counts per million for each of the annotated genes in each of the sequenced cells. We considered ten tissues that were also used for the construction of the Mouse ATAC Atlas (Cusanovich et al. 2018). We measured the expression breadth of each ORF corresponding to an annotated gene as the number of cell types in which expression could be detected in at least 1% of the cells assigned to a cell type. The homogeneity of expression across cell types was calculated as explained above for Hk, but considering the mean expression in each cell type where expression was detectable, rather than the mean expression across replicates.
Supplementary Material
Acknowledgments
J.L.P. acknowledges support from Swiss National Science Foundation (Grant No. PP00P3_170604). We thank Alejandro Cano and Macarena Toll-Riera for discussions.
References
- Abrusán G. 2013. Integration of new genes into cellular networks, and their structural maturation. Genetics 195(4):1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. 2014. An atlas of active enhancers across human cell types and tissues. Nature 507(7493):455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH, Wallbank RWR, Baxter SW, Pardo-Diaz C, Hanly JJ, Martin SH, Mallet J, Dasmahapatra KK, Salazar C, Joron M, et al. 2016. Evolutionary novelty in a butterfly wing pattern through enhancer shuffling. PLoS Biol. 14:e1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot C, Villar D, Horvath JE, Odom DT, Flicek P.. 2018. Complexity and conservation of regulatory landscapes underlie evolutionary resilience of mammalian gene expression. Nat Ecol Evol. 2(1):152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran E, Reinhardt JA, Wanjiru BM, Brant AT, Saelao P, Begun DJ, Jones CD.. 2013. De novo ORFs in Drosophila are important to organismal fitness and evolved rapidly from previously non-coding sequences. PLoS Genet. 9:e1003860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra JA, Pollard KS, Singh M.. 2010. Novel genes exhibit distinct patterns of function acquisition and network integration. Genome Biol. 11(12):R127.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carelli FN, Liechti A, Halbert J, Warnefors M, Kaessmann H.. 2018. Repurposing of promoters and enhancers during mammalian evolution. Nat Commun. 9(1):4066.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. 2001. Chance and necessity: the evolution of morphological complexity and diversity. Nature 409(6823):1102–1109. [DOI] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134(1):25–36. [DOI] [PubMed] [Google Scholar]
- Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, et al. 2012. Proto-genes and de novo gene birth. Nature 487(7407):370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catarino RR, Stark A.. 2018. Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription activation. Genes Dev. 32(3–4):202–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A, et al. 2011. The reality of pervasive transcription. PLoS Biol. 9(7):e1000625; discussion e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbran LL, Chen L, Capra JA.. 2019. Sequence characteristics distinguish transcribed enhancers from promoters and predict their breadth of activity. Genetics 211(4):1205–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L, Martins A, Danko C, Waters CT, Siepel A, Lis JT.. 2014. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 46(12):1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 107(50):21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, Achuthan P, Akanni W, Allen J, Amode MR, Armean IM, Bennett R, Bhai J, Billis K, Boddu S, et al. 2019. Ensembl 2019. Nucleic Acids Res. 47(D1):D745–D751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, Filippova GN, Huang X, Christiansen L, DeWitt WS, et al. 2018. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174(5):1309–1324.e1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby M, Rennie S, Andersson R.. 2018. FANTOM5 transcribed enhancers in mm10. Zenodo. doi:10.5281/zenodo.1411211.
- Davidson EH, Levine MS.. 2008. Properties of developmental gene regulatory networks. Proc Natl Acad Sci U S A. 105(51):20063–20066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G.. 2010. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 8(5):e1000384.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Lošo T, Brajković J, Tautz D.. 2007. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 23(11):533–539. [DOI] [PubMed] [Google Scholar]
- Emera D, Yin J, Reilly SK, Gockley J, Noonan JP.. 2016. Origin and evolution of developmental enhancers in the mammalian neocortex. Proc Natl Acad Sci U S A. 113(19):E2617–E2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiti F, Jindrich K, Fernandez-Valverde SL, Roper KE, Degnan BM, Tanurdžić M.. 2017. Landscape of histone modifications in a sponge reveals the origin of animal cis-regulatory complexity. eLife 6:e22194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V, Stark A.. 2018. Eukaryotic core promoters and the functional basis of transcription initiation. Nat Rev Mol Cell Biol. 19(10):621–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Chen C, Teng L, Tan K.. 2014. Global view of enhancer–promoter interactome in human cells. Proc Natl Acad Sci U S A. 111(21):E2191–E2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 39(3):311–318. [DOI] [PubMed] [Google Scholar]
- Ingolia NT. 2014. Ribosome profiling: new views of translation, from single codons to genome scale. Nat Rev Genet. 15(3):205–213. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne Gaëlle JS, Jackson SE, Wills MR, Weissman JS.. 2014. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 8(5):1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H. 2010. Origins, evolution, and phenotypic impact of new genes. Genome Res. 20(10):1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR.. 2007. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 8(6):413–423. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D.. 2002. The human genome browser at UCSC. Genome Res. 12(6):996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kherdjemil Y, Lalonde RL, Sheth R, Dumouchel A, de Martino G, Pineault KM, Wellik DM, Stadler HS, Akimenko M-A, Kmita M.. 2016. Evolution of Hoxa11 regulation in vertebrates is linked to the pentadactyl state. Nature 539(7627):89–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Mayer MG, Rödelsperger C, Witte H, Riebesell M, Sommer RJ.. 2015. The orphan gene dauerless regulates dauer development and intraspecific competition in nematodes by copy number variation. PLoS Genet. 11:e1005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465(7295):182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratochwil CF, Liang Y, Gerwin J, Woltering JM, Urban S, Henning F, Machado-Schiaffino G, Hulsey CD, Meyer A.. 2018. Agouti-related peptide 2 facilitates convergent evolution of stripe patterns across cichlid fish radiations. Science 362(6413):457–460. [DOI] [PubMed] [Google Scholar]
- Kryuchkova-Mostacci N, Robinson-Rechavi M.. 2015. Tissue-specific evolution of protein coding genes in human and mouse. PLoS One 10(6):e0131673.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ, Kamneva OK, Melo US, Barozzi I, Osterwalder M, Mannion BJ, Tissières V, Pickle CS, Plajzer-Frick I, Lee EA, et al. 2016. Progressive loss of function in a limb enhancer during snake evolution. Cell 167(3):633–642.e611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ, Kazmar T, Stampfel G, Yáñez-Cuna JO, Pagani M, Schernhuber K, Dickson BJ, Stark A.. 2014. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature 512(7512):91–95. [DOI] [PubMed] [Google Scholar]
- Levine M, Cattoglio C, Tjian R.. 2014. Looping back to leap forward: transcription enters a new era. Cell 157(1):13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Qing T, Zhu J, Wen Z, Yu Y, Fukumura R, Zheng Y, Gondo Y, Shi L.. 2017. A comprehensive mouse transcriptomic BodyMap across 17 tissues by RNA-seq. Sci Rep. 7:4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Yan Z, Lu L, Jiang H, Wang W.. 2014. Pleiotropy of the de novo-originated gene MDF1. Sci Rep. 4:7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Rosenfeld MG.. 2016. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 17(4):207–223. [DOI] [PubMed] [Google Scholar]
- Lizio M, Harshbarger J, Shimoji H, Severin J, Kasukawa T, Sahin S, Abugessaisa I, Fukuda S, Hori F, Ishikawa-Kato S, et al. 2015. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 16(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marletaz F, Firbas PN, Maeso I, Tena JJ, Bogdanovic O, Perry M, Wyatt CDR, de la Calle-Mustienes E, Bertrand S, Burguera D, et al. 2018. Amphioxus functional genomics and the origins of vertebrate gene regulation. Nature 564(7734):64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A, Hurst LD.. 2016. Open questions in the study of de novo genes: what, how and why. Nat Rev Genet. 17(9):567–578. [DOI] [PubMed] [Google Scholar]
- Medina-Rivera A, Santiago-Algarra D, Puthier D, Spicuglia S.. 2018. Widespread enhancer activity from core promoters. Trends Biochem Sci. 43(6):452–468. [DOI] [PubMed] [Google Scholar]
- Michel AM, Fox G, M. Kiran A, De Bo C, O’Connor PBF, Heaphy SM, Mullan JPA, Donohue CA, Higgins DG, Baranov PV.. 2014. GWIPS-viz: development of a ribo-seq genome browser. Nucl Acids Res. 42(D1):D859–D864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neme R, Tautz D.. 2013. Phylogenetic patterns of emergence of new genes support a model of frequent de novo evolution. BMC Genomics. 14(1):117.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neme R, Tautz D.. 2016. Fast turnover of genome transcription across evolutionary time exposes entire non-coding DNA to de novo gene emergence. eLife 5:e09977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi S, Arakawa T, Fukuda S, Furuno M, Hasegawa A, Hori F, Ishikawa-Kato S, Kaida K, Kaiho A, Kanamori-Katayama M, et al. 2017. FANTOM5 CAGE profiles of human and mouse samples. Sci Data 4(1):170112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder M, Barozzi I, Tissières V, Fukuda-Yuzawa Y, Mannion BJ, Afzal SY, Lee EA, Zhu Y, Plajzer-Frick I, Pickle CS, et al. 2018. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554(7691):239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partha R, Chauhan BK, Ferreira Z, Robinson JD, Lathrop K, Nischal KK, Chikina M, Clark NL.. 2017. Subterranean mammals show convergent regression in ocular genes and enhancers, along with adaptation to tunneling. eLife 6:pii: e25884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Davidson EH.. 2011. Evolution of gene regulatory networks controlling body plan development. Cell 144(6):970–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliner HA, Packer JS, McFaline-Figueroa JL, Cusanovich DA, Daza RM, Aghamirzaie D, Srivatsan S, Qiu X, Jackson D, Minkina A, et al. 2018. Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Mol Cell 71(5):858–871.e858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabh N, Rödelsperger C.. 2016. Are orphan genes protein-coding, prediction artifacts, or non-coding RNAs? BMC Bioinformatics 17:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud’homme B, Gompel N, Carroll SB.. 2007. Emerging principles of regulatory evolution. Proc Natl Acad Sci U S A. 104:8605–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscito JG, Sameith K, Parra G, Langer BE, Petzold A, Moebius C, Bickle M, Rodrigues MT, Hiller M.. 2018. Phenotype loss is associated with widespread divergence of the gene regulatory landscape in evolution. Nature Commun. 9:4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Orera J, Alba MM.. 2019. Translation of small open reading frames: roles in regulation and evolutionary innovation. Trends Genet. 35(3):186–198. [DOI] [PubMed] [Google Scholar]
- Ruiz-Orera J, Messeguer X, Subirana JA, Alba MM.. 2014. Long non-coding RNAs as a source of new peptides. eLife 3:e03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Orera J, Verdaguer-Grau P, Villanueva-Canas JL, Messeguer X, Alba MM.. 2018. Translation of neutrally evolving peptides provides a basis for de novo gene evolution. Nat Ecol Evol. 2(5):890–896. [DOI] [PubMed] [Google Scholar]
- Schmitz JF, Ullrich KK, Bornberg-Bauer E.. 2018. Incipient de novo genes can evolve from frozen accidents that escaped rapid transcript turnover. Nat Ecol Evol. 2(10):1626–1632. [DOI] [PubMed] [Google Scholar]
- Schwaiger M, Schonauer A, Rendeiro AF, Pribitzer C, Schauer A, Gilles AF, Schinko JB, Renfer E, Fredman D, Technau U.. 2014. Evolutionary conservation of the eumetazoan gene regulatory landscape. Genome Res. 24(4):639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Chomsky E, Pang K, Lara-Astiaso D, Gaiti F, Mukamel Z, Amit I, Hejnol A, Degnan BM, Tanay A.. 2018. Early metazoan cell type diversity and the evolution of multicellular gene regulation. Nat Ecol Evol. 2(7):1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Saudemont B, Chomsky E, Plessier F, Mailhe MP, Renno J, Loe-Mie Y, Lifshitz A, Mukamel Z, Schmutz S, et al. 2018. Cnidarian cell type diversity and regulation revealed by whole-organism single-cell RNA-Seq. Cell 173(6):1520–1534. [DOI] [PubMed] [Google Scholar]
- Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, Kodzius R, Watahiki A, Nakamura M, Arakawa T, et al. 2003. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci U S A. 100(26):15776–15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong E.. 2012. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 13(9):613–626. [DOI] [PubMed] [Google Scholar]
- Tabula Muris Consortium. 2018. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562:367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Domazet-Loso T.. 2011. The evolutionary origin of orphan genes. Nat Rev Genet. 12(10):692–702. [DOI] [PubMed] [Google Scholar]
- Teichmann SA, Babu MM.. 2004. Gene regulatory network growth by duplication. Nat Genet. 36(5):492–496. [DOI] [PubMed] [Google Scholar]
- Van Oss SB, Carvunis A-R.. 2019. De novo gene birth. PLoS Genet. 15(5):e1008160.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, Berthelot C, Aldridge S, Rayner Tim F, Lukk M, Pignatelli M, Park Thomas J, Deaville R, Erichsen Jonathan T, Jasinska Anna J, et al. 2015. Enhancer evolution across 20 mammalian species. Cell 160(3):554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnefors M, Eyre-Walker A.. 2011. The accumulation of gene regulation through time. Genome Biol Evol. 3:667–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner MS, Sieriebriennikov B, Prabh N, Loschko T, Lanz C, Sommer RJ.. 2018. Young genes have distinct gene structure, epigenetic profiles, and transcriptional regulation. Genome Res. 28(11):1675–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen A, Félez-Sánchez M, Bravo IG, Bapteste E.. 2019. Genome plasticity in papillomaviruses and de novo emergence of E5 oncogenes. Genome Biol Evol. 11(6):1602–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BA, Masel J.. 2011. Putatively noncoding transcripts show extensive association with ribosomes. Genome Biol Evol. 3:1245–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt E, Benjamin S, Svetec N, Zhao L.. 2019. Testis single-cell RNA-seq reveals the dynamics of de novo gene transcription and germline mutational bias in Drosophila. eLife 8:pii: e47138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Sharp PA.. 2013. Divergent transcription: a driving force for new gene origination? Cell 155(5):990–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chen J-Y, Shen QS, Zhou W-Z, Peng J, He BZ, Li Y, Liu C-J, Luan X, Ding W, et al. 2015. Emergence, retention and selection: a trilogy of origination for functional de novo proteins from ancestral lncRNAs in primates. PLoS Genet. 11:e1005391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ren Y, Yang T, Li G, Chen J, Gschwend AR, Yu Y, Hou G, Zi J, Zhou R, et al. 2019. Rapid evolution of protein diversity by de novo origination in Oryza. Nat Ecol Evol. 3(4):679–690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.