Abstract
Seven human induced pluripotent stem cell (iPSC) lines were generated from fibroblasts from three neonatal individuals using non-integrative reprogramming. Most control iPSCs are derived from adults, so these iPSCs meet the need for control iPSCs from young individuals. Donors were from different ethnicities and these lines provide unique genetic profiles. All iPSCs have normal karyotypes, express stem cell markers, and exhibit pluripotency, as assessed by capacity to differentiate into three germ layers. These lines are valuable to study human development, as age-matched controls for disorder-specific iPSCs, and as platforms for gene editing to control for age and ethnicity.
Keywords: Neonatal, unaffected, control iPSCs
Resource Table:
| Unique stem cell lines identifier | WC026i-5807–3 |
| WC027i-5807–5 | |
| WC028i-5807–6 | |
| WC029i-5907–1 | |
| WC030i-5907–2 | |
| WC031i-5907–6 | |
| WC032i-6007–1 | |
| Alternative names of stem cell lines | N/A |
| Institution | Waisman Center, University of Wisconsin-Madison, Madison, USA |
| Contact information of distributor | Anita Bhattacharyya; bhattacharyy@waisman.wisc.edu |
| Type of cell lines | iPSC |
| Origin | human |
| Cell Source | fibroblasts |
| Clonality | Clonal |
| Method of reprogramming | Episomal plasmid transfection |
| Multiline rationale | Multiple isogenic clones for both male and female neonatal donors from under-represented ethnicities |
| Gene modification | No |
| Type of modification | N/A |
| Associated disease | N/A |
| Gene/locus | N/A |
| Method of modification | N/A |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | 11/17/2017 |
| Cell line repository/bank | WiCell Research Institute, Madison, USA https://www.wicell.org/ |
| Ethical approval | University of Wisconsin-Madison Human Subjects Institutional Review Board Approval # 2016–0979. |
Resource utility
Human iPSCs hold great promise for basic research, disease modeling and regenerative medicine. iPSCs generated from neonates of varied ethnicities enable experimental control of age and ethnicity, providing a valuable resource for studies of human developmental biology, developmental disorders, and aging.
Resource details
iPSC clones were generated from fibroblasts of three unaffected neonatal individuals. Most iPSCs generated from control, healthy individuals are derived from adults and so these iPSC lines, and others (Trokovic, et al. 2015), are valuable lines for research in human developmental biology, developmental disorders, and aging. These lines enable age-matched control cell lines for disease research on childhood diseases. In addition, the young age of the donors may reduce the number of de novo genetic mutations or epigenetic modifications inherent in postnatal human development. The three donors were of different ethnicities, thus providing unique genetic profiles and ethnicity-matched control cell lines. Finally, by providing multiple characterized clones from two of the donors (one male and one female), these lines comprise a comprehensive toolset allowing researchers to rule out clonal variations in their studies. Thus, the establishment of these unique iPSC lines enable age-matched and ethnicity-matched control cell lines and platforms for gene editing.
Fibroblasts from neonatal donors were reprogrammed using the Yamanaka episomal reprogramming plasmids (via expression of OCT3/4, SOX2, KLF4, LIN28, and c-MYC) and delivered by electroporation (Okita, Nakagawa et al. 2008). Following clonal selection and amplification, visual characterization confirmed that these lines exhibited a characteristic iPSC morphology (Figure 1A, Supplementary Figure 1A). A large percentage (>95%) of cells within each clone express the self-renewal markers OCT4, SOX2 and NANOG, as assessed by immunofluorescence (Figure 1A, 1B and Supplementary Figure 1A, 1B). PCR amplification at passage 11 of the iPSCs verified the episomal reprogramming factors are not retained in these cells (Figure 1C; E1, E2, and E3 represent the three reprogramming plasmids). Second panel indicates positive genomic DNA was present in these samples. Karyotyping via G-band analysis on 20 metaphase spreads were analyzed for each sample. All iPSC lines and parental fibroblasts showed a normal karyotype of 46, XX or 46, XY (Figure 1A, Supplementary Figure 1A). Short Tandem Repeat (STR) analysis on 15 distinct genetic loci confirmed that the somatic cells and derivative iPSCs are derived from the same individual, and each line is clonal in nature without other human or murine feeder cell contamination (Available with the authors). Finally, the pluripotency of these cells was tested using a directed differentiation kit, followed by qPCR analysis for definitive germ layer markers. These experiments confirmed that each of the iPSC lines could successfully generate each of the three primary germ layers via the expression of PAX6 and NR2F2 for ectoderm, HAND1 and MIXL1 for mesoderm, and GATA6 and SOX17 for endoderm (Figure 1D, Supplementary Figure 1C). Data is presented as a fold change relative to undifferentiated iPSCs for each cell line. Asterisks indicate that fold-change is above 400. Taken together, we have characterized these seven neonatal iPSC lines and have found them sufficient to distribute to the research community. These cells are banked at the WiCell Research Institute (Madison, Wisconsin) and are ready for distribution.
Fig. 1.
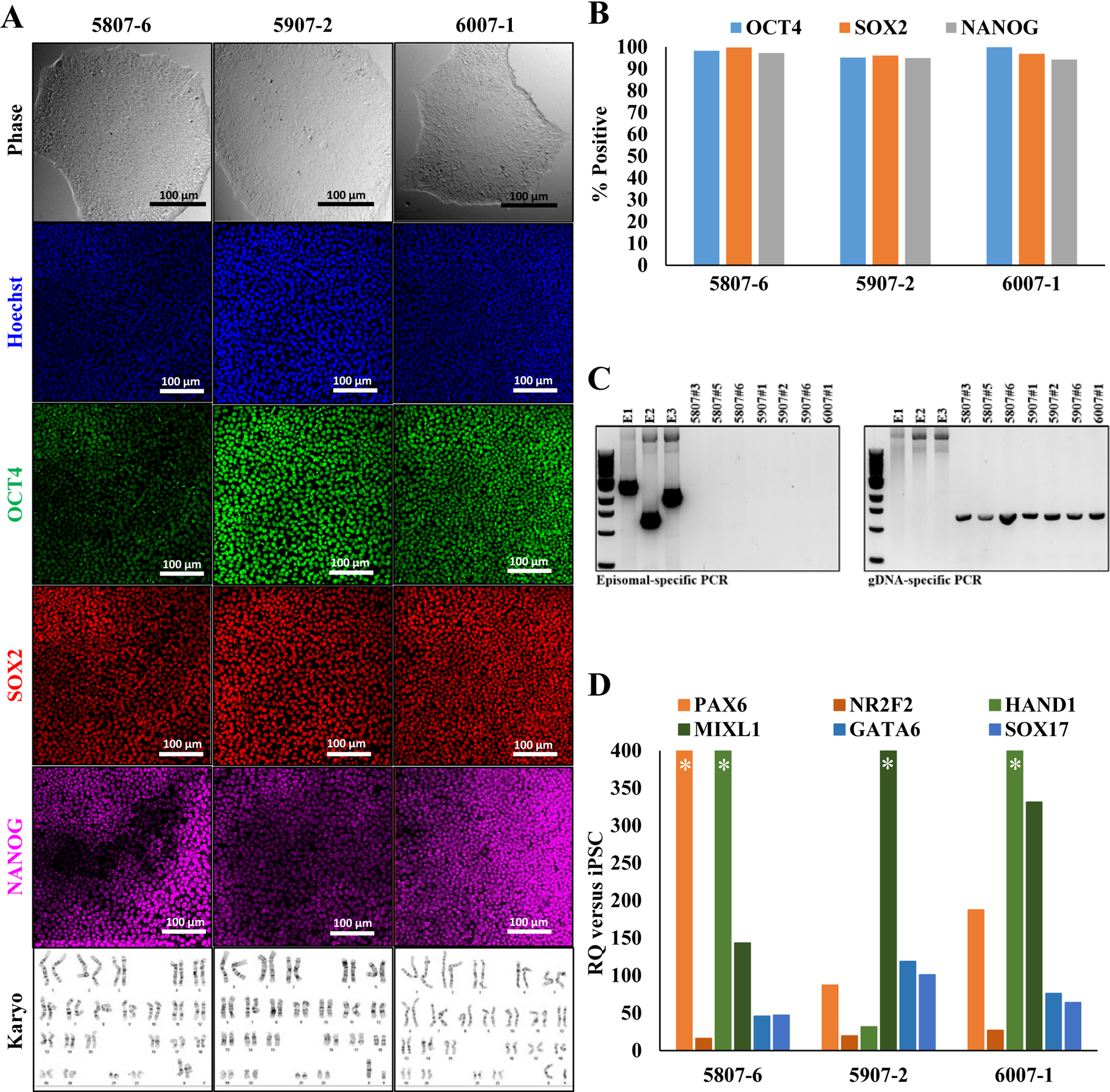
Characterization of induced pluripotent stem cells. A) Expression of self-renewal markers and normal karyotype of representative lines from each individual, B) Quantification of marker-positive cells. C) PCR amplification shows that episomal reprogramming factors are not retained in these cells. D) Pluripotency of these cells was tested using a directed differentiation kit, followed by qPCR analysis for definitive germ layer markers.
Materials and Methods:
Consent and sample collection:
Primary dermal fibroblasts were isolated from neonatal skin tissue provided by the International Institute for the Advancement of Medicine (IIAM) Neonatal Donor Program which provides non-transplantable, neonatal organs and tissues for medical research, education and diagnostic studies. IIAM follows standards established by the Center for Medicare/Medicaid Services and the United Network of Organ Sharing with approval from an internal Donor Family Advisory Committee and an External Review Committee. Tissue was acquired with approval from the University of Wisconsin-Madison Human Subjects Institutional Review Board (protocol #2016–0979).
Cell culture and iPSC generation:
Dermal fibroblasts were isolated and cultured in DMEM supplemented with 10% FBS (Life Technologies). Fibroblasts were reprogrammed by electroporation delivery of episomal vectors pCXLE-hOCT3/4-shp53-F (Addgene, 27077), pCXLE-hSK (Addgene, 27078) and pCXLE-hUL (Addgene, 27080). Three micrograms of each vector were delivered into 1 × 106 fibroblast cells using Amaxa 4D X 100-μl kit (Lonza) (Okita et al., 2011). After electroporation, cells were cultured on mouse embryonic fibroblast (MEF) feeder cells in a low oxygen incubator (5% O2, 5% CO2). Cells were fed with hESCM (DMEM-F12 media (Gibco) with 20% knock-out serum replacement (Gibco), 1X Non-Essential Amino Acids (Life Technologies), 0.5X GlutaMAX (Life Technologies), 0.1 mM 2-mercaptoethanol (Sigma), and 12 ng/mL bFGF (Waisman Biomanufacturing)). The iPSC colonies were manually picked between day 14–28 post-transfection. Following expansion, cells were transferred onto Matrigel (R&D) and cultured with mTeSR1 (Stemcell Technologies) for banking. iPSCs on MEF were passaged with dispase solution (Gibco) Split ratio is 1 to 6 every 5–7 days., and iPSCs on Matrigel were passaged with 0.5mM EDTA or ReLeSR (05872, Stemcell Technologies). iPSC colonies were passaged every 4–7 days at a 1:3 to 1:6 split ratio.
Episomal plasmid analysis:
To confirm loss of episomal reprogramming plasmids from the cells, PCR specific to these constructs was performed with Q5 polymerase (New England BioLabs). DNA extracts were prepared using QuickExtract DNA Extraction Solution 1.0 (Epicentre). Primer sequences can be found in Table 3. The episomal plasmid-specific pair of primers would be expected to amplify bands at approximately 2.8 kilobases for pCXLE-hSK (E1 in Figure 1C), 1.4 kilobases for pCXLE-hOCT3/4-shp53 (E2 in Figure 1C), and 2.1 kilobases for pCXLE-hUL (E3 in Figure 1C). The genomic DNA pair of primers would be expected to amplify a band at approximately 1.3 kilobases from genomic DNA samples, but not the episomal reprogramming plasmids. PCR conditions followed the standard Q5 polymerase protocol published by New England BioLabs, with the primer annealing temperature set at 63° C and the elongation temperature set at 72° C for 35 cycles.
Table 3:
Reagents details
| Antibodies used for immunocytochemistry/flow-cytometry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| Self-renewal Markers | Rabbit anti-NANOG | 1:200 | Stemgent Cat# 09–0020, RRID:AB_2298294 |
| Self-renewal Markers | Mouse anti-human OCT-3/4 | 1:1000 | Santa Cruz Biotechnology Cat# sc-5279, RRID:AB_628051 |
| Self-renewal Markers | Goat anti-human SOX2 | 1:1000 | R&D Systems Cat# AF2018, RRID:AB_355110 |
| Secondary antibodies | cy5 Donkey anti-rabbit | 1:500 | Jackson ImmunoResearch Labs Cat# 711–175-152, RRID:AB_2340607 |
| Secondary antibodies | 488 donkey anti-mouse | 1:500 | Molecular Probes Cat# A-21202, RRID:AB_141607 |
| Secondary antibodies | Cy3 donkey anti-goat | 1:500 | Jackson ImmunoResearch Labs Cat# 705–165-147, RRID:AB_2307351 |
| Primers | |||
| Target | Forward/Reverse primer (5′−3′) | ||
| Episomal plasmids | All episomal plasmids pCXLE-hSK 2.8 kB pCXLE-hUL 2.1 kB pCXLE-hOCT3/4-shp53 1.4 kB |
GTGACCGGCGGCTCTAGAGC/AAGCCATACGGGAAGCAATAGC | |
| gDNA control | Positive DNA control for episomal PCR (1.3 kB) | CATGCAGTCCTCCTTACCATC TCCTCTCTGGCTCCATCGTA | |
| Trilineage potential - experiment controls |
ACTIN B Loading Control GAPDH Loading Control |
Qiagen primer set number: PPH00073G PPH00150F |
|
| Trilineage potential – Ectodermal genes |
PAX6 NR2F2 |
PPH02598F PPH05883B |
|
| Trilineage potential – Mesodermal genes |
HAND1 MIXL1 |
PPH06879B PPH15840A |
|
| Trilineage potential – Endodermal genes |
GATA6 SOX17 |
PPH06943F PPH02451A |
|
Immunocytochemistry:
Cells were prepared for immunofluorescence following standard procedures. Antibody names and concentrations are found in Table 2. Cells were fixed in 4% paraformaldehyde for 20 min at room temperature, then permeabilized with 0.2% Triton X-100 (X100, Sigma-Aldrich) in phosphate buffered saline (PBS) for 20 minutes. Cells were blocked for 1 hour in 10% donkey serum and 0.2% Triton X-100 prepared in PBS before incubated with primary antibodies overnight at 4°C followed by incubation with fluorescent secondary antibodies. All antibodies were diluted in same blocking solution. Nuclei were stained with Hoechst (H3570, ThermoFisher) and cells on coverslips were mounted with Fluoromount-G Slide mounting medium (731604, Beckman Coulter). Samples without primary antibody served as negative controls. Images in Figure 1C were captured using a Nikon A1 laser confocal microscope, and quantitation results in Figure 1B were from a minimum of 10,000 cells plated on an optical 96-well plate and processed on a Perkin-Elmer Operetta high-content imaging system.
Table 2:
Characterization and validation
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | All lines exhibit normal morphology | Figure 1A, Supplementary Figure 1A |
| Phenotype | Qualitative analysis | All lines exhibit expression of self-renewal markers NANOG, OCT3/4, & SOX2 | Figure 1A, Supplementary Figure 1A |
| Quantitative analysis | All lines were greater than 90% positive for OCT4, SOX2 and NANOG | Figure 1B, Supplementary Figure 1B | |
| Genotype | G-banding Karyotype | 46,XX or 46,XY Normal Karyotype, resolution 425–600 nm | Figure 1A, Supplementary Figure 1A |
| Identity | STR analysis | STR analysis defines profile for each line, confirms clonality and purity to 95–98% confidence | Available with authors |
| 15 loci tested, all matched appropriate source fibroblasts | |||
| Mutation analysis (IF APPLICABLE) | Sequencing | Not Applicable | Not Applicable |
| Southern Blot OR WGS | Not Applicable | Not Applicable | |
| Microbiology and virology | Mycoplasma | Negative | Supplementary Figure 2 |
| Differentiation potential | Directed differentiation | Trilineage differentiation (StemCell Technologies) and qPCR verification, all lines express markers of each germ layer | Figure 1D, Supplementary Figure 1C |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping | N/A | N/A |
| HLA tissue typing | N/A | N/A |
Demonstration of trilineage differentiation capacity:
All iPSC lines at passage 11 were differentiated using the STEMdiff trilineage differentiation kit (Stemcell Technologies 05230) following the manufacturer’s protocol. Cells were harvested in Trizol and RNA was extracted using a Direct-zol RNA purification kit (Zymo Research). RNA was converted to cDNA using an iScript cDNA synthesis kit (BioRad), and analyzed using a custom RT2 profiler qPCR array (Qiagen) for the genes outlined in Table 3. The qPCR experiment was run on an Applied Biosystems Viia7 machine, using their standard run speed and a 2-step amplification protocol (95°C for 15 seconds, 60°C for 1 minute) for 40 cycles. The expression level of each gene was normalized to ACTIN and GAPDH within the differentiated sample and compared to these values from an iPSC sample from the same clone.
G-band karyotyping/STR analysis/Mycoplasma test:
Chromosomal G-band analyses were performed at WiCell Research Institute, according to the International System for Human Cytogenetic Nomenclature. All cell lines were tested at passage 10, with 20 cells in metaphase counted for the analysis. STR analysis was performed by the University of Wisconsin-Madison Department of Pathology TRIP Laboratory. Approximately 2 million cells were analyzed using the PowerPlex 16 HS System (Promega). MycoAlert assay (mycoplasma detection kit, Lonza LT07–218) was performed at WiCell Research Institute using manufacturer provided protocol to ensure all banked cells are mycoplasma free.
Supplementary Material
Table 1:
Summary of lines
| iPSC line names | Abbreviation in figures | Gender | Age | Ethnicity | Genotype of locus | Disease |
|---|---|---|---|---|---|---|
| WC026i-5807–3 | 5807–3 | female | neonatal | Caucasian | N/A | N/A |
| WC027i-5807–5 | 5807–5 | female | neonatal | Caucasian | N/A | N/A |
| WC028i-5807–6 | 5807–6 | female | neonatal | Caucasian | N/A | N/A |
| WC029i-5907–1 | 5907–1 | male | neonatal | African | N/A | N/A |
| WC030i-5907–2 | 5907–2 | male | neonatal | African | N/A | N/A |
| WC031i-5907–6 | 5907–6 | male | neonatal | African | N/A | N/A |
| WC032i-6007–1 | 6007–1 | male | neonatal | Asian | N/A | N/A |
Acknowledgments:
This study was supported by a core grant to the Waisman Center from the National Institute of Child Health and Human Development (U54 HD090256).
References
- Okita et al. (2011). “ A more efficient method to generate integration-free human iPS cells” Nature Methods 8, pages 409–412 [DOI] [PubMed] [Google Scholar]
- Trokovic R, Weltner J and Otonkoski T (2015). “Generation of iPSC line HEL24.3 from human neonatal foreskin fibroblasts.” Stem Cell Res 15(1): 266–268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


