Abstract
We compared nucleotide and deduced amino acid sequences of eight Japanese encephalitis virus (JEV) isolates derived from bats in China. We also compared the bat JEV isolates with other JEV isolates available from GenBank to determine their genetic similarity. We found a high genetic homogeneity among the bat JEVs isolated in different geographical areas from various bat species at different time periods. All eight bat JEV isolates belonged to genotype III. The mean evolutionary rate of bat JEV isolates was lower than those of isolates of other origin, but this difference was not statistically significant. Based on these results, we presume that the bat JEV isolates might be evolutionarily conserved. The eight bat JEV isolates were phylogenetically similar to mosquito BN19 and human Liyujie isolates of JEV. These results indicate that bats might be involved in natural cycle of JEV.
Keywords: West Nile Virus, Japanese Encephalitis Virus, Japanese Encephalitis, Amino Acid Mutation, Japanese Encephalitis Virus
Introduction
Japanese encephalitis (JE) is a severe zoonotic disease with a high fatality rate, ranging from 25 % to 50 % in humans. Nearly 50 % of survivors suffer from persistent neurological sequelae [22]. JE mainly occurs in China, India and Southeast Asia [12, 17, 35]. An estimated 67,900 cases occur globally each year (overall incidence: 1.8/100,000 people), of which only about 10 % are reported to the World Health Organization [4].
JE is caused by Japanese encephalitis virus (JEV), which belongs to the genus Flavivirus of the family Flaviviridae. JEV is an enveloped virus with a single-stranded, positive-sense RNA genome approximately 11 kb in length. The virus contains a single open reading frame (ORF) flanked by 5′ and 3′ nontranslated regions (NTRs). The 5′ one-third of the ORF encodes three structural proteins, named capsid protein (C), precursor membrane protein (PrM), and envelope protein (E), whereas seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) are encoded by the remaining 3′ region [38]. Five genotypes (GI -V) of JEV have been identified based on the nucleotide sequence of the envelope (E) gene [25, 42]. The predominant genotypes of JEV isolates exhibit geographical and temporal differences [6, 21, 29, 52]. GI includes isolates isolated in India, Southeast Asia, Australia, Korea and Japan from 1967 to the present, while GII includes isolates isolated from Korea, southern Thailand, Malaysia, Indonesia, Papua New Guinea and northern Australia between 1951 and 1999 [34]. In China, the dominant JEV isolates belong to GI and GIII. GIII JEV has been circulating in China since 1949, while GI began to replace GIII to become the dominant genotype in the 1980s [28, 47].
JEV can infect humans and a variety of vertebrate animals including pigs, horses, birds, sheep, dogs and monkeys [13, 28, 43]. However, only pigs and water birds are considered reservoirs of the virus [2, 22, 33, 43]. Bats are recognised as important reservoirs of a large number of zoonotic viruses [1, 3, 30, 45]. Some species of bats can maintain viruses for long periods of time [16, 37]. Previous studies have shown that JEV and/or serum antibody against JEV may exist in bats in Japan and China [10, 23, 28, 36, 48]. However, the role of bats in the JEV life cycle is unknown.
Limited information is currently available about bat-derived JEV isolates. The full-length nucleotide sequences of four JEV isolates (B58, GB30, HB49 and HN97) derived from bats in Yunnan Province, China, were determined [28, 48]. The B58 and GB30 isolates were isolated from a Rousettus leschenaultia bat in 1989 and a Murina aurata bat in 1997, respectively. The HB49 and HN97 isolates were isolated from a R. leschenaultia bat in 1990. These four bat JEV isolates belonged to GIII [28, 48].
In recent years, we collected four JEV isolates from bats captured in Guangdong, Hainan and Hunan provinces. The genetic relationship between the genomes of these bat-derived JEV isolates and the previously collected bat-derived isolates remains unknown. Here, we compared the genetic characteristics of eight bat JEV isolates and compared them to those of other original JEV isolates available from GenBank.
Materials and methods
Bats were sampled at four natural habitats in three regions in Guangdong, Hainan and Hunan provinces of southern China between July 2007 and August 2009. Bats were captured using mist nets at natural habitats of bats (e.g., caves or palm trees). The sampling method was followed as described previously [31]. Bat brain samples were taken in the laboratory, immediately placed into tubes containing 300 μl of RNAlater (QIAGEN, Hilden, Germany), and stored at −80 °C until used.
The supernatants from brain homogenates were used to inoculate baby hamster kidney (BHK-21) cells and were consecutively passed three times. The virus was isolated as described previously [48]. Four viruses were isolated and designated GD1, HN2, SY87 and YY158 isolates. Full-length genomic sequences were obtained from the GD1 and HN2 isolates, while only the sequence of the E gene was obtained for the SY87 and YY158 isolates. The GD1 isolate was obtained from a Myotis ricketti bat collected in Huizhou, Guangdong Province, in 2009, and the HN2 isolate was obtained from a Miniopterus schreibersii bat that was collected in Haikou, Hainan Province, in 2008. The SY87 isolate was obtained from a Rhinolophus affinis bat and the YY158 isolate was obtained from a M. schreibersi bat, both of which were collected in Yueyang, Hunan Province, in 2008.
A total of 105 full-length JEV genomic sequences were downloaded from GenBank, including four bat-derived JEV genomic sequences. Phylogenetic trees were constructed based on these 105 nucleotide sequences and two nucleotide sequences of JEV (GD1, HN2 isolates) determined in this study. Consequently, twenty-seven full-length genomic sequences of JEV isolates were selected from the phylogenetic tree based on region, isolation time, host and phylogenetic position. In addition, fourteen E gene sequences of JEV isolates were selected in the analyses, which included two sequences of the JEV E gene determined in this study. A total of 41 JEV isolates were used for constructing the phylogenetic trees, which contained isolates isolated from mosquitoes (n = 14), humans (n = 12), pigs (n = 3), vaccine (n = 2), midges (n = 2) and bats (n = 8) (Table 1). The JEV isolate that was first isolated from human brain in 1935 (Nakayama strain) was used as prototype strain in sequence comparisons.
Table 1.
Isolates of JEV used in the study
| Isolate name | Place of isolation | Year of isolation | Source | GenBank accession no. | Genotype |
|---|---|---|---|---|---|
| GD1† | China, Guangdong | 2009 | Bat | JN711458.1 | III |
| HN2† | China, Hainan | 2008 | Bat | JN711459.1 | III |
| SY87† | China Hunan | 2007 | Bat | JX050152.1* | III |
| YY158† | China Hunan | 2008 | Bat | JX093498.1 * | III |
| B58 | China, Yunnan | 1986 | Bat | FJ185036.1 | III |
| GB30 | China, Yunnan | 1997 | Bat | FJ185037.1 | III |
| HB49 | China, Yunnan | 1990 | Bat | JF706284.1 | III |
| HB97 | China, Yunnan | 1990 | Bat | JF706285.1 | III |
| Beijing-1 | China | 1949 | Human brain | L48961.1 | III |
| ChiangMai | Thailand | 1964 | Human | U70393.1* | III |
| Fj02-76 | China Fujian | 2002 | Human | JN381867.1 | III |
| FJ03-39 | China Fujian | 2003 | Human | JN381859.1 | III |
| Liyujie | China, Yunnan | 1979 | Human | FJ185039.1* | III |
| ML17-live | Taiwan | 1981 | Human | AY508812.1 | III |
| Nakayama | Japan | 1935 | Human brain | EF571853.1 | III |
| P3 | China | 1949 | Human brain | U47032.1 | III |
| P19-Br | Thailand | 1982 | Human | U70416.1* | I |
| Vellore | India | 1958 | Human brain | AF080251.1 | III |
| GP78 | India | 1978 | Human brain | AF075723.1 | III |
| 057434 | India | 2005 | Human | EF623988.1 | III |
| BN19 | China, Yunnan | 1982 | Mosquito | FJ185038.1* | III |
| DL04-45 | China Yunnan | 2004 | Mosquito | JN381854.1 | III |
| JaGAr01 | Japan | 1959 | Mosquito | AF069076.1 | III |
| JKT5441 | Indonesia | 1981 | Mosquito | U70406.1* | II |
| JKT7003 | Indonesia | 1981 | Mosquito | U70408.1* | IV |
| K94P05 | Korea | 1994 | Mosquito | AF045551.1 | I |
| K87P39 | South Korea | 1987 | Mosquito | AY585242.1 | III |
| M859 | Cambodia | 1967 | Mosquito | U70410.1* | I |
| SA14 | China | 1954 | Mosquito | U14163.1 | III |
| SH04-3 | China | 2004 | Mosquito | DQ404105.1* | III |
| VN118 | Vietnam | 1979 | Mosquito | U70420.1* | III |
| WTP-70-22 | Malaysia | 1970 | Mosquito | U70421.1* | II |
| YNJH04-18 | China | 2004 | Mosquito | DQ404146.1* | III |
| XZ0934 | China Tibet | 2009 | Mosquito | JF915894.1 | V |
| SA14-2-8 | China | NA | Vaccine | U15763.1 | III |
| SA14-14-2 | China | 1954 | Vaccine | AF315119.1 | III |
| B-2239 | Thailand | 1984 | Pig | U70391.1* | I |
| JEV/sw/Mie/40/2004 | Japan | 2004 | Swine | AB241118.1 | I |
| WHE | China | NA | Pig | EF107523.1 | III |
| YN83-Meng83-54 | China Yunnan | 1983 | Lasiohelea taiwana (Shiraki) | JF706282.1 | I |
| HLJ02-134 | China Heilongjiang | 2002 | Genus Culicoides | JF706276.1 | III |
NA, Not available in GenBank
† Isolates sequenced in this study
* Isolates for which E gene sequence information is available
Multiple sequence alignments were performed using MEGA 4.0 [40]. The percent identity within the nucleotide sequence alignment was determined using MegAlign (DNASTAR, Madison, WI, USA). Geneious 5.5.6 was used to show differences in the nucleotide and amino alignments.
Phylogenetic trees were constructed based on 41 E nucleotide sequences using the maximum-likelihood method in PHYLIP 3.9.6 [14]. The E gene of West Nile virus was used as an outgroup. In addition, the maximum-parsimony method in PHYLIP 3.9.6, the neighbor-joining method in Mega 4.0 [32], and the Bayesian method in BEAST 1.5.4 [11] were used in the analyses.
The rate of nucleotide substitutions per site was estimated using the Bayesian Markov chain Monte Carlo (MCMC) approach as implemented in the BEAST 1.5.4 package [11]. The analysis was performed by using the HKY substitution model under a coalescent model of constant population size. In each case, the relaxed molecular clock model was used. The resulting convergence was analyzed by using Tracer1.5. A 95 % high-probability density (HPD) was determined to ascertain the uncertainty in the parameter estimates.
Results
The eight bat JEV isolates (GD1, HN2, SY87, YY158, B58, GB30, HB49 and HB97) used were obtained at different times over two decades (1989-2009). Four of these isolates were isolated in Yunnan Province (Fig. 1), which has been a highly epidemic area for JE since the 1990s, with an average incidence of infection greater than 0.5/100,000 people [46, 48]. Two of the isolates were isolated in Hunan Province (Fig. 1), with an average incidence of infection between 0.2/100,000 and 0.5/100,000 people [46, 53]. The other two isolates were from Guangdong and Hainan Province (Fig. 1), respectively, which were once highly endemic areas for JE before the 1990s but are currently low-endemic areas with an incidence of less than 0.2/100,000 people annually [46, 53].
Fig. 1.
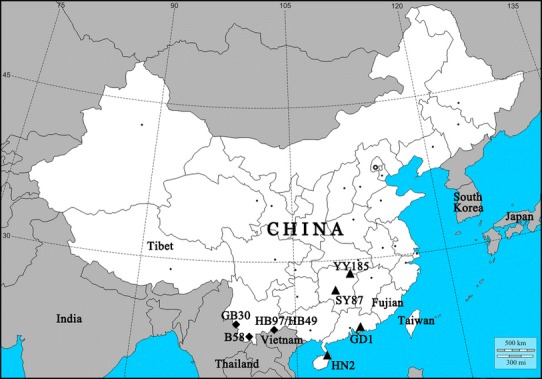
Geographic distribution of the eight bat JEV isolates in southern China. The filled triangles indicate locations of GD1, HN2, SY87 and YY158 (isolated in this study). The filled rhombuses indicate locations of GB30, B58, HB97 and HB49 (isolated in previous studies)
Nucleotide and amino acid sequence analysis
The six complete genomes of bat JEV isolates were analyzed, the length of which ranged from 10,975 to 10,977 nt. All of the isolates had a 95-nt 5′ nontranslated region (NTR). The HN2 isolate had a 581-nt 3′ NTR, while the other five isolates had a 582-nt 3′ NTR. The single ORF encoded a polyprotein of 3,432 amino acid residues, with the ATG start codon at 96-98 nt and the TAG stop codon at 10,392-10,394 nt. The genomes had similar guanine-cytosine content (51.42 % for the GD1 isolate, 51.41 % for the HN2 isolate, 51.44 % for the B58 isolate, 33.33 % for the GB30 isolate, 33.33 % for the HB49 isolate and 33.33 % for the HB97 isolate).
The diversity of the 27 full-length genomes and the 41 E genes at the nucleotide and amino acid level is shown in Table 2. The isolates generally shared high nucleotide and amino acid sequence identity. The amino acid sequence identities were higher than the corresponding nucleotide sequence identities. The full-length nucleotide sequences of bat JEV isolates shared identities from 99.4 % to 99.9 %, and the E gene sequences shared identities from 99.2 % to 99.9 %. However, all of isolates of full-length nucleotide sequences shared identities from 79.4 % to 99.9 %, and the identities of the E gene sequences ranged from 77.4 % to 99.9 %. When the comparison was restricted to the same isolation time period (1986 to 2009), there was 97.0-99.1 % identity in the nucleotide sequences and 96.7-99.7 % identity in the E gene sequences of JEVs isolated from humans. There was 79.6-97.2 % and 77.9-97.3 % identity in mosquito JEVs in the genomic and E gene sequence, respectively. The gene sequence homology of JEV isolates from bats was higher than those from other hosts (Table 2). When the comparison was restricted to GIII, the genetic homogeneity in the bat JEV isolates was likely higher than in those derived from humans and mosquitoes (data not shown).
Table 2.
Comparisons of nucleotide and amino acid sequence diversities among the JEV isolates isolated from the same source groups
| Isolates | Full nucleotide sequence | Full amino acid sequence | Isolates | E gene | E protein |
|---|---|---|---|---|---|
|
Bat isolates (n = 6) |
99.4 %-99.9 % | 99.4 %-99.9 % |
Bat isolates (n = 8) |
99.2 %-99.9 % | 99.0 %-99.9 % |
|
Mosquito isolates (n = 6) |
79.7 %-98.8 % | 91.5 %-99.7 % |
Mosquito isolates (n = 14) |
77.4 %-98.9 % | 89.8 %-99.8 % |
|
Human isolates (n = 9) |
95.7 %- 99.4 % | 98.6 %-99.9 % |
Human isolates (n = 12) |
87.3 %-99.9 % | 97.0 %-99.8% |
|
Pig isolates (n = 2) |
89.2 % | 98.1 % |
Pig isolates (n = 3) |
87.0 %-93.9 % | 96.4 %-98.4 % |
|
Midge isolates (n = 2) |
88.4 % | 97.7 % |
Midges isolates (n = 2) |
88.5% | 97.8 % |
|
All isolates (n = 25) |
79.4 %- 99.9 % | 91.1%- 99.9 % |
All isolates (n = 37) |
77.4 %-99.9 % | 89.6 %- 99.9 % |
We compared six bat JEV isolates (GD1, HN2, B58, GB30, HB49 and HB97) with the Nakayama strain on the basis of UTR variation (Table 3). The six bat JEVs shared the same nucleotide changes (C14 → T14 and T49 → C49) in the 5′ NTR (Table 3). Two nucleotide changes were found in the 3′ NTR of the bat isolates: G10434 → A10434 and G10448 → A10448 (Table 3). Two other nucleotide differences in the 3′ NTR of GD1 and HN2 were also revealed (Table 3). In addition, one nucleotide was absent in the 3′ NTR of the GD1 isolate, and two nucleotides were absent in that of the HN2 isolate (Table 3).
Table 3.
Comparison of nucleotide differences in the 3′ nontranslated region (NTR) and 5′ nontranslated region (NTR) between the JEV isolates isolated from bats and the Nakayama strain
| Isolate (origin) | 5′NTR | 3′NTR | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14 | 18 | 49 | 73 | 10408 | 10410 | 10434 | 10448 | 10496 | 10550 | 10554 | 10757 | 10802 | 10953 | 10977 | |
| GD1 (Bat) | T | G | C | G | G | G | A | A | A | A | A | G | § | G | T |
| HN2 (Bat) | T | G | C | G | G | G | A | A | A | A | A | § | § | G | C |
| GB30 (Bat) | T | G | C | G | A | A | A | A | A | A | A | G | C | T | T |
| B58 (Bat) | T | G | C | G | A | A | A | A | A | A | A | G | C | T | T |
| HB49 (Bat) | T | A | C | G | A | A | A | A | T | A | A | G | C | T | T |
| HB97 (Bat) | T | A | C | T | A | A | A | A | A | G | A | G | C | T | T |
| Nakayama (Human) | C | A | T | G | A | G | G | G | A | A | G | G | C | G | T |
§ Nucleotide sequence is missing
Differences were found in the comparisons of complete sequences between six bat-derived JEV isolates (GD1, HN2, B58, GB30, HB49 and HB97 isolates) and strain Nakayama (Fig. 2). The amino acid substitutions appeared in PrM/M, E, NS1, NS2A, NS2B, NS3, NS4A and NS5 (Table 4). The E protein from six of the bat isolates had 10 unique amino acid mutations. Fewer substitutions were observed in other proteins (Table 4). The six bat isolates had four identical amino acid mutations, which were in the E protein (E-83, K377 → E377; E-176, T470 → I470; E-290, R584 → K584) and the NS1 protein (NS1-8, A802 → I802) (Table 4).
Fig. 2.

Comparisons of genome and amino acid sequences of six bat JEV isolates (GD1, HN2, B58, GB30, HB49 and HB97) with the Nakayama (Nak) reference sequence. Single vertical black lines in the first line for each viral isolate indicate single nucleotide differences. Single vertical black lines in the second line for each viral isolate indicate single amino acid differences. Wider black boxes indicate larger regions of sequence differences, including areas of absent sequence. Dashes indicate gaps in sequence alignments
Table 4.
Amino acid sequence differences among bat JEV isolates and the Nakayama strain
| Protein | Amino acid position |
Isolates | ||||||
|---|---|---|---|---|---|---|---|---|
| GD1 | HN2 | GB30 | B58 | HB49 | HB97 | Nakayama | ||
| PreM/M | ||||||||
| 158 (M-31) | G | G | E | E | E | E | E | |
| 278 (M-151) | Q | Q | Q | H | Q | Q | Q | |
| E | ||||||||
| 377 (E-83) | E | E | E | E | E | E | K | |
| 417 (E-123) | S | S | R | R | R | R | S | |
| 440 (E-146) | T | T | A | A | A | A | T | |
| 470 (E-176) | I | I | I | I | I | I | T | |
| 503 (E-209) | R | R | K | K | K | K | - | |
| 570 (E-276) | N | N | S | S | S | S | N | |
| 584 (E-290) | K | K | K | K | K | K | R | |
| 712 (E-418) | A | A | V | V | V | V | A | |
| 747 (E-453) | F | F | F | F | L | F | F | |
| 759 (E-465) | G | G | G | G | A | G | G | |
| NS1 | ||||||||
| 802 (NS1-8) | I | I | I | I | I | I | A | |
| 971 (NS1-177) | D | G | D | D | D | D | D | |
| 1004 (NS1-210) | W | W | R | W | W | W | W | |
| 1014 (NS1-220) | I | I | V | V | V | V | V | |
| NS2A | ||||||||
| 1252 (NS2A-43) | V | V | A | V | V | V | V | |
| 1281 (NS2A-72) | A | A | A | A | T | A | A | |
| 1332 (NS2A-123) | K | K | R | R | R | R | R | |
| NS2B | ||||||||
| 1409 (NS2B-36) | A | A | A | A | E | A | A | |
| NS3 | ||||||||
| 1562 (NS3-58) | I | I | V | V | V | I | I | |
| 1988 (NS3-484) | G | G | S | S | S | S | S | |
| 2005 (NS3-501) | L | L | M | M | M | M | M | |
| 2022 (NS3-518) | F | F | S | S | S | S | L | |
| NS4A | ||||||||
| 2162 (NS4A-40) | T | T | A | A | A | A | A | |
| 2269 (NS4A-147) | V | V | V | V | V | V | M | |
| 2383 (NS4A-261) | F | F | F | F | F | F | V | |
| NS5 | ||||||||
| 2704 (NS5-177) | R | R | I | R | I | I | R | |
| 2865 (NS5-338) | V | V | A | A | A | A | A | |
| 2932 (NS5-405) | V | V | V | V | V | V | - | |
| 3036 (NS5-509) | V | V | V | V | G | V | V | |
| 3048 (NS5-521) | L | L | I | L | L | L | L | |
| 3292 (NS5-765) | - | Q | Q | Q | Q | Q | Q | |
| 3426 (NS5-899) | T | T | I | I | I | I | I | |
- Nucleotide sequence is missing
Phylogenetic analysis
Five genotypes were distinguished based on the E gene nucleotide sequences of the 41 selected JEV isolates (Fig. 3), which is consistent with the classification made by Chen and colleagues [6, 7]. The phylogenetic analysis demonstrated that all bat JEV isolates belonged to GIII. The isolates of GD1, HN2, SY87 and YY158 belonged to the same subgroup (Fig. 3). In addition, these isolates were similar to the GB30, B58, HB49 and HB97 isolates (Fig. 3). Notably, the BN19 isolate, which was isolated from a mosquito in Yunnan Province, China, in 1982 and the Liyujie isolate, which was obtained from a human in Yunnan Province, China, in 1979, were most closely related to the GD1 and HN2 isolates. Similar trees were produced by the neighbor-joining, maximum-parsimony, and Bayesian methods.
Fig. 3.
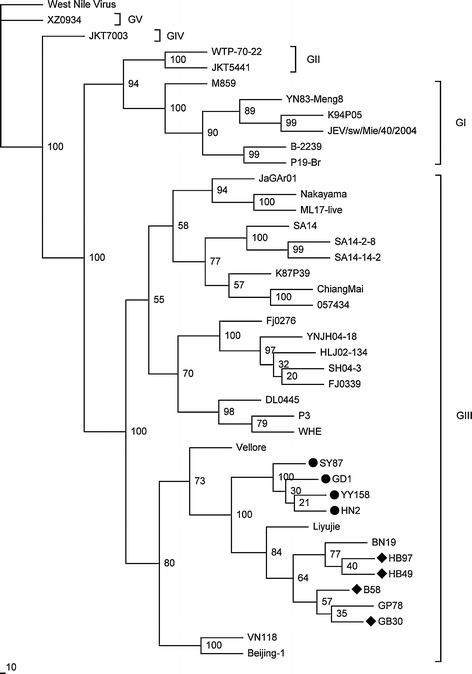
Phylogenetic tree generated based on the envelope (E) gene sequence using the maximum-likelihood method. Numbers above or below branches indicate neighbour-joining bootstrap values. West Nile virus was used as an outgroup. Genotypes are indicated on the right. The four bat JEV isolates sequenced in the present study are indicated with a circle on the left, and four other previously reported bat JEV isolates are indicated with a rhombus on the left. The scale bar indicates the number of nucleotide substitutions per site
Evolutionary analysis of eight E genes for bat JEV isolates showed that the mean evolutionary rate of bat JEV isolates was 1.44 × 10−4 (95 %HPD = 2.33 × 10−7 to 4.41 × 10−4) nucleotide substitutions per site per year. The mean evolutionary rate previously reported from an analysis of 35 full-length genomes derived from humans, pigs and mosquitoes was 4.35 × 10−4 (95 %HPD = 3.49 × 10−4 to 5.30 × 10−4) nucleotides substitutions per site per year [24].
Discussion
Bats are known to be reservoir hosts for many zoonotic viruses, such as SARS-coronavirus-like viruses of bats [19], Hendra virus [15] and Ebola virus [41]. JEV was isolated from naturally infected Miniopterus schreibersii [3]. There are some features of bats that might help explain the detection of JEV in bats. R. Affinis, M. ricketti and M. schreibersii can migrate hundreds of miles to their hibernation sites. Thus, bats have more opportunities to come into contact with humans or other animals at different geographical locations, which make it possible for interspecies transmission. Secondly, R. leschenaultia and M. schreibersii exhibit an exceptionally long lifespan, ranging up to 14 years. The long lifespan of bats may enhance the persistence of chronic infections [50]. In addition, some bat species also hibernate over the winter [49]. Sulkin et al. [37] found that infectious JEV was recovered from seropositive bats fifteen weeks after a shift in temperature. The reduced body temperature and metabolic rate may suppress immune responses and reduce the rate of virus replication, and therefore, JEV could persist for extensive periods without evidence of disease [37].
There are currently approximately 105 fully sequenced JEV isolates available from different hosts [28, 48]. Genetic variation has been reported among JEV isolates isolated from widely different time periods and geographical locations [6, 21, 28]. In the present study, we selected JEV isolates with genetic information available from GenBank based on their genotype, time period, geographic region and host from which they were isolated, and we used these reference isolates to compare the genetic variation of the eight bat-derived JEV isolates from China between 1986 and 2009. The isolates showed identities from 79.4 % to 99.9 % at the nucleotide level and identities from 91.1 % to 99.9 % at the amino acid level. Most of the differences were base substitutions and nucleotide changes that did not result in amino acid alterations (Fig. 2), which is consistent with previous findings [6]. The results indicate that most of the nucleotide mutations in the bat JEV isolates are silent.
Notably, the bat JEV isolates showed 99.4-99.9 % genetic homogeneity in the full-length nucleotide sequences and 99.2-99.9 % genetic homogeneity in the E gene sequences, which were higher than those from other hosts (Table 2). Also, the results of evolutionary analysis showed that bat JEV isolates probably had slower evolutionary rates than other original JEV isolates. The mean evolutionary rates of bat JEV isolates tended to be lower than those of isolates of other origin, but there was no statistically significant difference. This suggests that JEVs from bats might be more phylogenetically conserved than isolates from humans, swine and mosquitoes (Table 2). Moreover, according to the phylogenetic analysis, the GD1, HN2, SY87 and YY158 isolates were most closely related to the other four bat JEV isolates (B58, GB30, HB49 and HB97), showing a relatively high bootstrap value (Fig. 3). Eight bat JEV isolates were clustered into the same subgroup, although they were isolated from different bat species within separate regions and were originally isolated over the span of more than two decades. The reason for this phenomenon is unclear. It may be attributed to the host preference, with GIII JEVs having adapted to bats. Even though Van den Hurk et al. [44] performed laboratory-based infections on Pteropus alecto (Megachiroptera: Pteropididae) with JEV TS3306 (GII), there was no evidence that bats could harbor other JEV strains in nature except those belonging to GIII. It is unknown whether the other three genotypes of JEV circulate in bats in nature. In this study, the bats from which JEV isolates were isolated looked healthy, suggesting that the virus is not pathogenic to bats. Since sufficient nucleotide sequence information was not available about human or other host origins in the regions where bat JEVs were isolated, we could not determine the relationships between bat JEV isolates and isolates from other host origins in local areas. Further studies are needed to explore the role of bats in the natural cycle of JEV.
However, it is worth noting that the human Liyujie isolate and the mosquito-derived BN19 isolate from Yunnan Province in China were closely related to the six bat isolates (Fig. 3), with high amino acid similarities of 99.0 % to 99.6 % and 98.8 % to 99.8 %, respectively. This indicates that a relationship might exist between humans, mosquitoes and bats within the JEV transmission cycle.
Although no amino acid mutations were identified that were previously associated with viral phenotype alternation in the Nakayama isolate, which was isolated from a Japanese patient in 1935 (99.4 % to 99.9 % at the nucleotide level, 99.4 % to 99.5 % at the amino acid level, respectively), the comparison of the deduced amino acid sequences of bat JEV isolates and the Nakayama strain demonstrated that there were some variations in amino acid sequences (Fig. 2, Table 4). Amino acid changes in critical determinants of the viral proteins could cause alterations in viral propagation, virulence and neuroinvasion [5, 8, 9, 18, 20, 26, 27, 39, 51]. Even though no amino acid mutations were identified at key positions (E-102 [51], E-138 [39], E-300 [26], E-306 [52], E-308 [26], E-395 [26], C-52 [8, 18], C-109 [8], C-122 [8], NS3-109 [8] and NS3-122 [8]), we cannot exclude the possibility that a combination of these unique amino acids together would contribute to neurovirulence. The biological significance of the differences in amino acid sequences between the bat JEVs and other JEV isolates requires further study.
In conclusion, our study showed that eight bat JEV isolates (GD1, HN2, SY87, YY158, B58, GB30, HB49 and HB97) belonged to GIII of JEV and shared a high degree of genetic identity. We presume that bat JEV isolates might be more evolutionarily conserved than other original JEV isolates. In consideration of the bat JEV isolates being phylogenetically similar to the mosquito isolate (BN19) and the human isolate (Liyujie) from China and the Nakayama strain, bats might be involved in the JEV cycle in nature. However, we could not conclude whether bats are the hosts for JEV or are occasionally infected by JEV based on current evidence. Further virological and molecular epidemiologic studies of the bat JEVs are still needed.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 30972525), the National Natural Science Foundation of Guangdong Province (Grant No. 8151051501000056, 8151009101000005), the Natural Science Foundation of Hainan Province (Grant No. 311049), and the Guangdong Province 211 Project.
Footnotes
The GenBank ID: GD1, HN2, SY87 and YY158 JEV isolates nucleotide sequence are JN711458.1, JN711459.1, JX050152.1 and JX093498.1, respectively.
References
- 1.Banyard AC, Hayman D, Johnson N, McElhinney L, Fooks AR. Bats and lyssaviruses. Adv Virus Res. 2011;79:239–289. doi: 10.1016/B978-0-12-387040-7.00012-3. [DOI] [PubMed] [Google Scholar]
- 2.Buescher EL, Scherer WF, Mc CH, Moyer JT, Rosenberg MZ, Yoshii M, Okada Y. Ecologic studies of Japanese encephalitis virus in Japan. IV. Avian infection. Am J Trop Med Hyg. 1959;8:678–688. doi: 10.4269/ajtmh.1959.8.678. [DOI] [PubMed] [Google Scholar]
- 3.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell GL, Hills SL, Fischer M, Jacobson JA, Hoke CH, Hombach JM, Marfin AA, Solomon T, Tsai TF, Tsu VD, Ginsburg AS. Estimated global incidence of Japanese encephalitis: a systematic review. Bull World Health Organ. 2011;89:766–774. doi: 10.2471/BLT.10.085233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao JX, Ni H, Wills MR, Campbell GA, Sil BK, Ryman KD, Kitchen I, Barrett AD. Passage of Japanese encephalitis virus in HeLa cells results in attenuation of virulence in mice. J Gen Virol. 1995;76(Pt 11):2757–2764. doi: 10.1099/0022-1317-76-11-2757. [DOI] [PubMed] [Google Scholar]
- 6.Chen WR, Tesh RB, Rico-Hesse R. Genetic variation of Japanese encephalitis virus in nature. J Gen Virol. 1990;71(Pt 12):2915–2922. doi: 10.1099/0022-1317-71-12-2915. [DOI] [PubMed] [Google Scholar]
- 7.Chen WR, Rico-Hesse R, Tesh RB. A new genotype of Japanese encephalitis virus from Indonesia. Am J Trop Med Hyg. 1992;47:61–69. doi: 10.4269/ajtmh.1992.47.61. [DOI] [PubMed] [Google Scholar]
- 8.Chiou SS, Chen WJ. Mutations in the NS3 gene and 3’-NCR of Japanese encephalitis virus isolated from an unconventional ecosystem and implications for natural attenuation of the virus. Virology. 2001;289:129–136. doi: 10.1006/viro.2001.1033. [DOI] [PubMed] [Google Scholar]
- 9.Chung YJ, Nam JH, Ban SJ, Cho HW. Antigenic and genetic analysis of Japanese encephalitis viruses isolated from Korea. Am J Trop Med Hyg. 1996;55:91–97. doi: 10.4269/ajtmh.1996.55.91. [DOI] [PubMed] [Google Scholar]
- 10.Cui J, Counor D, Shen D, Sun G, He H, Deubel V, Zhang S. Detection of Japanese encephalitis virus antibodies in bats in Southern China. Am J Trop Med Hyg. 2008;78:1007–1011. [PubMed] [Google Scholar]
- 11.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolut Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erlanger TE, Weiss S, Keiser J, Utzinger J, Wiedenmayer K. Past, present, and future of Japanese encephalitis. Emerg Infect Dis. 2009;15:1–7. doi: 10.3201/eid1501.080311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan JM, Luo J, Chen L, Teng M, Bu D, Wang FY, Wang L, Wang CQ, Zhang GP. Genetic analysis of strains of Japanese encephalitis virus isolated from swine in central China. Virus Genes. 2010;40:357–361. doi: 10.1007/s11262-010-0464-9. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein J. Accuracy of coalescent likelihood estimates: do we need more sites, more sequences, or more loci? Mol Biol Evol. 2006;23:691–700. doi: 10.1093/molbev/msj079. [DOI] [PubMed] [Google Scholar]
- 15.Field H, de Jong C, Melville D, Smith C, Smith I, Broos A, Kung YH, McLaughlin A, Zeddeman A. Hendra virus infection dynamics in Australian fruit bats. PloS one. 2011;6:e28678. doi: 10.1371/journal.pone.0028678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.George DB, Webb CT, Farnsworth ML, O’Shea TJ, Bowen RA, Smith DL, Stanley TR, Ellison LE, Rupprecht CE. Host and viral ecology determine bat rabies seasonality and maintenance. Proc Natl Acad Sci USA. 2011;108:10208–10213. doi: 10.1073/pnas.1010875108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh D, Basu A. Japanese encephalitis-a pathological and clinical perspective. PLoS Negl Trop Dis. 2009;3:e437. doi: 10.1371/journal.pntd.0000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasegawa H, Yoshida M, Shiosaka T, Fujita S, Kobayashi Y. Mutations in the envelope protein of Japanese encephalitis virus affect entry into cultured cells and virulence in mice. Virology. 1992;191:158–165. doi: 10.1016/0042-6822(92)90177-Q. [DOI] [PubMed] [Google Scholar]
- 19.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S, Wang L-F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 20.Liu JJ, Tsai TH, Chang TJ, Wong ML. Cloning and sequencing of complete cDNA of Japanese encephalitis virus YL strain in Taiwan. Virus Genes. 2003;26:193–198. doi: 10.1023/A:1023443631659. [DOI] [PubMed] [Google Scholar]
- 21.Mangada MN, Takegami T. Molecular characterization of the Japanese encephalitis virus representative immunotype strain JaGAr 01. Virus Res. 1999;59:101–112. doi: 10.1016/S0168-1702(98)00130-0. [DOI] [PubMed] [Google Scholar]
- 22.Misra UK, Kalita J. Overview: Japanese encephalitis. Prog Neurobiol. 2010;91:108–120. doi: 10.1016/j.pneurobio.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Miura T, Kitaoka M. Viruses isolated from bats in Japan. Arch Virol. 1977;53:281–286. doi: 10.1007/BF01315626. [DOI] [PubMed] [Google Scholar]
- 24.Mohammed MA, Galbraith SE, Radford AD, Dove W, Takasaki T, Kurane I, Solomon T. Molecular phylogenetic and evolutionary analyses of Muar strain of Japanese encephalitis virus reveal it is the missing fifth genotype. Infect Genet Evol: J Mol Epidemiol Evol Genet Infect Dis. 2011;11:855–862. doi: 10.1016/j.meegid.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 25.Nga PT, del Carmen Parquet M, Cuong VD, Ma SP, Hasebe F, Inoue S, Makino Y, Takagi M, Nam VS, Morita K. Shift in Japanese encephalitis virus (JEV) genotype circulating in northern Vietnam: implications for frequent introductions of JEV from Southeast Asia to East Asia. J Gen Virol. 2004;85:1625–1631. doi: 10.1099/vir.0.79797-0. [DOI] [PubMed] [Google Scholar]
- 26.Ni H, Barrett AD. Molecular differences between wild-type Japanese encephalitis virus strains of high and low mouse neuroinvasiveness. J Gen Virol. 1996;77(Pt 7):1449–1455. doi: 10.1099/0022-1317-77-7-1449. [DOI] [PubMed] [Google Scholar]
- 27.Nitayaphan S, Grant JA, Chang GJ, Trent DW. Nucleotide sequence of the virulent SA-14 strain of Japanese encephalitis virus and its attenuated vaccine derivative, SA-14-14-2. Virology. 1990;177:541–552. doi: 10.1016/0042-6822(90)90519-W. [DOI] [PubMed] [Google Scholar]
- 28.Pan XL, Liu H, Wang HY, Fu SH, Liu HZ, Zhang HL, Li MH, Gao XY, Wang JL, Sun XH, Lu XJ, Zhai YG, Meng WS, He Y, Wang HQ, Han N, Wei B, Wu YG, Feng Y, Yang DJ, Wang LH, Tang Q, Xia G, Kurane I, Rayner S, Liang GD. Emergence of genotype I of Japanese encephalitis virus as the dominant genotype in Asia. J Virol. 2011;85:9847–9853. doi: 10.1128/JVI.00825-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paranjpe S, Banerjee K. Phylogenetic analysis of the envelope gene of Japanese encephalitis virus. Virus Res. 1996;42:107–117. doi: 10.1016/0168-1702(96)01306-8. [DOI] [PubMed] [Google Scholar]
- 30.Paterson BJ, Mackenzie JS, Durrheim DN, Smith D. A review of the epidemiology and surveillance of viral zoonotic encephalitis and the impact on human health in Australia. N S W Public Health Bull. 2011;22:99–104. doi: 10.1071/NB10076. [DOI] [PubMed] [Google Scholar]
- 31.Poon LL, Chu DK, Chan KH, Wong OK, Ellis TM, Leung YH, Lau SK, Woo PC, Suen KY, Yuen KY, Guan Y, Peiris JS. Identification of a novel coronavirus in bats. J Virol. 2005;79:2001–2009. doi: 10.1128/JVI.79.4.2001-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 33.Scherer WF, Moyer JT, Izumi T, Gresser I, Mc CJ. Ecologic studies of Japanese encephalitis virus in Japan. VI. Swine infection. The American journal of tropical medicine and hygiene. 1959;8:698–706. doi: 10.4269/ajtmh.1959.8.698. [DOI] [PubMed] [Google Scholar]
- 34.Schuh AJ, Li L, Tesh RB, Innis BL, Barrett AD. Genetic characterization of early isolates of Japanese encephalitis virus: genotype II has been circulating since at least 1951. J Gen Virol. 2010;91:95–102. doi: 10.1099/vir.0.013631-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Solomon T. Control of Japanese encephalitis—within our grasp? N Engl J Med. 2006;355:869–871. doi: 10.1056/NEJMp058263. [DOI] [PubMed] [Google Scholar]
- 36.Sulkin SE, Allen R, Miura T, Toyokawa K. Studies of arthropod-borne virus infections in chiroptera. VI. Isolation of Japanese B encephalitis virus from naturally infected bats. Am J Trop Med Hyg. 1970;19:77–87. doi: 10.4269/ajtmh.1970.19.77. [DOI] [PubMed] [Google Scholar]
- 37.Sulkin SE, Allen R. Virus infections in bats. Monogr Virol. 1974;8:1–103. [PubMed] [Google Scholar]
- 38.Sumiyoshi H, Mori C, Fuke I, Morita K, Kuhara S, Kondou J, Kikuchi Y, Nagamatu H, Igarashi A. Complete nucleotide sequence of the Japanese encephalitis virus genome RNA. Virology. 1987;161:497–510. doi: 10.1016/0042-6822(87)90144-9. [DOI] [PubMed] [Google Scholar]
- 39.Sumiyoshi H, Tignor GH, Shope RE. Characterization of a highly attenuated Japanese encephalitis virus generated from molecularly cloned cDNA. J Infect Dis. 1995;171:1144–1151. doi: 10.1093/infdis/171.5.1144. [DOI] [PubMed] [Google Scholar]
- 40.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 41.Taniguchi S, Watanabe S, Masangkay JS, Omatsu T, Ikegami T, Alviola P, Ueda N, Iha K, Fujii H, Ishii Y, Mizutani T, Fukushi S, Saijo M, Kurane I, Kyuwa S, Akashi H, Yoshikawa Y, Morikawa S. Reston Ebolavirus antibodies in bats, the Philippines. Emerg Infect Dis. 2011;17:1559–1560. doi: 10.3201/eid1708.101693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsarev SA, Sanders ML, Vaughn DW, Innis BL. Phylogenetic analysis suggests only one serotype of Japanese encephalitis virus. Vaccine. 2000;18(Suppl 2):36–43. doi: 10.1016/S0264-410X(00)00039-6. [DOI] [PubMed] [Google Scholar]
- 43.Misra UK, Kalita J. Overview: Japanese encephalitis. Prog Neurobiol. 2010;91:108–120. doi: 10.1016/j.pneurobio.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 44.van den Hurk AF, Smith CS, Field HE, Smith IL, Northill JA, Taylor CT, Jansen CC, Smith GA, Mackenzie JS. Transmission of Japanese Encephalitis virus from the black flying fox, Pteropus alecto, to Culex annulirostris mosquitoes, despite the absence of detectable viremia. Am J Trop Med Hyg. 2009;81:457–462. [PubMed] [Google Scholar]
- 45.Wacharapluesadee S, Boongird K, Wanghongsa S, Ratanasetyuth N, Supavonwong P, Saengsen D, Gongal GN, Hemachudha T. A longitudinal study of the prevalence of Nipah virus in Pteropus lylei bats in Thailand: evidence for seasonal preference in disease transmission. Vector Borne Zoonotic Dis. 2010;10:183–190. doi: 10.1089/vbz.2008.0105. [DOI] [PubMed] [Google Scholar]
- 46.Wang H, Li Y, Liang X, Liang G. Japanese encephalitis in mainland china. Jpn J Infect Dis. 2009;62:331–336. [PubMed] [Google Scholar]
- 47.Wang HY, Takasaki T, Fu SH, Sun XH, Zhang HL, Wang ZX, Hao ZY, Zhang JK, Tang Q, Kotaki A, Tajima S, Liang XF, Yang WZ, Kurane I, Liang GD. Molecular epidemiological analysis of Japanese encephalitis virus in China. J Gen Virol. 2007;88:885–894. doi: 10.1099/vir.0.82185-0. [DOI] [PubMed] [Google Scholar]
- 48.Wang JL, Pan XL, Zhang HL, Fu SH, Wang HY, Tang Q, Wang LF, Liang GD. Japanese encephalitis viruses from bats in Yunnan, China. Emerg Infect Dis. 2009;15:939–942. doi: 10.3201/eid1506.081525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang LF, Walker PJ, Poon LL. Mass extinctions, biodiversity and mitochondrial function: are bats ‘special’ as reservoirs for emerging viruses? Curr Opin Virol. 2011;1:649–657. doi: 10.1016/j.coviro.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilkinson GS, South JM. Life history, ecology and longevity in bats. Aging Cell. 2002;1:124–131. doi: 10.1046/j.1474-9728.2002.00020.x. [DOI] [PubMed] [Google Scholar]
- 51.Williams DT, Wang LF, Daniels PW, Mackenzie JS. Molecular characterization of the first Australian isolate of Japanese encephalitis virus, the FU strain. J Gen Virol. 2000;81:2471–2480. doi: 10.1099/0022-1317-81-10-2471. [DOI] [PubMed] [Google Scholar]
- 52.Zhang JS, Zhao QM, Zhang PH, Jia N, Cao WC. Genomic sequence of a Japanese encephalitis virus isolate from southern China. Arch Virol. 2009;154:1177–1180. doi: 10.1007/s00705-009-0421-x. [DOI] [PubMed] [Google Scholar]
- 53.Zheng Y, Li M, Wang H, Liang G. Japanese encephalitis and Japanese encephalitis virus in mainland China. Rev Med Virol. 2012;22:301–322. doi: 10.1002/rmv.1710. [DOI] [PubMed] [Google Scholar]


