Abstract
The efficacy of ginsenoside Rh2 (Rh2) in cancer therapy has been reported; however, its function in lung cancer remains unknown. To analyze the role of Rh2 in the inhibition of lung cancer cell proliferation in the present study, protein expression levels of E-cadherin, vimentin, β-catenin, Smo, Gli1, and α-catenin were assessed by western blotting, whilst mRNA expression levels of TCF7 FZD8, Smo, Gli1, Gli2, and Gli3 were determined by reverse transcription-quantitative PCR in the A549 cell line. Phosphorylation sites were detected by proteomic methods and cell proliferation was analyzed by MTT assay. The present study revealed that Rh2 treatment significantly inhibited cell proliferation. Western blotting indicated that the expression levels of E-cadherin were increased and vimentin was downregulated in Rh2-treated cells compared with control cells. Treatment of A549 cells with Rh2 suppressed phosphorylation of five distinct proteins and increased phosphorylation of nine proteins. Among them, the phosphorylation of α-catenin at S641 was significantly induced. Rh2 treatment suppressed the expression levels of key genes involved in Wnt (Wnt3, transcription factor 7 and frizzled class receptor 8) and hedgehog [smoothened, frizzled class receptor (Smo), GLI family zinc finger (Gli)1, Gli2, and Gli3] signaling. Immunoblotting results indicated that β-catenin, Smo and Gli1 protein expression levels were also suppressed by treatment with Rh2 compared with control treatment. Expression of α-catenin S641D, a phosphomimetic form of α-catenin, inhibited the accumulation of β-catenin and Gli1 and inhibited cell proliferation and invasion. Furthermore, knockdown of β-catenin (CTNNB1) or Gli1 with specific small interfering RNAs inhibited cell proliferation, whereas overexpression of these genes had an opposite effect. Additionally, overexpression of β-catenin or Gli1 activated cell proliferation, even in the presence of Rh2, suggesting that Rh2 affects A549 cell proliferation through inhibition of Wnt and hedgehog signaling by phosphorylation of α-catenin at S641. Together, these data suggested that Rh2 treatment may inhibit the proliferation of A549 lung cancer cells. Further exploration of the underlying mechanism by which Rh2 inhibits cell proliferation is warranted.
Keywords: ginsenoside Rh2, α-catenin, invasion, cell proliferation, lung cancer
Introduction
Lung cancer is a major cause of cancer-related death, and the 5-year survival rate remains very low, as more than half of patients are diagnosed too late for successful treatment (1). Advances in molecular translational research have resulted in greater understanding, diagnosis and management of lung cancer, improving patient survival rates (2). To date, efforts to develop innovative treatments have focused on targeting key signaling pathways involved in lung cancer growth and progression (3). Although considerable progress in targeted therapies has been achieved, further advances are required to improve prognosis and increase the overall life expectancy of patients with lung cancer (4). Distinct types of cancer result from the abnormal proliferation of different cells in the organism, and their properties and responses to treatment can vary substantially (5). Among the hallmarks of cancer cells, insensitivity to anti-growth signals, evading apoptosis and sustained angiogenesis are common in most, if not all, types of cancer (5,6). Accordingly, an effective strategy to treat cancer is the termination of out-of-control cell growth through activation of the intrinsic mechanisms of cell death (7).
Panax ginseng, the most common species of ginseng, is a herbal medicine that is widely applied in Asian countries (8). P. ginseng has been suggested to possess numerous beneficial properties, including anti-inflammatory, antioxidant and anticancer activity (9). Ginsenosides, a form of triterpene glycosides (saponins), are the major active components in ginseng and have been extensively used in traditional Chinese medicine as an anticancer agent (10). It has been suggested that ginseng extract blocks the proliferation of mammalian tumor cells by stimulating apoptosis (11). Ginsenoside Rh2 (Rh2) is characterized by low toxicity, low molecular weight and exhibits good solubility in lipids. Rh2 has been demonstrated to inhibit proliferation and migration of tumor cells, as well as angiogenesis. In addition, its inhibitory effect on angiogenesis in prostate cancer is mediated by regulating the expression of the metal cation transporter CNNM1(12). A previous study suggested that, in liver cancer cells, Rh2 is able to regulate the expression of a large number of non-coding RNAs (13), and an additional study in breast cancer cells suggested that Rh2 inhibits proliferation via epigenetic modifications of the cell-mediated immune pathway (14). Rh2 has additionally been suggested to inhibit the migration and invasion of lung cancer cells by modulation of tumor-induced macrophages (15). Pseudo-Rh2 has also been reported to induce apoptosis via the Ras/Raf/ERK/p53 pathway in the A549 adenocarcinoma cell line (16). Together, these findings suggest that Rh2 may exert anticancer activity through a range of diverse mechanisms.
Wnt signaling is essential during embryonic development and has a crucial role in the maintenance of the stem-like properties of tissue cells, including cancer cells (17). Hedgehog (Hh) signaling regulates diverse biological processes, among them the development of invertebrate and vertebrate organisms (18). The canonical Wnt signaling pathway, also known as the Wnt/β-catenin or β-catenin/T-cell factor pathway (19), performs its regulatory function by stabilizing the key transcription factor, β-catenin, which activates downstream gene expression (20-22). It is well documented that the activation of Wnt signaling is closely associated with the development of cancer in numerous types of tissue (23). Constitutive activation of Hh signaling affects the development and progression of cancer through several mechanisms (24). Aberrant activation of Hh signaling is required for almost all basal cell carcinomas, rhabdomyosarcomas, medulloblastomas and several other tumor types (18,25-27). The binding of Hh and protein patched homolog 1 molecules results in activation of the smoothened, frizzled class receptor (Smo) protein (26,28), which subsequently upregulates the expression of downstream transcriptional activator GLI-Kruppel family transcription factors to stimulate Hh signaling (28). GLI family zinc finger (Gli)1 has been demonstrated to function as a modulator of cancer cell properties controlled by E-cadherin/β-catenin signaling. Gli1 activates expression of the gel-forming mucin gene, MUC5AC, which in turn inhibits E-cadherin-dependent cell-to-cell adhesion, activating migration and invasion of pancreatic ductal adenocarcinoma cells (15). Together the available evidence indicates the involvement of Wnt and Hh signaling in cancer cell proliferation. However, whether a relationship exists between Rh2 and Wnt or Hh signaling remains to be determined.
The present study aimed to investigate the impact of Rh2 on the proliferation of A549 lung cancer cells, and on the expression of Wnt and Hh signaling markers. The relationship between the expression of β-catenin and Gli1 and the proliferation of A549 cells in the presence or absence of Rh2 was examined. The objective of this investigation was to establish the mechanism by which Rh2 regulates Wnt and Hh signaling and proliferation in A549 lung cancer cells.
Materials and methods
Cell culture and transfection assays
The human lung adenocarcinoma cell line A549 was obtained from the American Type Culture Collection. The cells were grown in Dulbecco's modified Eagle's medium, supplemented with glutamine, 10% fetal bovine serum, penicillin and streptomycin (Gibco; Thermo Fisher Scientific, Inc.). cDNA encoding β-catenin, α-catenin S641D and Gli1 were synthesized by Sangon Biotech. Co., Ltd. and pcDNA3.1 (+) (Invitrogen; Thermo Fisher Scientific, Inc.) was used to construct overexpression (OX) vectors. Small interfering RNA (siRNA) for β-catenin (ON-TARGETplus SMART pool; cat. no. L-004018), siRNA for Gli1 (ON-TARGETplus SMART pool; cat. no. J-041026-05) and ON-TARGETplus non-targeting pool (cat. no. D-001810) were purchased from GE Healthcare Dharmacon, Inc. A total of 1x106 cells were transfected with 2 µg β-catenin, α-catenin S641D or Gli1 OX plasmids, pcDNA3.1 (+) empty vector (OX control) and 30 nM siRNAs on day 0 using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) and Opti-MEM® I Reduced Serum Medium (Gibco; Thermo Fisher Scientific, Inc.), according to the manufacturers' protocol. Cells reached ~30% confluence 24 h post-transfection and the transfection media were replaced with full growth medium; cells were harvested for subsequent experiments and 72 h after transfection.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR) analysis
Total RNA was extracted using TRIzol® Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) from A549 cells treated with Rh2 (cat. no. 209058; Sigma-Aldrich; Merck KGaA) or transfected with β-catenin, α-catenin S641D and Gli1 OX plasmids, or β-catenin and Gli1 siRNAs for 72 h at 37˚C in an incubator with 5% CO2, according to manufacturer's protocol. A total of 2 µg RNA was reverse-transcribed using GoScript™ Reverse Transcription kit at 42˚C for 1 h (Promega Corporation), according to the manufacturer's instructions. SYBR® Green Master Mix (Bio-Rad Laboratories, Inc.) was used to perform qPCR in an Illumina Eco 3.0 Real-time PCR system (Illumina, Inc.). The thermocycling conditions consisted of an initial denaturation at 95˚C for 3 min, followed by 40 cycles of denaturation for 30 sec at 95˚C, annealing for 30 sec at 58˚C and extension at 72˚C for 30 sec, ending with a final extension at 72˚C for 5 min. The transcription levels were normalized against those of GAPDH using the 2-ΔΔCq method (29). The sequences of primers used are listed in Table I.
Table I.
Primer sequences.
| Primer | Sequences (5'-3') |
|---|---|
| Wnt3 F | ATCATAAGGGGCCGCCTGGCGAAGGCTGG |
| Wnt3 R | CTTGCAGGTGTGCACGTCGTAGA |
| TCF7 F | CTGCAGACCCCTGACCTCTCT |
| TCF7 R | ATCCTTGATGCTAGGTTCTGGTGT |
| FZD8 F | CTGGTGGAGATCCAGTGCTC |
| FZD8 R | TTGTAGTCCATGCACAGCGT |
| Smo F | ACCTATGCCTGGCACACTTC |
| Smo R | AGGAAGTAGCCTCCCACGAT |
| Gli1 F | CCAGAGTTCAAGAGCCTGG |
| Gli1 R | CCTCGCTCCATAAGGCTCAG |
| Gli2 F | GTTCCAAGGCCTACTCTCGCCTG |
| Gli2 R | 5'-CTTGAGCAGTGGAGCACGGACAT-3' |
| Gli3 F | GGGTGAACAGCATCAAAATGGAG |
| Gli3 R | CCGATAGCCATGTTGGTGG |
| β-catenin F | TCGCCAGGATGATCCCAGC |
| β-catenin R | GCCCATCCATGAGGTCCTG |
| GAPDH F | GACCTGCCGTCTAGAAAAAC |
| GAPDH R | CTGTAGCCAAATTCGTTGTC |
F, forward; FZD8, frizzled class receptor 8; Gli, GLI family zinc finger; R, reverse; Smo, smoothened, frizzled class receptor; TCF, transcription factor.
Western blot analysis
Both Rh2 treated and OX or siRNA construct transfected A549 cells were harvested in ice-cold lysis buffer (7 M urea, 2 M thiourea, 2% CHAPS, 40 mM Tris base, 40 mM dithiothreitol and 1% protease inhibitor) to obtain whole-cell extracts. Protein concentration was measured using a Bicinchoninic Acid assay kit (Merck KGaA). The total proteins were separated on a 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis gel at 100 V for 2 h following extraction followed by transferal onto Immobilon-P Transfer Membranes (EMD Millipore). The membranes were incubated in TBS containing 5% skimmed milk and 0.05% Tween-20 (EMD Millipore) at 25˚C for blocking for 1 h, followed by incubation with the following primary antibodies at 25˚C for overnight: Anti-vimentin (cat. no. ab193555; 1:1,000; Abcam), anti-E-cadherin (cat. no. ab194982; 1:1,000; Abcam), anti-Smo (cat. no. ab8969; 1:1,000; Abcam), anti-β-catenin (cat. no. ab16051; 1:2,000; Abcam), anti-Gli1 (cat. no. ab49314; 1:2,000; Abcam), anti-α-catenin (cat. no. 3240; 1:1,000; Cell Signaling Technology, Inc.), anti-phosphorylated (p)-α-catenin Ser641 antibody (cat no. 11330, 1:1,000; Signalway Antibody LLC) and anti-GAPDH (cat. no. ab8245; 1:2,000; Abcam). The membranes were washed twice with PBS and incubated with an anti-mouse or anti-rabbit horseradish peroxidase-conjugated secondary antibody (cat. no. 7074 and 7076; 1:2,000; Cell Signaling Technology, Inc.) for 1 h at 25˚C. Antigen-antibody complexes were visualized using an electrochemiluminescence kit (Beijing BioTrand, Inc.). Protein levels were normalized against GAPDH and protein expression was analyzed using ImageJ2 version 2.0 software (National Institutes of Health).
Phosphopeptide isolation and analysis
Total protein was extracted from control and Rh2-treated A549 cells and the protein concentration determined by bicinchoninic acid assay. A total of 1 mg extracted protein was digested overnight using trypsin (1:50 wt/wt) at 37˚C. Digested peptides were extracted and incubated at 25˚C for 15 min in 25 mM ammonium bicarbonate, and then for 15 min in 5% formic acid. Samples were desalted on a C18 column (cat no. IQLAALGABXFANUMBBD, Lancompare) according to the manufacturer's instructions and dried using a SpeedVac. Phosphopeptides were enriched according to a previously published protocol (30). Prior to binding phosphopeptides, the TiO2 beads were equilibrated with 200 µl 30 mg/ml 2,5-dihydroxybenzoic acid in 80% acetonitrile and 0.1% TFA, and the pH of the digested peptide lysate was adjusted to ≤1.9 with 1% TFA. The peptide mixture was then added to a 2-ml reaction tube containing 10 mg TiO2 beads, and each batch was incubated for 30 min at 25˚C with end-over-end rotation. Subsequently, the beads were spun down at 500 x g at 4˚C for 15 min and briefly washed once with 80% acetonitrile and 0.1% TFA, and once with 10% acetonitrile and 0.1% TFA. Finally, the bound peptides were eluted from the beads using 200 µl NH4OH in 30% acetonitrile (pH>10.0). The eluate was immediately neutralized with 5% TFA and dried.
The TiO2-enriched phosphopeptides (4 µl) were subjected to on-line nanoflow liquid chromatography (LC) using the EASY-Nano LC system (Proxeon Biosystems; Thermo Fisher Scientific, Inc.) with 10-cm capillary columns of an internal diameter of 75 µm filled with 3 µm Reprosil-Pur C18-A2 resin (Dr. Maisch HPLC GmbH). The sequence of gradients consisted of 10-30% (v/v) CAN in 0.1% (v/v) formic acid at the flow rate of 200 nl/min for 45 min, 30-100% (v/v) CAN in 0.1% (v/v) formic acid at a flow rate of 200 nl/min for 1 min, and 100% CAN in 0.1% formic acid at a flow rate of 200 nl/min for 10 min. The elution was electrosprayed using a Proxeon nanoelectrospray ion source by electrospray ionization (ESI). The ESI-tandem mass spectrometry (MS/MS) analysis was performed using a Thermo Fisher LTQ Velos Pro (Thermo Fisher Scientific, Inc.) using full ion scan mode over the m/z range of 200-1,800. Collision-induced dissociation (CID) was performed in the linear ion trap using a 4.0-Th isolation width and 35% normalized collision energy with helium as the collision gas. Five independent MS/MS scans were performed on each ion using dynamic exclusion. Additionally, the precursor ion selected for CID was dynamically excluded from further MS/MS analysis for 30 sec. The MS/MS spectra were processed using Proteome Discoverer (Version 1.3; Thermo Fisher Scientific, Inc.), and the database search was performed using the Mascot search engine (Mascot 2.3; Matrix Science) against a concatenated target decoy approach. The Uniprot-KB/Swiss-Prot protein sequence database (release 54.5; https://web.expasy.org/docs/swiss-prot_guideline.html) was searched, with corresponding taxonomy selection for different samples. The following search parameters were applied: Mass error tolerance for the precursor ions, 1 Da; mass error tolerance for the fragment ions, 0.8 Da; fixed modifications, carbamidomethylation (C); variable modifications, oxidation (M), phosphorylation (S, T, Y); number of missed cleavages, 1; significance threshold, P<0.05; type of instrument, ESI-TRAP. Protein identifications were validated only if they met the following three requirements: i) Their score was significant (P<0.05) with cut-off criteria; ii) one peptide had a score >15; iii) proteins were identified in at least two out of the three runs. Proteins identified by a set or subset of peptides used for identification of another protein were not considered.
Cell proliferation analysis
To determine cell proliferation rate, cells were plated in a volume of 150 µl at a density of 2,000 cells/well in 96-well plates. Cell proliferation was analyzed following treatment with 50 or 100 µM Rh2, or transfections with siRNAs and OX plasmids using a CCK-8 kit (Dojindo Molecular Technologies, Inc.) as previously described (31). At each indicated time point (0, 24, 48, 72 and 96 h), MTT solution (Beyotime Insitute of Biotechnology) was added to each well to a final concentration of 5 mg/ml followed by incubation at 37˚C for 4 h. A total of 100 µl acidic isopropanol (10% SDS, 5% isopropanol and 0.01 M HCl) was then added into each well to stop the reaction and the plates were incubated at 37˚C overnight.
Cell invasion assay
The invasive properties of A549 cells treated with 100 µM Rh2 or transfected with α-catenin S641D were assessed using a 96-well 3D spheroid cell invasion assay (cat. no. 3500-096-K; Trevigen Inc.; Bio-Techne), according to the manufacturer's instructions.
Statistical analysis
Statistical analysis was performed with Prism 5 software package (GraphPad Software, Inc.). The data are presented as the mean ± SEM with at least three experimental replicates. Comparisons between two groups and the determination of statistical significance was done by Student's t-test. Comparisons between more than two groups were performed using one-way ANOVA, followed by Bonferroni's multiple comparison test.
Results
Rh2 inhibits the proliferation of A549 lung cancer cells
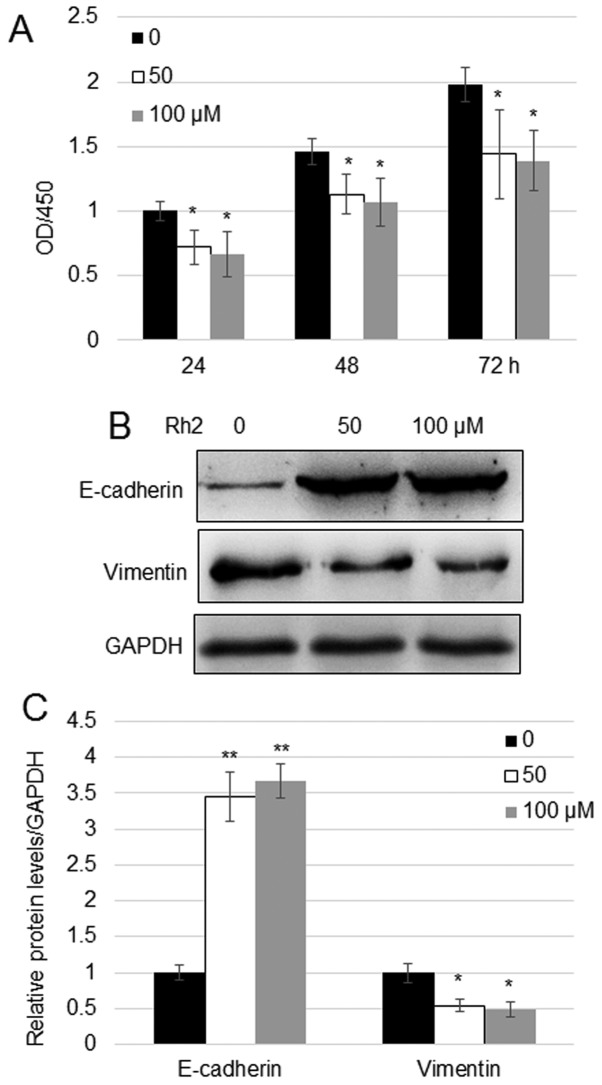
Rh2 is well established as an anticancer molecule and is widely used in cancer therapy in China, but its function in lung cancer cells remains unclear (10,11). Cell proliferation is a key process of spreading of cancer in human tissues and is directly associated with the severity of the disease (5). To analyze the function of Rh2 in lung cancer cells, the A549 cells were treated with Rh2 for 24, 48 and 72 h. In comparison with untreated cells, exposure to 50 or 100 µM Rh2 led to significant inhibition of the proliferation of A549 cells (Fig. 1A). Since E-cadherin and vimentin levels are related to the migration of cancer cells, the impact of Rh2 on these proteins was analyzed. Western blotting indicated that treatment with 50 and 100 µM Rh2 increased the expression levels of E-cadherin ~3.5-fold and reduced the expression levels of vimentin by ~50% compared with untreated cells (Fig. 1B and C).
Figure 1.
Rh2 treatment inhibits the proliferation of A549 lung cancer cells. (A) MTT assay was performed to analyze the effects of Rh2 on A549 cell proliferation at 0, 24 and 48 h after treatment with 50 and 100 µM Rh2. Data represent the mean values ± SE of 20 replicates. *P<0.05 vs. Control. (B) Western blot analysis was performed to assess the protein expression levels of E-cadherin and vimentin. GAPDH was used as an internal control. (C) Densitometry of bands shown in (B). Data represent the mean values ± SE (n=3). *P<0.05 and **P<0.01 vs. Control. OD, optical density; Rh2, ginsenoside Rh2.
Rh2 reduces the expression of Wnt and Hh signaling genes in A549 lung cancer cells
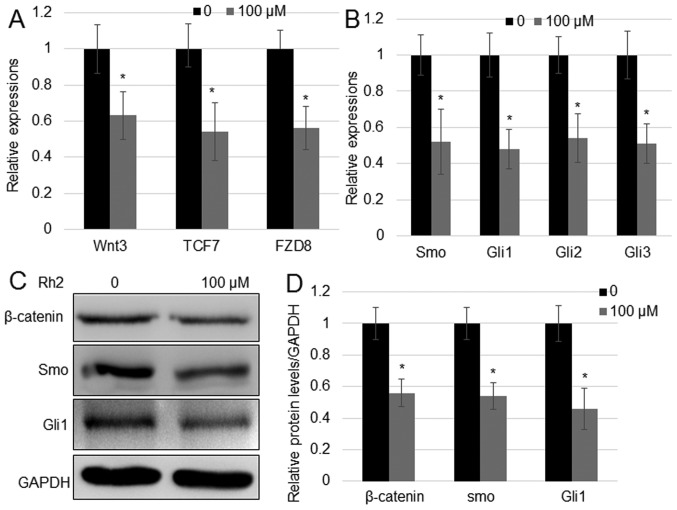
Wnt and Hh signaling serve a critical role in the proliferation of hepatocellular carcinoma (32). To analyze whether Rh2 regulates Wnt and Hh signaling genes in A549 cells, the expression levels of key genes implicated in the two signaling pathways were analyzed. The mRNA expression levels of Wnt signaling genes [Wnt3, transcription factor 7 (TCF7) and frizzled class receptor 8 (FZD8)] and Hh signaling genes (Smo, Gli1, Gli2, and Gli3) were measured by RT-qPCR. The results indicated that, in comparison with untreated cells, Rh2 reduced the expression of both Wnt signaling genes (Wnt3, TCF7 and FZD8) and Hh signaling genes (Smo, Gli1, Gli2 and Gli3) (Fig. 2A and B). In addition, the protein expression levels of the key Wnt signaling regulator β-catenin and Hh signaling regulator Smo, and the expression of Gli1 were analyzed by western blotting. In agreement with the results of RT-qPCR, the protein expression levels of β-catenin, Smo and Gli1 were reduced with Rh2 treatment compared with untreated cells (Fig. 2C and D).
Figure 2.
Rh2 suppresses the expression of genes implicated in Wnt and Hh signaling. (A) Expression of Wnt signaling genes Wnt3, TCF7 and FZD8, and (B) Hh signaling genes Smo, Gli1, Gli2 and Gli3 in A549 cells after treatment with 100 µM Rh2. GAPDH was used as an internal control to normalize expression levels. Error bars indicate mean ± SE (n=3). (C) β-catenin, Smo and Gli1 protein levels in the presence or absence of 100 µM Rh2 examined by western blot analysis. GAPDH was used as the loading control. (D) Relative levels of proteins shown in (C). *P<0.05 vs. Control. FZD8, frizzled class receptor 8; GLI, Gli family zinc finger; Hh, hedgehog; Rh2, ginsenoside Rh2; Smo, smoothened, frizzled class receptor; TCF7, transcription factor 7.
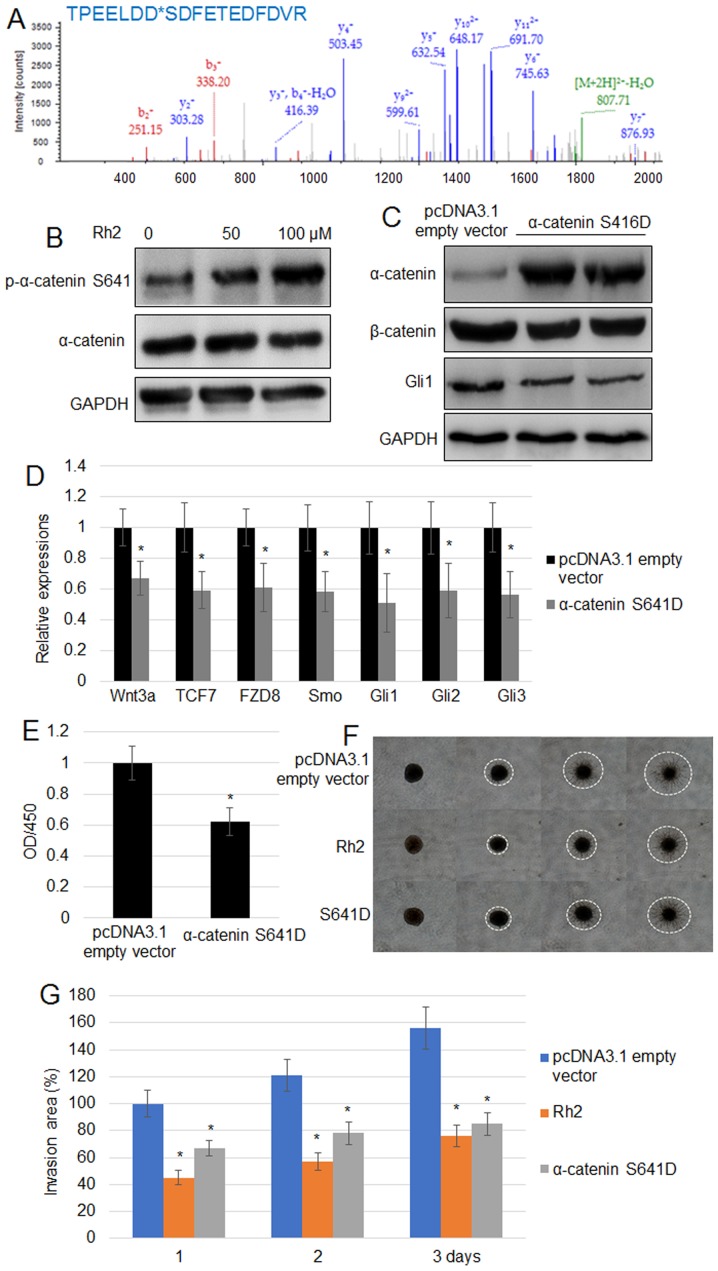
Rh2 induces the phosphorylation of α-catenin at S641
To further analyze the mechanisms implicated in the effects of Rh2, phosphopeptides were analyzed using the TiO2-enrichment method. Significant changes in the levels of phosphorylation were identified in 14 phosphopeptides. Among them, the phosphorylation level of five proteins was reduced, whereas the phosphorylation of nine proteins was induced by Rh2 (Tables II and III). The five proteins displaying reduced phosphorylation levels were ACTA, ITGA5, RACK1, ARHGEF6 and FAM129B, while the nine with higher levels of phosphorylation were PPP1R12A, FERMT2, α-catenin, CCDC6, LIMCH1, GRHPR, STEAP3, PPP3CA and HS1BP3. Given the suppression of Wnt signaling by Rh2, the phosphorylation of α-catenin was evaluated further. LC/MS data for histogram of p-peptide showed that α-catenin at the S641 residue exhibited the highest peak (691.70), suggesting that α-catenin phosphorylation may be at the S641 residue (Fig. 3A). To verify further, the phosphorylation level of α-catenin at the S641 residue, western blotting using a specific p-α-catenin S641 antibody confirmed that Rh2 treatment significantly increased the levels of p-α-catenin S641 without changing the levels of total α-catenin (Fig. 3B). Expression of α-catenin S641D, a phosphomimetic form of α-catenin S641, reduced the accumulation of β-catenin and Gli1 in A549 cells (Fig. 3C). Additionally, RT-qPCR indicated that the expression levels of Wnt signaling genes (Wnt3, TCF7 and FZD8) and Hh signaling genes (Smo, Gli1, Gli2 and Gli3) in A549 cells were suppressed by the expression of α-catenin S641D (Fig. 3D). Furthermore, α-catenin S641D expression significantly inhibited cell proliferation (Fig. 3E). The results of a cell invasion assay suggested that α-catenin S641D expression or treatment with 100 µM Rh2 significantly reduced the invasiveness of A549 cells. White dashed circles mark the borders of cell invasion area in each cell (Fig. 3F and G).
Table II.
Proteins with phosphorylation suppressed by Rh2.
| Accession number | Gene name | Description | Mr | pI | Phosphorylated peptides | Ion score | E-value | Ion precursor | Ion charge | Phosphosite |
|---|---|---|---|---|---|---|---|---|---|---|
| P68133 | ACTA | Actin, α skeletal muscle | 42366 | 5.23 | K.CDIDIRKDLYANNVMSGGTTMYPGIADR.M | 35 | 0.0072 | 1108.5046 | 3 | Y296 |
| P08648 | ITGA5 | Integrin α-5 | 115605 | 5.5 | R.LLESSLSSSEGEEPVEYK.S | 72 | 7.60E-06 | 1032.2283 | 2 | S127 |
| P63244 | RACK1 | Guanine nucleotide-binding protein subunit β-2-like 1 | 35511 | 7.6 | M.TEQMTLR.G | 35 | 0.034 | 528.0834 | 2 | T2, T6 |
| Q15052 | ARHGEF6 | Rho guanine nucleotide exchange factor 6 | 88698 | 5.79 | R.MSGFIYQGK.I | 52 | 0.00074 | 555.6702 | 2 | S488 |
| Q96TA1 | FAM129B | Niban-like protein 1 | 84598 | 5.82 | R.GLLAQGLRPESPPPAGPLLNGAPAGESPQPK.A | 48 | 0.0005 | 1033.0134 | 3 | S665, S652 |
Mr, Peptide molecular weight score.
Table III.
Proteins with phosphorylation increased by Rh2.
| Accession number | Gene name | Description | Mr | pI | Phosphorylated peptides | Ion score | E-value | Ion precursor | Ion charge | Phosphosite |
|---|---|---|---|---|---|---|---|---|---|---|
| O14974 | PPP1R12A | Protein phosphatase 1 regulatory subunit 12A | 115610 | 5.31 | R.RSTQGVTLTDLQEAEK.T | 64 | 8.4-5 | 928.7751 | 2 | T696 |
| Q96AC1 | FERMT2 | Fermitin family homolog 2 | 78438 | 6.26 | K.KLDDQSEDEALELEGPLITPGSG SIYSSPGLYSK.T | 74 | 3.60E-05 | 1226.3796 | 3 | S159 |
| P35221 | CTNNA1 | Catenin α-1 | 100693 | 5.95 | R.TPEELDDSDFETEDFDVR.S | 98 | 9.50E-09 | 1120.2859 | 2 | S641 |
| Q16204 | CCDC6 | Coiled-coil domain-containing protein 6 | 53429 | 6.87 | K.LDQPVSAPPSPR.D | 38 | 0.0045 | 712.4462 | 2 | S240, S244 |
| Q9UPQ0 | LIMCH1 | LIM and calponin homology domains-containing protein 1 | 122818 | 6.1 | K.SPEPEATLTFPFLDK.M | 52 | 0.0031 | 886.3337 | 2 | S718 |
| Q9UBQ7 | GRHPR | Glyoxylate reductase/hydroxypyruvate reductase | 36045 | 7.01 | R.GDVVNQDDLYQALASGK.I | 38 | 0.019 | 976.8146 | 2 | Y255 |
| Q658P3 | STEAP3 | Metalloreductase STEAP3 | 55079 | 8.86 | R.ESNAEYLASLFPTCTVVK.A | 36 | 0.0031 | 1095.3168 | 2 | S128, Y132 |
| Q08209 | PPP3CA | Serine/threonine-protein phosphatase 2B catalytic subunit α isoform | 59335 | 5.58 | R.IITEGASILR.Q | 31 | 0.023 | 617.1566 | 2 | T66, S70 |
| Q53T59 | HS1BP3 | HCLS1-binding protein 3 | 42868 | 4.89 | K.GEDAEESLEEEEALDPLGIMR.S | 30 | 0.015 | 1206.755 | 2 | S194 |
Figure 3.
Rh2 treatment increases the level of p-α-catenin S641 in A549 cells. (A) Phosphopeptide diagram for α-catenin showed phosphorylation of the S641 residue. (B) p-α-catenin S641 and α-catenin levels were analyzed by western blotting following treatment with 0, 50 and 100 µM Rh2. GAPDH was used as the loading control. (C) α-catenin, β-catenin and GLI1 levels were examined by western blot analysis in pcDNA3.1 empty vector transformed and α-catenin S641D-overexpressing A549 cells. GAPDH was used as the internal control. (D) Expression levels of Wnt signaling genes (Wnt3, TCF7 and FZD8) and Hh signaling genes (Smo, Gli1, Gli2 and Gli3) were assessed in the pcDNA3.1 empty vector transformed and α-catenin S641D-expressing A549 cells. *P<0.05 vs. pcDNA3.1 empty vector. (E) Cell proliferation rate was measured in the pcDNA3.1 empty vector transformed and α-catenin S641D-expressing A541 cells. *P<0.05 vs. pcDNA3.1 empty vector. (F) Cell invasion assay using A549 cells transformed with the pcDNA3.1 empty vector, expressing α-catenin S641D or treated with Rh2 (100 µM) for 3 days. The images of cells invading into the surrounding matrix were acquired at days 0, 1, 2 and 3. White dashed circles indicate the area invaded by the cells. Bar, 500 µm. (G) Measurement of cell invasion areas depicted in (F). Data are presented as the mean values ± SE of 10 replicates. *P<0.05 vs. pcDNA3.1 empty vector. FZD8, frizzled class receptor 8; GLI, GLI family zinc finger; OD, optical density; p, phosphorylated; Rh2, ginsenoside Rh2; Smo, smoothened, frizzled class receptor; TCF7, transcription factor 7.
β-catenin and Gli1 positively regulate lung cancer cell proliferation
As Rh2 treatment appeared to reduce the expression of key genes involved in Wnt and Hh signaling, the role of these pathways in A549 cell proliferation was analyzed. β-catenin and Gli1 levels were reduced by siRNA or overexpressed post-transfection with OX plasmids compared with control levels. RT-qPCR results revealed that transfection with the siRNA duplex and OX plasmid significantly suppressed or induced, respectively, the expression of β-catenin (Fig. 4A) and Gli1 (Fig. 4C). Comparison of cell proliferation among the control, siRNA and OX groups indicated that β-catenin knockdown by siRNA inhibited proliferation, whereas OX enhanced proliferation of A549 cells (Fig. 4B). Similar results were obtained with Gli1 knockdown and OX (Fig. 4D).
Figure 4.
Role of β-catenin and Gli1 in A549 cell proliferation. (A) β-catenin expression was analyzed in the pcDNA3.1 empty vector, NC siRNA, β-catenin siRNA and β-catenin OX cells. Error bars indicate the mean ± SE (n=3). *P<0.05 and **P<0.01 vs. NC siRNA. (B) Cell proliferation rate was analyzed in the pcDNA3.1 empty vector, NC siRNA, β-catenin siRNA and β-catenin OX cells. *P<0.05 vs. NC siRNA. (C) Gli1 expression was analyzed in the pcDNA3.1 empty vector, NC siRNA, Gli1 siRNA and Gli1 OX cells. Error bars indicate the mean ± SE (n=3). *P<0.05 and **P<0.01 vs. pcDNA3.1 empty vector. (D) Cell proliferation rate in the pcDNA3.1 empty vector, NC siRNA, Gli1 siRNA and Gli1 OX cells. Data represent the mean ± SE of 6 replicates. *P<0.05 vs. pcDNA3.1 empty vector. GLI, GLI family zinc finger; NC, non-targeting control; OD, optical density; OX, overexpression; siRNA, small interfering RNA.
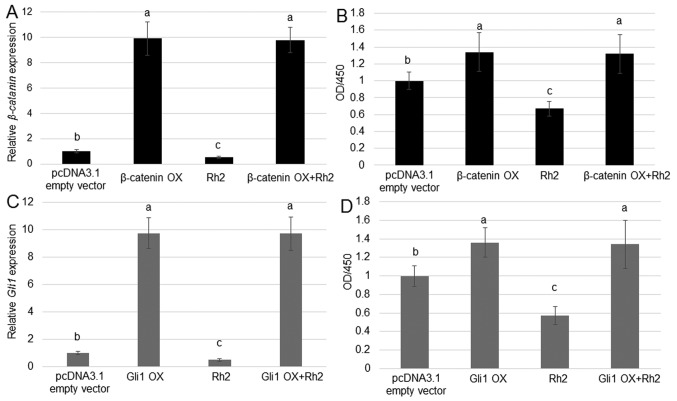
β-catenin or Gli1 OX blocks the inhibition of A549 cell proliferation by ginsenoside Rh2
As Rh2 reduced the expression of β-catenin and Gli1, and levels of these proteins were positively associated with A549 cell proliferation, whether OX of β-catenin or Gli1 could counteract the inhibitory effect of Rh2 on A549 cell proliferation was tested. RT-qPCR results suggested that β-catenin and Gli1 were highly expressed in cells transfected with OX plasmids compared with the control group (Fig. 5A and C). The treatment with Rh2 inhibited the rate of cell proliferation, whereas OX of β-catenin and Gli1 had an opposite effect. Furthermore, OX of β-catenin and Gli1 reversed the inhibitory effect of Rh2, accelerating the proliferation of A549 cells (Fig. 5B and D).
Figure 5.
Effects of β-catenin or Gli1 OX on the impact of Rh2 on the proliferation of A549 cells. (A) β-catenin expression was analyzed in A549 cells treated with the β-catenin OX plasmid, 100 µM Rh2, or Rh2 together with the β-catenin OX plasmid. pcDNA3.1 empty vector transformed cells were used as control. (B) Cell proliferation in A549 cells treated with the pcDNA3.1 empty vector, β-catenin OX plasmid, 100 µM Rh2, or a combination of Rh2 and β-catenin OX plasmid. (C) Gli1 expression was analyzed in A549 cells treated with the Gli1 OX plasmid, 100 µM Rh2, or a combination of Rh2 and the Gli1 OX plasmid. (D) Cell proliferation in A549 cells treated with the pcDNA3.1 empty vector, 100 µM Rh2, Gli1 OX plasmid, or a combination of Rh2 and Gli1 OX plasmid. P<0.0.5 at 'a' vs. 'b', 'b' vs. 'c', and P<0.01 at 'a' vs. 'c' Data are presented as the GLI, GLI family zinc finger; OD, optical density; OX, overexpression; Rh2, ginsenoside Rh2.
Discussion
The majority of lung cancer cases worldwide, are diagnosed as non-small-cell lung cancer; accounting for 85% of lung cancer-related deaths (33). The identification of an effective therapeutic approach to lung cancer is therefore an urgent issue. Rh2 has been suggested to act as an anticancer molecule that affects diverse types of cancer cells. However, the mechanism by which it regulates the proliferation of lung cancer cells is not yet known.
The present study sought to determine whether Rh2 could regulate the proliferation of A549 cells and to identify the molecular mechanism involved. Treatment with Rh2 significantly inhibited cell proliferation, upregulated E-cadherin expression and downregulated vimentin compared with in untreated cells, suggesting that this molecule affects the behavior of A549 lung cancer cells. Subsequent experiments elucidated the role of Wnt and Hh signaling in mediating the effects of Rh2. This was an essential question, since the Wnt (23) and Hh signaling pathways are known to be associated with cancer development and progression (24). Rh2 treatment suppressed key Wnt signaling genes, Wnt3, TCF7 and FZD8, and Hh signaling genes, Smo, Gli1, Gli2 and Gli3. Moreover, it decreased the protein expression levels of β-catenin (Wnt signaling), Smo (Hh signaling) and GLI1 (Hh signaling) in A549 cells, suggesting that Rh2 suppresses both signaling pathways. The phosphoproteomic study revealed that the phosphorylation of 14 distinct peptides was significantly altered by treatment with Rh2. Among them, phosphorylation levels of five peptides were suppressed, and phosphorylation levels of nine peptides were increased. Further analysis demonstrated that phosphorylation of the α-catenin S641 residue was significantly increased and that the expression of α-catenin S641D, a phosphomimetic of α-catenin S641, markedly reduced the levels of β-catenin and Gli1. α-catenin S641D expression additionally significantly suppressed key Wnt signaling genes, Wnt3, TCF7 and FZD8, as well as Hh signaling genes, Smo, Gli1, Gli2 and Gli3. These findings indicated that the expression of α-catenin S641D may inhibit Wnt and Hh signaling in A549 cells. Previously, α-catenin was reported as a tumor suppressor acting as an inhibitor of the inflammatory response in breast cancer cells (34). However, α-catenin phosphorylation signaling has not been extensively investigated. The present results indicated that Rh2 stimuli may activate the phosphorylation of α-catenin, which, in turn, inhibits the accumulation of β-catenin and Gli1. In addition, expression of α-catenin S641D severely attenuated the proliferation and invasion of A549 cells, resembling the effect of Rh2 on lung cancer cells. The roles of Wnt and Hh signaling in A549 cell proliferation were also investigated. The data suggested that suppression of the expression of β-catenin and Gli1 by siRNAs inhibited cell proliferation, whereas OX of these genes promoted cell proliferation, indicating that Wnt and Hh signaling cascades exert positive control on the proliferation rate of A549 cells. To test the possibility that Rh2 might suppress Wnt and Hh signaling to inhibit lung cancer cell proliferation, β-catenin and Gli1 were overexpressed in A549 cells. Administration of Rh2 did not change β-catenin and Gli1 levels in overexpressing cells but downregulated the expression of these genes in untransfected A549 cells. This finding suggested that Rh2 might control activities of the promoters of β-catenin and Gli1 genes. It will be of interest to further dissect the regulatory mechanism by which Rh2 affects the transcription of β-catenin and Gli1. These data indicated that β-catenin and Gli1 promoted proliferation not only of control cells, but also of cells treated with Rh2, suggesting that β-catenin and Gli1 may function downstream of Rh2 signaling in the control of lung cancer cell proliferation.
The obtained results indicated that Rh2 suppressed both Wnt and Hh signaling in the A549 lung cancer cell line. A previous study on skin fibroblasts reported that β-catenin complex directly activates two Hh signaling genes, Smo and Gli1, via promoter binding (35). Thus, it may be hypothesized that, in lung cancer cells, Rh2 regulates Wnt directly and it is the modification of Wnt signaling that affects the activity of genes involved in Hh signaling. It is apparent that further experiments are needed to define the regulatory model. In conclusion, the present study documented that Rh2 may suppress Wnt and Hh signaling via the activation of α-catenin S641 phosphorylation, inhibiting lung cancer cell proliferation and invasion. These findings expand our understanding of the regulatory mechanism controlled by Rh2 and its anticancer activity.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
GZ and ZM designed the experiments. GZ, LH, JC and BX performed the experiments. GZ, LH, JC, BX and ZM analyzed the data. GZ and ZM wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent for publication
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Chan BA, Hughes BG. Targeted therapy for non-small cell lung cancer: Current standards and the promise of the future. Transl Lung Cancer Res. 2015;4:36–54. doi: 10.3978/j.issn.2218-6751.2014.05.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts PJ, Stinchcombe TE, Der CJ, Socinski MA. Personalized medicine in non-small-cell lung cancer: Is KRAS a useful marker in selecting patients for epidermal growth factor receptor-targeted therapy? J Clin Oncol. 2010;28:4769–4777. doi: 10.1200/JCO.2009.27.4365. [DOI] [PubMed] [Google Scholar]
- 4.de Looff M, de Jong S, Kruyt FA. The role of Src in TRAIL signaling in non-small cell lung cancer cells. Proceedings: AACR Annual Meeting 2018. April 14-18, 2018. Cancer Res 78: 4377, 2018. [Google Scholar]
- 5.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 6.Arbiser JL, Bonner MY, Gilbert LC. Targeting the duality of cancer. doi: 10.1038/s41698-017-0026-x. NPJ Precision Oncol 1: pii: 23, 017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfeffer CM, Singh ATK. Apoptosis: A target for anticancer therapy. Int J Mol Sci. 2018;19(448) doi: 10.3390/ijms19020448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rastogi V, Santiago-Moreno J, Doré S. Ginseng: A promising neuroprotective strategy in stroke. Front Cell Neurosci. 2015;8(457) doi: 10.3389/fncel.2014.00457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiefer D, Pantuso T. Panax ginseng. Am Fam Physician. 2003;68:1539–1542. [PubMed] [Google Scholar]
- 10.Helms S. Cancer prevention and therapeutics: Panax ginseng. Altern Med Rev. 2004;9:259–274. [PubMed] [Google Scholar]
- 11.Fei XF, Wang BX, Tashiro S, Li TJ, Ma JS, Ikejima T. Apoptotic effects of ginsenoside Rh2 on human malignant melanoma A375-S2 cells. Acta Pharmacol Sin. 2002;23:315–322. [PubMed] [Google Scholar]
- 12.Huang Y, Huang H, Han Z, Li W, Mai Z, Yuan R. Ginsenoside Rh2 inhibits angiogenesis in prostate cancer by targeting CNNM1. J Nanosci Nanotechnol. 2019;19:1942–1950. doi: 10.1166/jnn.2019.16404. [DOI] [PubMed] [Google Scholar]
- 13.Chen WW, Huang YF, Hu ZB, Liu YM, Xiao HX, Liu DB, Zhuang YZ. Microarray analysis of altered long non-coding RNA expression profile in liver cancer cells treated by ginsenoside Rh2. J Asian Nat Prod Res. 2019;21:742–753. doi: 10.1080/10286020.2018.1490273. [DOI] [PubMed] [Google Scholar]
- 14.Lee H, Lee S, Jeong D, Kim SJ. Ginsenoside Rh2 epigenetically regulates cell-mediated immune pathway to inhibit proliferation of MCF-7 breast cancer cells. J Ginseng Res. 2018;42:455–462. doi: 10.1016/j.jgr.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Huang N, Zhu W, Wu J, Yang X, Teng W, Tian J, Fang Z, Luo Y, Chen M, Li Y. Modulation the crosstalk between tumor-associated macrophages and non-small cell lung cancer to inhibit tumor migration and invasion by ginsenoside Rh2. BMC Cancer. 2018;18(579) doi: 10.1186/s12885-018-4299-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Xu H, Lu Z, Yu X, Lv C, Tian Y, Sui D. Pseudo-ginsenoside Rh2 induces A549 cells apoptosis via the Ras/Raf/ERK/p53 pathway. Exp Ther Med. 2018;15:4916–4924. doi: 10.3892/etm.2018.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bushman W. Hedgehog signaling in development and cancer. In: Prostate cancer Springer, pp107-118, 2007. [Google Scholar]
- 19.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: Diseases and therapies. Nat Rev Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 21.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 22.Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, Rando TA. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 23.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 24.Xin M, Ji X, De La Cruz LK, Thareja S, Wang B. Strategies to target the Hedgehog signaling pathway for cancer therapy. Med Res Rev. 2018;38:870–913. doi: 10.1002/med.21482. [DOI] [PubMed] [Google Scholar]
- 25.Beauchamp EM, Ringer L, Bulut G, Sajwan KP, Hall MD, Lee YC, Peaceman D, Ozdemirli M, Rodriguez O, Macdonald TJ, et al. Arsenic trioxide inhibits human cancer cell growth and tumor development in mice by blocking Hedgehog/GLI pathway. J Clin Invest. 2011;121:148–160. doi: 10.1172/JCI42874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K, Pan L, Che X, Cui D, Li C. Sonic Hedgehog/GLI1 signaling pathway inhibition restricts cell migration and invasion in human gliomas. Neurol Res. 2010;32:975–980. doi: 10.1179/016164110X12681290831360. [DOI] [PubMed] [Google Scholar]
- 27.Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: Conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 2006;133:3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- 28.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Larsen MR, Thigholm TE, Jensen ON, Roepstorff P, Jørgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Xuan YH, Huang BB, Tian HS, Chi LS, Duan YM, Wang X, Zhu ZX, Cai WH, Zhu YT, Wei TM, et al. High-glucose inhibits human fibroblast cell migration in wound healing via repression of bFGF-regulating JNK phosphorylation. PLoS One. 2014;9(e108182) doi: 10.1371/journal.pone.0108182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tripathy A, Thakurela S, Sahu MK, Uthanasingh K, Behera M, Ajay AK, Kumari R. The molecular connection of histopathological heterogeneity in hepatocellular carcinoma: A role of Wnt and Hedgehog signaling pathways. PLoS One. 2018;13(e0208194) doi: 10.1371/journal.pone.0208194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neal RD, Hamilton W, Rogers TK. Lung cancer. BMJ. 2014;349(g6560) doi: 10.1136/bmj.g6560. [DOI] [PubMed] [Google Scholar]
- 34.Piao HL, Yuan Y, Wang M, Sun Y, Liang H, Ma L. α-catenin acts as a tumour suppressor in E-cadherin-negative basal-like breast cancer by inhibiting NF-κB signalling. Nat Cell Biol. 2014;16:245–254. doi: 10.1038/ncb2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Lin P, Wang Q, Zheng M, Pang L. Wnt3a-regulated TCF4/β-catenin complex directly activates the key Hedgehog signalling genes Smo and GLI1. Exp Ther Med. 2018;16:2101–2107. doi: 10.3892/etm.2018.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.