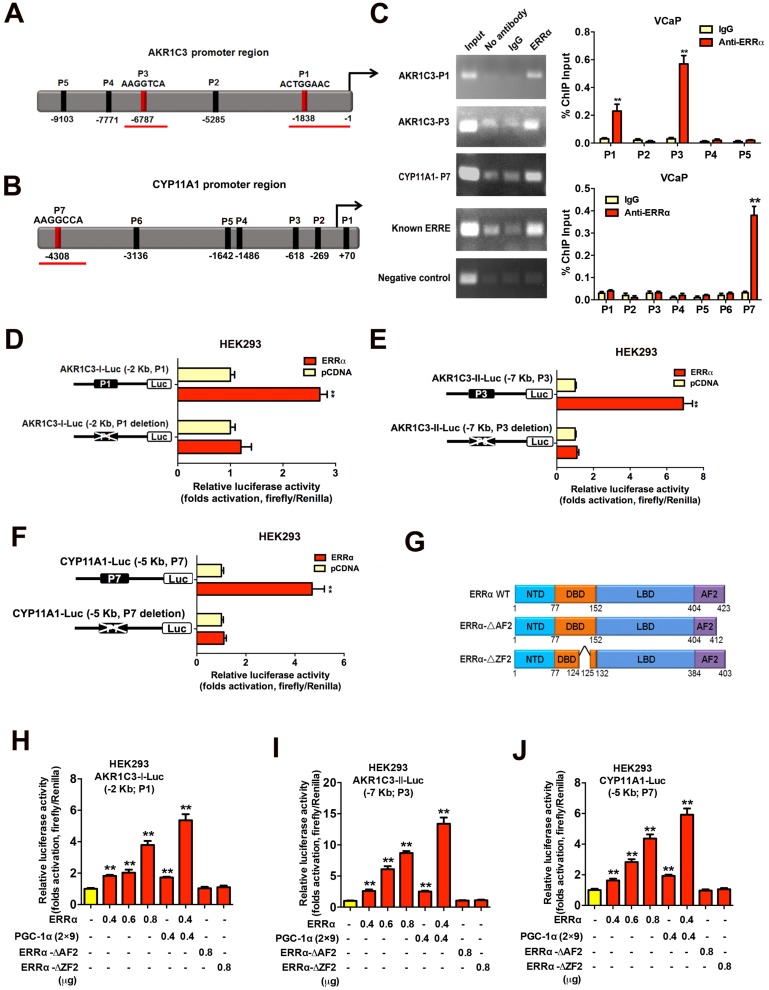
Figure 5.
Direct transactivation of AKR1C3 and CYP11A1 genes by ERRα in prostate cancer cells. (A and B) Schematic diagrams depict the putative ERRα-binding sites (ERREs), as predicted by the online program MatInspector (https://www.genomatrix.de), located in the (A) AKR1C3 (P1-P5) and (B) CYP11A1 (P1-P7) gene promoter/regulatory regions. (C) ChIP-PCR assay of AKR1C3 and CYP11A1 gene promoters performed in VCaP prostate cancer cells. Results validated that two ERRE sites (P1 and P3) located respectively at 1.8 kb and 6.8 kb upstream of the transcription start site of AKR1C3 promoter (upper graph), and one ERRE site (P7) located at 4.3 kb upstream of the CYP11A1 promoter (lower graph), were enriched of ERRα. **, P < 0.01 versus non-immune IgG-treated DNA. (D-F) Luciferase reporter assay of AKR1C3-Luc and CYP11A1-Luc reporters performed in ERRα-transfected HEK293 cells. The AKR1C3-Luc and CYP11A1-Luc reporter constructs, containing inserts of ERRα promoter/regulatory regions, could be significantly transactivated by the transfected ERRα. Deletion of identified ERREs in the reporters (P1 and P3 in AKR1C3-Luc; P7 in CYP11A1-Luc) abolished or significantly reduced the ERRα-induced transactivation. (G) Schematic diagram shows the functional domains of wild-type ERRα protein and two of its truncated mutants generated. (H-J) Luciferase reporter assays of AKR1C3-Luc and CYP11A1-Luc reporters performed in HEK293 cells. The AKR1C3-Luc and CYP11A1-Luc reporters could be dose-dependently transactivated by ERRα and further potentiated by co-transfection with co-regulator PGC-1α (2×9), but not by ERRα-ΔAF2 and ERRα-ΔZF2 truncated mutants. *, P < 0.05; **, P < 0.01 versus empty vector pcDNA3.1. Data are presented as mean ± SD (n = 3) and analyzed by Students' t-test.