Abstract
Despite multiple sexual exposures to HIV-1 virus, some individuals remain HIV-1 seronegative. Although several genetic factors have been related to HIV-1 resistance, the homozygosity for a mutation in CCR5 gene (the 32-bp deletion, i.e., CCR5-Delta32 allele) is presently considered the most relevant one. The C-type lectins, DC-SIGN (present on dendritic cells and macrophages) and DC-SIGNR (present on endothelial cells in liver and lymph nodes) efficiently bind and transmit HIV-1 to susceptible cell in trans, thereby augmenting the infection. A potential association of the DC-SIGN and DC-SIGNR neck domain repeat polymorphism and risk of HIV-1 infection is currently under debate. To determine the influence of host genetic factors on HIV-1 resistance, we conducted genetic risk association study in HIV-1-exposed seronegative (n = 47) individuals, HIV-1 seronegative (n = 262) healthy control, and HIV-1-infected seropositive patients (n = 168) for polymorphism in neck domain of DC-SIGN and DC-SIGNR genes. The DC-SIGN and DC-SIGNR genotypes were identified by polymerase chain reaction method in DNA extracted from peripheral blood and confirmed by sequencing. Fisher exact or χ 2 test was used for static analysis. DC-SIGN genotype and allele distribution was fairly similar in HIV-1-exposed seronegative, HIV-1 seropositive, and HIV-1 seronegative control. There was no statistical significance in the differences in the distribution of DC-SIGN genotypes. A total of 13 genotypes were found in DC-SIGNR neck repeat region polymorphism. Among all the genotypes, only 5/5 homozygous showed significant reduced risk of HIV-1 infection in HIV-1-exposed seronegative individuals (p = 0.009). A unique genotype 8/5 heterozygous was also found in HIV-1 seropositive individual, which is not reported elsewhere.
Keywords: DC-SIGN, DC-SIGNR, HIV-1, tandem repeats, genetic association, gene polymorphism
Introduction
Host genetic factors play an important role in susceptibility to HIV-1 infection and progression to AIDS. Repeated sexual exposure to HIV-1 does not always result in infection. Although several factors have been related to HIV-1 infection resistance, the possible genetic mechanism underlying this resistance presently remains elusive [1, 2]. The most investigated genetic factor associated with HIV-1 infection resistance is the homozygous presence of a 32-bp deletion in CCR5 gene (CCR5 delta 32) [3], i.e., the main co-receptor used by the macrophage (M)-tropic strain of the virus to infect peripheral blood mononuclear cells.
The dendritic cell receptor, DC-SIGN (dendritic cell-specific ICAM-3 grabbing non-integrin, encoded by CD209) and the closely related DC-SIGNR (DC-SIGN Related also called as L-SIGN for liver/lymph node-specific ICAM-3 grabbing non-integrin, encoded by CLEC4M) are members of the C-type lectin receptor family. They can act as both cell-adhesion receptors and pathogen-recognition receptors and shares 77% amino acid identity [4]. They appear to have related and perhaps overlapping abilities to bind mannose, GlcNAc, and fucose-containing ligands. These sugar-binding proteins have been ascribed multiple roles in the innate and adaptive immune response [5]. Both DC-SIGN and DC-SIGNR have been of considerable interest because of their ability to enhance infection of T cells by the HIV-1 and because of their interactions with glycoproteins found on the surface of other enveloped viruses [6]. As pathogen-recognition receptors, these lectins have been shown to recognize a vast range of microbes, some of which are of major public health importance. Indeed, DC-SIGN captures bacteria such as Mycobacterium tuberculosis, Helicobacter pylori, and certain Klebsiella pneumonia strains; viruses such as HIV-1, Ebola virus, cytomegalovirus, hepatitis C virus, dengue virus, and SARS-coronavirus; and parasites like Leismania pifanoi and Schistosoma mansoni. With regard to DC-SIGNR, studies to date have shown an interaction with a variety of viruses, including HIV, hepatitis C virus [18], Ebola virus [19] and coronavirus, as well as with the parasite S. mansoni [7].
Both DC-SIGN and DC-SIGNR are organized into three domains: an N-terminal cytoplasmic region, a neck region containing seven tandem repeats of the 23 amino acid sequence, and a C-terminal domain with homology to C-type lectins. This neck region presents high nucleotide identity between repeats, both within each molecule and between DC-SIGN and DC-SIGNR. It has been shown that this region plays a crucial role in the oligomerization and support of the carbohydrate-recognition domain, thus, influencing the pathogen-binding properties of these two receptors [8–10]. The neck region of DC-SIGN and DC-SIGNR may be important in determining the orientation of the carbohydrate-binding C-type lectin domain and may therefore have an impact on ligand specificity [5]. DC-SIGN may be important in facilitating intrauterine vertical transmission of HIV [11]. Recent data demonstrate the capacity of both DC-SIGN and DC-SIGNR to mediate cellular entry of Ebola virus both in cis and in trans [12]. In this context, the efficiency of the DC-SIGN and DC-SIGNR in pathogen recognition and subsequent processing may have important consequences for the quality of host immune responses and consequent pathogen control and/or clearance.
The tandem repeats in the neck region are variable in both DC-SIGN and DC-SIGNR. In contrast to DC-SIGN, DC-SIGNR has considerable polymorphism in the neck region (tandem repeat domain of exon 4) which consists of three to nine repeats of a 69-base pair segment, with seven repeats being predominant in the general population [13]. Recent studies have provided evidence linking polymorphism in the neck of DC-SIGN and DC-SIGNR with susceptibility to HIV-1 infection [15]. Heterozygous DC-SIGN individuals were shown to be at reduced risk of HIV-1 infection [14]. In some recent studies, DC-SIGNR homozygous 7/7 repeat in the neck region was associated with an increased risk of HIV-1 infection, whereas DC-SIGNR heterozygous 7/5 repeat was correlated with resistance to HIV-1 infection [15], while earlier studies could not detect any association between DC-SIGNR repeat region polymorphism and HIV-1 infection [17]. Homozygosity for DC-SIGNR/L-SIGN repeat region was also associated with protective role during SARS infection [16].
As there is a large ethnic variation in the prevalence of the described polymorphisms and studies on the role of DC-SIGN and DC-SIGNR in HIV-1 disease transmission have yielded conflicting results, we sought to investigate the effect of DC-SIGN and DC-SIGNR neck domain repeat polymorphism on HIV-1 susceptibility among North Indian HIV-1 seronegative, HIV-1 seropositive, and HIV-1-exposed seronegative individuals.
Material and Methods
Patient Selection
One hundred sixty-eight HIV-1 seropositive patients (HSP) were enrolled from the outpatients attending the clinics of Sanjay Gandhi Post Graduate Institute of Medical Sciences, India from January 2004 to November 2006. Most of the patients were from the state of Uttar Pradesh in North India. Two hundred sixty-two age-matched controls were healthy staff members of institute with HIV-1 seronegative status. Forty-seven HIV-1 seronegative individuals with history of repeated sexual intercourse (twice a week) without any protection with HIV-1-infected partners for at least 1 year were recruited in HIV-1-exposed seronegative group (HES). HIV-1 seronegative status of the HES subjects was confirmed by Western blot at the regular interval of 3 months until 1 year. Demographic profiles of the study groups are given in Table I. All the subjects with similar ethnicity were randomly selected from same source population. After an informed consent, 5-ml blood sample was taken in EDTA for analysis of DNA.
Table I.
Demographic Profile of the Study Groups
| Groups | Age (Median, Range) | Sex (M/F ratio) |
|---|---|---|
| HIV-1 seropositive (HSP; n = 168) | 58 years (42–74) | 8/1 |
| HIV-1 seronegative (HSN; n = 262) | 56 years (38–60) | 6/1 |
| HIV-1-exposed seronegative (HES; n = 47) | 59 years (35–55) | 7/1 |
n Number of individuals
Determination of HIV-1 Status
All individuals were screened for their HIV-1 status by primarily screening with enzyme-linked immunosorbent assay (Vironostika, HIV Uni-FormII Ag/Ab, Biomerieux, Netherlands) and subsequently confirmed with Western blot (LAV Blot I, BioRad, France).
Genomic DNA Isolation
The genomic DNA samples were obtained from 0.2–0.3 ml peripheral whole blood using QIAamp Blood kit (Qiagen, CA, USA) according to the protocols supplied by the manufacturer. Usually, ∼0.1 μg genomic DNA was used for the genotyping studies.
Genotyping of CCR5 delta 32 Mutation
CCR5 delta 32 genotype was determined by sizing polymerase chain reaction (PCR) amplicons that include the entire region of the deletion. PCR was conducted in a 25-μl reaction containing 50 ng of genomic DNA, 5 pmol of each primer, 175 μM dNTPs, 1.5 mM MgCl2, 10X PCR buffer and 0.5 U of Taq polymerase (Roche). Thermocycling procedure (PerkinElmer 9600) consisted of initial denaturation at 94°C for 4 min followed by 35 cycles of 94°C for 30 s, 52°C for 45 s and 72°C for 1 min, and final extension of 72°C for 7 min. Amplicons were visualized by ultraviolet transillumination in 2% agarose gel containing ethidium bro-mide. The sense primer was 5-TGTTTGCGTCTCCCAG-3 and antisense was 5-CAC AGC CCT GTG CCT CTT-3, which result in a 233-bp product for the wild-type amplicon and 201 bp for the deletion product.
Genotyping of DC-SIGN Neck Repeat (Exon 4)
The DC-SIGN repeat region in exon 4 was amplified from genomic DNA with the following pair of primer: forward: 5′-CCA CTT TAG GGC AGG AC-3′ and reverse: 5′-AGC AAA CTC ACA CCA CAC AA-3′ with some modifications as mentioned by Liu et al. [11]. PCR amplification was performed in a volume of 25 μl containing 0.25 mmol/l dNTPs, 1.0 μmol/l each primer, 1 μl glycerol, 2.5 mmol/l MgCl2, and 1.0 U Taq polymerase (Roche) in a 1X reaction buffer. The cycle conditions were 5 min at 94°C followed by 30 cycles of 15 s at 95°C, 7 s at 61°C, 30 s at 72°C, and then a single cycle of 7 min at 72°C. Wild-type alleles (7/7) yielded 852 bp PCR products. Alleles were differentiated on the basis of 3% agarose gel and ethidium-bromide staining.
Genotyping of DC-SIGNR Neck Repeat (Exon 4)
The DC-SIGNR repeat region was genotyped by use of the following pair of primer: forward: 5′-TGT CCA AGG TCC CCA GCT CCC-3′ and reverse: 5′-GAA CTC ACC AAA TGC AGT CTT CAA ATC-3′ with some modifications [12]. PCR amplification was performed in a volume of 25 μl containing 0.25 mmol/l of each dNTP, 1.0 μmol/l of each primer, and 1.0 U of Taq enzyme in 1X reaction buffer. The cycle conditions were as follows: 94°C for 5 min followed by 35 cycles of 94°C for 5 s and 70°C for 1 min, and then one cycle of 70°C for 10 min. Alleles were differentiated on the basis of 3% agarose gel and ethidium-bromide staining. Gel documentation was done by Alphaimager™ 1220, Alpha Innotech Corporation (San Leandro, CA, USA).
DNA Sequencing
PCR-amplified fragments of both DC-SIGN and DC-SIGNR alleles were randomly selected to be cloned and sequenced according to the manufacturers protocols (Applied Biosystems/Perkin Elmer Foster City, CA, USA) using the dye terminator cycle sequencing kit in ABI PRISM 310 genetic analyzer.
Statistical Analysis
Statistical analysis was done by SPSS software version 11.5 (SPSS, Chicago, IL, USA). Direct gene counting method was used to determine the frequency of genotypes and alleles. The Fisher exact or χ 2 test was used to determine differences in allele/genotype frequencies of DC-SIGN and DC-SIGNR polymorphism. Odds ratios (OR) and its 95% confidence interval (CI) were obtained to describe the strength of association. A p value <0.05 was considered to be statistically significant.
Results
We examined DC-SIGN and DC-SIGNR exon 4 repeats polymorphism in neck region among 262 HIV-1 seronegative healthy controls, 168 HIV-1 seropositive, and 47 HIV-1-exposed seronegative. Genotyping of CCR5delta 32 mutation was done to rule out any possibility of protection/resistance conferred by CCR5delta32 mutation in HIV-1-exposed seronegative group. None of the 47 HIV-1-exposed seronegative individuals has this mutation. Striking differences were observed between the degree of polymorphism of two genes, i.e., DC-SIGN and DC-SIGNR. The strong difference in the neck region lengths between the two genes was consequently visible in the heterozygosity values: DC-SIGN exhibited an overall heterozygosity of only 0.38%, whereas DC-SIGNR presented a value of 48.85% (Table II). Our findings agree with previous studies of Barrerio et al. [7] suggesting that DC-SIGN has been under strong selective constraints that prevent accumulation of any amino acid changes overtime, whereas DC-SIGNR variability has most likely been driven by the action of balancing selection in North Indians also.
Table II.
Statistical Analysis of DCSIGNR Homozygosity/Heterozygosity
| HSP (%) | HSN (%) | HES (%) | P value | Odds ratio | 95% Confidence Interval | |
|---|---|---|---|---|---|---|
| Heterozygotes (%) | 82 (48.88) | 128 (48.85) | 1.000 | 1.002 | 0.680–1.476 | |
| Homozygotes (%) | 86 (51.19) | 134 (51.14) | 1.000 | 0.998 | 0.677–1.471 | |
| Total | 168 | 262 | ||||
| Heterozygotes (%) | 82 (48.88) | 24 (51.06) | 0.869 | 1.094 | 0.573–2.090 | |
| Homozygotes (%) | 86 (51.19) | 23 (48.93) | 0.869 | 0.914 | 0.478–1.745 | |
| Total | 168 | 47 |
Number in parentheses gives the data in percentage.
HSP HIV-1 seropositive, HSN HIV-1 seronegative, HES HIV-1-exposed seronegative
Genotyping of DC-SIGN Exon 4 Tandem Repeat Polymorphism
In the studied North Indian population, the genetic polymorphism in DC-SIGN neck region was rare. Allele 7 was most common, and its frequency was 99.62%. Among all the North Indian individuals genotyped, only one individual from HSN group was found to be heterozygous 8/7 mutant, as shown in Fig. 1. There were no statistically significant differences of DC-SIGN genotype/allele between different groups studied (data not shown).
Fig. 1.
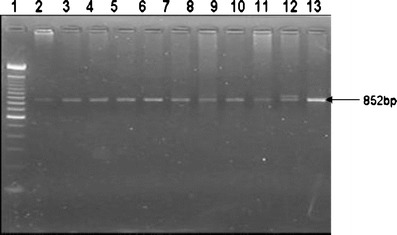
Agarose gel (3%) photograph of amplified DC-SIGN exon 4 repeat region. Lane 1–100 bp Marker (Genetix); Lane 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 13–7/7 repeat (wild homozygous); Lane 12–8/7 repeat (heterozygous).
Genotyping of DC-SIGNR Exon 4 Tandem Repeat Polymorphism
The frequency of DC-SIGNR genotypes and alleles in all the study groups is summarized in Tables III and IV, respectively. A total of 13 genotypes in the DC-SIGNR repeat region based on numbers of repeats (ranging from 4 to 9) were found, as shown in Fig. 2. The allele frequencies found in HSN healthy control North Indian populations were 1.14% for allele 4, 24.42% for allele 5, 4.96% for allele 6, 63.7% for allele 7, and 5.72% for allele 9. The frequency of homozygous 5/5 genotype was lower in HSP (3.57%) than in HSN (8.39%, p = 0.070). When homozygous 5/5 genotype was compared between HSP (3.57%) and HES (12.7%), a significant difference was found (p = 0.009, OR = 0.212, CI = 0.067–0.664). When allele 5 was compared between HSP (19.04%) and HES (30.85%), then also a significant difference were found (p = 0.016, OR = 0.527, CI = 0.315–0.883). Although heterozygous genotype 7/5 was found to be lowest in HSP (23.81%) when compared with HSN (29%, p = 0.266) and HES (34.04%, p = 0.188), no significant differences were found. The difference in genotype frequency of homozygous 7/7 between HSP (47.61%) and HSN (41.22%, p = 0.197), HES (36.17%, p = 0.135) was not statistically significant. Comparison of heterozygous 9/7 genotype between HSP (11.90%) and HSN (9.16%, p = 0.415) and HES (6.38%, p = 0.423) also showed no significant differences. Of the 13 genotypes identified, seven genotypes were present at frequencies of <1%, thereby limiting the intergroup comparability of 9/9, 9/6, 9/5, 8/7, 8/5, 7/4, 4/4 genotypes.
Table III.
Frequency Distribution of DC-SIGNR Genotypes in HIV-1 Seropositive, HIV-1 Seronegative and HIV-1-Exposed Seronegative North Indian Individuals
| Genotypes | HSP (%) | HSN (%) | HES (%) | P value | Odds ratio | 95% Confidence Interval |
|---|---|---|---|---|---|---|
| 4/4 | 0 | 2 (0.76) | – | – | – | |
| 5/5 | 6 (3.57) | 22 (8.39) | 0.070 | 0.40 | 0.160–1.018 | |
| 6/5 | 6 (3.57) | 8 (3.05) | 0.786 | 1.176 | 0.401–3.451 | |
| 7/5 | 40 (23.81) | 76 (29.00) | 0.266 | 0.765 | 0.491–1.192 | |
| 7/6 | 6 (3.57) | 16 (6.11) | 0.272 | 0.569 | 0.218–1.486 | |
| 7/7 | 80 (47.61) | 108 (41.22) | 0.197 | 1.296 | 0.878–1.915 | |
| 7/4 | 2 (1.19) | 2 (0.76) | – | – | – | |
| 8/5 | 2 (1.19) | 0 | – | – | – | |
| 8/7 | 2 (1.19) | 0 | – | – | – | |
| 9/5 | 4 (2.38) | 0 | – | – | – | |
| 9/6 | 0 | 2 (0.76) | – | – | – | |
| 9/7 | 20 (11.90) | 24 (9.16) | 0.415 | 1.640 | .715–2.511 | |
| 9/9 | 0 | 2 (0.76) | – | – | – | |
| Total | 168 | 262 | ||||
| 4/4 | 0 | 0 | – | – | – | |
| 5/5 | 6 (3.57) | 6 (12.7) | 0.009 | 0.212 | 0.067–0.664 | |
| 6/5 | 6 (3.57) | 1 (2.12) | 1.000 | 1.704 | 0.200–14.512 | |
| 7/5 | 40 (23.81) | 16 (34.04) | 0.188 | 0.605 | 0.301–1.219 | |
| 7/6 | 6 (3.57) | 2 (4.25) | 0.687 | 0.833 | 0.163–4.270 | |
| 7/7 | 80 (47.61) | 17 (36.17) | 0.135 | 1.761 | 0.897–3.460 | |
| 7/4 | 2 (1.19) | 1 (2.12) | 0.525 | 0.554 | 0.049–6.249 | |
| 8/5 | 2 (1.19) | 0 | – | – | – | |
| 8/7 | 2 (1.19) | 1 (2.12) | 0.525 | 0.554 | 0.049–6.249 | |
| 9/5 | 4 (2.38) | 0 | – | – | – | |
| 9/6 | 0 | 0 | – | – | – | |
| 9/7 | 20 (11.90) | 3 (6.38) | 0.423 | 1.982 | 0.563–6.982 | |
| 9/9 | 0 | 0 | – | – | – | |
| Total | 168 | 47 |
Number in parentheses gives the data in percentage.
HSP HIV-1 seropositive, HSN HIV-1 seronegative, HES HIV-1-exposed seronegative
Table IV.
Allelic Frequency Distribution of DC-SIGNR in Different Groups
| Allele Frequency | HSP (%) | HSN (%) | HES (%) | P value | Odds ratio | 95% Confidence Interval |
|---|---|---|---|---|---|---|
| 4 | 2 (0.59) | 6 (1.14) | – | – | – | |
| 5 | 64 (19.04) | 128 (24.42) | 0.066 | 0.728 | 0.519–1.020 | |
| 6 | 12 (3.57) | 26 (4.96) | 0.397 | 0.709 | 0.353–1.426 | |
| 7 | 230 (68.4) | 334 (63.7) | 0.163 | 1.234 | 0.923–1.651 | |
| 8 | 4 (1.19) | 0 | – | – | – | |
| 9 | 24 (7.14) | 30 (5.72) | 0.472 | 1.267 | 0.727–2.207 | |
| Total no. of alleles | 336 | 524 | ||||
| 4 | 2 (0.59) | 1 (1.06) | – | – | – | |
| 5 | 64 (19.04) | 29 (30.85) | 0.016 | 0.527 | 0.315–0.883 | |
| 6 | 12 (3.57) | 3 (3.19) | 1.000 | 1.123 | 0.310–4.066 | |
| 7 | 230 (68.4) | 57 (60.63) | 0.173 | 1.408 | 0.877–2.261 | |
| 8 | 4 (1.19) | 1 (1.06) | 1.000 | 1.120 | 0.124–10.146 | |
| 9 | 24 (7.14) | 3 (3.19) | 0.229 | 2.333 | 0.687–7.925 | |
| Total no. of alleles | 336 | 94 |
Number in parentheses gives the data in percentage.
HSP HIV-1 seropositive, HSN HIV-1 seronegative, HES HIV-1-exposed seronegative
Fig. 2.
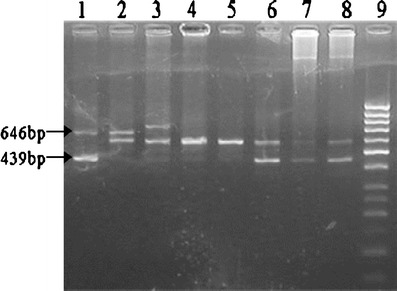
Agarose gel (3%) photograph of amplified DC-SIGNR exon 4 repeat region. Lane 1–8/5; Lane 2–;8/7; Lane 3–;9/7; Lane 4, 5–;7/7; Lane 6, 7, 8–;7/5; Lane 9–;Marker (50 bp DNA ladder, #SM1133 Fermentas).
When level of DC-SIGNR heterozygosity was compared between HSP individuals and HSN (95%CI 0.680–1.476; p = 1.000) and HES (95%CI 0.573–2.090; p = 0.869) individuals, no significant association for HIV-1 susceptibility have been found (Table II). No significant differences in the distribution of DC-SIGNR genotype frequencies were found when compared between HIV-1 seropositive, HIV-1 seronegative and HIV-1-exposed seronegative individuals after stratifying them on gender basis (data not shown).
Discussion
Humans demonstrate significant variability in their susceptibility to HIV-1 infection. Genetic epidemiological cohort studies have shown that polymorphism in the genes encoding chemokine receptor CCR5 (i.e., CCR5 delta 32) is associated with resistance against acquiring HIV infection in HIV-1-exposed seronegative individuals. Homozygous CCR5 delta 32 accounts for the resistance to HIV-1 infection in a small proportion of HIV-1-exposed seronegative individuals, whereas major part of the resistance conferred to these individuals is still unknown and to be elucidative. The existence of exposed yet uninfected individuals suggests that natural and acquired immunity to HIV exists and is a major determinant of clinical outcome. Variation in human genes having innate antiviral function may contribute to these alterations in gene expression or protein function. Understanding the effects of polymorphism in innate antiviral factor on HIV-1 susceptibility has provided essential insights in novel variants in DC-SIGN and DC-SIGNR exon 4 neck domain gene sequence. The DC-SIGN and DC-SIGNR lectins function as a trans-receptor for HIV-1. The exon 4 of these lectins comprises of variable number of 69 base pair tandem repeats encoding for parts of the extracellular neck domain. It has been shown that polymorphism in DC-SIGN and DC-SIGNR neck domain has significant effects in the transmission of HIV-1 infection. However, studies of genetic polymorphism effect on protection against HIV-1 infection have been less substantial.
In the present study, we analyzed the impact of the tandem repeat polymorphism in exon 4 of the DC-SIGN and DC-SIGNR gene with respect to the inter-individual transmission of HIV-1 infection. All HIV-1-exposed seronegative individuals who were found to be CCR5 delta 32 negative were enrolled to study the effect of DC-SIGN and DC-SIGNR exon 4 variants against HIV-1 susceptibility. We found no significant differences with regard to the overall DC-SIGN allele/genotype frequencies in a group of HIV-1 seropositive (HSP) compared to HIV-1 seronegative (HSN) and HIV-1-exposed seronegative (HES) individuals. Thus, the DC-SIGN genotype does not seem to influence the host’s susceptibility to the HIV-1 infection in North Indians. Whereas, Liu et al. [14] reported that heterozygous DC-SIGN reduced the risk of HIV-1 infection (3.2% in exposed seronegative individuals vs 0.0% in HIV-1 seropositive individuals; p = 0.011). A likely explanation for this discrepancy may be because of variation in number of sample studied.
The results of the present study suggest that individuals with the DC-SIGNR homozygous 5/5 genotype in the neck region have a reduced risk of HIV-1 infection (p = 0.009). Lichterfeld et al., in their comparison of 134 healthy volunteers and 391 HIV-1 seropositive individuals, did not detect any association between DC-SIGNR repeat-region polymorphism and HIV-1 transmission and clinical progression to AIDS [19]. The probable explanation for Lichterfeld’s failure to detect any association between DC-SIGNR repeat-region polymorphisms and HIV-1 transmission is that the study did not include HIV-1-exposed seronegative groups as also suggested by Liu et al. [15]. Impact of DC-SIGNR repeat-region polymorphisms on clinical progression of HIV-1 seropositive patients to AIDS could not be done in the present study, as patients could not be classified on the basis of number of years of known seroprevalence without antiretroviral therapy and viral load (data not available). Recently, Liu et al. [15] reported that DC-SIGNR homozygous 7/7 repeat was associated with an increased risk of HIV-1 infection (17.5% in high-risk HIV-1 seronegative individuals vs 28.5% in HIV-1 seropositive individuals; p = 0.0015), whereas the DC-SIGNR heterozygous 7/5 repeat tended to be correlated with resistance to HIV-1 infection (35.5% in high-risk HIV-1 seronegative individuals vs 27.6% in HIV-1 seropositive individuals; p = 0.0291). Whereas in the present study, although homozygous 7/7 individuals were more frequent in HIV-1 seropositive (47.61%) than HSN group (41.2%, p = 0.197) and HES (36.17%, p = 0.135), no significant association could be established. This suggests the need for larger number of individuals to be studied. We also did not find any significant differences in heterozygous 7/5 repeat between different group studies. The discrepancy between these results may be partially explained by the number of individuals included in the studies and/or by the relative impact of the different populations.
DC-SIGNR homozygous 7/7 repeat associated with an increased risk of HIV-1 infection [15] can be partially explained on the basis that extended neck regions (7/7 repeat) stabilizes tetramers of the DC-SIGNR, directing these receptors away from glycans on glycoprotein in the same cell membrane and toward pathogen surfaces [5] whereas in the present study, individuals having DC-SIGNR homozygous 5/5 repeat have reduced risk of HIV-1 infection can be justified as short DC-SIGNR receptors will not be easily accessible for HIV-1 binding [5]. Moreover, five repeat in the neck region can, although, form stable tetramers, but with the least affinity to target molecule HIV-1 gp120 [22, 23]. The affinity enhancement displayed towards oligosaccharide is due, in part, to multiple binding modes at the primary calcium ion site, which provide both additional contacts and statistical enhancement of binding [21]. Gramberg et al. [20] showed that DC-SIGNR alleles with five or six repeat units in the neck domain are expressed efficiently and augment viral infection similarly to wild DC-SIGNR which contains seven repeat units [20]. There might be indirect mechanism, which accounts for an association of DC-SIGNR homozygous 5/5 repeat genotype with reduced risk of HIV-1 infection in North Indian individuals. Nonetheless, the mechanism whereby DC-SIGNR repeat region polymorphisms affect HIV-1 transmission remains an area for future study.
Differential susceptibility to HIV infection in males and females with respect to DC-SIGNR neck repeat polymorphism have been recently reported [24], whereas no significant association was found when we stratified our data on gender basis likely due to differences in sample size of females studied and ethnicity.
To the best of our knowledge, the present study is the first to investigate the allelic distribution of the genetic variants DC-SIGN and DC-SIGNR in a group of North Indian HIV-1 seropositive, HIV-1 seronegative, and HIV-1-exposed seronegative. Although potential limitations of our study must be considered, our results showed a protective effect of DC-SIGNR homozygous 5/5 repeat region in the North Indian HIV-1-exposed seronegative population. This necessitates further molecular studies involving larger cohort sizes with longitudinal follow-up to confirm and fully describe the influence of DC-SIGN and DC-SIGNR host gene polymorphism on HIV-1 susceptibility, transmission, and disease progression in the North Indian population.
Acknowledgments
We thank Mr. Pradeep K. Singhal, Professional Biotech Pvt. Ltd., New Delhi for unwavering support and for insightful advice. We gratefully thank Mr. Santosh Pandey, Department of Microbiology, SGPGIMS, Lucknow for assistance in coordinating the clinical samples and all of the individuals who participated in the study. The Senior Research Fellowship provided by Indian Council of Medical Research (ICMR, New Delhi) to Mr. Anurag Rathore (first author) is greatly acknowledged.
References
- 1.O’Brien SJ, Nelson GW. Human genes that limit AIDS. Nat Genet. 2004;36(6):565–574. doi: 10.1038/ng1369. [DOI] [PubMed] [Google Scholar]
- 2.Hogan CM, Hammer SM. Host determinants in HIV infection and disease. Part 2: genetic factors and implications for antiretroviral therapeutics. Ann Intern Med. 2001;134(10):978–996. doi: 10.7326/0003-4819-134-10-200105150-00012. [DOI] [PubMed] [Google Scholar]
- 3.Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86(3):367–377. doi: 10.1016/S0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 4.Soilleux EJ, Barten R, Trowsdale J. DC-SIGN; a related gene, DC-SIGNR; and CD23 from a cluster on 19p13. J Immunol. 2000;165:2937–2942. doi: 10.4049/jimmunol.165.6.2937. [DOI] [PubMed] [Google Scholar]
- 5.Feinberg H, Mitchell DA, Drickamer K, Weis WI. Extended neck regions stabilize tetramers of the receptors DC-SIGN and DC-SIGNR. J Biol Chem. 2005;280:1327–1335. doi: 10.1074/jbc.M409925200. [DOI] [PubMed] [Google Scholar]
- 6.Geijtenbeek TB, Kwon DS, Torensma R, van Vliet SJ, van Duijnhoven GC, Middel J, et al. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–589. doi: 10.1016/S0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 7.Barreiro LB, Patin E, Nyrolles O, Cann HM, Gicquel B. The heritage of pathogen Pressure and Ancient Demography in the Human Innate Immunity CD 209/CD 209 L Region. Am J Hum Genet. 2005;77:869–886. doi: 10.1086/497613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchell DA, Fadden AJ, Drickamer K. A novel mechanism of carbohydrate recognition by the C-type lectins DC-SIGN and DC-SIGNR: subunit organization and building to multivalent ligands. J Biol Chem. 2001;276:28939–28945. doi: 10.1074/jbc.M104565200. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg H, Mitchell DA, Drickamer R, Weis WI. Structural basis for selective recognition of Oligosaccharide by DC-SIGNR. Science. 2001;294:2163–2166. doi: 10.1126/science.1066371. [DOI] [PubMed] [Google Scholar]
- 10.Guo Y, Feinberg H, Electrophoresis C, Mitchell DA, Alvarez R, Blixt O, et al. Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat Struct Mol Biol. 2004;11:591–598. doi: 10.1038/nsmb784. [DOI] [PubMed] [Google Scholar]
- 11.Soilleux EJ, Morris LS, Lee B, Pohlmann S, Trowsdale J, Doms RW, et al. Placental expression of DC-SIGN may mediate intrauterine vertical transmission of HIV. J Pathol. 2001;195:586–592. doi: 10.1002/path.1026. [DOI] [PubMed] [Google Scholar]
- 12.Alvarez CP, Lasala F, Carrillo J, Muniz O, Corbi AL, Delgado R. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J Virol. 2002;76:6841–6844. doi: 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bashirova AA, Geijtenbeek TB, Van Duijnhoven GC, Van Vliet SJ, Eilering JB, Martin MP, et al. A dendritic cell specific intracellular adhesion molecule 3 grabbing nonintegrin (DC-SIGN) related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV-1 infection. J Exp Med. 2001;193:671–678. doi: 10.1084/jem.193.6.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H, Hwangbo Y, Holte S, Lee J, Wang C, Kaupp N, et al. Analysis of genetic polymorphisms in CCR5, CCR2, stromal cell-derived factor-1, RANTES, and dendritic cell-specific intercellular adhesion molecular-3-grabbing nonintegrin in seronegative individual repeatedly exposed to HIV-1. J Infect Dis. 2004;190:1055–1058. doi: 10.1086/423209. [DOI] [PubMed] [Google Scholar]
- 15.Liu H, Carrington M, Wang C, Holte S, Lee J, Greene B, et al. Repeat-region polymorphisms in the gene for the dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin-related molecule: effects on HIV susceptibility. J Infect Dis. 2006;193:698–702. doi: 10.1086/499820. [DOI] [PubMed] [Google Scholar]
- 16.Chan VSF, Chan KYK, Chen Y, Poon LLM, et al. Homozygous L-SIGN (CLEC4M) plays a protective role in SARS Corona virus infection. Nat Genet. 2006;38:38–46. doi: 10.1038/ng1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lichterfeld M, Nischalke HD, van Lunzen J, et al. The tandem-repeat polymorphism of the DC-SIGNR gene does not affect the susceptibility to HIV infection and the progression to AIDS. Clin Immunol. 2003;107:55–59. doi: 10.1016/S1521-6616(02)00050-5. [DOI] [PubMed] [Google Scholar]
- 18.Pohlmann S, Zhang J, Baribaud F, Chen Z, et al. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J Virol. 2003;77:4070–4080. doi: 10.1128/JVI.77.7.4070-4080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simmons G, Reeves JD, Grogan CC, Vandenberghe LH, Baribaud F, Whitbeck JC, et al. DC-SIGN and DC-SIGNR bind Ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology. 2003;305:115–123. doi: 10.1006/viro.2002.1730. [DOI] [PubMed] [Google Scholar]
- 20.Gramberg T, Zhu T, Chaipan C, et al. Impact of polymorphisms in the DC-SIGNR neck domain on the interaction with pathogens. Virology. 2006;347:354–363. doi: 10.1016/j.virol.2005.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feinberg H, CAstelli R, Drickmer K, Seeberger PH, Weis WI. Multiple modes of binding enhance the affinity of DC-SIGN for high mannose N-linked glycans found on viral glycoproteins. J Biol Chem. 2006;286(6):4202–4209. doi: 10.1074/jbc.M609689200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snyder GA, Ford J, Torabi-Parizi P, et al. Characterization of DC-SIGN/R interaction with human immunodeficiency virus type 1 gp120 and ICAM molecules favors the receptor’s role as an antigen-capturing rather than an adhesion receptor. J Virol. 2005;79:4589–4598. doi: 10.1128/JVI.79.8.4589-4598.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y, Atkinson CE, Taylor ME, Drickmer K. All but the shortest polymorphic forms of the viral receptor DC-SIGNR assemble into homo- and heterotetramers. J Biol Chem. 2006;281:16794–16798. doi: 10.1074/jbc.M602430200. [DOI] [PubMed] [Google Scholar]
- 24.Wichukchinda N, Kitamura Y, Rojanawiwat A, Nakayama EE, Song H, et al. The polymorphism in DC-SIGNR affect susceptibility to HIV type 1 infection. AIDS Res Hum Retrovir. 2007;23:686–692. doi: 10.1089/aid.2006.0212. [DOI] [PubMed] [Google Scholar]


