Abstract
Canine enteric coronaviruses (CCoVs) are important enteric pathogens of dogs. CCoVs with different variations are typically pantropic and pathogenic in dogs. In this study, we isolated a CCoV, designated HLJ-073, from a dead 6-week-old male Pekingese with gross lesions and diarrhea. Interestingly, sequence analysis suggested that HLJ-073 contained a 350-nt deletion in ORF3abc compared with reference CCoV isolates, resulting in the loss of portions of ORF3a and ORF3c and the complete loss of ORF3b. Phylogenetic analysis based on the S gene showed that HLJ-073 was more closely related to members of the FCoV II cluster than to members of the CCoV I or CCoV II cluster. Furthermore, recombination analysis suggested that HLJ-073 originated from the recombination of FCoV 79-1683 and CCoV A76, which were both isolated in the United States. Cell tropism experiments suggested that HLJ-073 could effectively replicate in canine macrophages/monocytes and human THP-1 cells. This is the first report of the isolation of strain HLJ-073 in China, and this virus has biological characteristics that are different from those of other reported CCoVs.
Electronic supplementary material
The online version of this article (10.1007/s00705-019-04296-9) contains supplementary material, which is available to authorized users.
Keywords: Canine coronavirus, ORF3abc, Recombination, Pantropic, Viral cell tropism
Introduction
Canine coronavirus (CCoV) is a member of virus family Coronaviridae, order Nidovirales. CCoV is single-stranded positive-sense RNA viruses with a genome that is between 28 and 31 kb in length [1]. Based on antigenic and genetic relationships, CCoV consists of two distinct genotypes, CCoV- I and CCoV- II [7, 14]. CCoV was first recognized as an enteric pathogen of dogs in 1971, when it was observed to cause clinical signs including anorexia, lethargy, vomiting, and mild-to-severe diarrhea [3]. The clinical signs usually lasted 1-2 weeks and were followed by recovery, or occasionally by dehydration that led to death, mainly in puppies [6].
In recent years, researchers have reported an increasing number of cases of virulent CCoV infection in dogs and the emergence of CCoV variants with recombinant S genes. Recombination occurs when different CCoV types infect the same host simultaneously [9, 16, 20, 22]. Therefore, CCoV has become a cause of concern as an emerging pathogen in dogs. In the present study, we report the emergence and molecular characterization of an, FCoV-like recombinant CCoV-HLJ-073, which was isolated from a fecal sample from a dead dog that exhibited enteritis.
Materials and methods
Clinical case
During the spring of 2015, a dead dog, a privately owned 6-week-old male Pekingese, was submitted for laboratory investigation. The clinical symptoms were lethargy, inappetence, fluid diarrhea, vomiting and dehydration with death after 2 days. Necropsy of the dog showed hemorrhagic enteritis and severe lesions in the liver, spleen and lung. Hemorrhages of the liver and spleen were observed on their surfaces. Multiple areas of congestion were found in the lung.
Isolation, identification, and purification of the virus
A fecal sample was collected and subjected to virological investigations. Rapid kits were employed to identify common canine viral pathogens, including canine distemper virus (CDV), canine parvovirus (CPV), canine adenovirus-1 (CAV-1), CAV-2 and CCoV (Bionote, Hwaseong-si, Gyeonggi-do, South Korea). The primers P-F and P-R (Table S1) were employed to confirm the above results [5].
The CRFK cell line was used for virus isolation. After three rounds of purification by plaque assay [23], the virus was propagated and titrated and then harvested by one cycle of freezing and thawing. Viral RNA was extracted from the original fecal sample and from CRFK cells infected with CCoV using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). Fifteen primer pairs were designed based on the conserved regions of CCoV strain TN-449 (Table S1). The RT-PCR assay was performed as described previously [13].
Electron microscopy
The protocol for electron microscopy was described previously [13].
Indirect immunofluorescence assay (IFA)
An indirect immunofluorescence assay (IFA) was conducted using a standard procedure. An anti-CCoV antibody (VMRD, WA, USA) and FITC-conjugated rabbit anti-dog IgG (Sigma, CA, USA) were used as primary and secondary antibodies, respectively. A CCoV-N polyclonal antibody was prepared as follows [15]: Briefly, the complete N gene was amplified using a forward primer (5’ TTTGGATCCATGGCCAACCAGGGACAACGC3’) and a reverse primer (5’ TTTGCGGCCGCTTAGTTCGTTACCTCATCAAT3’). Then, the products were cloned into the vector pGEX6p-1. Purified GST-N recombinant protein was used as an antigen to inject female BALB/c mice. After three immunizations, serum was collected and stored at -80 °C.
Quantitative real-time RT-PCR assay
RNA was extracted from fecal and organ samples using a previously established TaqMan-based real-time RT-PCR assay for rapid detection and quantification of CCoV RNA [10].
Detection of subgenomic (sg) ORF3abc mRNAs by RT-PCR
??The primers?? DF2F and ORF3CR (Table S1) [2] were used to detect sg mRNAs from the ORF3abc region by RT-PCR, using a One-Step RT-PCR Kit (abm, Richmond, BC, Canada).
Sequence alignment and phylogenetic analysis
A phylogenetic tree based on the sequences of the structural proteins was constructed using the neighbor-joining (NJ) method of molecular evolutionary genetics analysis (MEGA) software (version 4.0). Bootstrap values were estimated for 1,000 replicates. Simplot 3.5.1 was used for nucleotide sequence comparison of HLJ-073 to the reference FCoV strains and CCoV strains. The HLJ-073 sequence obtained in this study was assembled and submitted to the GenBank database under accession number KY063618.
Cell tropism experiments
Canine blood monocytes originating from specific-pathogen-free (SPF) dogs were isolated [2]. Phorbol 12-myristate 13-acetate (PMA) was purchased from Sigma-Aldrich (St. Louis, USA). The human monocytic cell line THP-1 was provided by the National Institute for Communicable Disease Control and Prevention, Chinese Centers for Disease Control and Prevention (Beijing, China). THP-1 cells were induced to differentiate into a mature-macrophage-like state by stimulation with 50 ng of PMA per mL for 48 h before treatment.
Results and discussion
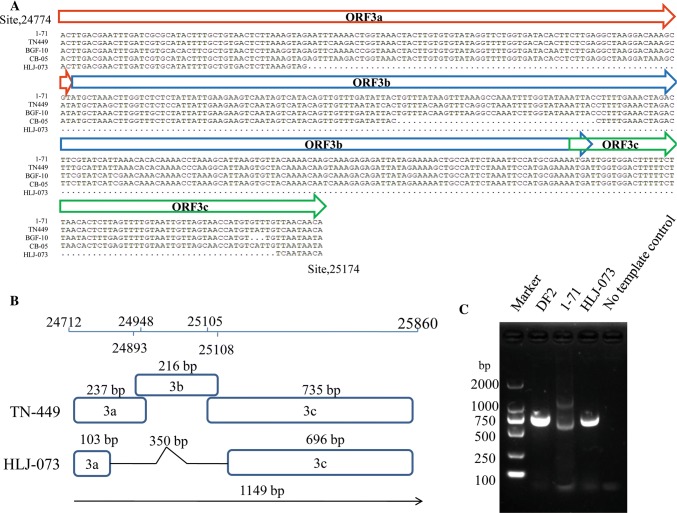
We isolated a canine coronavirus with a deletion in the ORF3abc region from a dead dog in China. Real-time RT-PCR assays were carried out to determine the viral load in the faeces, intestine, lung, liver, spleen, kidney and heart of the dead dog. All of the organs except the kidney were found to be positive for CCoV (Table S2), indicating that this strain is a pantropic CCoV strain. Next, after three rounds of plaque assay screening, a novel isolate, HLJ-073, was successfully isolated and purified. The presence of virus particles was confirmed by electron microscopy of negatively stained samples (Fig. S2). Sequence analysis suggested that there was a 350-nt deletion in ORF3abc when compared to isolates 1-71, TN-449 and BGF-10. This deletion resulted in truncations in ORF3a (103 nt) and ORF3c (696 nt) and the complete loss of ORF3b (Fig. 1A and B). To investigate the stability of the ORF3abc region in the HLJ-073 genomic RNA, the virus was propagated in CRFK cells for nine generations. Total RNA was extracted from HLJ-073-infected CRFK cells and from the original faecal sample. Analysis by PCR confirmed that the ORF3abc deletion was present not only in the purified virus but also in the original faecal sample (Fig. S1), indicating that the deletion was not due to viral adaptation during cell culture passage. Examination of subgenomic ORF3abc mRNAs showed that the genome of HLJ-073 contained a truncated ORF3abc (Fig. 1C). Previous studies have demonstrated that this region is variable, and a natural CB/05 strain that causes fatal infections and long-lasting lymphopenia has also been shown to contain a deletion in ORF3b [4, 12, 8]. Strains 450/07 and NA/09 are closely related to the prototype strain CB/05, which causes diarrhea and acute lymphopenia [11, 17]. Furthermore, strain BGF10 also has a deletion in the ORF3abc region [21]. These results demonstrate that the ORF3abc region may be prone to being lost during the evolution of the virus.
Fig. 1.
(A) Multiple sequence alignment of the ORF3abc region of CCoV strains (B) Schematic diagram of ORF3a, 3b and 3c of CCoVs. (C) Identification of mutant strains by RT-PCR
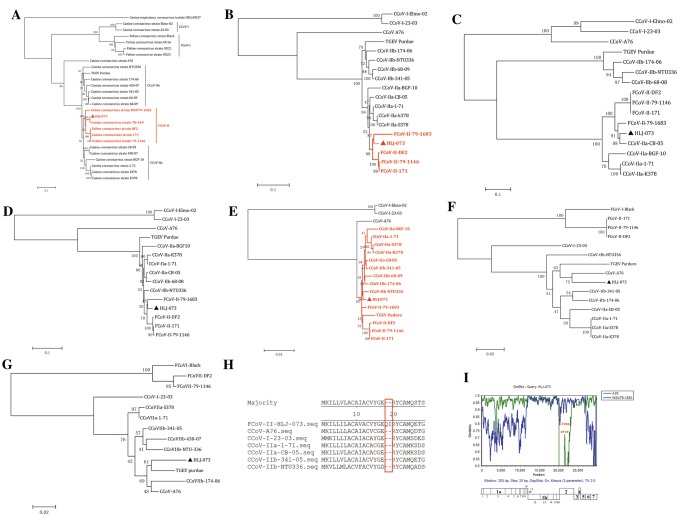
Recombination plays a vital role in the evolution of CCoV, allowing the emergence of new strains with altered virulence and immunogenicity. In recent years, CCoVs with different genotypes and subgenotypes have been detected. CCoV-II is the oldest genotype, which has been known since 1971 [3]. More recently, recombination between CCoV and TGEV has been detected, which has led to the emergence of CCoV-IIb isolates in which the N-terminus of the S gene is very similar to that of TGEV, while the rest of the genome resembles that of the reference CCoV-II isolate [9]. A CCoV strain (UCD-1) that had potentially recombined with TGEV was identified in the late 1990s [24]. Genetic analysis of several CCoVs circulating in Italy first revealed a new canine genetic cluster in which recombination events within the S gene had increased the similarity of CCoVs to the feline homolog [19]. On the basis of their genetic relationship to FCoV-I, the new genotype was initially designated “FCoV-like CCoVs”. Recently, an additional ORF, named ORF3, located between the end of the S gene and the ORF3a gene, was also detected [14]. In this study, phylogenetic analysis based on the S protein revealed that HLJ-073 is closely related to members of the FCoV-II cluster and is distinct from CCoVs (Fig. 2A). Analysis of the S1 (receptor-binding) domain showed that it clustered closely with FCoV-II 79-1683 (Fig. 2B), while the S2 (fusion) domain clustered with CCoV- II, FCoV-II and TGEV (Fig. 2E). The coronavirus S1 protein consists of two independent subdomains, the N-terminal domain (NTD) and the C-terminal domain (C-domain) [18]. The NTD clustered closely with CB/05 and 79-1683 (Fig. 2C), and the C-domain clustered with 79-1683 (Fig. 2D). In contrast, the M gene clustered with A76 and had a 2-amino-acid deletion compared with A76, CB/05, 23/03 (Fig. 2H), and the E gene cluster with TGEV and did not clustered with CCoV and FCoV (Fig. 2G). These data indicate that HLJ-073 is probably a recombinant of TGEV, FCoV-II and CCoV-I/II, with recombination sites located between the S, E and M genes. Simplot analysis was carried out to identify possible recombination events and breakpoints in the complete nucleotide sequence of the HLJ-073 genome. The analysis confirmed the probably occurrence of recombination in the S, E and M genes of HLJ-073, which was probably the result of recombination between FCoV 79-1683 and CCoV A76.
Fig. 2.
(A-G) Phylogenetic analysis of the spike protein (S), S1, S1-NTD, S1-C-domain, S2, membrane (M) and envelope (E) genome regions of HLJ-073 and the most closely related strains in GenBank for which whole genome sequences were available. Neighbor-joining was used for the construction of the phylogenetic tree with bootstrap values of 1000 replicates shown at the branches. The scale bar represents the p-distance. (H) Sequence alignment of the N-terminal membrane region of CCoV strains analyzed in the present study with CCoV strains A76, 23-03, 1-71, CB-05, 341-05, and NTU336. (I) The crossover region in the HLJ-073 genome was detected by Simplot 3.5.1. The y-axis shows the percentage of permuted trees employing a sliding window of 200 nucleotides (nt) and a step size of 20 nt. The other parameters used included the Kimura (2-parameter) distance model, 2.0 Ts/Tv ratio, neighbor-joining tree model, and 1000 bootstrap replicates
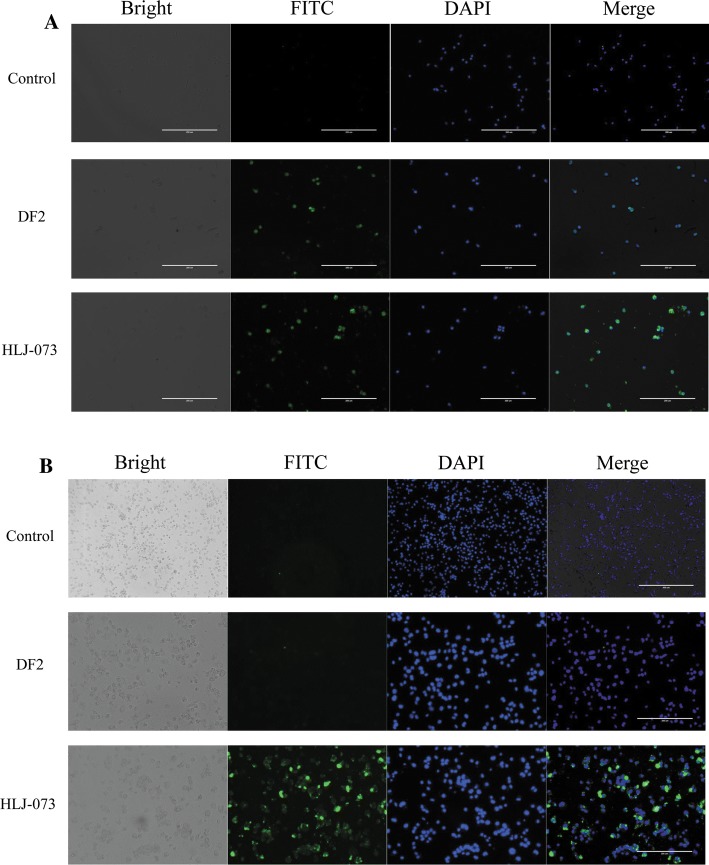
The ORF3abc region of feline coronavirus has been shown to affect viral cell tropism. Truncated ORF3abc of FIPV DF-2 has been shown to effectively replicate in macrophages/monocytes, while infection with FCoVs with the complete ORF3abc region was confined to the intestinal tract [2]. In the present study, we found that HLJ-073 and FIPV DF-2 could also effectively replicate in canine macrophages/monocytes (Fig. 3A). However, HLJ-073 could replicate in THP-1 cells, while DF-2 could not (Fig. 3B). These results suggest that HLJ-073 has the ability to alter its tissue tropism from the intestinal tract to systemic infection and to alter its viral cell tropism from dogs to humans, which should be regarded as a potential threat to domestic dogs. Accordingly, epidemiologic studies are required to determine whether domestic CCoV isolates have similar characteristics.
Fig. 3.
Indirect immunofluorescence detection of CCoVs in canine macrophage/monocyte (A) and THP-1 cells (B). The cells were infected with HLJ-073 or DF2 (MOI = 1) and detected using CCoV N protein-positive serum
Conclusion
We report for the first time that CCoV HLJ-073, with an ORF3abc deletion, has been isolated in China. HLJ-073 represents a recombinant coronavirus with distinct host cell tropism.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 Fig. S1 (A) Testing of the fecal samples by PCR. (B) Testing of the genetic stability (24778F and 25440R) by PCR (TIFF 3087 kb)
Supplementary material 2 Fig. S2 (A) Control (uninfected) CRFK cells. (B) Cytopathic effect (CPE) induced by the HLJ-073 strain in CRFK cells. (C) Ultrathin sections of CRFK cells infected with HLJ-073. (D) Electron micrograph of HLJ-073 negatively stained with 2% phosphotungstic acid. The scale bar represents 500 nm. (E-F) Indirect immunofluorescence detection of CCoVs in mock-infected or HLJ-073-infected CRFK cells using CCoV-positive serum. (G-H) Plaque assay using mock-infected and HLJ-073-infected in CRFK cells (TIFF 4430 kb)
Acknowledgements
This work was supported by the State Key Laboratory of Veterinary Biotechnology Foundation (SKLVBP2018002).
Abbreviations
- CCoV
Canine coronavirus
- TGEV
Transmissible gastroenteritis virus
- CDV
Canine distemper virus
- CPV
Canine parvovirus
- CAV-1
Canine adenovirus 1
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiaoliang Hu, Email: huxiaoliang@caas.cn.
Liandong Qu, Email: quliandong@caas.cn.
References
- 1.Enjuanes L, Spaan W, Snijder E, Cavanagh D. Virus taxonomy, 6th report of the International Committee on Taxonomy of Viruses. Arch Virol Suppl. 1995;10:1–586. [PubMed] [Google Scholar]
- 2.Balint A, Farsang A, Zadori Z, Hornyak A, Dencso L, Almazan F, Enjuanes L, Belak S. Molecular characterization of feline infectious peritonitis virus strain DF-2 and studies of the role of ORF3abc in viral cell tropism. J Virol. 2012;86:6258–6267. doi: 10.1128/JVI.00189-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binn LN, Lazar EC, Keenan KP, Huxsoll DL, Marchwicki RH, Strano AJ. Recovery and characterization of a coronavirus from military dogs with diarrhea. Proc Annu Meet US Anim Health Assoc. 1974;78:359–366. [PubMed] [Google Scholar]
- 4.Buonavoglia C, Decaro N, Martella V, Elia G, Campolo M, Desario C, Castagnaro M, Tempesta M. Canine coronavirus highly pathogenic for dogs. Emerg Infect Dis. 2006;12:492–494. doi: 10.3201/eid1203.050839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canio B, Nicola D, Vito M, Gabriella E, Marco C, Costantina D, Massimo C, Maria T. Canine coronavirus highly pathogenic for dogs. Emerg Infect Dis. 2006;12:492–494. doi: 10.3201/eid1203.050839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carmichael LE, Binn LN. New enteric viruses in the dog. Adv Vet Sci Comp Med. 1981;25:1–37. [PubMed] [Google Scholar]
- 7.Decaro N, Buonavoglia C. An update on canine coronaviruses: viral evolution and pathobiology. Vet Microbiol. 2008;132:221–234. doi: 10.1016/j.vetmic.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decaro N, Elia G, Martella V, Campolo M, Mari V, Desario C, Lucente MS, Lorusso E, Kanellos T, Gibbons RH. Immunity after natural exposure to enteric canine coronavirus does not provide complete protection against infection with the new pantropic CB/05 strain. Vaccine. 2010;28:724–729. doi: 10.1016/j.vaccine.2009.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decaro N, Mari V, Campolo M, Lorusso A, Camero M, Elia G, Martella V, Cordioli P, Enjuanes L, Buonavoglia C. Recombinant canine coronaviruses related to transmissible gastroenteritis virus of Swine are circulating in dogs. J Virol. 2009;83:1532–1537. doi: 10.1128/JVI.01937-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decaro N, Pratelli A, Campolo M, Elia G, Martella V, Tempesta M, Buonavoglia C. Quantitation of canine coronavirus RNA in the faeces of dogs by TaqMan RT-PCR. J Virol Methods. 2004;119:145–150. doi: 10.1016/j.jviromet.2004.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decaro N, Mari V, von Reitzenstein M, Lucente MS, Cirone F, Elia G, Martella V, King VL, Di Bello A, Varello K, Zhang S, Caramelli M, Buonavoglia C. A pantropic canine coronavirus genetically related to the prototype isolate CB/05. Vet Microbiol. 2012;159:239–244. doi: 10.1016/j.vetmic.2012.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Decaro N, Martella V, Elia G, Campolo M, Desario C, Cirone F, Tempesta M, Buonavoglia C. Molecular characterisation of the virulent canine coronavirus CB/05 strain. Virus Res. 2007;125:54–60. doi: 10.1016/j.virusres.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu X, Jr, Li N, Jr, Tian Z, Jr, Yin X, Jr, Qu L, Qu J. Molecular characterization and phylogenetic analysis of transmissible gastroenteritis virus HX strain isolated from China. BMC Vet Res. 2015;11:72. doi: 10.1186/s12917-015-0387-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Poder S. Feline and canine coronaviruses: common genetic and pathobiological features. Adv Virol. 2011;2011:1–11. doi: 10.1155/2011/609465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, Lyu D. Preparation and characterization of mouse polyclonal antibody against conserved region of human FOXO3. Xi bao yu fen zi mian yi xue za zhi = Chin J Cell Mol Immunol. 2017;33:838–844. [PubMed] [Google Scholar]
- 16.Licitra BN, Duhamel GE, Whittaker GR. Canine enteric coronaviruses: emerging viral pathogens with distinct recombinant spike proteins. Viruses. 2014;6:3363–3376. doi: 10.3390/v6083363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ntafis V, Xylouri E, Mari V, Papanastassopoulou M, Papaioannou N, Thomas A, Buonavoglia C, Decaro N. Molecular characterization of a canine coronavirus NA/09 strain detected in a dog’s organs. Arch Virol. 2012;157:171–175. doi: 10.1007/s00705-011-1141-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng G, Sun D, Rajashankar KR, Qian Z, Holmes KV, Li F. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc Natl Acad Sci USA. 2011;108:10696–10701. doi: 10.1073/pnas.1104306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pratelli A, Martella V, Decaro N, Tinelli A, Camero M, Cirone F, Elia G, Cavalli A, Corrente M, Greco G, Buonavoglia D, Gentile M, Tempesta M, Buonavoglia C. Genetic diversity of a canine coronavirus detected in pups with diarrhoea in Italy. J Virol Methods. 2003;110:9–17. doi: 10.1016/S0166-0934(03)00081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Regan AD, Millet JK, Tse LP, Chillag Z, Rinaldi VD, Licitra BN, Dubovi EJ, Town CD, Whittaker GR. Characterization of a recombinant canine coronavirus with a distinct receptor-binding (S1) domain. Virology. 2012;430:90–99. doi: 10.1016/j.virol.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez-Morgado JM, Poynter S, Morris TH. Molecular characterization of a virulent canine coronavirus BGF strain. Virus Res. 2004;104:27–31. doi: 10.1016/j.virusres.2004.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Li C, Guo D, Wang X, Wei S, Geng Y, Wang E, Wang Z, Zhao X, Su M, Liu Q, Zhang S, Feng L, Sun D. Co-circulation of canine coronavirus I and IIa/b with high prevalence and genetic diversity in heilongjiang province, Northeast China. PLoS One. 2016;11:e0146975. doi: 10.1371/journal.pone.0146975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuchiya K, Kasaoka T, Azetaka M, Takahashi E, Konishi S. Plaque assay for canine coronavirus in CRFK cells. Nihon Juigaku Zasshi. 1987;49:571–573. doi: 10.1292/jvms1939.49.571. [DOI] [PubMed] [Google Scholar]
- 24.Wesley RD. The S gene of canine coronavirus, strain UCD-1, is more closely related to the S gene of transmissible gastroenteritis virus than to that of feline infectious peritonitis virus. Virus Res. 1999;61:145–152. doi: 10.1016/S0168-1702(99)00032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 Fig. S1 (A) Testing of the fecal samples by PCR. (B) Testing of the genetic stability (24778F and 25440R) by PCR (TIFF 3087 kb)
Supplementary material 2 Fig. S2 (A) Control (uninfected) CRFK cells. (B) Cytopathic effect (CPE) induced by the HLJ-073 strain in CRFK cells. (C) Ultrathin sections of CRFK cells infected with HLJ-073. (D) Electron micrograph of HLJ-073 negatively stained with 2% phosphotungstic acid. The scale bar represents 500 nm. (E-F) Indirect immunofluorescence detection of CCoVs in mock-infected or HLJ-073-infected CRFK cells using CCoV-positive serum. (G-H) Plaque assay using mock-infected and HLJ-073-infected in CRFK cells (TIFF 4430 kb)