Abstract
Platelets are the key cellular components of blood primarily contributing to formation of stable hemostatic plugs at the site of vascular injury, thus preventing excessive blood loss. On the other hand, excessive platelet activation can contribute to thrombosis. Platelets respond to many stimuli that can be of biochemical, cellular, or physical origin. This drives platelet activation kinetics and plays a vital role in physiological and pathological situations. Currently used bulk assays are inadequate for comprehensive biomechanical assessment of single platelets. Individual platelets interact and respond differentially while modulating their biomechanical behavior depending on dynamic changes that occur in surrounding microenvironments. Quantitative description of such a phenomenon at single‐platelet regime and up to nanometer resolution requires methodological approaches that can manipulate individual platelets at submicron scales. This review focusses on principles, specific examples, and limitations of several relevant biophysical methods applied to single‐platelet analysis such as micropipette aspiration, atomic force microscopy, scanning ion conductance microscopy and traction force microscopy. Additionally, we are introducing a promising single‐cell approach, real‐time deformability cytometry, as an emerging biophysical method for high‐throughput biomechanical characterization of single platelets. This review serves as an introductory guide for clinician scientists and beginners interested in exploring one or more of the above‐mentioned biophysical methods to address outstanding questions in single‐platelet biomechanics.
Keywords: biophysics, cytoskeleton, force microscopy, mechanobiology, platelets, single‐cell analysis
Essentials.
Platelets form hemostatic plugs and under pathological conditions contribute to thrombosis.
Platelets sense and mechanotransduce physicochemical cues and generate biomechanical forces.
Biophysical tools can quantify single‐platelet biomechanics during hemostasis and thrombosis.
Platelet biomechanical properties may serve as useful biomarkers of platelet dysfunction.
1. INTRODUCTION
Platelets are discoidal, anucleate, multifunctional cellular fragments (1‐3 µm in diameter) generated from bone marrow megakaryocytes and are released into the blood circulation.1 Circulating platelets are essential for hemostasis. They “survey” the integrity of the vascular system. Upon vascular injury, platelets adhere to the exposed extracellular matrix (ECM) and form a hemostatic plug to seal the wound. However, platelets may also contribute to thrombosis, in case of pathological clot formation. Platelets function as complex biological units that sense and mechanotransduce physicochemical cues and stimuli from their surrounding environment (ie, outside‐in signaling via ligand receptor–mediated interactions) and actively respond through mechanotransduction events (ie, inside‐out signaling triggering platelet adhesion, activation, spreading and contraction).2 Platelet activation at interfaces is associated with spreading and involves the formation of lamellipodial and filopodial protrusions. These processes result in generation of biomechanical contractile forces through interactions between platelet cytoskeletal components (eg, actin, myosin, tubulin, and several other proteins), which regulate, from the internal side of platelets, distribution of surface receptors (eg, glycoprotein Ib‐V‐IX complex, integrin α2β1 and αIIbβ3) that recognize the ligands (eg, von Willebrand factor), exposed ECM (eg, collagen), and activated platelets.3 Apart from their protective function, platelets contribute to pathophysiological conditions and onset of diseases as they interact with cells of the vasculature, a wide variety of cells from the adaptive and innate immune system, and pathogens of viral and bacterial origins.4 These dynamic interactions occur at length scales from nanometers to several microns and at time scales of milliseconds up to minutes mediated by specific receptors. Little is known about the longer‐term functions of platelets, that are incorporated into a thrombus. Currently, in platelet research a wide array of methods are practiced routinely to decipher such interactions and their outcomes for research as well as for diagnostic purposes in clinical practice. Frequently used in vitro and ex vivo assays are platelet aggregometry in platelet‐rich plasma or in whole blood by impedance aggregometry and rheological techniques (require relatively large sample volumes); flow cytometry (requires small sample volumes but is often restricted to end‐point readout); and real‐time thrombus formation in parallel plate flow chambers (which allows insights into clot formation).5 Although these approaches have significantly improved our understanding of hemostasis and thrombosis from a biochemical and cell biological perspective, much less is known about underlying biomechanical properties and biophysical forces generated, which govern protective functions such as hemostasis and pathophysiological origins of thrombosis. Within this context, several of these methods are not sufficiently informative to assess directly biomechanical properties of single platelets and how these in turn influence platelet function and how they drive the complex spatiotemporal dynamics during hemostasis and thrombosis.
1.1. Why should we investigate biomechanical properties of single platelets using single‐cell biophysical techniques, and what does this add to our current knowledge of platelets in both hemostasis and thrombosis?
Given the extent of cell‐to‐cell variations, rare subpopulations, and intrinsic fluctuations within biological systems, bulk population‐level experiments often conceal and average out vital causal relationships underlying the biological phenomena.6 Single‐cell biophysical techniques and their sensitivity have enabled quantification and unveiling of previously unknown relationships and fundamental paradigms in platelet biology and their underlying biomechanical principles with a resolution up to single‐molecule level.7, 8, 9, 10 However, many of these single‐cell techniques are still limited in their ability to scale to measure a suitably large number of individual cells and independent samples.11 In this review, we focus on biophysical methods suitable to assess and quantify biomechanical characteristics such as elastic modulus, stiffness (or deformability), forces generated during adhesion, and spreading and contraction of single platelets in health and disease states. We also provide some insights into limitations and highlight improvements of the methods described herein. Most of the methods described in this review rely on application of external mechanical forces such as stretch, compression, and shear (Figure 1) to quantify biomechanical properties of single platelets. This review is specifically directed toward clinician scientists and beginners who are interested in exploring applications of single‐cell–based biophysical approaches in unraveling the role of platelet biomechanics in hemostasis and thrombosis research.
Figure 1.
Simplified schematics and glossary of terms used for defining mechanical properties of a deformable biological viscoelastic material (A) with a defined area A, initial length L0 and width W0 under applied shear force undergoing sample elongation ΔL along the direction of the shear (B). The same deformable viscoelastic material can be either stretched (ie, material undergoes elongation) (C) or compressed (ie, material undergoes deformation) under the application of external force perpendicular to the surface area A, resulting in changes in length Ln and width Wn, thereby allowing determination of several mechanical parameters including the Young’s modulus E. (Figure 1 was adapted and modified from Wu et al.11)
2. TECHNIQUES FOR MEASURING THE BIOMECHANICAL PROPERTIES OF SINGLE PLATELETS
2.1. Micropipette aspiration
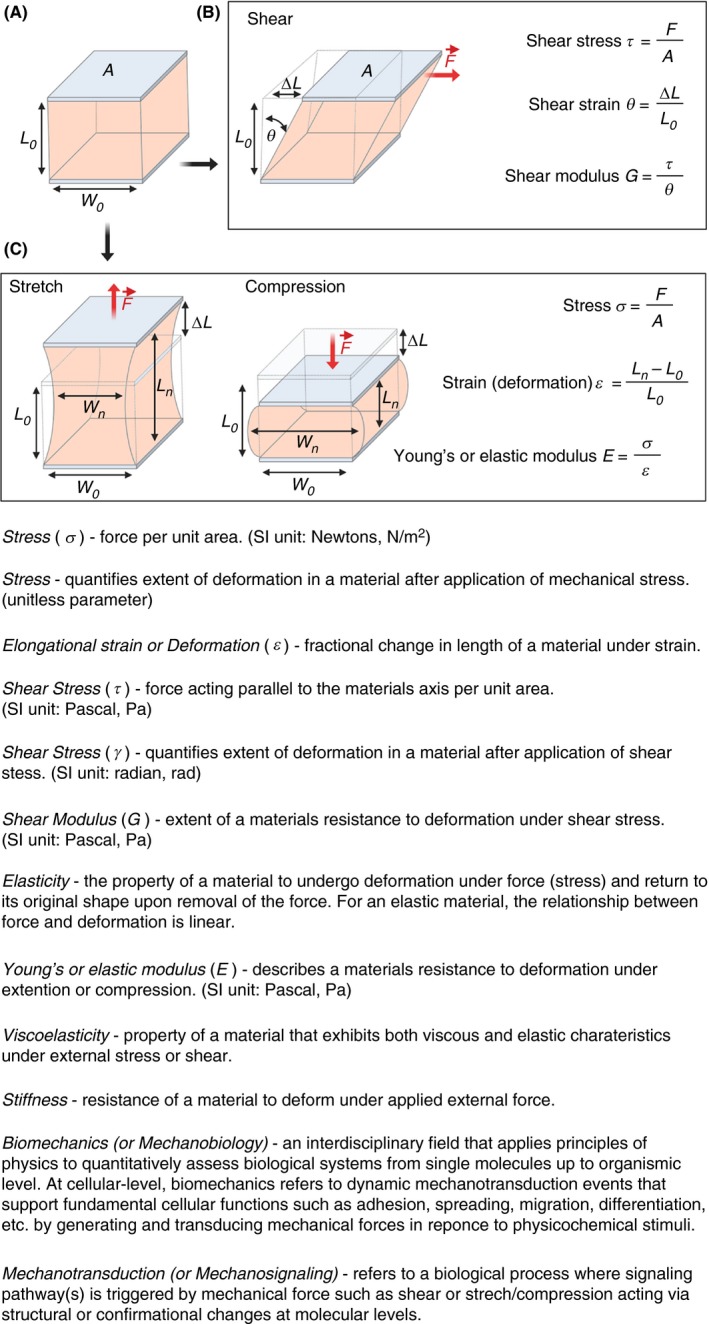
Micropipette aspiration has been indispensable for membrane biophysicists interested in quantifying phase behavior, elasticity, and rupture tension of lipid bilayers.12, 13, 14 When applied to single cells, micropipette aspiration allows for measuring the biomechanical properties of single cells by observing cellular deformation upon application of defined suction pressure.15 It is one of the earliest biophysical tools used in single‐platelet manipulation and quantification of platelet biomechanics.16 Micropipette aspiration (Figure 2A), as the name suggests, relies on suction of part of the single‐platelet membrane into a borosilicate glass micropipette (inner diameter of 0.5‐1.5 µm) connected to a micromanipulator by applying negative pressure in a stepwise manner. The subsequent change in the length of the platelet membrane aspirated into the micropipette over time is tracked by video microscopy (Figure 2B).15, 17 The data obtained from this type of experiment is then used to characterize material properties of a deforming cell using the Law of Laplace, which gives the relationship between the surface tension and pressure within a fluid drop that has a membrane with surface tension in it (Figure 2C). Depending on the instrument setup, suction pressures from 0.1 pN/μm2 up to 101 325 N/m2 (ie, atmospheric) can be applied and membrane tension forces between 10 pN up to 104 nN can be measured with a membrane edge detection accuracy of ±25 nm.18 Using micropipette aspiration viscoelastic and biomechanical changes in single platelets induced by soluble antithrombotic drugs (eg, acetylsalicylic acid), platelet agonists (eg, ADP, thrombin, and the calcium ionophore A23187) and influence of cytoskeleton destabilizing drugs (eg, vincristine, colchicine, taxol, and cytochalasin D) on platelet cytoskeleton have been comprehensively assessed.16, 19, 20 Micropipette aspiration measurements show the Young’s modulus of resting platelets is about 1.7 ± 0.63 × 103 dyn cm−2 with a viscous modulus of 1.0 ± 0.5 × 104 dyn s cm−2.21 In addition, the mechanistic effect of low‐temperature–induced (platelets cooled to 4°C and rewarmed to 37°C) platelet deformation was shown to be directly dependent on microtubule integrity.22 Furthermore, the capacity of platelets from patients with Bernard–Soulier syndrome, gray platelet syndrome, and MYH9 disorders to undergo membrane deformation based on their size have been characterized by micropipette aspiration.23 In particular, these measurements revealed that platelets from Bernard–Soulier syndrome required application of lower suction pressure thresholds during aspiration, showed longer membrane protrusions within the micropipette, and were highly deformable in comparison to normal platelets. Apart from single platelets, recently, micropipette aspiration has been also used to investigate the megakaryocyte cytoskeletal biomechanics and its influence on pro‐platelet formation.24
Figure 2.
A, Schematic diagram of micropipette aspiration setup of a single resting platelet. The micropipette movement immersed in platelet suspension is controlled by a precision micromanipulator, ΔPa is the aspiration pressure applied parallel to the z‐axis through the aspirator, Ri and Ro‐inner and outer radius of the micropipette respectively and L pro is the length of the protrusion of platelet body into the micropipette. Change in L pro is recorded continuously with a high‐speed camera. B, Micropipette aspiration is a dynamic procedure with three distinct stages: In Stage I (initial), radius of protrusion R pro > L pro, the length of protrusion. During Stage II (critical) R pro = L pro and in Stage III (final) R pro < L pro. In Stage II when L pro/R pro = 1, the Law of Laplace can be applied to determine the surface tension n (SI units pN/μm). C, Simplified schematics of the Law of Laplace, which provides a relationship between the radius R of a membrane of a spherical cell subjected to the pressure difference ΔP = P int − P out, where P int and P out are the pressures inside and outside the cell, respectively. The membrane force F mem = n 2πR is the result of the surface tension n acting on the cell membrane along the circumference C = 2PR as shown in the free body diagram of a spherical cell cut in half. It is in equilibrium with the forces F pre resulting from the pressure difference ΔP acting on the cell area A = PR 2. Combining these two we arrive at the Law of Laplace, which gives the relationship between cell wall and its curvature 1/R in terms with the surface tension n. Applying this to the critical Stage II during micropipette aspiration where R pro = L pro for radius of protrusion and that of cell we can effectively determination the surface tension n of the cell. (Figure modified from Gonzalez‐Bermudez et al.15)
Even though there are several elegant examples of the use of micropipette aspiration to elucidate fundamental biophysical properties of platelets, this method is seldom used for single‐platelet analysis. This is mainly due to its low throughput, and it is technically highly demanding. Recent developments in miniaturization and automation to biomechanically characterize a few hundred cells in a single experiment by using parallel arrays of serial micropipettes assembled into a microfluidic device may potentially be able to also facilitate investigation of platelets.25, 26 Additionally, such devices would have the advantage over rapid exchange of soluble agonists/antagonists around the cells within millisecond time scales, thus facilitating measurement of kinetics of biomechanical changes in real time.
2.2. Atomic force microscopy
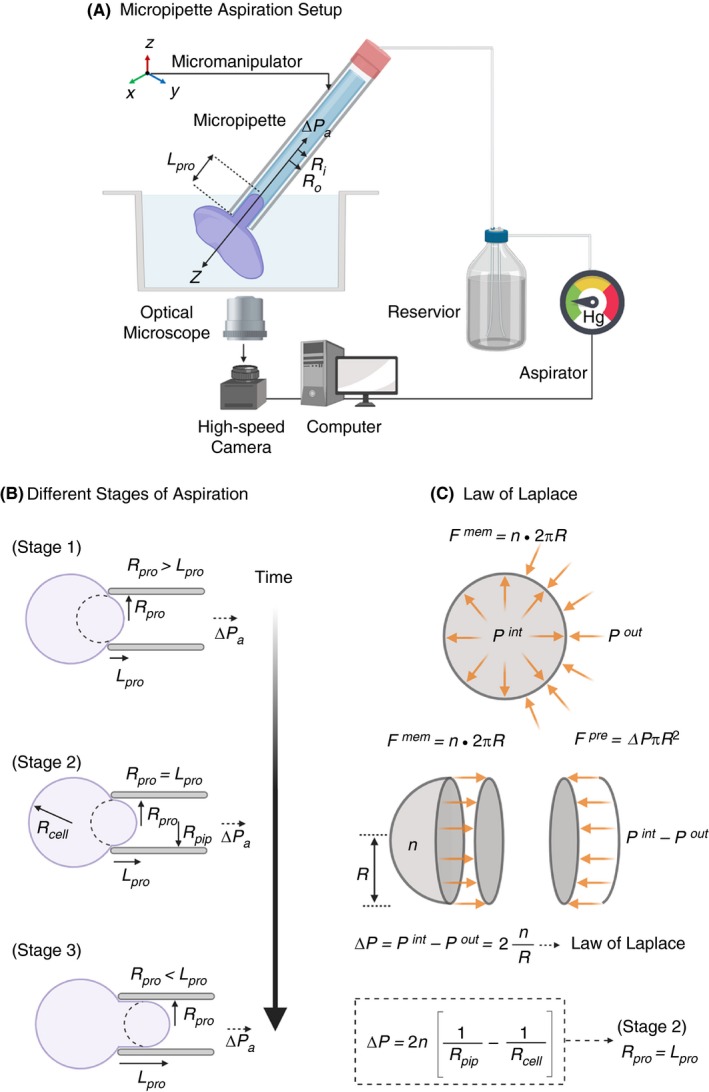
The technique of atomic force microscopy (AFM), also known as scanning force microscopy, provides the capability to simultaneously visualize and quantify mechanical properties of both organic and inorganic materials in 3 dimensions on a nanometer (nm) scale at ambient atmospheric conditions, and in solution, including (biological) buffers, beyond the light diffraction limit.27 AFM can achieve a lateral resolution up to 1 nm, or below with a subatomic vertical resolution limit of <0.1 nm while simultaneously allowing direct visualization of dynamic molecular events at millisecond time scales.28, 29 One of the most important features of AFM is its ability to manipulate single molecules and cells, thus facilitating measurement of intermolecular forces generated by biological systems down to sub‐picoNewton (pN) regimes. These powerful features have made AFM the tool of choice for biologists, chemists, and physicists alike. The typical AFM setup combines a sensitive octapoled piezoelectric transducer for x‐y scanning, and the third dimension, z, correlates to height movement in the piezo linked to an optical lever detection system. Scanning is performed with a thin flexible silicon nitride (Si3N4) cantilever, 100‐200 µm in length, with an integrated pyramidal probe tip 4 to 8 µm tall with a radius of curvature of 5 to 10 nm at tip apex (Figure 3A). The cantilever is moved over the sample (x‐ and y‐axis) and deflected (z‐axis) according to the height of the sample (ie, scanning mode). The cantilever deflection (d) is registered onto a quadrant photo diode from reflected laser and provides a quantitative signal for the applied force (F). This information can be now used to calculate F based on Hooke’s law: F = −k d, where k is the calibrated spring constant of the cantilever. Alternatively, the cantilever can be pressed on the sample (ie, tapping mode), and the forces needed to deflect it are measured (Figure 3B, 3, and 3), or it can be used to pull on the sample using an adhesive tip. Determination of the spring constant k of the AFM cantilever is the most critical step of setting up an AFM experiment. This can be measured by applying classical cantilever‐beam theory (or Euler–Bernoulli beam theory) to determine the spring constant of the cantilever under investigation with cantilever tips shaped as a sphere, cone, or a pyramid and taking into account the geometric constrains of the cantilever (Figure 3A). Furthermore, after data acquisition, it is also necessary to apply suitable theoretical models such as Hertz, Sneddon, Derjaguin–Müller–Toporov, and Johnson–Kendall–Roberts (JKR) models to extract mechanical properties of biological systems.30
Figure 3.
A, Schematic representation of a typical rectangular cantilever with a pyramidal sharp tip used for AFM and the equation for determination of cantilever spring constant k based on beam theory; C, 3D topography (height) and D, Young’s modulus E of a single platelet spreading on fibrinogen passivated surface obtained by force‐volume imaging by acquiring force‐distance curves in x‐y plane. (B, C, and D adapted and modified from Sorrentino et al.44) E, Schematic diagram of single platelet force spectroscopy using flat cantilever to assess biomechanical forces generated during single platelet adhesion at the tip of the cantilever. Single platelet firmly adhering on the lower side of a collagen‐coated tipless cantilever (fplatelet labeled with plasma membrane dye DiI and imaged with confocal florescence microscopy) lowered along the z‐axis (Step 1) and allowed to approach and contact briefly with collagen coated substrate (Step 2) to facilitate adhesion. Next, the cantilever is then retracted along z‐axis in (Step 3) to allow the platelet to detach from the adhesive substrate (Step 4). These steps are repeated several hundreds of times in x‐, y‐, and z‐axis to scan a defined area to obtain precise quantification of single platelet adhesion and spreading forces (g) on different substrates. (E, F, and G are adapted and modified from Nguyen et al.46)
Since its development in the early 1990’s, AFM has accelerated our understanding of biophysical properties governing hemostasis at the molecular and cellular levels.31, 32, 33, 34, 35, 36, 37, 38, 39, 40 With respect to surface characterization of platelets using AFM in scanning mode, Fritz et al41, 42 were first to report on detailed topography maps of human platelets upon adhesion and spreading on glass substrates. Subsequent tapping‐mode AFM tip‐based indentation imaging studies revealed the elastic modulus of adherent human platelets to be in the range of 1 to 50 kPa.43 It was found that platelet granulomere was the softest part of platelets (1.5‐4 kPa), and areas surrounding the granulomere were heterogeneous in their stiffness (10‐40 kPa), while platelet edges were the stiffest (up to 53 kPa). Since the subcortical architecture of actin cytoskeleton influences elastic modulus of cells, the observed increased stiffness at platelet edges is a result of shorter, densely packed, and homogeneous distribution actin filaments.43 These observations were recently confirmed by Sorrentino et al,44 who investigated the mechanical stiffness of thrombin‐activated single platelets spreading on fibrinogen surface by force‐volume mapping by acquiring force‐distance curves in x‐y plane (Figure 3B‐D) revealing Young’s modulus of platelet granulomere to be 32 kPa, and the peripheral regions showed higher stiffness of ~224 kPa.
Using bulk methods, it has been shown previously that elasticity of platelet‐rich clots is 10‐fold greater (~600 Pa) than the elasticity of a clot devoid of platelets (~70 Pa). This is mainly due to high contractile forces generated by platelet actomyosin machinery on the fibrin network. To investigate these processes at single‐platelet level, Lam et al9, 45 and Chaudhuri et al9, 45used a custom‐designed AFM setup that simultaneously allowed single‐platelet manipulation and fluorescence imaging from the sides. A single platelet was suspended between a flat AFM cantilever passivated with fibrinogen and a fibrinogen‐coated planar surface. The subsequent adhesion and spreading of the platelet between both fibrinogen surfaces generates adhesive and contractile forces that induce cantilever bending, which was the readout for quantifying the generated forces. This approach for the first time revealed that single platelets undergo instantaneous contact activation on fibrinogen while generating average maximum contractile and adhesive forces of 29 nN and 70 nN, respectively. These forces peaked at 2 to 3 minutes after contact with fibrinogen and were sustained beyond 15 minutes.9 Furthermore, when platelets were held under an isometric clamp, they exerted higher force (2‐fold increase) and became stiffer and more adhesive. This implies that platelets are able to modulate their contractile forces depending of the mechanical properties of the microenvironment. Recently, Nguyen et al,46 using a similar approach investigated adhesive and rupture forces generated by single platelets (Figure 3E‐G) and between 2 single platelets adhered to ECM proteins. Single‐platelet interaction assays showed that human platelets produce an adhesion force of 23 ± 5 nN and ≥35 ± 4 nN on fibronectin and collagen type 1, respectively. However, when 2 individual platelets were brought into contact, the platelet‐platelet interaction forces were found to be much lower, 1.50 ± 0.05 nN, when platelets adhered on collagen and 2.01 ± 0.05 nN when they adhered on fibronectin. These observations suggest that single platelets generate differential adhesion forces on different ECM substrates, which may depend on platelet activation states.
One of the most complex sets of multiple biomolecular direct and indirect interactions occurs when platelets engage with Gram‐negative and Gram‐positive bacteria.47, 48 These interactions often lead to platelet activation and in part contribute to the pathophysiology of infective endocarditis and disseminated intravascular coagulopathy. So far, little is known about how these interactions come into play and how this affects platelet activation from a biophysical perspective. Using AFM imaging, Xu and Siedlecki49 showed that platelet adhesion to Staphylococcus epidermidis is mainly promoted by fibrinogen and fibronectin, which induces platelet activation, resulting in bacteria/platelet aggregates. These observations may further explain recently observed platelet‐bacteria interactions leading to bacterial capture and their killing. Biomechanical properties of platelets are important, as bacterial killing by platelets is dependent on the integrity of the platelet acto‐myosin complex.50, 51 Additionally, bacterial secret products such as pore‐forming toxins: α‐hemolysin (from Staphylococcus aureus) and streptolysin O (from Streptococcus pyogenes), which are known to permeabilize platelet membranes.52 AFM made it possible to obtain detailed dynamic maps and kinetics of elasticity changes in the presence of streptolysin O on the platelet plasma membrane.53 Streptolysin O induced a gradual increase in platelet elasticity starting from the granulomere, then spreading to the periphery. Upon stabilization of the F‐actin cytoskeleton by phalloidin, streptolysin O–treated platelets undergo a marked decrease in their stiffness. These intriguing observations highlight the role of major cytoskeletal proteins to stabilize plasma membrane elasticity in platelets.
Although AFM has emerged as an excellent tool for precise measurements of single‐platelet biomechanics, there are still many limitations mainly related to tip scan speed (temporal resolution for profiling rapid changes in platelet shape) and scan area, thereby restricting the throughput to a few adherent platelets. Additionally, single‐platelet force spectroscopy experiments require immobilization of platelets to measure forces, either on the cantilever or on the solid phase. This immobilization procedure may cause undesirable platelet preactivation, may affect adhesion kinetics, and potentially introduce additional artifacts including operator bias. However, such artifacts can be minimized by carefully selecting immobilizing substrates depending on the scientific objectives.46 Recently, automation and parallelization combined with positioning of cells on prepatterned adhesive substrates have achieved higher‐throughput measurements of cell mechanics using AFM.54, 55 However, these advanced setups are not widely available yet.
2.3. Scanning ion‐conductance microscopy
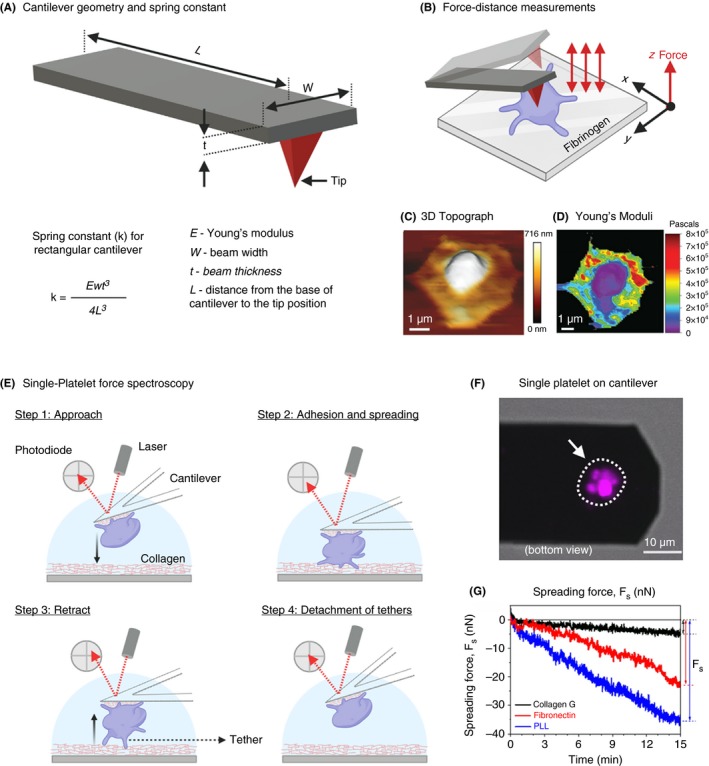
Scanning ion‐conductance microscopy (SICM) is a label‐free, nonforce contact, noninvasive, and high‐resolution topography imaging technique suitable for biophysical characterization of biological samples from several micrometers down to molecular resolution of few nanometers.56, 57, 58 The resolution limit in SICM is mainly determined by the nanopipette inner radius, which is typically in the range of 8 nm up to 2 µm.56, 59, 60 In addition, SICM is also capable of monitoring surface charge and ion flux across membranes during topography imaging of biological samples.61, 62 The working principle of SICM is based on precise measurement of ionic current that flows between a quasi‐reference counter electrode (QRCE) inside a borosilicate glass nanopipette and a second QRCE immersed in an electrolyte solution connected to a feedback control system that maintains the pipette‐sample distance along a vertical axis (z) during the lateral scanning process (Figure 4A).63 Topography imaging is then performed by immersing the nanopipette in the bath, where the ion current is limited initially by the resistance of the pipette. As the pipette gradually approaches the surface (eg, platelet membrane), the ion current (IL) reduces. This reduction in ion current is used for distance feedback control and high‐resolution noncontact topographical imaging. In order to map the local biomechanical properties such as elasticity, SICM uses application of hydrostatic pressure (0.1‐150 kPa) through the nanopipette aperture at the surface of a cell membrane (Figure 4B). The deformation of membrane triggers the distance feedback control, adjusting the nanopipette position, thereby keeping the reduction in ion current at a constant value. Since the pipette position z (Figure 4A and 4) is a function of applied hydrostatic pressure and corresponding response of the mechanical properties of the cell beneath the pipette, one can noninvasively locally indent the cell membrane.64, 65 This facilitates simultaneous noncontact true topography imaging and biomechanical characterization of platelets (Figure 4C and 4).66 Recently, SICM has attracted considerable attention among platelet biologists and is fast emerging as the method of choice for high‐speed morphometric and biomechanical characterization of single platelets.67 Using SICM, Rheinlaender et al66 elegantly demonstrated simultaneous imaging of morphodynamics at submicrometer resolution and showed that the mean elastic modulus of single platelets during spreading increased 5‐fold from 3 to 15 kPa within 20 minutes. In addition, in the presence of thrombin, the elastic modulus of platelets decreased significantly from 12.4 kPa (resting platelet) to 7.5 kPa (thrombin activation) with a mean softening time of 7.1 minutes. Surprisingly, in platelets treated with cytochalasin D, mean elastic modulus decreased from 11.1 kPa to 6.5 kPa with a mean softening time of 2.7 minutes. These findings revealed platelet activation by thrombin and cytoskeleton depolymerization by cytochalasin D result in differential spatial distributions of the stiffening and softening regions on platelets, respectively. Subsequent SICM investigations of contribution of motor proteins such as dynein in the presence of inhibitors ciliobrevin D and erythro‐9‐(2‐hydroxy‐3‐nonyl)‐adenine revealed decreased contact‐induced platelet spreading activity in thrombin‐treated platelets, whereas blebbistatin (myosin II inhibitor), Y‐27632 (ROCK inhibitor), nocodazole (microtubule polymerization inhibitor), and aurintricarboxylic acid‐ATA (kinesin ATPase inhibitor) showed no significant effects.68 Recently, in a clinically relevant study, SICM was found to be a useful label‐free morphometric biophysical tool for obtaining fast 3‐dimensional (3D) topographic images to unravel a critical role of platelet‐derived high‐mobility group box 1 (HMGB1) in trauma and hemorrhagic shock, linking inflammation and microvascular thrombosis.69 Platelet‐derived HMGB1 was instrumental on acceleration of adhesion speed and increased spreading area of platelets on collagen and von Willebrand factor (VWF). Similarly, 3D topographic measurements using SICM have shown that platelets deficient in anaphylatoxin receptor C3aR exhibit a reduced spreading area on fibrinogen after thrombin stimulation compared to their normal platelets.70 These observations indicate a role of complement activation fragment C3a and C3aR for platelet function during thrombus formation.
Figure 4.
A, Schematics of scanning ion conductance microscopy (SICM) used for high‐resolution topography imaging and biomechanical characterization of adherent platelets. The SICM setup consists of a pressure‐controlled borosilicate glass nanopipette with a Ag/AgCl electrode and a second electrode connects the electrolyte‐filled culture dish with the nanopipette. The culture dish is mounted on a sample scanner that drive in x and y‐axis (for lateral movement) and z‐piezo motors (for vertical scanning). An applied voltage between 2 electrodes induces an ionic leakage current IL through the electrolyte‐filled nanopipette, which depends on the distance d between pipette and sample. A controller records this ion current and drives the xy‐ and z‐piezo. For cell biological applications the SICM can be easily integrated with an optical microscope. B, Theoretical modeling using in silico finite element mechanics (FEM) simulation and calculations showing resulting deformation of an elastic sample as a function of the vertical SICM nanopipette position upon application of fluid flow induced by the pressure p0 applied to the upper pipette end.; C, Representative SICM 3D topography and Young’s modulus mapping of spreading platelets. (Figure adapted and modified from Rheinlaender and Schaffer65 and Rheinlaender et al.66)
Even though SICM has attracted a broader attention of biologists, most SICM instrument setups do not allow fast and large‐area scanning of live biological samples. For those interested in simultaneous mapping of platelet topography and elastic properties, the time factor is of key importance, since platelet morphodynamics occur within seconds in response to agonists. Recent technical improvements in SICM instrument design have specifically addressed these challenges and facilitate imaging of a 20 × 20 μm2 scan area within 10 seconds.71 It is foreseen that adaptation of these technical improvements will generate a broader appeal of SICM and pave the way for new discoveries in platelet biology.
3. TRACTION FORCE MEASUREMENTS
Platelets exert traction (or contractile) forces. These are generated by the actomyosin‐motor protein complex in activated platelets upon specific receptor‐ligand interactions on exposed subendothelial ECM during blood clot and thrombus formation.72, 73 Broadly categorized under traction force microscopy (TFM), several methods are currently available to precisely measure and quantify cellular traction forces at a single or multicellular level in 2D and 3D.74 In principle, these techniques rely on quantitative light microscopy to measure substrate displacements, which are then converted into forces. However, substrate preparation methods, implementation, modeling and data analysis of TFM varies between different approaches.75, 76 To measure traction forces produced by platelets, displacement tracking by TFM on hydrogel substrates and bending of elastomeric micropillar arrays are most commonly used.
3.1. TFM on hydrogel substrates
TFM on hydrogel substrates is a versatile and perturbation‐free approach to access cellular forces from single cells up to tissue levels. In its simplest implementation, TFM on hydrogel substrates measures cell‐exerted traction forces from the spatial images of substrate stress by tracking the displacement of fiducial markers such as fluorescent tracer beads embedded in a continuum soft elastic substrate that undergoes linear deformations (Figure 5A and 5).77 Traction forces are then computed by applying appropriate assumptions/constraints, taking into account substrate stiffness and its deformation in response to traction force and boundary conditions such as cell geometry. The resolution of measured force relies on substrate stiffness, density of tracer beads, image acquisition parameters, and the mathematical algorithm applied for data assessment.76, 78, 79
Figure 5.
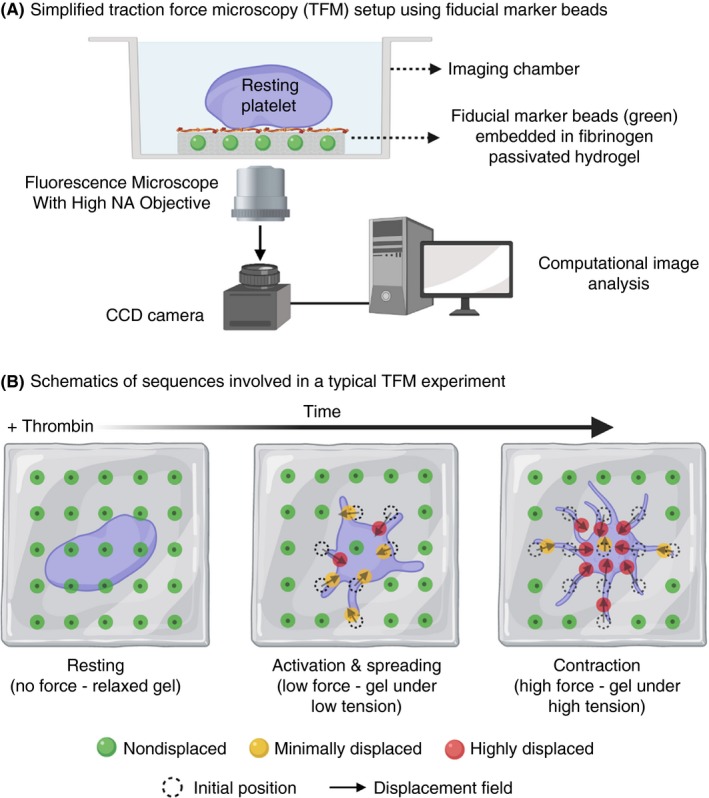
A, Schematic diagram showing set‐up of a traction force microscopy experiment using fibrinogen passivated hydrogel of known stiffness embedded with of fiducial marker microbeads; B, Resting platelets are seeded low densities on the fibrinogen passivated hydrogel surface. Upon thrombin stimulation, activated platelets adhere, spread, and ultimately contract resulting in generation of “traction forces” over the entire process. These forces exert mechanical tension on the hydrogel that leads to displacement of embedded microbeads. These displacements can be precisely imaged and tracked overtime with a fluorescence microscope. Post‐image acquisition, image‐processing algorithms are used to compute displacement fields that provide spatial and temporal dynamics of platelet generated traction forces
Blood clots are mainly composed of platelets, red blood cells, and a dense network of fibrin fibers. During hemostasis, the terminal stage of blood clot maturation by platelet‐driven clot retraction and fibrin matrix remodeling is one of the physiologically most relevant mechanisms where integrin αIIbβ3‐mediated fibrin interactions and contractile force generation by the actomyosin complex acting through talin comes into play.80 Primarily, this allows for reestablishing nonobstructed blood flow and normal hemodynamics past otherwise obstructive thrombi within a blood vessel.81 Although the biochemical nature of this process is well understood, the mechanobiological principles have remained elusive. Using TFM, Schwarz Henriques et al82 showed for the first time temporal evolution of contractile forces in thrombin‐activated platelets spreading on fibrinogen functionalized polyacrylamide gel with a physiologically relevant elastic modulus of 4 kPa. They observed, upon reaching steady state after 25 minutes, that single platelets were able to exert a traction field (ie, displacement field of gel surface) as large as ≈3.3 × 103 Pa with total forces in the range of ≈34 nN. Using polyacrylamide gels with a tunable elastic modulus, Qiu et al83 observed that fibrinogen functionalized stiffer gels (elastic modulus of 5.0 kPa and 50 kPa) promoted platelet adhesion, spreading, and activation, while on soft gels (0.5 kPa) platelet function was abrogated and was dependent on Rac1 and actomyosin activity. Subsequent TFM experiments on substrates of higher elastic modulus (from 19 kPa up to 83 kPa) revealed that traction forces generated by fully activated platelets were independent of the matrix stiffness.84 Although platelets generated isotropic contractile traction forces, at the steady state, assessment of force localization showed that these were largest at the periphery of platelets, while the traction force was focused near the platelet granulomere. From a broader perspective, single‐platelet TFM experiments have provided meaningful biophysical insight into how isotropic contractile traction and subsequent mechanotransduction events driven by the actomyosin complex may play an important role in engagement of platelet αIIbβ3 on fibrin during clot retraction in hemostasis.80 Recently, quantitative fluorescence microscopy provided mechanistic insights into how transmission of contractile forces generated by single platelets occurs via filopodial extensions, which bend and shorten fibrin fibers, thereby causing clot volume shrinkage and alterations at the macro level.85 Additionally, single‐platelet contractile force measurements also explain the role of mechanotransduction in innate immune responses of platelets such as those involving FcγRIIA‐mediated recognition, sequestration, and killing of immunoglobulin G opsonized bacteria under the platelet granulomere.51
TFM has gained wide popularity due to its high sensitivity and relative simplicity. However, traditional TFM suffers from low measurement throughput and artifacts related to variations in hydrogel mechanical properties. To address these drawbacks, a chip‐based high‐throughput platelet contraction cytometer was developed recently.86 The chip‐based contraction cytometer uses microfabrication technology to fabricate a large array of fibrinogen passivated microdot pairs on a hydrogel substrate with varied stiffness to assess the nanomechanics of hundreds of individual platelets under physiologically relevant conditions. The readout of contractile force output is based on measurement of lateral displacement of 2 adjacent fibrinogen microdots by a single activated platelet. Using a platelet contraction cytometer, Myers et al86 revealed that platelet‐generated contractile force during clot formation requires both biochemical (eg, thrombin) and mechanical (eg, substrate stiffness) inputs and the mechanosensitive contraction is highly dependent on the Rho/ROCK pathway. Furthermore, assessment of platelet contractile forces from patients with Wiskott‐Aldrich syndrome and MYH9 disorder (nonmuscle myosin IIa mutations) on a platelet contraction cytometer showed that up to 30% of these platelets from patients with cytoskeleton‐related platelet disorders exhibit the near‐zero contractile forces on stiffer substrates, while this was limited to only 6% in healthy controls. These findings are significant, as they point toward the critical role of platelet cytoskeletal machinery at a single‐platelet level in achieving a mechanically stable hemostatic plug.
3.2. Deformable elastomeric micropillar and microbeam arrays
As an alternative to the continuum substrates used in TFM, deformable micropillar (also called as micropost) arrays are uniformly spaced, compliant, vertical elastomeric cylindrical cantilevers made of polydimethylsiloxane (PDMS) of defined dimensions (diameter and height) and stiffness (linearly elastic).87 In contrast to TFM, the ligand functionalized tips of the micropillars serve as adhesive surfaces.88 Upon cell adhesion, the displacements of each micropillar in an array is tracked by video microscopy, and the applied force on the cantilever can be calculated from force‐displacement relationship for pure bending of an elastic cylindrical beam using beam theory (Figure 6A and 6).89 Micropillar arrays have been used in a wide variety of contexts, not only to measure cell‐generated forces (as low as 1 nN), but also to analyze the relationship between substrate rigidity and single‐cell responses as well as at the tissue level, thus making it a highly versatile biophysical tool.90, 91 Liang et al92 adapted micropillar arrays spaced at 9 µm apart and passivated with fibrinogen to quantify contractile forces produced by platelets in a microthrombus. The average contractile force produced by a single platelet in microthrombi on micropillars was found to be 2.1 ± 0.1 nN that was dependent on thrombin concentration, while the contractile force increased steadily overtime as the microthrombus grew. These results demonstrated quantitatively for the first time that each platelet acts as an active contractile force–producing unit in the microthrombus and subsequent recruitment of platelets leads to incremental augmentation of total contractile force. Follow‐up investigations by Feghhi et al93 using fibronectin passivated micropillars revealed that the generation of contractile forces by platelets in microthrombi is regulated by nonmuscle myosin IIA ATPase activity through Rho kinase (ROCK) and myosin light‐chain kinase. Additionally, elastomeric micropillars coated with VWF have been found to be useful in assessing the role of integrin αIIbβ3 and GPIb‐IX‐V complex in mechanotransduction of forces through cytoskeleton at the single‐platelet level.94 Currently, deformable micropillar arrays are fabricated using a photolithographic technique followed by replica molding using PDMS. However, this labor‐intensive and time‐consuming process limits rapid prototyping necessary for optimizing substrate deformation to increase measurement sensitivity. In addition, fabrication of complex geometries to measure cellular traction forces on 3D substrates is impossible to achieve via replica molding. To address these issues our laboratory has adopted a maskless technique based on direct laser writing using 2‐photon polymerization (2PP) in 3D of photosensitive resists to print high‐resolution deformable micro‐ and nanostructures in 3D.95 Using 2PP we have 3D printed deformable micropillars as wells as suspended horizontal microbeams that can be functionalized with platelet adhesive ligand of choice (Figure 6D and 6).96 Quantification of micropillar and microbeam deformation upon platelet adhesion showed single platelets generate a mean traction force of 46.3 nN on micropillars of aspect ratio of 1:10 (diameter 700 nm and height 7 µm) and 31.9 nN on microbeams (diameter 400 nm, length 25 µm) functionalized with fibrinogen. We assume these differences in measured forces arise as a result of differential platelet adhesion and spreading that is unique to substrate geometry and depends on ligand‐functionalized surface area (Figure 5C and 5). We are currently further investigating these observations to better understand whether platelets sense and modulate traction forces in response to changes in topography and the chemical nature of their microenvironment.
Figure 6.
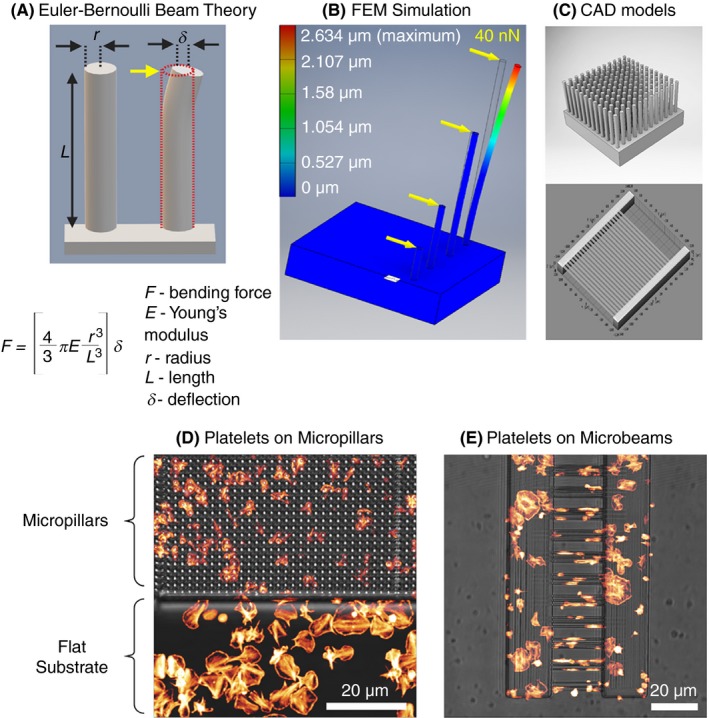
A, Schematics showing Euler‐Bernouli beam theory as applied to calculation of lateral force (yellow arrow) acting on the free end of a micropillar attached to a solid base leading to its deflection of the vertical micropillar. B, In silico finite element mechanics (SEM) simulation of bending of micropillars upon application of a lateral force of 40 nN acting on the free end of the micropillar causing a maximum defection of 2.62 μm. C, Models of computer‐assisted design (CAD) used for fabrication of deformable elastomeric micropillar (D) and microbeam array microstructures using 2‐photon polymerization (2PP) and 3D printing functionalized with fibrinogen showing comparative differences in platelet spreading and cytoskeletal organization on different substrate geometries
4. REAL‐TIME DEFORMIBILITY CYTOMETRY
Real‐time deformability cytometry is a rapidly emerging mechanophenotyping approach that combines the technique of flow cytometry with measuring cell stiffness using microfluidic technology. As discussed in previous sections, currently available biophysical techniques for biomechanical characterization of platelets are labor intensive, suffer from measurement throughput, and most importantly cannot be performed in complex fluids such as whole blood. These limitations have restricted the adoption of such techniques to investigate platelet biology and function testing for diagnostic purposes in clinical and translational scenarios. Real‐time deformability cytometry (RT‐DC) has shown to be capable of probing single‐cell deformation at >1000 cells per second in real time in whole blood thereby providing comprehensive biomechanical signatures of individual cell types in blood in a short period of time.97, 98 RT‐DC uses application of hydrodynamic shear stress to induce cell deformation as the cells pass through a narrow microfluidic channel (15‐30 µm in cross section). During this passage, the sample is illuminated with a high‐power light‐emitting diode (LED) as a light source and imaged concurrently with a high‐speed camera synchronized with the LED at millisecond intervals and cell contour images are computationally analyzed on the fly, providing rapid insights into cellular physiological states based on their stiffness contributed by cytoskeletal components (Figure 7A‐C). This approach facilitates extraction of additional quantitative parameters such as cell size and morphology, thus typifying cells and simultaneously characterizing cell deformability at single‐cell resolution without prior separation, enrichment, and labeling procedures.99 Due to these capabilities, RT‐DC is a promising method with potential relevance to several fields of life sciences.100 To demonstrate the potential of RT‐DC as a high‐throughput biophysical method for mechanophenotyping platelets, as a proof of principle we exposed human platelets to agonists and cytoskeleton destabilizing chemical agents for 10 minutes and subsequently performed RT‐DC measurements on live platelets (Figure 7D, unpublished results). Thrombin receptor activator for peptide 6 (TRAP‐6) treated platelets deformed less, that is, platelets became stiffer (median deformation, 0.0475, n = 2813 single platelets), whereas cytochalasin D and blebbistatin showed increased deformation, that is, platelets became softer (0.166, n = 3651; and 0.176, n = 3479, respectively), in comparison to resting platelets (0.139, n = 4013). Our preliminary results are encouraging since they hint at the role of the platelet cytoskeleton contributing to intrinsic biomechanical deformation characteristics of platelets depending on their physiological state. We are currently evaluating the suitability of RT‐DC to understand the relationship between platelet activation status and corresponding biomechanical characteristics as a novel approach for platelet function testing, quality control of platelets during storage, and diagnostics of inherited platelet diseases related to the cytoskeleton.
Figure 7.
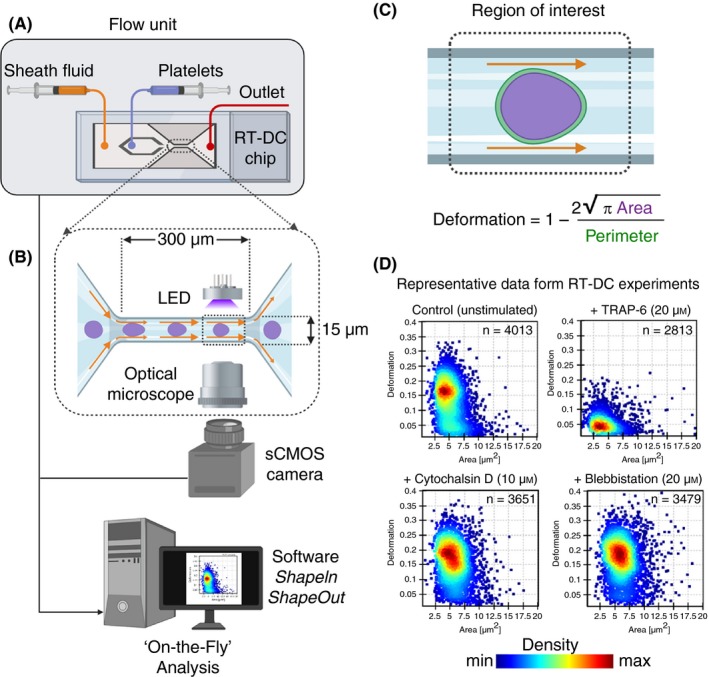
The real‐time deformability cytometry (RT‐DC) microfluidic chip connected to a flow unit (A) consist of dedicated inlets for sheath fluid and for cells/platelets, which combine into to a channel with narrow constriction zone (B) where cell/platelets undergo deformation as a result of hydrodynamic compression brought about by sheath fluid. A high‐powered light‐emitting diode (LED) is used to illuminate the samples. Images are recorded by a scientific complementary metal–oxide–semiconductor (sCMOS) camera working at high frame rates (≈2000 images/s) that is synchronized with the frequency of illumination time. This combined together with microfluidic flow, RT‐DC allows for mechanoprofiling of >100 unique cells/platelets per second in a contact‐ and label‐free manner. Platelet deformation (C) is determined on the fly in real time by computational image processing algorithm that taking into account the area and perimeter of the deformed platelet as it passes through the region of interest. Results of a typical RT‐DC measurements involving washed platelets (D) show that unstimulated deform more (ie, softer), while those treated with agonist (TRAP‐6) deform less (ie, stiffer; upper panels). Platelets preincubated with cytochalasin D and blebbistatin lead to increased deformation (ie, softer; lower panels). Data show density maps of deformation against area (μm2) from n ≥ 2500 individual platelets from a single experiment
5. CONCLUSIONS
Current knowledge of fundamental biochemical and cell biological processes governing thrombosis and hemostasis have been steadily advanced as result of continued development and implementation methods, which vary widely in their readouts, sensitivities, and spatiotemporal resolution on a macroscale. Building on this, using cutting‐edge biophysical methods to manipulate single platelets, one can gain novel insights into biomechanical principles influencing hemostasis and thrombus formation at subnanoscale regimes. Adaptation of the appropriate biophysical platform will mainly depend on the primary research question and on availability and suitability of one or more of the methods described here (Table 1).
Table 1.
Comparison of currently available biophysical tools suitable for biomechanical assessment of platelets
| Biophysical method | Biomechanical properties | Restrictions | Throughput | Technical know‐how | Commercial vendors |
|---|---|---|---|---|---|
| Micropipette aspiration | Elastic and viscoelastic | Nonadherent/In suspension and adherent |
Low 1‐5 platelets/h |
Highly skilled | NA |
| Atomic force microscopy | Elastic, viscoelastic, and adhesion forces | Adherent |
Low 1‐5 platelets/h |
Highly skilled | Yes |
| Scanning ion conductance microscopy | Elastic and viscoelastic | Adherent |
Low 1‐10 platelets/h |
Highly skilled | Yes |
| Traction force microscopy on hydrogel substrates | Adhesion and contraction | Adherent | Medium to high | Skilled | NA |
| Micropillars and deformable 3D substrates | Adhesion and contraction | Adherent | Medium | Skilled | Yes |
| Real‐time deformability cytometry | Deformability | Nonadherent/In suspension |
Very high 100‐1000 platelets/sec |
Skilled | Yes |
Due to the highly interdisciplinary nature of these investigations, clinicians and basic scientists are encouraged to collaborate extensively to maximize the potential of many of such biophysical methods to address outstanding questions in platelet biology.
RELATIONSHIP DISCLOSURE
The authors report nothing to disclose.
AUTHOR CONTRIBUTIONS
RP and LS performed literature review, composition of the primary manuscript, preparation of figures, and editing. AG and CD contributed to the primary text and participated in revision of the manuscript. RP, LS, and CD performed the experiments, analyzed the results, and prepared the RT‐DC, micropost, and micropillar data presented in the review. RP and CD acquired the funding. All authors contributed to revision of the final version of the manuscript.
ACKNOWLEDGMENTS
The authors thank Prof. Dr Markus Münzenburg, Interface and Surface Physics, and Dr Oliver Otto, Biomechanics, Center for Innovation Competence at University of Greifswald, Germany, for providing access instrumentation and facilities. The authors also thank Prof. Dr Ohad Medalia, Department of Biochemistry, University of Zurich, Switzerland, and Prof. Dr Tilman E. Schäffer, Institute of Applied Physics, University of Tübingen, Germany, for providing high‐resolution, representataive AFM and SICM images of biomechanical charaterization of single platelets, respectively.
Sachs L, Denker C, Greinacher A, Palankar R. Quantifying single‐platelet biomechanics: An outsider’s guide to biophysical methods and recent advances. Res Pract Thromb Haemost. 2020;4:386–401. 10.1002/rth2.12313
Handling Editor: Yotis Senis.
Funding information
LS is funded by the Deutsche Forschungsgemeinschaft (DFG, German Research foundation) Project Number 374031971 – TRR 240, Project A06 granted to RP and Forschungsverbund für Molekulare Medizin, Universitätsmedizin Greifswald (Research Network Molecular Medicine of University of Medicine Greifswald) FOVB‐2017‐07 to CD and RP.
REFERENCES
- 1. Machlus KR, Italiano JE Jr. The incredible journey: from megakaryocyte development to platelet formation. J Cell Biol. 2013;201:785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Qiu Y, Ciciliano J, Myers DR, Tran R, Lam WA. Platelets and physics: how platelets "feel" and respond to their mechanical microenvironment. Blood Rev. 2015;29:377–86. [DOI] [PubMed] [Google Scholar]
- 3. Hansen CE, Qiu Y, McCarty OJT, Lam WA. Platelet mechanotransduction. Annu Rev Biomed Eng. 2018;20:253–75. [DOI] [PubMed] [Google Scholar]
- 4. Yeaman MR. Platelets: at the nexus of antimicrobial defence. Nat Rev Microbiol. 2014;12:426–37. [DOI] [PubMed] [Google Scholar]
- 5. Cardigan R, Turner C, Harrison P. Current methods of assessing platelet function: relevance to transfusion medicine. Vox Sang. 2005;88:153–63. [DOI] [PubMed] [Google Scholar]
- 6. Taheri‐Araghi S, Brown SD, Sauls JT, McIntosh DB, Jun S. Single‐cell physiology. Annu Rev Biophys. 2015;44:123–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ju L, Chen Y, Xue L, Du X, Zhu C. Cooperative unfolding of distinctive mechanoreceptor domains transduces force into signals. Elife. 2016;5:e15447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Butera D, Passam F, Ju L, Cook KM, Woon H, Aponte‐Santamaria C, et al. Autoregulation of von Willebrand factor function by a disulfide bond switch. Sci Adv. 2018;4:eaaq1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lam WA, Chaudhuri O, Crow A, Webster KD, Li TD, Kita A, et al. Mechanics and contraction dynamics of single platelets and implications for clot stiffening. Nat Mater. 2011;10:61–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen Y, Ju LA, Zhou F, Liao J, Xue L, Su QP, et al. An integrin αIIbβ3 intermediate affinity state mediates biomechanical platelet aggregation. Nat Mater. 2019;18:760–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu P‐H, Aroush DR‐B, Asnacios A, Chen W‐C, Dokukin ME, Doss BL, et al. A comparison of methods to assess cell mechanical properties. Nat Methods. 2018;15:491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Evans E, Needham D. Physical properties of surfactant bilayer membranes: thermal transitions, elasticity, rigidity, cohesion and colloidal interactions. J Phys Chem. 1987;91:4219–28. [Google Scholar]
- 13. Dimova R. Recent developments in the field of bending rigidity measurements on membranes. Adv Colloid Interface Sci. 2014;208:225–34. [DOI] [PubMed] [Google Scholar]
- 14. Longo ML, Ly HV. Micropipet aspiration for measuring elastic properties of lipid bilayers. Methods Mol Biol. 2007;400:421–37. [DOI] [PubMed] [Google Scholar]
- 15. Gonzalez‐Bermudez B, Guinea GV, Plaza GR. Advances in micropipette aspiration: applications in cell biomechanics, models, and extended studies. Biophys J. 2019;116:587–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. White JG, Burris SM, Tukey D, Smith C 2nd, Clawson CC. Micropipette aspiration of human platelets: influence of microtubules and actin filaments on deformability. Blood. 1984;64:210–4. [PubMed] [Google Scholar]
- 17. Kee YS, Robinson DN. Micropipette aspiration for studying cellular mechanosensory responses and mechanics. Methods Mol Biol. 2013;983:367–82. [DOI] [PubMed] [Google Scholar]
- 18. Hochmuth RM. Micropipette aspiration of living cells. J Biomech. 2000;33:15–22. [DOI] [PubMed] [Google Scholar]
- 19. Burris SM, Smith CM 2nd, Tukey DT, Clawson CC, White JG. Micropipette aspiration of human platelets after exposure to aggregating agents. Arteriosclerosis. 1986;6:321–5. [DOI] [PubMed] [Google Scholar]
- 20. Burris SM, Smith CM 2nd, Rao GH, White JG. Aspirin treatment reduces platelet resistance to deformation. Arteriosclerosis. 1987;7:385–8. [DOI] [PubMed] [Google Scholar]
- 21. Haga JH, Beaudoin AJ, White JG, Strony J. Quantification of the passive mechanical properties of the resting platelet. Ann Biomed Eng. 1998;26:268–77. [DOI] [PubMed] [Google Scholar]
- 22. Burris SM, Smith CM 2nd, Tukey D, Clawson CC, White JG. Micropipette aspiration of human platelets: influence of rewarming on deformability of chilled cells. J Lab Clin Med. 1986;107:238–43. [PubMed] [Google Scholar]
- 23. White JG, Burris SM, Hasegawa D, Johnson M. Micropipette aspiration of human blood platelets: a defect in Bernard‐Soulier's syndrome. Blood. 1984;63:1249–52. [PubMed] [Google Scholar]
- 24. Chen Y, Aardema J, Kale S, Whichard ZL, Awomolo A, Blanchard E, et al. Loss of the F‐BAR protein CIP4 reduces platelet production by impairing membrane‐cytoskeleton remodeling. Blood. 2013;122:1695–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mak M, Erickson D. A serial micropipette microfluidic device with applications to cancer cell repeated deformation studies. Integr Biol (Camb). 2013;5:1374–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Myrand‐Lapierre ME, Deng X, Ang RR, Matthews K, Santoso AT, Ma H. Multiplexed fluidic plunger mechanism for the measurement of red blood cell deformability. Lab Chip. 2015;15:159–67. [DOI] [PubMed] [Google Scholar]
- 27. Patel AN, Kranz C. (Multi)functional atomic force microscopy imaging. Annu Rev Anal Chem (Palo Alto Calif). 2018;11:329–50. [DOI] [PubMed] [Google Scholar]
- 28. Gross L, Schuler B, Pavlicek N, Fatayer S, Majzik Z, Moll N, et al. Atomic force microscopy for molecular structure elucidation. Angew Chem Int Ed Engl. 2018;57:3888–908. [DOI] [PubMed] [Google Scholar]
- 29. Ando T. Directly watching biomolecules in action by high‐speed atomic force microscopy. Biophys Rev. 2017;9:421–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krieg M, Fläschner G, Alsteens D, Gaub BM, Roos WH, Wuite GJL, et al. Atomic force microscopy‐based mechanobiology. Nat Rev Phys. 2019;1(1):41–57. [Google Scholar]
- 31. Hussain MA, Siedlecki CA. The platelet integrin alpha(IIb) beta(3) imaged by atomic force microscopy on model surfaces. Micron. 2004;35:565–73. [DOI] [PubMed] [Google Scholar]
- 32. Carvalho FA, Connell S, Miltenberger‐Miltenyi G, Pereira SV, Tavares A, Ariens RA, et al. Atomic force microscopy‐based molecular recognition of a fibrinogen receptor on human erythrocytes. ACS Nano. 2010;4:4609–20. [DOI] [PubMed] [Google Scholar]
- 33. Tobimatsu H, Nishibuchi Y, Sudo R, Goto S, Tanishita K. Adhesive forces between A1 domain of von Willebrand factor and N‐terminus domain of glycoprotein ibalpha measured by atomic force microscopy. J Atheroscler Thromb. 2015;22:1091–9. [DOI] [PubMed] [Google Scholar]
- 34. Soman P, Rice Z, Siedlecki CA. Measuring the time‐dependent functional activity of adsorbed fibrinogen by atomic force microscopy. Langmuir. 2008;24:8801–6. [DOI] [PubMed] [Google Scholar]
- 35. Aponte‐Santamaria C, Huck V, Posch S, Bronowska AK, Grassle S, Brehm MA, et al. Force‐sensitive autoinhibition of the von Willebrand factor is mediated by interdomain interactions. Biophys J. 2015;108:2312–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Posch S, Obser T, Konig G, Schneppenheim R, Tampe R, Hinterdorfer P. Interaction of von Willebrand factor domains with collagen investigated by single molecule force spectroscopy. J Chem Phys. 2018;148:123310. [DOI] [PubMed] [Google Scholar]
- 37. Kang I, Raghavachari M, Hofmann CM, Marchant RE. Surface‐dependent expression in the platelet GPIb binding domain within human von Willebrand factor studied by atomic force microscopy. Thromb Res. 2007;119:731–40. [DOI] [PubMed] [Google Scholar]
- 38. Marchant RE, Lea AS, Andrade JD, Bockenstedt P. Interactions of von Willebrand factor on mica studied by atomic force microscopy. J Colloid Interface Sci. 1992;148:261–72. [Google Scholar]
- 39. Zhmurov A, Protopopova AD, Litvinov RI, Zhukov P, Weisel JW, Barsegov V. Atomic structural models of fibrin oligomers. Structure. 2018;26(6):857–868.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brown AE, Litvinov RI, Discher DE, Weisel JW. Forced unfolding of coiled‐coils in fibrinogen by single‐molecule AFM. Biophys J. 2007;92:L39–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fritz M, Radmacher M, Gaub HE. In vitro activation of human platelets triggered and probed by atomic force microscopy. Exp Cell Res. 1993;205:187–90. [DOI] [PubMed] [Google Scholar]
- 42. Fritz M, Radmacher M, Gaub HE. Granula motion and membrane spreading during activation of human platelets imaged by atomic force microscopy. Biophys J. 1994;66:1328–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Radmacher M, Fritz M, Kacher CM, Cleveland JP, Hansma PK. Measuring the viscoelastic properties of human platelets with the atomic force microscope. Biophys J. 1996;70:556–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sorrentino S, Studt JD, Horev MB, Medalia O, Sapra KT. Toward correlating structure and mechanics of platelets. Cell Adh Migr. 2016;10:568–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chaudhuri O, Parekh SH, Lam WA, Fletcher DA. Combined atomic force microscopy and side‐view optical imaging for mechanical studies of cells. Nat Methods. 2009;6:383–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nguyen TH, Palankar R, Bui VC, Medvedev N, Greinacher A, Delcea M. Rupture forces among human blood platelets at different degrees of activation. Sci Rep. 2016;6:25402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hamzeh‐Cognasse H, Damien P, Chabert A, Pozzetto B, Cognasse F, Garraud O. Platelets and infections ‐ complex interactions with bacteria. Front Immunol. 2015;6:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fitzgerald JR, Foster TJ, Cox D. The interaction of bacterial pathogens with platelets. Nat Rev Microbiol. 2006;4:445–57. [DOI] [PubMed] [Google Scholar]
- 49. Xu L‐C, Siedlecki CA. Effects of plasma proteins on staphylococcus epidermidis RP62A adhesion and interaction with platelets on polyurethane biomaterial surfaces. J Biomater Nanobiotechnol. 2012;03:487–98. [Google Scholar]
- 50. Gaertner F, Ahmad Z, Rosenberger G, Fan S, Nicolai L, Busch B, et al. Migrating platelets are mechano‐scavengers that collect and bundle bacteria. Cell. 2017;171(6):1368–1382.e23. [DOI] [PubMed] [Google Scholar]
- 51. Palankar R, Kohler TP, Krauel K, Wesche J, Hammerschmidt S, Greinacher A. Platelets kill bacteria by bridging innate and adaptive immunity via platelet factor 4 and FcgammaRIIA. J Thromb Haemost. 2018;16:1187–97. [DOI] [PubMed] [Google Scholar]
- 52. Flaumenhaft R. Platelet Permeabilization In: Gibbins JM, Mahaut‐Smith MP, editors. Platelets and Megakaryocytes: Volume 2: Perspectives and Techniques. Totowa, NJ: Humana Press, 2004; 365–78. [Google Scholar]
- 53. Walch M, Ziegler U, Groscurth P. Effect of streptolysin O on the microelasticity of human platelets analyzed by atomic force microscopy. Ultramicroscopy. 2000;82:259–67. [DOI] [PubMed] [Google Scholar]
- 54. Favre M, Polesel‐Maris J, Overstolz T, Niedermann P, Dasen S, Gruener G, et al. Parallel AFM imaging and force spectroscopy using two‐dimensional probe arrays for applications in cell biology. J Mol Recognit. 2011;24:446–52. [DOI] [PubMed] [Google Scholar]
- 55. Wang Z, Liu L, Wang Y, Xi N, Dong Z, Li M, et al. A fully automated system for measuring cellular mechanical properties. J Lab Autom. 2012;17:443–8. [DOI] [PubMed] [Google Scholar]
- 56. Schaffer TE. Nanomechanics of molecules and living cells with scanning ion conductance microscopy. Anal Chem. 2013;85:6988–94. [DOI] [PubMed] [Google Scholar]
- 57. Happel P, Thatenhorst D, Dietzel ID. Scanning ion conductance microscopy for studying biological samples. Sensors (Basel). 2012;12:14983–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shevchuk AI, Frolenkov GI, Sanchez D, James PS, Freedman N, Lab MJ, et al. Imaging proteins in membranes of living cells by high‐resolution scanning ion conductance microscopy. Angew Chem Int Ed Engl. 2006;45:2212–6. [DOI] [PubMed] [Google Scholar]
- 59. Rheinlaender J, Schaffer TE. Lateral resolution and image formation in scanning ion conductance microscopy. Anal Chem. 2015;87:7117–24. [DOI] [PubMed] [Google Scholar]
- 60. Korchev YE, Milovanovic M, Bashford CL, Bennett DC, Sviderskaya EV, Vodyanoy I, et al. Specialized scanning ion‐conductance microscope for imaging of living cells. J Microsc. 1997;188:17–23. [DOI] [PubMed] [Google Scholar]
- 61. Page A, Perry D, Young P, Mitchell D, Frenguelli BG, Unwin PR. Fast nanoscale surface charge mapping with pulsed‐potential scanning ion conductance microscopy. Anal Chem. 2016;88:10854–9. [DOI] [PubMed] [Google Scholar]
- 62. Chen CC, Zhou Y, Morris CA, Hou J, Baker LA. Scanning ion conductance microscopy measurement of paracellular channel conductance in tight junctions. Anal Chem. 2013;85:3621–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hansma PK, Drake B, Marti O, Gould SA, Prater CB. The scanning ion‐conductance microscope. Science. 1989;243:641–3. [DOI] [PubMed] [Google Scholar]
- 64. Sánchez D, Johnson N, Li C, Novak P, Rheinlaender J, Zhang Y, et al. Noncontact measurement of the local mechanical properties of living cells using pressure applied via a pipette. Biophys J. 2008;95:3017–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rheinlaender J, Schäffer TE. Mapping the mechanical stiffness of live cells with the scanning ion conductance microscope. Soft Matter. 2013;9:3230–6. [Google Scholar]
- 66. Rheinlaender J, Vogel S, Seifert J, Schachtele M, Borst O, Lang F, et al. Imaging the elastic modulus of human platelets during thrombin‐induced activation using scanning ion conductance microscopy. Thromb Haemost. 2015;113:305–11. [DOI] [PubMed] [Google Scholar]
- 67. Kraus MJ, Seifert J, Strasser EF, Gawaz M, Schaffer TE, Rheinlaender J. Comparative morphology analysis of live blood platelets using scanning ion conductance and robotic dark‐field microscopy. Platelets. 2016;27:541–6. [DOI] [PubMed] [Google Scholar]
- 68. Seifert J, Rheinlaender J, Lang F, Gawaz M, Schaffer TE. Thrombin‐induced cytoskeleton dynamics in spread human platelets observed with fast scanning ion conductance microscopy. Sci Rep. 2017;7:4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vogel S, Bodenstein R, Chen Q, Feil S, Feil R, Rheinlaender J, et al. Platelet‐derived HMGB1 is a critical mediator of thrombosis. J Clin Invest. 2015;125:4638–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sauter RJ, Sauter M, Reis ES, Emschermann FN, Nording H, Ebenhoch S, et al. A functional relevance of the anaphylatoxin receptor C3aR for platelet function and arterial thrombus formation marks an intersection point between innate immunity and thrombosis. Circulation. 2018;138(16):1720–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhukov A, Richards O, Ostanin V, Korchev Y, Klenerman D. A hybrid scanning mode for fast scanning ion conductance microscopy (SICM) imaging. Ultramicroscopy. 2012;121:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Carr ME Jr, Zekert SL. Measurement of platelet‐mediated force development during plasma clot formation. Am J Med Sci. 1991;302:13–8. [DOI] [PubMed] [Google Scholar]
- 73. Carr ME Jr. Development of platelet contractile force as a research and clinical measure of platelet function. Cell Biochem Biophys. 2003;38:55–78. [DOI] [PubMed] [Google Scholar]
- 74. Roca‐Cusachs P, Conte V, Trepat X. Quantifying forces in cell biology. Nat Cell Biol. 2017;19:742–51. [DOI] [PubMed] [Google Scholar]
- 75. Ribeiro AJ, Denisin AK, Wilson RE, Pruitt BL. For whom the cells pull: Hydrogel and micropost devices for measuring traction forces. Methods. 2016;94:51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Schwarz US, Soine JR. Traction force microscopy on soft elastic substrates: a guide to recent computational advances. Biochim Biophys Acta. 2015;1853:3095–104. [DOI] [PubMed] [Google Scholar]
- 77. Plotnikov SV, Sabass B, Schwarz US, Waterman CM. High‐resolution traction force microscopy. Methods Cell Biol. 2014;123:367–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zundel M, Ehret AE, Mazza E. Factors influencing the determination of cell traction forces. PLoS ONE. 2017;12:e0172927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Style RW, Boltyanskiy R, German GK, Hyland C, MacMinn CW, Mertz AF, et al. Traction force microscopy in physics and biology. Soft Matter. 2014;10:4047–55. [DOI] [PubMed] [Google Scholar]
- 80. Osdoit S, Rosa JP. Fibrin clot retraction by human platelets correlates with alpha(IIb)beta(3) integrin‐dependent protein tyrosine dephosphorylation. J Biol Chem. 2001;276:6703–10. [DOI] [PubMed] [Google Scholar]
- 81. Muthard RW, Diamond SL. Blood clots are rapidly assembled hemodynamic sensors: flow arrest triggers intraluminal thrombus contraction. Arterioscler Thromb Vasc Biol. 2012;32:2938–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Schwarz Henriques S, Sandmann R, Strate A, Koster S. Force field evolution during human blood platelet activation. J Cell Sci. 2012;125:3914–20. [DOI] [PubMed] [Google Scholar]
- 83. Qiu Y, Brown AC, Myers DR, Sakurai Y, Mannino RG, Tran R, et al. Platelet mechanosensing of substrate stiffness during clot formation mediates adhesion, spreading, and activation. Proc Natl Acad Sci U S A. 2014;111:14430–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hanke J, Probst D, Zemel A, Schwarz US, Koster S. Dynamics of force generation by spreading platelets. Soft Matter. 2018;14:6571–81. [DOI] [PubMed] [Google Scholar]
- 85. Kim OV, Litvinov RI, Alber MS, Weisel JW. Quantitative structural mechanobiology of platelet‐driven blood clot contraction. Nat Commun. 2017;8:1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Myers DR, Qiu Y, Fay ME, Tennenbaum M, Chester D, Cuadrado J, et al. Single‐platelet nanomechanics measured by high‐throughput cytometry. Nat Mater. 2017;16:230–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Gupta M, Kocgozlu L, Sarangi BR, Margadant F, Ashraf M, Ladoux B. Micropillar substrates: a tool for studying cell mechanobiology. Methods Cell Biol. 2015;125:289–308. [DOI] [PubMed] [Google Scholar]
- 88. Sniadecki NJ, Han SJ, Ting LH, Feghhi S. Micropatterning on micropost arrays. Methods Cell Biol. 2014;121:61–73. [DOI] [PubMed] [Google Scholar]
- 89. Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc Natl Acad Sci U S A. 2003;100:1484–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Han SJ, Bielawski KS, Ting LH, Rodriguez ML, Sniadecki NJ. Decoupling substrate stiffness, spread area, and micropost density: a close spatial relationship between traction forces and focal adhesions. Biophys J. 2012;103:640–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Song J, Shawky JH, Kim Y, Hazar M, LeDuc PR, Sitti M, et al. Controlled surface topography regulates collective 3D migration by epithelial‐mesenchymal composite embryonic tissues. Biomaterials. 2015;58:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Liang XM, Han SJ, Reems JA, Gao D, Sniadecki NJ. Platelet retraction force measurements using flexible post force sensors. Lab Chip. 2010;10:991–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Feghhi S, Tooley WW, Sniadecki NJ. Nonmuscle myosin IIA regulates platelet contractile forces through rho kinase and myosin light‐chain kinase. J Biomech Eng. 2016;138:1045061–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Feghhi S, Munday AD, Tooley WW, Rajsekar S, Fura AM, Kulman JD, et al. Glycoprotein Ib‐IX‐V complex transmits cytoskeletal forces that enhance platelet adhesion. Biophys J. 2016;111:601–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Klein F, Striebel T, Fischer J, Jiang Z, Franz CM, von Freymann G, et al. Elastic fully three‐dimensional microstructure scaffolds for cell force measurements. Adv Mater. 2010;22:868–71. [DOI] [PubMed] [Google Scholar]
- 96. Palankar R, Denker C, Varma M, Münzenberg M, Greinacher A. Deformable nano and microstructures for quantifying platelet contractile forces and platelet cytoskeletal mechanics. Hamostaseologie. 2018;38:A2–A91. [Google Scholar]
- 97. Otto O, Rosendahl P, Mietke A, Golfier S, Herold C, Klaue D, et al. Real‐time deformability cytometry: on‐the‐fly cell mechanical phenotyping. Nat Methods. 2015;12:199–202, 4 p following. [DOI] [PubMed] [Google Scholar]
- 98. Otto O, Rosendahl P, Golfier S, Mietke A, Herbig M, Jacobi A, et al. Real‐time deformability cytometry as a label‐free indicator of cell function. Conf Proc IEEE Eng Med Biol Soc. 2015;2015:1861–4. [DOI] [PubMed] [Google Scholar]
- 99. Mietke A, Otto O, Girardo S, Rosendahl P, Taubenberger A, Golfier S, et al. Extracting cell stiffness from real‐time deformability cytometry: theory and experiment. Biophys J. 2015;109:2023–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Urbanska M, Rosendahl P, Krater M, Guck J. High‐throughput single‐cell mechanical phenotyping with real‐time deformability cytometry. Methods Cell Biol. 2018;147:175–98. [DOI] [PubMed] [Google Scholar]


