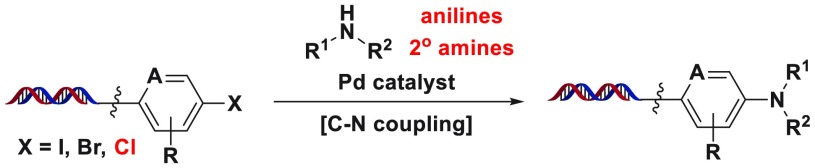
Abstract
DNA-encoded chemical library (DECL) screens are a rapid and economical tool to identify chemical starting points for drug discovery. As a robust transformation for drug discovery, palladium-catalyzed C–N coupling is a valuable synthetic method for the construction of DECL chemical matter; however, currently disclosed methods have only been demonstrated on DNA-attached (hetero)aromatic iodide and bromide electrophiles. We developed conditions utilizing an N-heterocyclic carbene–palladium catalyst that extends this reaction to the coupling of DNA-conjugated (hetero)aromatic chlorides with (hetero)aromatic and select aliphatic amine nucleophiles. In addition, we evaluated steric and electronic effects within this catalyst series, carried out a large substrate scope study on two representative (hetero)aryl bromides, and applied this newly developed method within the construction of a 63 million-membered DECL.
Introduction
In recent years, DNA-encoded chemical libraries (DECLs) have emerged as a powerful hit generation tool for discovery campaigns.1−5 DECLs are large collections of small molecules covalently tagged with a unique DNA sequence which enables multiplexed affinity- or activity-based screens of libraries against biological targets with subsequent decoding by high-throughput DNA sequencing.6,7 Although many forms of the underlying DNA-tag concept have been reported,8−18 the solution-phase DNA-recorded synthesis variant has proven to be a versatile platform for affinity-based screens as library compounds may be built directly with inventory chemical building blocks,19 expensive large-scale synthetic DNA stocks may be reused between DECLs,9 novel million-member DECLs can routinely be produced at scales for thousands of screens, and affinity-based target screens of billions of compounds may be performed, sequenced and analyzed in as little as 2 weeks.20 However, although there are robust methods for the manipulation of the encoding DNA tags,21 only a limited set of chemical transformations have been reported for DECL small molecule synthesis.22−35 DECL DNA-chemical conjugates generally require partially aqueous media to remain in solution, and DNA is sensitive to some conditions (e.g., strong acids, high temperatures, oxidants);25 thus, adaptation of common synthetic transformations and bond-disconnect strategies to DNA-compatible processes is needed to further expand DECL small molecule chemical space.
Palladium-catalyzed C–N coupling holds a prominent place in the chemist’s toolbox as a widely used reaction in fine chemical synthesis and medicinal chemistry efforts.36,37 In particular, the palladium-catalyzed C–N coupling of (hetero)aromatic or aliphatic amines with (hetero)aromatic electrophiles has been utilized in the creation of screening compound collections,38 for diversification of hit series in structure–activity studies,39 and as a synthetic disconnect in large-scale pharmaceutical productions.40 C–N coupling is particularly useful in combinatorial approaches as very large and diverse collections of multifunctional (hetero)aryl electrophiles and amino nucleophiles are commercially available.38 This, along with the advent of specialized phosphine ligands for palladium-catalyzed C–N coupling41,42 and innovations in the rapid generation of active palladium species,43,44 has spurred development of a variety of mild C–N coupling conditions, including in water or aqueous solvent mixtures.45 However, extension of metal-catalyzed C–N coupling to solution-phase DECL synthesis is challenging as not only must it be applicable for thousands to millions of sterically and electronically distinct substrate pairs but also working in multiwell plate format in the presence of DNA and water introduces additional challenges: (a) ready decomposition of catalysts from aerobic or aqueous decomposition pathways; (b) palladium- or heat-promoted degradation of DNA or molecular linkages particularly in the case of prolonged reaction times; (c) a basicity limit in the system due to the presence of water; (d) dehalogenation pathways arising from palladium species produced from water or β-hydride transfer; (e) poor catalyst solubility in aqueous–organic mixtures.
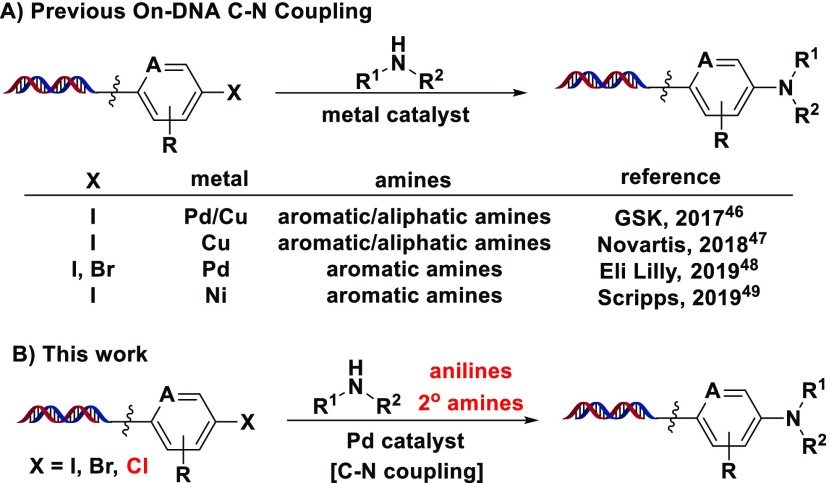
Despite these challenges, several groups have reported procedures for metal-catalyzed C–N coupling on DNA–chemical conjugates for DECL applications: (1) Lu et al. disclosed the palladium-catalyzed coupling of different aromatic amines and a copper-catalyzed coupling of aliphatic amines on a p-iodobenzamide substrate;46 (2) Ruff et al. reported a copper-catalyzed system for Ullmann-type couplings of a variety of primary and secondary aliphatic amines with different (hetero)aromatic iodides;47 (3) de Pedro Beato et al. described a palladium-catalyzed method for the coupling of different (hetero)aryl bromides with aromatic amines;48 (4) Flood et al. published a solid-phase nickel-catalyzed electrochemical amination of aliphatic and aromatic amines on a p-iodobenzamide substrate (Scheme 1A).49 These conditions were demonstrated on iodide and bromide electrophiles; however, many halogenated building blocks from commercial or historical collections may only be readily available in brominated or chlorinated form, limiting the input DECL building block diversity. Having previously utilized C–N coupling successfully within several internal DECL productions,26 we sought to develop alternate solution-phase palladium-catalyzed conditions that might couple a variety of (hetero)aryl bromides and chlorides with aromatic amines and if possible select aliphatic amine nucleophiles (Scheme 1B). In addition, we sought to validate these conditions through a wide study of coupling partners and functional group tolerance, investigation on substrates with library-relevant DNA tags, and demonstration within a DECL build with DNA sequencing analysis for ultimate application in target-based “selection”50 campaigns.
Scheme 1. Metal-Catalyzed C–N Coupling.
Results and Discussion
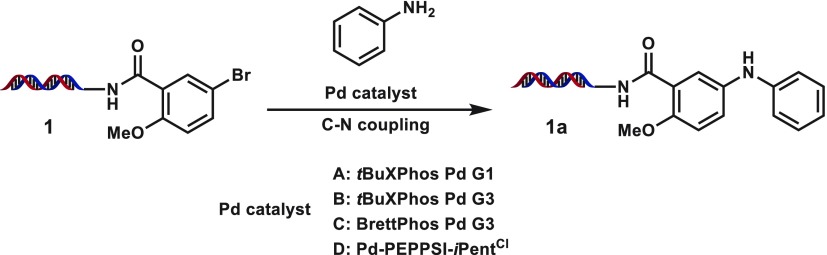
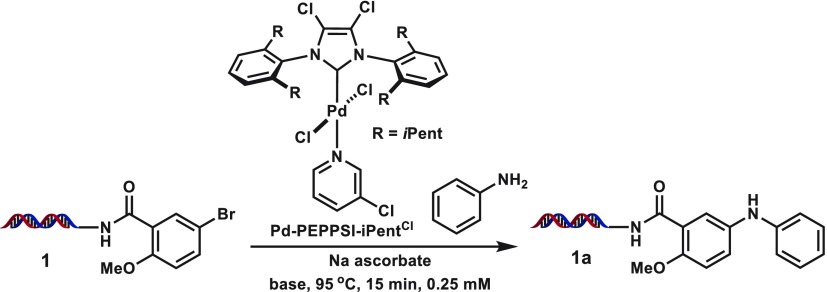
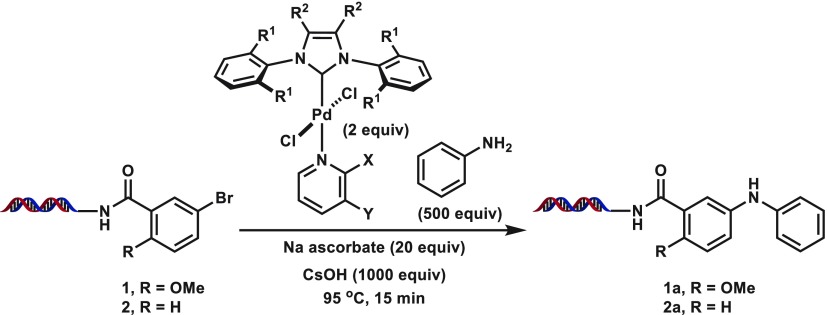
We began our studies by investigating previously disclosed palladium-based conditions46,48 on the coupling of 17-bp dsDNA-linked, methoxy-substituted aryl bromide 1 with aniline (Table 1; see Supporting Information for full structural information on conjugate). Although the conditions from Lu et al. could produce 1a in good yield (entry 1, Table 1), DNA decomposition was detected due to the harsh reaction conditions (hydroxide base for 3 h at 100 °C). Application of the system from de Pedro Beato et al.48 or from a recent DECL synthesis we reported26 provided the coupled product in poor conversions (entries 2 and 3, Table 1), and attempts to further optimize BrettPhos Pd G3 were not successful (e.g., entries 4 and 5, Table 1). PEPPSI (pyridine-enhanced precatalyst preparation stabilization and initiation) catalysts are a series of stable Pd(+2) complexes that include a N-heterocylic carbene (NHC) ligand and a fast-dissociating pyridyl ligand and have been widely used in palladium-catalyzed cross-coupling.51−53 Although use of Pd-PEPPSI-iPentCl with 1-methoxy-2-isopropanol cosolvent failed to provide 1a, upon switching to dimethylacetamide cosolvent a very modest amount of product was observed (entries 6 and 7, Table 1; see Table 2 for catalyst structure). Hypothesizing that the low conversion could stem from inefficient formation of the active palladium species, we were pleased to observe fair conversions to 1a when sodium ascorbate was included as a reducing agent/oxygen scavenger (entries 8 and 9, Table 1).47 Encouraged by this result, we began a screen of reaction parameters with the PEPPSI-iPentCl catalyst (Table 2). From this initial condition, increasing the amount of catalyst loading or equivalents of sodium ascorbate used had no beneficial effect (entries 1–3, Table 2), whereas lowering the amount of catalyst hurt the conversion (entries 4–6, Table 2).
Table 1. General Screening of Palladium Catalyst Systems.
| entry | catalyst | base | time | solvent | additive | conversiona |
|---|---|---|---|---|---|---|
| 1b | A | CsOH | 100 °C, 3 h | DMA | N/A | 75% |
| 2c | B | NaOH | 60 °C, 2 h | DMA | N/A | 30% |
| 3d | C | CsOH | 95 °C, 25 min | MIPO | N/A | 33% |
| 4e | C | CsOH | 95 °C, 25 min | MIPO | N/A | 0% |
| 5e | C | CsOH | 95 °C, 25 min | DMA | N/A | 6% |
| 6f | D | CsOH | 95 °C, 25 min | MIPO | N/A | 0% |
| 7f | D | CsOH | 95 °C,25 min | DMA | N/A | 14% |
| 8g | D | CsOH | 95 °C, 25 min | DMA | Na ascorbate | 57% |
| 9g | D | CsOH | 95 °C, 15 min | DMA | Na ascorbate | 63% |
The conversion was determined by LC-MS.
Condition reported by GSK46.
Condition reported by Eli Lilly48.
Condition reported by BCM26.
Reaction conditions: 1 (5 nmol), 250 equiv of aniline (400 mM), 2 equiv of catalyst (5 mM), 500 equiv of CsOH (1000 mM), H2O (2.5 μL), solvent (4.9 μL).
Reaction conditions: 1 (5 nmol), 250 equiv of aniline (400 mM), 2 equiv of catalyst (5 mM), 500 equiv of CsOH (1000 mM), H2O (2.5 μL), solvent (4.9 μL).
Reaction conditions: 1 (5 nmol), 250 equiv of aniline (400 mM), 2 equiv of catalyst (5 mM), 500 equiv of CsOH (1000 mM), 20 equiv of sodium ascorbate (40 mM), DMA (4.9 μL). DMA = dimethylacetamide. MIPO = 1-methoxy-2-isopropanol.
Table 2. Optimization of Parameters.
| entry | aniline (equiv) | Pd (equiv) | ascorbate (equiv) | base (equiv) | conversiona |
|---|---|---|---|---|---|
| 1 | 250 | 2 | 20 | CsOH (500) | 63% |
| 2 | 250 | 2 | 40 | CsOH (500) | 63% |
| 3 | 250 | 5 | 50 | CsOH (500) | 59% |
| 4 | 250 | 1 | 5 | CsOH (500) | 0% |
| 5 | 250 | 1 | 10 | CsOH (500) | 20% |
| 6 | 250 | 1 | 20 | CsOH (500) | 53% |
| 7 | 250 | 2 | 20 | Cs2CO3 (500) | 32% |
| 8 | 250 | 2 | 20 | pH 9.5 Borate (500) | 0% |
| 9b | 250 | 2 | 20 | CsOH (500) | 0% |
| 10c | 250 | 2 | 20 | CsOH (500) | 63% |
| 11d | 250 | 2 | 20 | CsOH (500) | 56% |
| 12e | 250 | 2 | 20 | CsOH (500) | 0% |
| 13 | 250 | 2 | 20 | CsOH (750) | 66% |
| 14 | 250 | 2 | 20 | CsOH (1000) | 67% |
| 15 | 500 | 2 | 20 | CsOH (1000) | 80% |
The conversion was determined by LC-MS.
The condition was carried out at 80 °C.
The reaction was run for 5 min.
The reaction was run for 60 min.
The reaction concentration was 0.05 mM.
Attempts to use weaker bases such as carbonate or borate (e.g., entries 7 and 8, Table 2) led to lower conversion. No coupling was observed when the reaction was run at 80 °C, similar conversions were observed for 5 and 60 min reaction times, and the reaction was sensitive to further dilution (entries 8–11, Table 2). However, upon increasing the amounts of building block and base, boosts in conversion were observed, with 500 equiv of building block and 1000 equiv of CsOH as the optimal amount within practical solubility limits (entries 13–15, Table 2).
Previous studies into PEPPSI-type catalysts have shown coupling yields can significantly be affected by the degree of substitution at the ortho positions of the NHC phenyl groups and the electron density at the palladium center, particularly when applied to challenging coupling partners.54 To assess these effects we investigated the coupling of two m-bromobenzamide substrates 1 and 2 with aniline under our optimized conditions (entry 15, Table 2) with a variety of differentially substituted PEPPSI-type catalysts (Table 3). Commercially available PEPPSI catalysts Pd-PEPPSI-iPr, Pd-PEPPSI-SIPr, Pd-PEPPSI-iPent, and Pd-PEPPSI-iHept—a series that features increasingly bulky ortho positions on a simple NHC backbone—all failed to provide any of the cross-coupled product (entries 1–4, Table 3). Surprisingly, even dichlorinated NHC analog Pd-PEPPSI-iPrCl was not able to provide any product; only the combination of high steric bulk and backbone chlorination in catalyst Pd-PEPPSI-iPentCl could promote the cross-coupling within this system. Substitution effects at the labile pyridyl ligand could affect the initiation rate and ultimate success of the coupling, and PEPPSI complexes with various pyridine ligands have been reported.52 However, no significant difference was seen upon switching 3-chloropyridine to 2-picoline or unsubstituted pyridine (entries 6–8, Table 3). Finally, increasing the ortho substitution from i-pentyl to i-heptyl within this chlorinated NHC series did not improve the conversion, and the catalyst suffered from increased initial insolubility (entry 9, Table 3). Ultimately, Pd-PEPPSI-iPentCl-pyr was chosen as the optimal catalyst to progress to further studies due to its high reactivity and as it does not produce halogenated pyridyl byproducts that might be capable of oxidative addition.
Table 3. Investigation of PEPPSI Catalystsa.
| entry | NHC catalyst | R1 | R2 | X | Y | 1a | 2a |
|---|---|---|---|---|---|---|---|
| 1 | Pd-PEPPSI-iPr | i-Pr | H | H | Cl | 0% | 0% |
| 2 | Pd-PEPPSI-iPent | i-Pent | H | H | Cl | 0% | 0% |
| 3 | Pd-PEPPSI-iHept | i-Hept | H | H | Cl | 0% | 0% |
| 4 | Pd-PEPPSI-SIPr | i-Pr | Hb | H | Cl | 0% | 0% |
| 5 | Pd-PEPPSI-iPrCl | i-Pr | Cl | H | Cl | 0% | 0% |
| 6 | Pd-PEPPSI-iPentCl | i-Pent | Cl | H | Cl | 80% | 92% |
| 7 | Pd-PEPPSI-iPentCl-o-picoline | i-Pent | Cl | Me | H | 69% | 91% |
| 8 | Pd-PEPPSI-iPentCl-pyr | i-Pent | Cl | H | H | 78% | 91% |
| 9 | Pd-PEPPSI-iHeptCl | i-Hept | Cl | H | Cl | 80% | 90% |
The conversion was determined by LC-MS.
Pd-PEPPSI-SIPr has a saturated NHC ring.
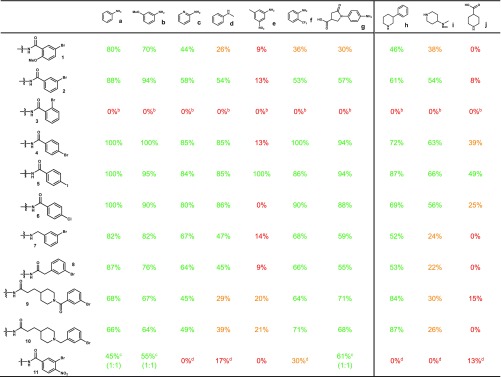
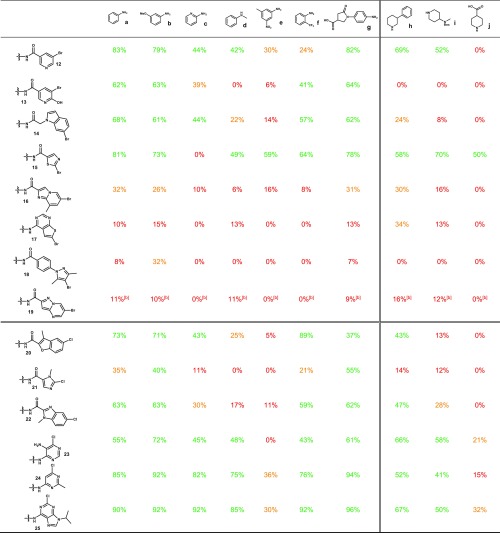
With an optimized catalyst and reaction condition in hand, we next turned to exploring the substrate scope of a set of 10 representative amino nucleophiles on (hetero)aryl electrophiles attached to 17-bp dsDNA constructs (Table 4). In general, good to excellent conversions were observed with simple aromatic amines such as aniline a, 3-methoxyaniline b, 2-aminopyridine c, 2-trifluoromethylaniline f, or carboxylated aniline g with a variety of halide-coupling partners. Notably, similar results were obtained for these amines with privileged 4-iodo, -bromo, and -chloro benzamide congeners 4–6. However, more challenging aromatic amines, such as N-methyl aniline d and 3-methyl-5-nitroaniline e, suffered from lowered conversions. The presence of nearby coordinating functionality may influence reaction success; although meta- and para-bromo benzamide 2 and 4 underwent efficient coupling, ortho-bromo 3 universally failed to provide coupled products, presumably due to the formation of an intramolecular palladacycle-type interaction.55 Although a nearby basic amine did not affect the coupling (e.g., 9 vs 10), the presence of an ortho-nitro group in bromide 11 diminished its conversions. We next looked to evaluate these substrates on more challenging cyclic, secondary aliphatic amines with phenyl amine h, N-Boc diamine i, and amino acid j chosen as representative examples. Reduced conversions were generally observed utilizing this amine class, particularly in the case of negatively charged amine j. Despite this, many substrate combinations coupled in synthetically useful conversion, suggesting that use of these conditions with cyclic secondary aliphatic amines could be a tractable DECL synthetic process. Unfortunately, extension of these conditions to the coupling of acyclic secondary aliphatic amines and primary aliphatic amines with (hetero)aromatic iodides, bromides, and chlorides generally provided the coupled products in low conversion. These results may stem from an enhanced ability of these amines to initiate β-hydride transfer processes and may require further optimization of this catalyst system. Representative results for these type of aliphatic amines, as well as indoles, are presented in the Supporting Information.
Table 4. C–N Coupling between Phenyl Halides and Various Anilines and Secondary Aminesa.
Conversion determined by LC-MS.
No desired product was observed.
The product was observed as a 1:1 ratio of reduced to unreduced nitro group.
All components were observed with a fully reduced nitro group.
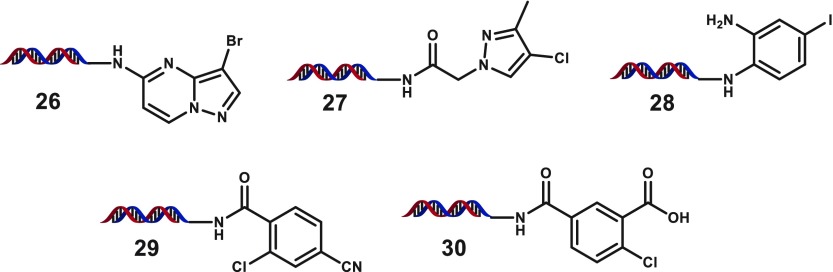
To further evaluate these conditions for DECL production, we prepared an expanded set of heterocyclic bromides and chlorides (selected examples, Table 5). Effects from small changes could be observed, for example, the oxygenation added to 13 relative to nicotinate bromide 12 lowered conversions for anilines a–g and completely suppressed coupling to aliphatic amines h and i. Although indole 14 and azole 15 were satisfactory substrates, complex heterocyclic bromides 16–19 could not form C–N coupling products in synthetically useful conversions. Heterocyclic chlorides represent a particularly valuable class of substrates to leverage for diverse bifunctional building blocks from historical collections. Fortunately, extension to complex heterocyclic chlorides was fruitful as acyl-functionalized chlorides 20–22, and electron-deficient chlorides 23–25 generally provided the desired C–N products in good to excellent yields. Additionally, some heterocyclic substrates prepared within this set failed to provide any C–N products with a variety of amine partners, which may derive from the presence of metal coordination motifs (selected examples, Figure 1). Taken together, these results highlight the need to evaluate building block sets against representative coupling partner panels to inform building block input and keep C–N coupling DECL productions within desired conversion ranges.
Table 5. C–N Coupling Screening on Various Heteroaryl Bromidesa.
Conversion determined by LC-MS.
Mostly dehalogenation or addition of dimethylamine was observed.
Figure 1.
Examples of halide substrates that failed to give any desired C–N coupling products.
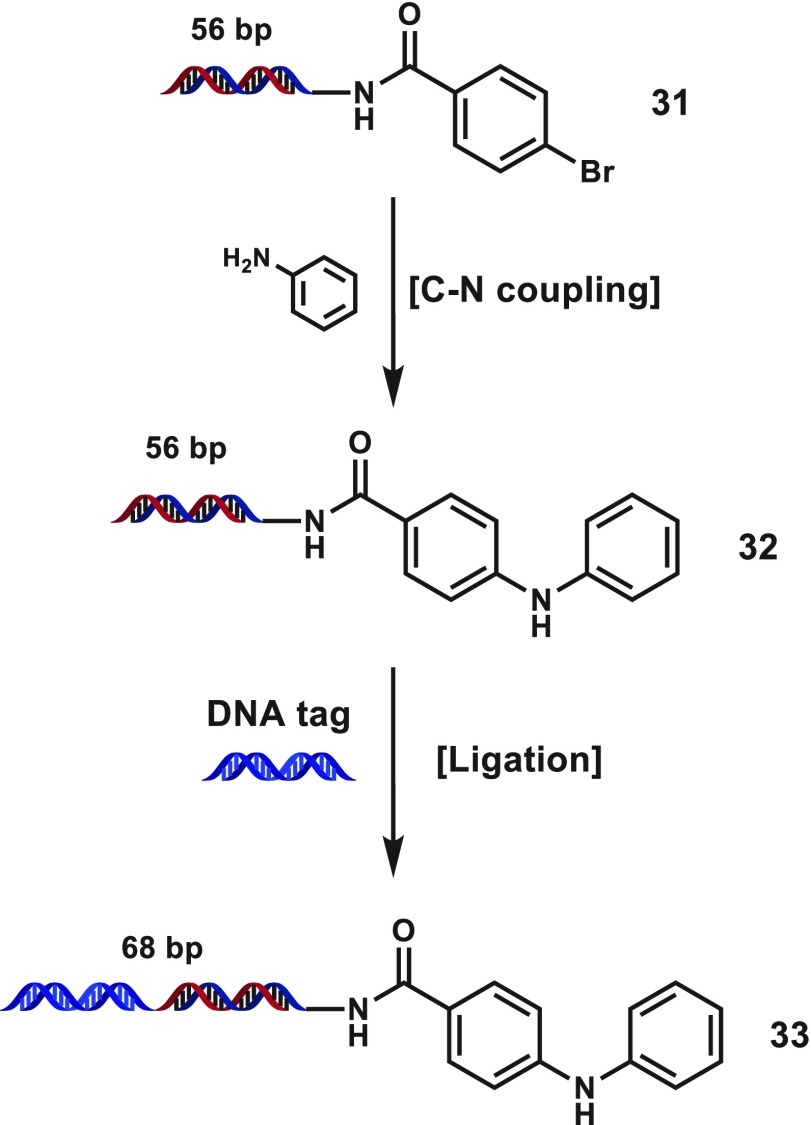
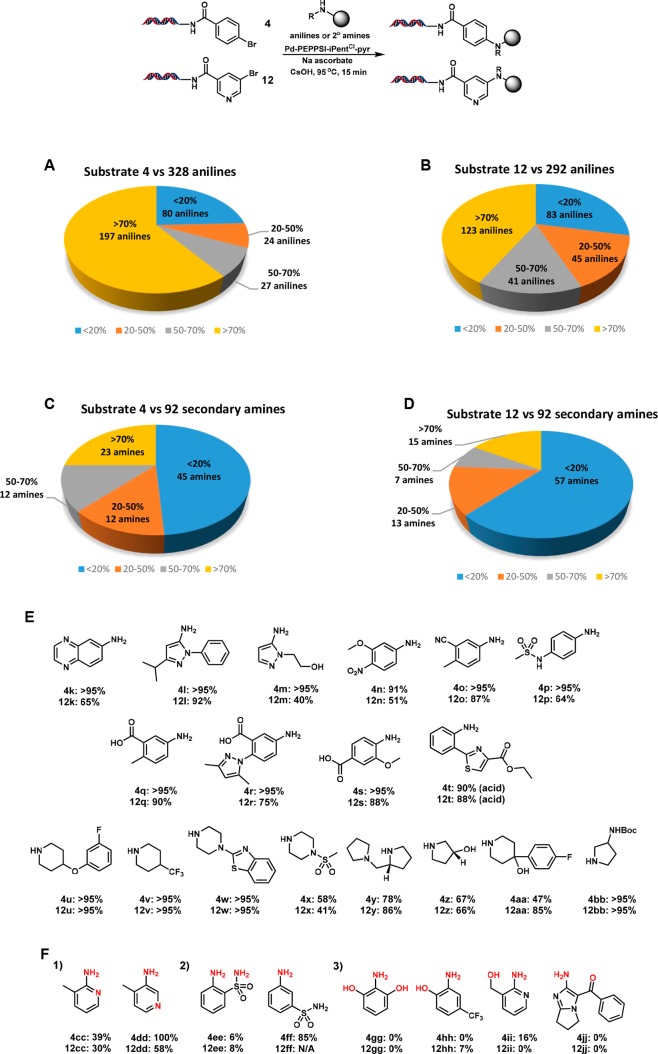
To assess the effect of DNA length on the outcome of a late-stage C–N coupling during a DECL production, we prepared 56-bp DNA substrate 31 from 4 via the ligation of two 39-mer oligonucleotides. Application of our optimized Pd-PEPPSI-iPentCl-pyr system to substrate 31 successfully provided biphenyl amine 32 in comparable conversion, and 32 subsequently underwent efficient DNA ligation (Scheme 2, see Supporting Information for reaction, LC-MS and gel electrophoresis details). With a library-relevant C–N coupling condition in hand, we sought to assess amine substitution effects across a diverse collection of ∼300 aromatic amines and 92 cyclic secondary aliphatic amines against two representative DNA-conjugated substrates, 4-bromo benzoate 4, and 5-bromo nicotinate 12 (Figure 2, see Supporting Information for a full list of building block structures and coupling results). Although this amine set had not been filtered for reaction compatibility, a majority of screened (hetero)aromatic amines coupled in >50% conversion for both substrates and many cyclic secondary aliphatic amines coupled in good conversion. Selected examples showing the tolerance for heterocyclic cores as well as distal nitro, cyano, or sulfonamide substitution are shown (3k–3p, 12k–12p, Figure 2). Notably, a variety of aromatic amines with distal carboxyl substitution were well tolerated (3q–3t, 12q–12t, Figure 2), potentially a versatile source of bifunctional building blocks for DECL productions. Separately, a variety of piperadinyl, pyrrolidinyl, and piperazinyl building blocks coupled in good to excellent conversions (3u–3bb, 12u–12bb, Figure 2), although cyclic secondary amines with α-substitution often did not couple significantly. (Hetero)aromatic amines featuring iodide, bromide, or chloride substitution generally failed, presumably due to competing oxidative addition to the building block. In addition, amines that featured a proximal coordinating group generally suffered lowered conversion that was separate from electronic effects (3cc/12cc vs 3dd/12dd or 3ee vs 3ff, Figure 2), and coupled products with amines containing a phenolic anion were generally not observed (3gg, 3hh, 12gg, 12hh, Figure 2).
Scheme 2. DNA Stability and Ligation Test.
Figure 2.
Amine validation with 4 and 12. (A) Summary of aniline validation with 4. (B) Summary of aniline validation with 12. (C) Summary of secondary amine validation with 4. (D) Summary of secondary amine validation with 12. (E) Validation results of selected anilines and secondary amines. (F) Representative comparison of the effect from neighboring group.
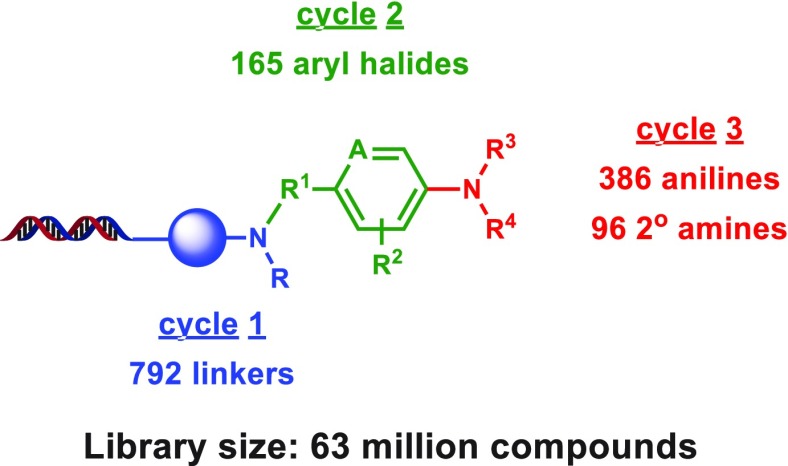
After determining the scope of this amination, we sought to further evaluate our C–N coupling conditions through the synthesis of a test-scale, three-cycle “split-and-pool” DECL. This type of DECL consists of iterative cycles of splitting a single or preassembled collection of DNA-appended substrates into individual wells, the attachment of a unique building block, and the concurrent ligation of a unique encoding DNA segment within each well and then a global pooling of all wells.
Thus, three-cycle DECLs with hundreds to thousands of compounds per cycle may produce collections of millions of compounds for screening efforts. In this DECL (Figure 3), a series of N-protected bifunctional amines was incorporated in cycle 1, and after an N-deprotection, a collection of bifunctional cores containing a (hetero)aryl iodide, bromide, or chloride was attached through nucleophilic substitution, reductive amination, or acylation in cycle 2. In cycle 3, a collection of approximately 500 (hetero)aromatic and cyclic secondary aliphatic amines was attached through C–N coupling to provide a DECL of ∼63 million compounds. Postcycle 3 recovery was satisfactory (estimated 72% recovery), and a “naïve” sample of the library (i.e., a DECL sample that has not been subjected to target-based affinity screens) exhibited normal PCR amplification consistent to other DECLs of identical DNA architecture. After sequencing of the naïve DECL sample, all expected sequences were detected within narrow, Gaussian-like distributions, altogether suggesting a limited degradation or imbalance of DNA-encoding segments during the DECL production. In addition, analysis of the percentage of perfect sequences observed during naïve sequencing was close to our historical average observed over dozens of libraries (see Supporting Information for DECL synthesis, sequencing details, and sequencing analysis).
Figure 3.
Use of the developed C–N coupling condition in a three-cycle DECL.
Conclusions
In conclusion, we developed a mild and general C–N condition that may be applied to DNA-conjugated (hetero)aryl iodides, bromides, and chlorides with (hetero)aromatic amines and cyclic secondary aliphatic amines in good to excellent conversions with negligible DNA decomposition. Within this study, we investigated the substitution needed within the PEPPSI catalyst series and additives required to promote effective coupling, demonstrated the broad substrate scope, and further validated the condition within the synthesis and subsequent use within a DECL. This report builds upon the work of others by extending DNA-compatible C–N coupling to chloride substrates, which should enable the synthesis of more diverse DECLs by expanding the available sourcing of (hetero)aryl electrophiles within current building block collections. However, further work is needed to extend this class of palladium catalysts broadly to the coupling of primary aliphatic amines and acyclic secondary aliphatic amines. Catalyst modifications designed to increase the catalyst room-temperature solubility in mixed aqueous solvent as well as alter the initiation mechanism may improve catalyst performance and are currently underway.
Experimental Procedures
Materials and Instrumentation
The central dsDNA oligonucleotides with chemically modified phosphates that end in an amine terminus and encoding 5′-phosphorylated oligonucleotides were purchased from LGC Biosearch Technologies. All DNA oligonucleotides were assessed through the general analytical procedure for purity. DNA sequences were designed to maximize sequence reads and minimize close similarity while sequencing. DNA oligomers in each codon-duplexed pair were designed to feature a divergent mass greater than 5 Da for efficient quality control analysis. A 10-mer DNA oligomer (spike-in), featuring a cholesterol tag and amine terminus, was obtained from Sigma-Aldrich and charged in a library pool to monitor chemical reactions. High-concentration T4 DNA ligase was obtained from Enzymatics (Qiagen). The oligomer ligation test with DTSU was incorporated to determine the ligase activity before use. DNA working solutions were prepared using DNase/RNase-free ultrapure water (Invitrogen), HPLC-grade acetonitrile (Fisher), or high-purity absolute ethanol (Koptec). LC/MS running solvents were made from Optima LC/MS-grade water (Fisher), Optima LC/MS-grade methanol (Fisher), hexafluoroisopropanol (99+% purity, Sigma-Aldrich), and HPLC-grade triethylamine (Fisher). All listed buffers and ionic solutions, including HEPES 10× ligation buffer (300 mM 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid, 100 mM MgCl2, 100 mM dithiothreitol, 10 mM adenosine triphosphate, pH 7.8), aq. NaOH, aq. NaCl (5 M), and basic borate buffer (250 mM sodium borate/boric acid, pH 9.5), were prepared in house. Chemical building blocks and reagents were purchased from various vendor sources and used without further purification. Building blocks were generally used from aliquots dissolved in acetonitrile (MeCN), methoxyisopropanol (MIPO), dimethylacetamide (DMA), or mixed aqueous acetonitrile and stored in 2D barcoded tubes (Phenix) with septacaps at −80 °C. Barcoded tubes were read using a SampleScan 96 scanner (BiomicroLab) and decoded using Vortex software (Dotmatics). Solutions were transferred utilizing Biotix brand or Fisher brand pipet tips and Biotix reservoirs. Reactions and library transformations were generally performed in polypropylene PCR tubes (Genemate), polypropylene tubes (Eppendorf), 96-well polypropylene PCR plates (Phenix or ThermoFisher), or 96-well, deep-well plates (USA Scientific). Plates were sealed for incubation with AlumaSeal II sealing films (Excel Scientific), and large-volume DNA precipitations were performed in polypropylene 250 mL screw-cap bottles (various vendors) or centrifuge tubes. Heated reactions were performed in benchtop heating blocks (ThermoFisher), Mastercycler nexus gradient (Eppendorf), a TS-DW deep-well plate themoshaker (Grant), or laboratory ovens (Fisher). Solutions were centrifuged in 5424R centrifuges (Eppendorf), Allegra X-15R centrifuges (Beckman-Coulter), or Lynx 4000 centrifuges (ThermoFisher). Optical density measurements were made using a Biophotometer (Eppendorf). A Vanquish UHPLC system was integrated with a LTQ XL ion trap mass spectrometer (ThermoFisher Scientific) for LC/MS analysis of oligonucleotides. DNA was visualized with a Molecular Imager Gel Doc XR system (BIO-RAD) after staining in an ethidium bromide solution.
Data Analysis
Samples were analyzed on a Thermo Vanquish UHPLC system coupled to an electrospray LTQ ion trap mass spectrometer. An oligonucleotide column (Thermo DNAPac RP, 2.1 × 50 mm, 4 μm) was equipped with ion-pairing mobile phase (15 mM TEA/100 mM HFIP in a water/methanol solvent system) for all of the separations. All mass spectra were acquired in the full scan negative-ion mode over the m/z range of 500–2000. Data analysis was performed by exporting the raw instrument data (.RAW) to an automated biomolecule deconvolution and reporting software (ProMass) which uses a novel algorithm (ZNova) to produce artifact-free mass spectra. Deconvoluted mass spectra were standardized against cocurrently run samples of DNA standards to account for any drift from theoretical mass during deconvolution.
Ethanol Precipitation and DNA Reconstitution
To a DNA reaction aqueous mixture was added 4% (v/v) 5 M NaCl solution and 3 times of the reaction volume of absolute ethanol. The mixture was mixed thoroughly before being stored at −20 °C overnight for DNA precipitation. The slurry was then centrifuged at 4000g for 1 h, followed by decanting the supernatant. The pellet was washed with 75% chilled ethanol, and the pellet was centrifuged at 4000g for another hour. The DNA pellet was dried in air after the supernatant was decanted. Water was added to reconstitute the DNA to the needed concentration. Ethanol precipitation was generally performed after each chemical reaction and ligation. Additional ethanol precipitation can be applied if residual reagent was observed after the first purification. Dilution with 2–4 times the reaction volume of water may be added before purification, while a higher percentage of water-miscible solvent was used, such as DMSO.
General Procedure for DNA-Compatible PEPPSI C–N Coupling
To a solution of DNA-conjugated aryl halide substrate (10 μL, 10 nmol, 1.0 mM in water) was added DMA (3.5 μL), 1000 equiv of CsOH (5 μL, 2000 mM in water), 500 equiv of amines (12.5 μL, 400 mM in DMA), and 20 equiv of freshly prepared sodium ascorbate (5 μL, 40 mM in water). The mixture was thoroughly mixed before adding 2 equiv of freshly prepared Pd-PEPPSI-iPentCl-pyr (4 μL, 5 mM in DMA). The reaction mixture was heated at 95 °C for 15 min, followed by addition of 100 equiv of the scavenger sodium l-cysteine (5 μL, 200 mM in water) and then heating at 80 °C for another 15 min. The mixture was then cooled to room temperature before assessing with LC-MS or purification by EtOH precipitation. PEPPSI catalysts used in this study were synthesized via known procedures and/or purchased from either Total Synthesis Ltd. or Sigma-Aldrich and used without further purification. While conducting a subsequent ligation, insurance of maximum removal of residual building blocks or salts before ligation was necessary as a large amount of residue may influence the ligation efficiency.
Acknowledgments
This work was supported by the Welch Foundation (Grant Q-0042), a Core Facility Support Award (RP160805) from the Cancer Prevention Research Institute of Texas (CPRIT), grant OPP1160866 from The Bill and Melinda Gates Foundation, and National Institutes of Health grant P01HD087157 from The Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.bioconjchem.9b00863.
Details of experimental procedures, DNA structures, DECL construction, and selection experiments (PDF)
Author Present Address
† G.M.: Arvinas, Incorporated, 5 Science Park, 395 Winchester Avenue, New Haven, Connecticut 06511, United States.
Author Present Address
‡ N.S.: Janssen Research & Development, 3210 Merryfield Row, San Diego, California 92121, United States.
Author Contributions
Y.-C.C. optimized the reaction conditions, prepared substrates, explored the substrate scope, conducted ligation tests, cobuilt the DECL, and composed the draft manuscript. J.C.F. performed cheminformatic analysis of the DECL. A.K. validated the DECL through resynthesis of chemical hits. G.M. conducted DECL naïve sequencing experiments. K.R. performed bioinformatic analysis of the DECL. K.M.B. validated the DECL through testing of synthesized hits. M.N.U. designed building block input for the DECL. M.M.M. advised experiments. Z.Y. validated the DECL through successful target selection campaigns. N.S. conducted initial PEPPSI investigations, optimized reaction conditions, cobuilt the DECL, advised experiments, and composed the draft manuscript. All authors contributed to and reviewed the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Goodnow R. A. Jr.; Dumelin C. E.; Keefe A. D. (2017) DNA-encoded chemistry: enabling the deeper sampling of chemical space. Nat. Rev. Drug Discovery 16, 131–147. 10.1038/nrd.2016.213. [DOI] [PubMed] [Google Scholar]
- Neri D.; Lerner R. A. (2018) DNA-encoded chemical libraries: A selection system based on endowing organic compounds with amplifiable information. Annu. Rev. Biochem. 87, 479–502. 10.1146/annurev-biochem-062917-012550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimbacher M.; Zhang Y.; Mannocci L.; Stravs M.; Geppert T.; Scheuermann J.; Schneider G.; Neri D. (2012) Discovery of small-molecule interleukin-2 inhibitors from DNA-encoded chemical library. Chem. - Eur. J. 18, 7729–7737. 10.1002/chem.201200952. [DOI] [PubMed] [Google Scholar]
- Harris P. A.; King B. W.; Bandyopadhyay D.; Berger S. B.; Campobasso N.; Capriotti C. A.; Cox J. A.; Dare L.; Dong X.; Finger J. N.; et al. (2016) DNA-encoded library screening identifies Benzo[b][1,4]oxazepin-4-ones as highly potent and monoselective receptor interacting protein 1 kinase inhibitor. J. Med. Chem. 59, 2163–2178. 10.1021/acs.jmedchem.5b01898. [DOI] [PubMed] [Google Scholar]
- Harris P. A.; Berger S. B.; Jeong J. U.; Nagilla R.; Bandyopadhyay D.; Campobasso N.; Capriotti C. A.; Julie A. C.; Dare L.; Dong X.; et al. (2017) Discovery of a first-in-class receptor interacting protein 1 (RIP1) kinase specific clinical candidate (GSK2982772) for the treatment of inflammatory diseases. J. Med. Chem. 60, 1247–1261. 10.1021/acs.jmedchem.6b01751. [DOI] [PubMed] [Google Scholar]
- Neri D. (2017) Twenty-five years of DNA-encoded chemical libraries. ChemBioChem 18, 827–828. 10.1002/cbic.201700130. [DOI] [PubMed] [Google Scholar]
- Satz A. L. (2018) What Do You Get from DNA-Encoded Libraries?. ACS Med. Chem. Lett. 9, 408–410. 10.1021/acsmedchemlett.8b00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrenn S. J.; Weisinger R. M.; Halpin D. R.; Harbury P. B. (2007) Synthetic ligands discovery by in vitro selection. J. Am. Chem. Soc. 129, 13137–13143. 10.1021/ja073993a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. A.; Acharya R.; Arico-Muendel C. C.; Belyanskaya S. L.; Benjamin D. R.; Carlson N. R.; Centrella P. A.; Chiu C. H.; Creaser S. P.; Cuozzo J. W.; et al. (2009) Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat. Chem. Biol. 5, 647–654. 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]
- MacConnell A. B.; McEnaney P. J.; Cavett V. J.; Paegel B. M. (2015) DNA-encoded solid-phase synthesis: Encoding language design and complex oligomer library synthesis. ACS Comb. Sci. 17, 518–534. 10.1021/acscombsci.5b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusemark C. J.; Tilmans N. P.; Brown P. O.; Harbury P. B. (2016) Directed chemical evolution with an outsized genetic code. PLoS One 11, e0154765 10.1371/journal.pone.0154765. [DOI] [PMC free article] [PubMed] [Google Scholar]; Halpin D. R.; Harbury P. B. (2004) DNA display I. Sequence-encoded routing of DNA populations. PLoS Biol. 2, e173 10.1371/journal.pbio.0020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse B. N.; Snyder T. M.; Shen Y.; Liu D. R. (2008) Translation of DNA into a library of 13000 synthetic small-molecule macrocycles suitable for in vitro selection. J. Am. Chem. Soc. 130, 15611–15626. 10.1021/ja805649f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M. H.; Blakskjær P.; Petersen L. K.; Hansen T. H.; Højfeldt J. W.; Gothelf K. V.; Hansen N. J. V. (2009) A yoctoliter-scale DNA reactor for small-molecule evolution. J. Am. Chem. Soc. 131, 1322–1327. 10.1021/ja808558a. [DOI] [PubMed] [Google Scholar]
- Gartner Z. J.; Tse B. N.; Grubina R.; Doyon J. B.; Snyder T. M.; Liu D. R. (2004) DNA-templated organic synthesis and selection of a library of macrocycles. Science 305, 1601–1605. 10.1126/science.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrenn S. J.; Harbury P. B. (2007) Chemical evolution as a tool for molecular discovery. Annu. Rev. Biochem. 76, 331–349. 10.1146/annurev.biochem.76.062205.122741. [DOI] [PubMed] [Google Scholar]
- Gartner Z. J.; Liu D. R. (2001) The generality of DNA-templated synthesis as a basis for evolving non-natural small molecules. J. Am. Chem. Soc. 123, 6961–6963. 10.1021/ja015873n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann J.; Neri D. (2015) Dual-pharmacophore DNA-encoded chemical libraries. Curr. Opin. Chem. Biol. 26, 99–103. 10.1016/j.cbpa.2015.02.021. [DOI] [PubMed] [Google Scholar]
- Daguer J. P.; Ciobanu M.; Alvarez S.; Barluenga S.; Winssinger N. (2011) DNA-templated combinatorial assembly of small molecule fragments amenable to selection/amplification cycles. Chem. Sci. 2, 625–632. 10.1039/c0sc00574f. [DOI] [Google Scholar]
- Satz A. L.DNA-compatible chemistry. In A handbook for DNA-encoded chemistry: theory and applications for exploring chemical space and drug discovery; Goodnow R. A., Eds.; Wiley, 2014; pp 99–121. [Google Scholar]
- Decurtins W.; Wichert M.; Franzini R. M.; Buller F.; Stravs M. A.; Zhang Y.; Neri D.; Scheuermann J. (2016) Automated screening for small organic ligands using DNA-encoded chemical libraries. Nat. Protoc. 11, 764–780. 10.1038/nprot.2016.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannocci L.; Zhang Y.; Scheuermann J.; Leimbacher M.; De Bellis G.; Rizzi E.; Dumelin C.; Melkko S.; Neri D. (2008) High-throughput sequencing allows the identification of binding molecules isolated from DNA-encoded chemical libraries. Proc. Natl. Acad. Sci. U. S. A. 105, 17670–17675. 10.1073/pnas.0805130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buller F.; Mannocci L.; Zhang Y.; Dumelin C. E.; Scheuermann J.; Neri D. (2008) Design and synthesis of a novel DNA-encoded chemical library using Diels-Alder cycloadditions. Bioorg. Med. Chem. Lett. 18, 5926–5931. 10.1016/j.bmcl.2008.07.038. [DOI] [PubMed] [Google Scholar]
- Luk K.-C., and Satz A. L.. DNA-compatible chemistry. In A handbook for DNA-encoded chemistry: theory and applications for exploring chemical space and drug discovery; Goodnow R. A., Eds.; Wiley, 2014; pp 69–76. [Google Scholar]
- Satz A. L.; Cai J.; Chen Y.; Goodnow R.; Gruber F.; Kowalczyk A.; Petersen A.; Naderi-Oboodi G.; Orzechowski L.; Strebel Q. (2015) DNA Compatible Multistep Synthesis and Applications to DNA Encoded Libraries. Bioconjugate Chem. 26, 1623–1632. 10.1021/acs.bioconjchem.5b00239. [DOI] [PubMed] [Google Scholar]
- Malone M. L.; Paegel B. M. (2016) What is a “DNA-Compatible” Reaction?. ACS Comb. Sci. 18, 182–187. 10.1021/acscombsci.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H.-C.; Simmons N.; Faver J. C.; Yu Z.; Palaniappan M.; Riehle K.; Matzuk M. M. (2019) A mild, DNA-compatible nitroreduction using B2(OH)4. Org. Lett. 21, 2194–2199. 10.1021/acs.orglett.9b00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H.-C.; Bangs M. C.; Simmons N.; Matzuk M. M. (2019) Multistep synthesis of 1,2,4-oxadiazoles via DNA-conjugated aryl nitrile substrates. Bioconjugate Chem. 30, 1304–1308. 10.1021/acs.bioconjchem.9b00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Gabriele E.; Samain F.; Favalli N.; Sladojevich F.; Scheuermann J.; Neri D. (2016) Optimized reaction conditions for amide bond formation in DNA-encoded combinatorial libraries. ACS Comb. Sci. 18, 438–443. 10.1021/acscombsci.6b00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y.; Clark M. A. (2015) Robust Suzuki-Miyaura cross-coupling on DNA-linked substrates. ACS Comb. Sci. 17, 1–4. 10.1021/co5001037. [DOI] [PubMed] [Google Scholar]
- Yuen L.; Franzini R. M. (2017) Stability of oligonucleotide-Small molecule conjugates to DNA-deprotection conditions. Bioconjugate Chem. 28, 1076–1083. 10.1021/acs.bioconjchem.7b00005. [DOI] [PubMed] [Google Scholar]
- Geigle S. N.; Petersen A. C.; Satz A. L. (2019) Development of DNA-compatible Van Leusen three-component imidazole synthesis. Org. Lett. 21, 9001–9004. 10.1021/acs.orglett.9b03406. [DOI] [PubMed] [Google Scholar]
- Phelan J. P.; Lang S. B.; Sim J.; Berritt S.; Peat A. J.; Billings K.; Fan L.; Molander G. A. (2019) Open-air alkylation reaction in photoredox-catalyzed DNA-encoded library synthesis. J. Am. Chem. Soc. 141, 3723–3732. 10.1021/jacs.9b00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Lundberg H.; Asai S.; Martin-Acosta P.; Chen J. S.; Brown S.; Farrell W.; Dushin R.; O’Donnell C. J.; Ratnayake A. S.; et al. (2018) Kinetically guided radical-based synthesis of C(sp3)-C(sp3) linkages on DNA. Proc. Natl. Acad. Sci. U. S. A. 115, E6404–E6410. 10.1073/pnas.1806900115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai P.; Yang G.; Zhao L.; Wan J.; Li J.; Liu G. (2019) Synthesis of C3-Alkylated Indoles on DNA via Indolyl Alcohol Formation Followed by Metal-Free Transfer Hydrogenation. Org. Lett. 21, 6633–6637. 10.1021/acs.orglett.9b02132. [DOI] [PubMed] [Google Scholar]
- Kunig V. B. K.; Ehrt C.; Dömling A.; Brunschweiger A. (2019) Isocyanide multicomponent reactions on solid-phase coupled DNA oligonucleotides for encoded library synthesis. Org. Lett. 21, 7238–7243. 10.1021/acs.orglett.9b02448. [DOI] [PubMed] [Google Scholar]
- Fischer C.; Koenig B. (2011) Palladium- and copper-mediated N-aryl bond formation reactions for the synthesis of biological active compounds. Beilstein J. Org. Chem. 7, 59–74. 10.3762/bjoc.7.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Castillo P.; Buchwald S. L. (2016) Applications of palladium-catalyzed C–N cross-coupling reactions. Chem. Rev. 116, 12564–12649. 10.1021/acs.chemrev.6b00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roughley S. D.; Jordan A. M. (2011) The medicinal chemist’s toolbox: An analysis of reactions used in the pursuit of drug candidates. J. Med. Chem. 54, 3451–3479. 10.1021/jm200187y. [DOI] [PubMed] [Google Scholar]
- Uehling M. R.; King R. P.; Krska S. W.; Cernak T.; Buchwald S. L. (2019) Pharmaceutical diversification via palladium oxidative addition complexes. Science 363, 405–408. 10.1126/science.aac6153. [DOI] [PubMed] [Google Scholar]
- Magano J.; Dunetz J. R. (2011) Large-Scale Applications of Transition Metal-Catalyzed Couplings for the Synthesis of Pharmaceuticals. Chem. Rev. 111, 2177–2250. 10.1021/cr100346g. [DOI] [PubMed] [Google Scholar]
- Surry D. S.; Buchwald S. L. (2008) Biarylphosphane ligands in palladium-catalyzed amination. Angew. Chem., Int. Ed. 47, 6338–6361. 10.1002/anie.200800497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surry D. S.; Buchwald S. L. (2011) Dialkylbiaryl phosphines in Pd-catalyzed amination: a user’s guide. Chem. Sci. 2, 27–50. 10.1039/C0SC00331J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forero-Cortés P. A.; Haydl A. M. (2019) The 25th anniversary of the Buchwald-Hartwig amination: Development, applications, and outlook. Org. Process Res. Dev. 23, 1478–1483. 10.1021/acs.oprd.9b00161. [DOI] [Google Scholar]
- Fors B. P.; Krattiger P.; Strieter E.; Buchwald S. L. (2008) Water-mediated catalyst preactivation: An efficient protocol for C–N cross-coupling reactions. Org. Lett. 10, 3505–3508. 10.1021/ol801285g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas A. S.; Gothelf K. V. (2005) Effect of water on the palladium-catalyzed amidation of aryl bromides. J. Org. Chem. 70, 3321–3323. 10.1021/jo0500176. [DOI] [PubMed] [Google Scholar]
- Lu X.; Roberts S. E.; Franklin G. J.; Davie C. P. (2017) On-DNA Pd and Cu promoted C–N cross-coupling reactions. MedChemComm 8, 1614–1617. 10.1039/C7MD00289K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff Y.; Berst F. (2018) Efficient copper-catalyzed amination of DNA-conjugated aryl iodides under mild aqueous conditions. MedChemComm 9, 1188–1193. 10.1039/C8MD00185E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pedro Beato E.; Priego J.; Gironda-Martínez A.; González F.; Benavides J.; Blas J.; Martín-Ortega M. D.; Toledo M. Á.; Ezquerra J.; Torrado A. (2019) Mild and efficient palladium-mediated C–N cross-coupling reaction between DNA-conjugated aryl bromides and aromatic amines. ACS Comb. Sci. 21, 69–74. 10.1021/acscombsci.8b00142. [DOI] [PubMed] [Google Scholar]
- Flood D. T.; Asai S.; Zhang X.; Wang J.; Yoon L.; Adams Z. C.; Dillingham B. C.; Sanchez B. B.; Vantourout J. C.; Flanagan M. E.; et al. (2019) Expanding reactivity in DNA-encoded library synthesis via reversible binding of DNA to an inert quaternary ammonium support. J. Am. Chem. Soc. 141, 9998–10006. 10.1021/jacs.9b03774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- “Selections” are experiments designed to identify or enrich compounds of interest, typically binders to a biological target, from a DECL. Successful “selection” campaigns with this DECL will be described in a future publication.
- Organ M. G.; Abdel-Hadi M.; Avola S.; Dubovyk I.; Hadei N.; Kantchev E. A. B.; O’Brien C. J.; Sayah M.; Valente C. (2008) Pd-catalyzed aryl amination mediated by well defined, N-heterocyclic carbene (NHC)–Pd precatalysts, PEPPSI. Chem. - Eur. J. 14, 2443–2452. 10.1002/chem.200701621. [DOI] [PubMed] [Google Scholar]
- Nasielski J.; Hadei N.; Achonduh G.; Kantchev E. A. B.; O’Brien C. J.; Lough A.; Organ M. G. (2010) Structure–activity relationship analysis of Pd–PEPPSI complexes in cross-couplings: A close inspection of the catalytic cycle and the precatalyst activation model. Chem. - Eur. J. 16, 10844–10853. 10.1002/chem.201000138. [DOI] [PubMed] [Google Scholar]
- Pompeo M.; Farmer J. L.; Froese R. D. J.; Organ M. G. (2014) Room-temperature amination of deactivated aniline and aryl halide partners with carbonate base using a Pd-PEPPSI-IPentCl-o-picoline catalyst. Angew. Chem., Int. Ed. 53, 3223–3226. 10.1002/anie.201310457. [DOI] [PubMed] [Google Scholar]
- Valente C.; Pompeo M.; Sayah M.; Organ M. G. (2014) Carbon–Heteroatom Coupling Using Pd-PEPPSI Complexes. Org. Process Res. Dev. 18, 180–190. 10.1021/op400278d. [DOI] [Google Scholar]
- The failure of these substrates have been previously observed, see ref (48).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.