Abstract
Background: Hepatocellular carcinoma (HCC) recurrence appears commonly after liver transplantation (LT), and it severely affected the long-term survival of patients. Previous studies have proved that Rap1A is involved in hepatocarcinogenesis and metastasis, and demonstrated the significant association between Rap1A gene rs494453 polymorphism and HCC. However, the relationship between Rap1A rs494453 polymorphism and HCC recurrence after LT remained unclear.
Methods: A total of 74 HCC patients who underwent LT from July 2005 to June 2015 was analyzed. The genotypes of both donors and recipients had been confirmed as Rap1A rs494453. The independent risk factors that associated with HCC recurrence were investigated with univariate and multivariate logistic regression analysis. The recurrence-free (RFS) and overall survival (OS) were calculated with Cox regression analysis. The Rap1A rs494453 genotype frequencies were determined using the Χ² test and the minor allele frequencies (MAFs) of Rap1A rs494453 genotypes were calculated by Hardy-Weinberg equilibrium.
Results: We found that the donor Rap1A rs494453 polymorphism was profoundly associated with HCC recurrence after LT. Moreover, the Milan criteria, microvascular invasion and donor Rap1A rs494453 genotype were proved to be independent risk factors for HCC recurrence. Patients with donor AG/GG genotypes had a distinct lower RFS and OS than AA genotype. The TNM stage, Milan criteria, microvascular invasion, and donor Rap1A rs494453 genotype were independent factors for the RFS of LT patients.
Conclusions: Donor Rap1A rs494453 is a potential predictive marker for HCC recurrence risk after LT.
Keywords: Rap1A polymorphism, hepatocellular carcinoma, recurrence, liver transplantation
Introduction
Hepatocelluar carcinoma (HCC) is one of the major cancer-related death causes 1. The main features of HCC include rapid progression, poor prognosis, and frequent tumor recurrence. Among novel therapies, the orthotopic liver transplantation (LT) is still considered the best treatment for HCC patients having cirrhosis syndrome without local or distant metastasis 2.
However, the recurrence after LT remains frequently among 1.3% to 44.9%, which depends on individual series 3,4. Abundant studies have concluded the main causes for high HCC recurrence after surgery, including microvascular invasion, poor tumor grade, larger tumor diameter, ischemia time, vascular invasion and elevated AFP 5-7. These factors are viewed to be significant prediction of HCC recurrence by famous institute, such as the Milan, University of California San Francisco (UCSF), and Hangzhou criteria 8,9.
Rap1A is a member of Ras oncogene family of small G protein, which regulates different cellular processes, including proliferation, adhesion and cancer progression. It was reported that Rap1A promoted ovarian cancer tumorigenesis and metastasis via activating ERK, MAPK, Notch pathways 10. Decreasing Rap1A suppressed cell proliferation, adhesion, and invasion in prostate cancer and several other cancer types 11-13. A recent study further demonstrated that Rap1A expression abolished the tumor-suppressive effects of EYA4 in HCC cells, which could promote the proliferation and recurrence 14. In clinical studies, Rap1A is not only overexpressed in a cohort of Oral Cavity Squamous Cell Carcinoma (OCSCC) specimens but also correlated with the clinical characteristics of the advanced tumor stage 13. As Rap1A plays important role in malignancies, it is worthy to understand the relationship between Rap1A expression and HCC recurrence after LT.
Patients who underwent LT were divided into recurrence group and nonrecurrence group for comparation, to identify the clinicopathological factors associated with the poor prognosis. Rap1A genotypes were observed to have function on HCC relapse. Hence, we endeavoured to uncover whether donor or recipient HCC susceptibility gene (Rap1A) variations contribute to the HCC recurrence after LT in a Han Chinese population.
Materials and Methods
Patients
We collected 74 HCC patients among Han Chinese undergoing LT from July 2005 to June 2015 at the department of liver transplantation Surgery, Shanghai General Hospital, Shanghai, China. The median age was 47.3 years (range from 32-66 years), including 65 males and 9 females. The mean follow-up period was 49.2 months (range from 2.3-120 months). The LT criteria for patients with HCC included the absence of extrahepatic malignancies, macroscopic tumor thrombosis, and extrahepatic metastasis of HCC.
Patients were treated with standard immunosuppression regimen consisting of the triple drugs combination of cyclosporin (CsA) or tacrolimus (FK506), mycophenolate (MMF) and prednisone. Lamivudine and hepatitis B immune globulin (HBIG) were used for HBV DNA positive patients to prevent hepatitis B recurrence. The antineoplastic prophylaxis of chemotherapy and targeted therapies was used for certain patients. All patients were asked to monitor tumor recurrence or metastasis by undergoing the AFP test, ultrasonography, chest X-ray, and emission computed tomography every three months for the first two years and semiannually thereafter.
Ethics statement
Organ donation and transplantation were approved by the Institutional Review Board, Shanghai General Hospital, and carried out strictly under the guidelines of the Ethics Committee of the hospital, and Declaration of Helsinki. All patients provided written informed consent.
Data collection
Clinicopathological data such as age, sex, hepatitis B status, cirrhosis, histologic grade (well differentiated, moderately differentiated and poorly differentiated), Milan criteria, TNM stage, tumor size, multinodular status, microvascular invasion, and pre-LT serum AFP level were recorded. The pre-transplant data were all collected within 24h before transplantation.
DNA Isolation and Polymorphism Genotyping
LT patients were divided into two groups, patients with HCC recurrence and nonrecurrence control groups. These samples were previously stored at -80oC before being genotyped. DNA was isolated from EDTA-anticoagulated whole blood or liver tissues of donors and recipients using the AllPrep DNA/RNA Mini kit (Qiagen, Venlos, the Netherlands). PCR reaction system (10 ul) included 1xGC buffer, 3.0 mM Mg2+, 0.3 mM dNTP, 1 U HotStarTaq polymerase, 1 ul templates DNA and 1 ul PCR primer pair: 5' CAGAAGCTGGTGGAGTGGG 3' and 5' ACCTGGATAGACGCTGGCC 3'. The conditions of PCR amplification and the mass extension reaction were available on request. Genotyping was performed in a blind way. To assess the reliability of genotyping, we randomly choose more than 10% of the samples to sequence twice. Single nucleotide polymorphism (SNP) genotyping was executed using a Sequenom Mass ARRAY SNP genotyping platform (Sequenom, San Diego, CA) 15.
Statistical analysis
Statistical analysis was performed with SPSS statistical software version 21.0 (SPSS Inc., Chicago, IL, USA). Quantitative variables were denoted as medians (±SD) and compared using Student's t-test or the Wilcoxon signed-rank test. Categorical variables were expressed as values (percentages) and compared by Pearson's Χ² test or Fisher's exact test. The allele and genotype frequencies were determined using the Χ² test. SHEsis Online Version (http://analysis.bio-x.cn/myAnalysis.php) was used to analyze Hardy-Weinberg equilibrium. Risk factors for HCC recurrence were evaluated by logistic regression analysis. Recurrence-free survival (RFS) and overall survival (OS) were calculated using Kaplan-Meier analyiss. Statistical significance was defined as P < 0.05.
Results
Patient characteristics
A total of 74 HCC patients was divided into the recurrence group (n=33, 44.6%) and the nonrecurrence group (n=41, 55.4%), as shown in Table 1, under the criteria of the disease states after LT. The median follow-up time was 26.1 months (range from 2.3 to 91 months) in the recurrence group and 71.2 months (range from 8 to 120 months) in the nonrecurrence group, and 28 deaths (37.8%) were recorded. There was significant difference between the two groups (P<0.01) analyzed by Student's t-test, indicating a close association of the follow-up time and OS with carcinoma recurrence. Moreover, Chi-squared test or Fisher's exact test revealed that the Milan criteria, TNM stage, tumor size, nodules number and microvascular invasion were significant risk factors that involved in recurrence (P<0.05). However, other factors such as age, sex, hepatitis B, cirrhosis, Child-Pugh grading, tumor differentiation, pre-LT serum AFP level, and antineoplastic prophylaxis after LT failed to influence patient recurrence events (P > 0.05).
Table 1.
Clinicopathological characteristics and two groups of the HCC patients
| Parameter | Recurrence group (n=33) |
Nonrecurrence group (n=41) | P value |
|---|---|---|---|
| Recipient age | 47(33-64) | 49(32-66) | 0.689 |
| Recipient sex | |||
| Male | 30(90.9%) | 35(85.4%) | |
| Female | 3(9.1%) | 6(14.6%) | 0.468 |
| Follow up time(month) | 26.1(2.3-91) | 71.2(8-120) | <0.001 |
| Hepatitis B | |||
| Yes | 29(87.9%) | 39(95.1%) | |
| No | 4(12.1%) | 2(4.9%) | 0.257 |
| Cirrhosis | |||
| Yes | 24(72.7%) | 35(85.4%) | |
| No | 9(27.3%) | 6(14.6%) | 0.179 |
| Child-Pugh grading | |||
| A | 20(60.6%) | 27(65.9%) | |
| B+C | 13(39.4%) | 14(34.1%) | 0.641 |
| Histologic grade | |||
| Well+moderately | 27(81.8%) | 37(90.2%) | |
| Poorly | 6(18.2%) | 4(9.8%) | 0.292 |
| Milan criteria | |||
| Within | 4(12.1%) | 25(61.0%) | |
| Beyond | 29(87.9%) | 16(39.0%) | <0.001 |
| TNM stage | |||
| 1-2 | 4(12.1%) | 26(63.4%) | |
| 3-4 | 29(87.9%) | 15(36.6%) | <0.001 |
| Tumor size(cm) | |||
| <5 | 13(39.4%) | 31(75.6%) | |
| ≥5 | 20(60.6%) | 10(24.4%) | 0.002 |
| Number of nodules | |||
| <3 | 19(58.8%) | 33(79.6%) | |
| ≥3 | 14(41.2%) | 8(20.4%) | 0.032 |
| Microvascular invasion | |||
| Yes | 19(58.8%) | 3(7.3%) | |
| No | 14(41.2%) | 38(92.7%) | <0.001 |
| Pre-LT serum AFP level | |||
| ≤400(ng/ml) | 25(75.8%) | 28(68.3%) | |
| >400(ng/ml) | 8(24.2%) | 13(31.7%) | 0.479 |
| Antineoplastic prophylaxis after LT | |||
| Yes | 8(24.2%) | 3(7.3%) | |
| No | 25(75.8%) | 38(92.7%) | 0.120 |
HCC: hepatocellular carcinoma; AFP: alpha-fetoprotein; LT: liver transplantation.
Rap1A genotype distribution and its association with HCC recurrence
The minor allele frequencies (MAFs) of Rap1A rs494453 genotypes in both donors and recipients were more than 0.01, which were calculated by Hardy-Weinberg equilibrium. When comparing the recipients and donors carring Rap1A rs494453 polymorphisms, the latter presented distinct distributions in recurrence and nonrecurrence group while the former did not show significance, as shown in Table 2. It is of note that patients who received donor Rap1A rs494453 AG/GG genotype experienced a significantly higher recurrence rate after LT than those received AA genotype (78.8% vs. 21.2%, P=0.001). However, the Rap1A rs494453 genotypes of recipients did not influence the incidence rates of HCC recurrence after LT.
Table 2.
Donor and recipient Rap1A genotype distribution and the association with HCC recurrence
| Genotype distribution, n (%) | P value | HWE value | ||
|---|---|---|---|---|
| Recurrence(n=33) | Nonrecurrence(n=41) | |||
| Donor rs494453 | ||||
| GG | 8(24.3%) | 3(7.3%) | 0.056 | |
| AG | 18(54.5%) | 12(29.3%) | 0.497 | |
| AA | 7 (21.2%) | 26 (63.4%) | 0.003 | |
| Dominant model | ||||
| AA | 7(21.2%) | 26(63.4%) | ||
| AG/GG | 26(78.8%) | 15(36.4%) | 0.001 | |
| Recessive model | ||||
| GG | 8(24.2%) | 3(7.3%) | ||
| AG/AA | 25(75.8%) | 38(92.7%) | 0.201 | |
| Additive model | ||||
| AG | 18(54.5%) | 12(29.3%) | ||
| AA/GG | 15(45.5%) | 29(70.7%) | 0.062 | |
| Recipient rs494453 | ||||
| GG | 7(21.2%) | 8(19.5%) | 0.853 | |
| AG | 22(66.7%) | 27(65.9%) | 0.904 | |
| AA | 4(12.1%) | 6(14.6%) | 0.993 | |
| Dominant model | ||||
| AA | 4(12.1%) | 6(14.6%) | ||
| AG/GG | 29(87.9%) | 35(85.4%) | 0.988 | |
| Recessive model | ||||
| GG | 7(21.2%) | 8(19.5%) | ||
| AG/AA | 26(78.8%) | 33(80.5%) | 0.998 | |
| Additive model | ||||
| AG | 22(66.7%) | 27(65.9%) | ||
| AA/GG | 11(33.3%) | 14(34.1%) | 0.956 | |
HCC: hepatocellular carcinoma; HWE: Hardy-Weinberg equilibrium.
Risk factors for HCC recurrence after LT by logistic regression analysis
As previous studies reported that AFP level and histologic grade also reflect the recurrence rate, we included the two predictive factors into the logistic regression analysis. Univariate logistic regression analysis was carried out. We found that the Milan criteria, TNM stage, tumor size, nodules numbers, microvascular invasion and donor Rap1A rs494453 genotypes turn out to be significant determinants for HCC recurrence after LT. To avoid collinearity, the tumor size and nodule numbers were excluded and the remaining variables indeed had no collinearity (tolerance>0.1). Multivariate logistic analysis revealed that TNM stage (OR=6.444, 95% CI 2.671-26.679; P=0.007), microvascular invasion (OR=20.283, 95% CI 3.277-125.527; P=0.001), Milan criteria (OR=5.924, 95% CI 1.379-25.446; P=0.017) and donor Rap1A rs494453 genotypes (OR=12.014, 95% CI 2.351-61.401; P=0.003) were associated with HCC recurrence (Table 3).
Table 3.
Univariate and Multivariate logistic regression analysis of risk factors associated with HCC recurrence
| Variables | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| Odds ratio(95%CI) | P value | Odds ratio(95%CI) | P value | ||
| Recipient age(0 ≤50,1>50) | 1.21(0.51-2.77) | 0.609 | |||
| Recipient sex (0 female,1 male) | 1.66(0.37-7.65) | 0.421 | |||
| Hepatitis B(0 No, 1 Yes) | 0.51(0.16-2.55) | 0.367 | |||
| Cirrhosis(0 No,1 Yes) | 0.53(0.16-1.58) | 0.201 | |||
| Child-Pugh grade (0 Grade A, 1 Grade B+C) | 1.21(0.52-2.77) | 0.733 | |||
| Histologic grade (0 well+moderately, 1 poorly) | 3.01(0.81-10.55) | 0.123 | |||
| Pre-LT serum AFP level (0 ≤400ng/ml,1>400ng/ml) | 0.744(0.25-2.23) | 0.566 | |||
| TNM stage(0 stage1-2, 1 stage3-4) | 8.33(2.38-24.88) | 0.003 | 6.444(2.671-26.679) | 0.007 | |
| Tumor size(cm) (0<5, 1 ≥5) | 4.54(1.76-11.09) | 0.011 | |||
| Number of nodules (0<3, 1 ≥3) | 2.54(1.11-7.01) | 0.042 | |||
| Microvascular invasion(0 No, 1 Yes) | 14.12(4.54-47.12) | <0.001 | 20.283(3.277-125.527) | 0.001 | |
| Milan criteria(0 within, 1 beyond) | 7.44(2.32-23.01) | 0.002 | 5.924(1.379-25.446) | 0.017 | |
| Donor Rap1A rs494453 genotype (0 AA, 1 AG/GG) | 2.211(1.114-5.898) | 0.009 | 12.014(2.351-61.401) | 0.003 | |
CI: confidence interval.
Association between donor rs494453 polymorphisms and survival rates
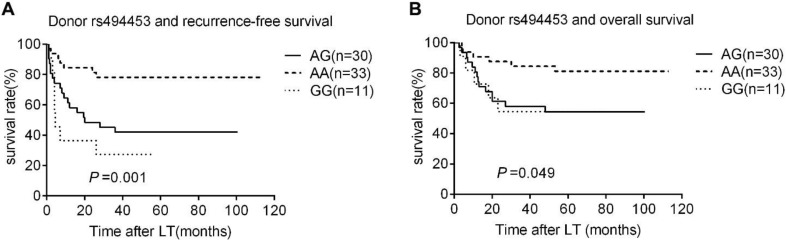
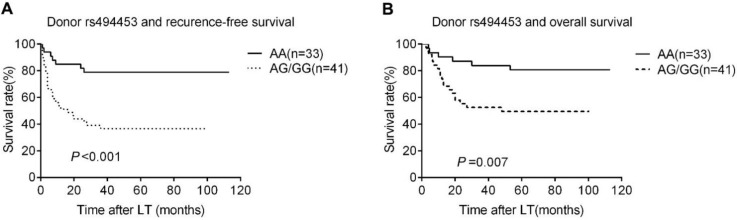
The Rap1A rs494453 genotype polymorphisms of donors, but not the recipients mainly contributed to the recurrence differences in both RFS and OS by univariate Cox regression analysis. The RFS and OS of recipients carring donor's AA, AG and GG alleles were further analysed by Kaplan-Meier survival and the log-rank test to distinguish the effect of different donor rs494453 polymorphism on prognosis. Distinctions of the AG, AA and GG genotypes of donors were observed on RFS and OS of recipients (P=0.001 and P=0.049, respectively; Fig. 1A and 1B). In addition, the RFS and OS of recipients with AG/GG genotypes of donor group were dramatically decreased compared to those with AA genotype (P<0.001 and P=0.007, respectively; Fig. 2A and 2B). However, recipient rs494453 genotypes did not show the influence on RFS and OS differentially (S1 A and B and S2 A and B).
Figure 1.
Kaplan-Meier survival estimates of recurrence-free survival (RFS) (A) and overall survival (OS) (B) among different donor genotypes (AA, GG, and AG). Patients carrying donors genotype GG and AG had the most lower RFS and OS.
Figure 2.
Kaplan-Meier survival estimates of recurrence-free survival (RFS) (A) and overall survival (OS) (B) between different donor genotypes (AA vs. AG/GG). Patients carrying donor homozygous AG/GG had a significantly lower RFS and OS than AA patients.
Risk factors for recurrence-free survival rate and overall survival rate by Cox regression analysis
We selected more impressive factors to perform multivariate Cox regression analysis, including pre-LT serum AFP level (>400ng/ml), histologic grade (poorly differentiated), TNM stage (stages 3-4), tumor size (≥5cm), number of nodules (≥3), Milan criteria (beyond), microvascular invasion and donor rs1927914 AG/GG genotype, and once again excluded tumor size and the number of nodules to prevent collinearity, as mentioned above. The remaining variables showed no collinearity (tolerance>0.1). The Milan criteria (HR=3.936, 95% CI 1.228-12.614; P=0.021), TNM stage (HR=16.004, 95% CI 2.047-125.110; P=0.008), microvascular invasion (HR=3.046, 95% CI 1.394-6.657; P=0.005), and donor rs494453 AG/GG genotype (HR=3.430, 95% CI 1.474-7.981; P=0.004) were found to be independent risk factors for RFS (Table 4). Additionally, for overall survival, the TNM stage (HR=11.490, 95% CI 1.432-92.192; P=0.022), microvascular invasion (HR=3.638, 95% CI 1.532-8.638; P=0.003), and donor rs494453 AG/GG genotype (HR=2.816, 95% CI 1.104-7.181; P=0.030) were also considered as independent risk factors (Table 5).
Table 4.
Cox regression analysis of the risk factors for recurrence-free survival rate
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| Hazard ratio (95% CI) |
P value | Hazard ratio (95% CI) |
P value | |
| Pre-LT serum AFP level (>400ng/ml) | 0.812(0.355-1.867) | 0.821 | ||
| Histologic grade (poorly) |
1.621(0.668-3.930) | 0.285 | ||
| TNM stage (stage3-4) | 7.669(2.681-21.940) | <0.001 | 16.004(2.047-125.110) | 0.008 |
| Tumor size (≥5cm) | 3.633(1.777-7.429) | <0.001 | ||
| Number of nodules (≥3) | 2.206(1.103-4.409) | 0.025 | ||
| Milan criteria (beyond) | 6.754(2.363-19.301) | <0.001 | 3.936(1.228-12.614) | 0.021 |
| Microvascular invasion | 5.950(2.919-12.129) | <0.001 | 3.046(1.394-6.657) | 0.005 |
| Donor rs494453 AG/GG genotype | 2.596(1.034-6.515) | 0.002 | 3.430(1.474-7.981) | 0.004 |
HCC: hepatocellular carcinoma; AFP: alpha-fetoprotein.
Table 5.
Cox regression analysis of the risk factors for overall survival rate
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| Hazard ratio (95% CI) |
P value | Hazard ratio (95% CI) |
P value | |
| Pre-LT serum AFP level (>400ng/ml) | 1.177(0.511-2.976) | 0.605 | ||
| Histologic grade (poorly) |
1.209(0.443-3.23) | 0.787 | ||
| Milan criteria(beyond) | 4.182(1.432-12.207) | 0.013 | ||
| Tumor size(≥5cm) | 2.606(1.168-5.814) | 0.019 | ||
| Number of nodules(≥3) | 2.794(1.271-6.139) | 0.011 | ||
| TNM stage(stage3-4) | 23.862(3.220-176.823) | 0.002 | 11.490(1.432-92.192) | 0.022 |
| Microvascular invasion | 7.797(3.396-17.901) | <0.001 | 3.638(1.532-8.638) | 0.003 |
| Donor rs494453 AG/GG genotype |
2.460(1.155-5.240) | 0.020 | 2.816(1.104-7.181) | 0.030 |
CI: confidence interval.
Discussion
Rap1A is a GTPase that belongs to the Ras-associated protein (Rap) family, which is similar to Ras mostly 16, and associated with cancer initiation and progression 10. Rap1A plays a molecular switch role by converting the inactive GDP-bound state to an active GTP-bound state 17. In previous studies, Rap1A gene over-expression had been reported in various cancers, including ovarian cancer 10, breast cancer 18, and OCSCC 13. The relationship between SNP genotype and HCC recurrence after LT has been reported in VEGF gene 19, IL-15 gene 20 and HPSE gene 21, signifying that the SNP genotype of hosts, including recipients and donors, has a significant role in the progression of HCC recurrence after LT.
In this study, we explored whether the genetic variations of Rap1A were associated with the HCC recurrence after LT. Our results demonstrated that polymorphisms of donor Rap1A rs494453 were remarkably significant in the prediction of HCC recurrence after LT by Kaplan-Meyer curve analysis. Additionally, the increased risk of HCC recurrence was accompanied with AG/GG genotype of donors. Multivariate analysis showed that TNM stage, Milan criteria, microvascular invasion and the donor Rap1A rs494453 genotype AG/GG were considered as independent risk factors for HCC recurrence and RFS. In addition, the TNM stage, microvascular invasion and the donor Rap1A rs494453 genotype AG/GG were independent risk factors for OS. These results were consistent with those of previous studies 7,22,23.
However, the mechanism about how the genetic variation of Rap1A increased the recurrence risk remains unknown. Rap1A encoded protein regulates signaling pathways that affect cell proliferation and adhesion, which plays a role in tumor malignancy. MO et al. found that TNF-a activated NF-kB pathway promotes Rap1A protein over-expression then further increases the HCC progression and metastasis 14. Tao et al. found that Rap1A activates the MAPK signaling pathway to develop breast cancer 24. Other studies also showed that Rap1A promotes cell proliferation and metastasis by activating the extracellular signal-regulated kinase (ERK) and regulating the integrin-mediated cellular activities 25,26. Regarding our results, the microvascular invasion influenced mostly on HCC recurrence, which may be explanded by the circulating tumor cells from microvascular tumor thrombus 27,28.
The limitations of our study were embodied in the two respects. The first limitation was the little patient sample amount with 80.9% of it having HBV related liver disease. To obtain more reliable results, it is needed to expand the patient's sample amount and take in more HCC samples caused by different pathogenic factors, except for HBV. Besides, although we have concluded that the variant genotype influenced HCC recurrence of patients after LT surgery, there is a need to investigate the function and mechanism of Rap1A rs494453 in HCC recurrence.
In conclusion, this is the first investigation on the relationship of Rap1A gene rs494453 polymorphisms in HCC patients treated with LT. These results concluded the positive relationship between donor Rap1A rs494453 polymorphisms with HCC recurrence and OS of LT patients. Furthermore, Rap1A rs494453, microvascular invasion and Milan criteria actively predicted the risk of HCC recurrence after liver LT. These findings could be used for the clinical prognosis of liver transplantation.
Supplementary Material
Supplementary figures.
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China under grant numbers 81401956 and 81702341, and by the Medical Engineering Crossing Project Grant from Shanghai Jiao Tong University (grant number: YG2016QN24). The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. Mar 1. 2015;136(5):E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Lee KK, Kim DG, Moon IS, Lee MD, Park JH. Liver transplantation versus liver resection for the treatment of hepatocellular carcinoma. J Surg Oncol. Jan 1. 2010;101(1):47–53. doi: 10.1002/jso.21415. [DOI] [PubMed] [Google Scholar]
- 3.Lee S, Hyuck David Kwon C, Man Kim J. et al. Time of hepatocellular carcinoma recurrence after liver resection and alpha-fetoprotein are important prognostic factors for salvage liver transplantation. Liver Transpl. Sep. 2014;20(9):1057–1063. doi: 10.1002/lt.23919. [DOI] [PubMed] [Google Scholar]
- 4.Andreou A, Bahra M, Schmelzle M. et al. Predictive factors for extrahepatic recurrence of hepatocellular carcinoma following liver transplantation. Clin Transplant. Jul. 2016;30(7):819–827. doi: 10.1111/ctr.12755. [DOI] [PubMed] [Google Scholar]
- 5.Kondili LA, Lala A, Gunson B. et al. Primary hepatocellular cancer in the explanted liver: outcome of transplantation and risk factors for HCC recurrence. Eur J Surg Oncol. Sep. 2007;33(7):868–873. doi: 10.1016/j.ejso.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Lai Q, Avolio AW, Lerut J. et al. Recurrence of hepatocellular cancer after liver transplantation: the role of primary resection and salvage transplantation in East and West. J Hepatol. Nov. 2012;57(5):974–979. doi: 10.1016/j.jhep.2012.06.033. [DOI] [PubMed] [Google Scholar]
- 7.Nagai S, Yoshida A, Facciuto M. et al. Ischemia time impacts recurrence of hepatocellular carcinoma after liver transplantation. Hepatology. Mar. 2015;61(3):895–904. doi: 10.1002/hep.27358. [DOI] [PubMed] [Google Scholar]
- 8.Mazzaferro V, Chun YS, Poon RT. et al. Liver transplantation for hepatocellular carcinoma. Ann Surg Oncol. Apr. 2008;15(4):1001–1007. doi: 10.1245/s10434-007-9559-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Xu X, Wu J. et al. The stratifying value of Hangzhou criteria in liver transplantation for hepatocellular carcinoma. PLoS One. 2014;9(3):e93128. doi: 10.1371/journal.pone.0093128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu L, Wang J, Wu Y, Wan P, Yang G. Rap1A promotes ovarian cancer metastasis via activation of ERK/p38 and notch signaling. Cancer Med. Dec. 2016;5(12):3544–3554. doi: 10.1002/cam4.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang J, Bian C, Wang H, Huang S, Wu D. MiR-203 down-regulates Rap1A and suppresses cell proliferation, adhesion and invasion in prostate cancer. J Exp Clin Cancer Res. Jan 31. 2015;34:8. doi: 10.1186/s13046-015-0125-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sayyah J, Bartakova A, Nogal N, Quilliam LA, Stupack DG, Brown JH. The Ras-related protein, Rap1A, mediates thrombin-stimulated, integrin-dependent glioblastoma cell proliferation and tumor growth. J Biol Chem. Jun 20. 2014;289(25):17689–17698. doi: 10.1074/jbc.M113.536227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen CH, Chuang HC, Huang CC. et al. Overexpression of Rap-1A indicates a poor prognosis for oral cavity squamous cell carcinoma and promotes tumor cell invasion via Aurora-A modulation. Am J Pathol. Feb. 2013;182(2):516–528. doi: 10.1016/j.ajpath.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 14.Mo SJ, Hou X, Hao XY. et al. EYA4 inhibits hepatocellular carcinoma growth and invasion by suppressing NF-kappaB-dependent RAP1 transactivation. Cancer Commun (Lond) Apr 3. 2018;38(1):9. doi: 10.1186/s40880-018-0276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. Jan; 2009. Chapter 2:Unit 2 12. [DOI] [PubMed] [Google Scholar]
- 16.Bos JL, de Rooij J, Reedquist KA. Rap1 signalling: adhering to new models. Nat Rev Mol Cell Biol. May. 2001;2(5):369–377. doi: 10.1038/35073073. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Mitra RS, Henson BS. et al. Rap1GAP inhibits tumor growth in oropharyngeal squamous cell carcinoma. Am J Pathol. Feb. 2006;168(2):585–596. doi: 10.2353/ajpath.2006.050132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmed SM, Theriault BL, Uppalapati M. et al. KIF14 negatively regulates Rap1a-Radil signaling during breast cancer progression. J Cell Biol. Dec 10. 2012;199(6):951–967. doi: 10.1083/jcb.201206051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu LM, Xie HY, Zhou L, Yang Z, Zhang F, Zheng SS. A single nucleotide polymorphism in the vascular endothelial growth factor gene is associated with recurrence of hepatocellular carcinoma after transplantation. Arch Med Res. Oct. 2009;40(7):565–570. doi: 10.1016/j.arcmed.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 20.Zhang T, Liu Y, Peng X, Fan J, Peng Z. Association between Recipient IL-15 Genetic Variant and the Prognosis of HBV-Related Hepatocellular Carcinoma after Liver Transplantation. Dis Markers. 2017;2017:1754696. doi: 10.1155/2017/1754696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu L, Zhang X, Zhai Y. et al. Association of polymorphisms in the heparanase gene (HPSE) with hepatocellular carcinoma in Chinese populations. Genet Mol Biol. Oct-Dec. 2017;40(4):743–750. doi: 10.1590/1678-4685-GMB-2014-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen D, Liu S, Chen S. et al. Donor interleukin 6 gene polymorphisms predict the recurrence of hepatocellular carcinoma after liver transplantation. Int J Clin Oncol. Dec. 2016;21(6):1111–1119. doi: 10.1007/s10147-016-1001-1. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Shi ZL, Yang X, Yin ZF. Targeting of circulating hepatocellular carcinoma cells to prevent postoperative recurrence and metastasis. World J Gastroenterol. Jan 7. 2014;20(1):142–147. doi: 10.3748/wjg.v20.i1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang T, Jiang K, Zhu X. et al. miR-433 inhibits breast cancer cell growth via the MAPK signaling pathway by targeting Rap1a. Int J Biol Sci. 2018;14(6):622–632. doi: 10.7150/ijbs.24223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorn A, Zoellner A, Follo M. et al. Rap1a deficiency modifies cytokine responses and MAPK-signaling in vitro and impairs the in vivo inflammatory response. Cell Immunol. Mar-Apr. 2012;276(1-2):187–195. doi: 10.1016/j.cellimm.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 26.Pizon V, Baldacci G. Rap1A protein interferes with various MAP kinase activating pathways in skeletal myogenic cells. Oncogene. Dec 7. 2000;19(52):6074–6081. doi: 10.1038/sj.onc.1203984. [DOI] [PubMed] [Google Scholar]
- 27.Kelley RK, Magbanua MJ, Butler TM. et al. Circulating tumor cells in hepatocellular carcinoma: a pilot study of detection, enumeration, and next-generation sequencing in cases and controls. BMC Cancer. Mar 31. 2015;15:206. doi: 10.1186/s12885-015-1195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang YL, Wang RC, Cheng K, Ring BZ, Su L. Roles of Rap1 signaling in tumor cell migration and invasion. Cancer Biol Med. Feb. 2017;14(1):90–99. doi: 10.20892/j.issn.2095-3941.2016.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.