Abstract
Adenoviruses are common pathogens that are responsible for a wide variety of infectious syndromes. The objectives of this study were to identify and characterize members of different adenovirus species at the molecular level and to describe the correlation between viruses and clinical syndromes during a period of 4 years. Between 2002 and 2006, 45 of 512 respiratory specimens (8%) from patients with acute respiratory tract infection tested positive for adenovirus. Four adenovirus isolates from samples sent for enterovirus isolation were also analyzed. This research identified 49 confirmed cases of human adenovirus infection by PCR and/or viral culture. The most common diagnosis was upper respiratory infection (44%). Human adenovirus D was the major species found (59%), followed by Human adenovirus C (36%) and Human adenovirus B (4%). Human adenovirus 5 was the major serotype found producing bronchiolitis, followed by human adenovirus 6. In patients with upper respiratory infection, the major serotype found was human adenovirus 17. Viruses of the species Human adenovirus D were identified in seven (77%) cases of acute febrile syndrome. Four isolates from clinical materials obtained from patients with encephalitis, acute flaccid paralysis and meningoencephalitis were identified as belonging to the species Human adenovirus D. Our data demonstrate a surprising result about the identification of an unusual association of viruses of the species Human adenovirus D with different clinical syndromes. This observation could be evaluated as a possible indicator of the emergence of a novel strain but further studies are required.
Keywords: Meningoencephalitis, Hemorrhagic Cystitis, Adenovirus Infection, Acute Flaccid Paralysis, Human Adenovirus
Introduction
Human adenoviruses (HAdVs) are members of the family Adenoviridae, whose members infect hosts across a broad spectrum of vertebrates. There are 51 serotypes of HAdV in the genus Mastadenovirus, distributed in six species, A–F (formerly subgroups or subgenera) on the basis of their physicochemical, biological and genetic properties. A new serotype isolated recently (52) has been proposed to represent a new species G [21].
Human adenoviruses are associated with sporadic infection, and community and institutional outbreaks. These viruses cause a variety of clinical manifestations, such as conjunctivitis, pneumonia, gastroenteritis, and hemorrhagic cystitis. Some serotypes can occasionally infect tissues of the central nervous system (CNS) and cause aseptic meningitis, meningoencephalitis, and encephalitis. They can cause especially severe disease in infants, young children, immunocompromised persons, and transplant recipients [2, 5, 14, 15, 25, 30, 31].
Serosurveys suggest that virtually all people are exposed to HAdV during childhood [22, 35]. They can remain in an asymptomatic carrier state until at least young adulthood [18], and virus may be actively shed long after symptomatic infection [16].
Over the past decades, neutralization tests, ELISA, and virus isolation had been used for the detection and identification of adenovirus serotypes. However, these methods are relatively complicated, labour-intensive, and time-consuming, and they have low sensitivity. [26, 39, 42]. These disadvantages have limited their use. Amplification of the viral genome by PCR has been introduced as a convenient and powerful alternative for molecular diagnosis. Additionally, genome amplification allows further characterization of the adenovirus serotype by sequence analysis [1, 7, 34].
The objectives of this study were the identification and molecular characterization of different HAdV isolates and to describe the correlation between viruses and clinical syndromes during a period of 4 years.
Methods
Clinical samples
Between October 2002 and September 2006, 512 respiratory specimens (nasopharyngeal swabs and pharyngeal washes) from patients with acute respiratory tract infection (ARTI) were sent specifically to the National Reference Laboratory of Respiratory Viruses for testing respiratory viruses, including influenza virus A, B and C, human respiratory syncitial virus (HRSV), human parainfluenza virus 1–4, HAdV, human coronavirus, human rhinovirus and human metapneumovirus (HMPV). Routine virological testing for respiratory pathogens was performed using a combination of direct immunofluorescence, isolation in cell culture and PCR assays [9, 10, 27]. In addition, four samples (three stools and one cerebrospinal fluid) from the enterovirus (EV) laboratory with previous viral isolation without identification were analyzed. Nasopharyngeal swabs and pharyngeal washes were collected in 3 ml of virus transport medium (MEM, Gibco-BRL, Life Technologies, Paisley, Scotland; penicillin 200 U/ml, and streptomycin 200 μg/ml, BioWhittaker, MA; mycostatin 200 U/ml, Sigma; bovine serum albumin 0.25%, Merck, Darmstadt, Germany) The specimens were frozen and stored at –70°C until the analysis was carried out.
Cell culture isolation
A human embryonic fibroblast cell line was used for primary isolation of EV. Tubes with 80% confluent monolayers were inoculated with 0.2 ml of homogenized samples. Cells were fed with 2 ml of 2% fetal calf serum in Basal Medium Eagle and visualized for cytopathic effect (CPE) twice a week. When a CPE that was not typical of EV was observed, the monolayer was scraped and tested for adenovirus antigen by immunofluorescence with a specific monoclonal antibody (Chemicon, Temecula, CA).
Nucleic acid extraction
Total viral RNA/DNA from 200-μl aliquots from clinical samples or supernatant of infected cell culture was extracted using the guanidinium thiocyanate method as described previously by Casas et al. [6]. The lysis buffer included 100 copies of the cloned, amplified product of the internal control described by Coiras et al. [9]. It was used for checking the extraction process, the amplification efficiency, and the presence of inhibitors in the clinical specimens. After processing, the dried pellet was resuspended in 15 μl of RNAse-free sterile water. Negative controls consisting of RNase-free sterile water (Sigma) were treated following the same procedure. For each assay, known positive controls, derived from infected viral cells, were added.
Detection of viral genomes
In all cases, nucleic acid extracts (5 μl) were examined for influenza virus A, B and C, HAdV, HRSV A and B [9], human parainfluenza virus 1–4, coronaviruses, rhinoviruses, and EV [10], using nRT-PCR. In addition, a multiplex nPCR assay for human herpesvirus was used in the case of a patient with encephalitis [40]. The protocols employed have been published previously and used for clinical diagnosis. Three different biosafety cabinets were used for master mix preparation, sample handling and primary reaction product handling. Different laboratory wear and coats were used for each cabinet. Amplicon detection was done in a different room.
DNA sequencing and phylogenetic analysis
Specimens that were positive for HAdV were processed by two independent nested reactions with 20 pmol of ADHEX2F, nt 20485 to 20503; (5′CCCITTYAACCACCACCG3′) and ADHEX1R, nt 20836 to 20854; (5′KATGGGGTARAGCATGTT3′) or 20 pmol of ADHEX1F, nt 20380 to 20400; (5′CAACACCTAYGASTACATGAA3′) and ADHEX2R, nt 20632 to 20652; (5′ACATCCTTBCKGAAGTTCCA3′) degenerate primers as described previously [7]. These nested primer pairs were designed to bind inside the hexon protein coding region of the adenovirus genome [1]. The PCR products were purified with QIAquick PCR purification kit (Qiagen) according to the manufacturers’s protocol and were sequenced in both directions using the primer pairs described above.
Sequences were obtained using an automatic DNA sequencer (ABI Prism 3700; Applied Biosystems) and a Big Dye Terminator Cycle Sequencing kit version 3.1 (Applied Biosystems). The fragment size sequenced corresponded to 122 amino acids, from amino acid position 540–662 of the hexon protein. This fragment is located at the external surface L2 of the hexon protein monomer but does not overlap with the HRV7 domain [11].
The nucleotide sequences obtained were first analysed using the CHROMAS software (version 1.3). The forward and reverse sequences were combined using BioEdit and were compared and aligned with the corresponding previously published sequences of prototype viruses available from GenBank, using the CLUSTAL X (version 1.83) program. Specimens that yielded identity scores ≥90%, were considered to be good genotype matches. Phylogenetic trees were inferred using programs from the MEGA package (version 4) and reconstructed using the neighbour-joining method. The evolutionary distances were estimated using Kimura’s two-parameter method [24]. The statistical significance of a particular tree topology was evaluated by 1,000 replicates of boostrap re-sampling. The criteria for serotype assignation were the shortest distance from a prototype strain, as deduced from the phylogenetic tree (Fig. 1), and the highest similarity value obtained after sequence comparisons.
Fig. 1.
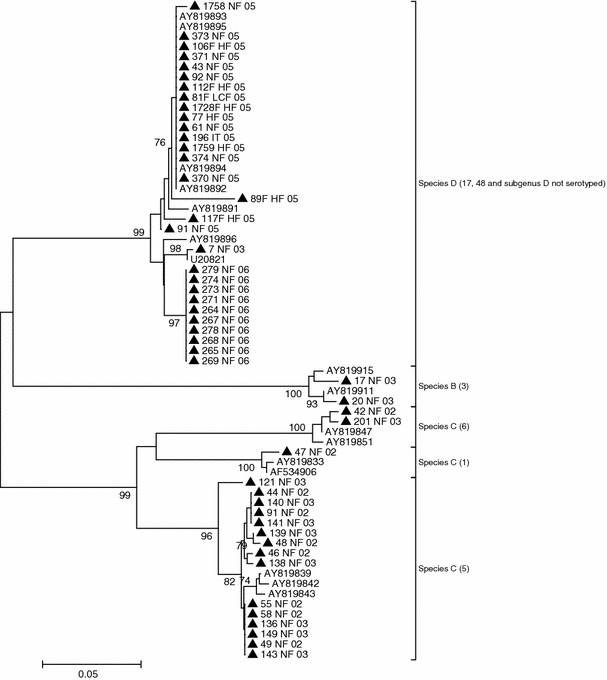
Phylogenetic analysis based on sequence alignment of HAdV amplicons
Nucleotide sequence accession numbers
The GenBank accession numbers of the nucleotide sequences presented in this study are: EU179755–EU179763, EU179765, EU179768–EU179785 and EU439569–EU439589.
Results
PCR and clinical features of patients
In the present report, the nested PCR method used was able to detect different HAdVs in clinical samples and supernatant culture with a sensitive internal control system to assure the quality of reaction conditions in each individual tube.
In this investigation, 49 confirmed cases of HAdV infection were identified by PCR and/or viral culture (Table 1).
Table 1.
Clinical features of patients with adenoviral infection
| Samplea | Typing result | Syndrome | Type of specimen/months of collection | Patient age | Epidemiology |
|---|---|---|---|---|---|
| 42/02 | Ad 6 | Bronchiolitis/Hospitalized | NPS/June | 2 Months | Sporadic |
| 44/02 | Ad 5 | Bronchiolitis/Hospitalized | NPS/June | 5 Months | Sporadic |
| 46/02 | Ad 5 | Bronchiolitis/Hospitalized | NPS/June | 4 Months | Sporadic |
| 47/02 | Ad 1 | Bronchiolitis/Hospitalized | NPS/June | 5 Months | Sporadic |
| 48/02 | Ad 5 | Bronchiolitis/Hospitalized | NPS/June | 4 Months | Sporadic |
| 49/02 | Ad 5 | Bronchiolitis/Hospitalized | NPS/July | 3 Months | Sporadic |
| 55/02 | Ad 5 | Bronchiolitis/Hospitalized | NPS/July | 5 Months | Sporadic |
| 58/02 | Ad 5 | Bronchiolitis/Hospitalized | NPS/July | 6 Months | Sporadic |
| 91/02 | Ad 5 | Bronchiolitis/Hospitalized | NPS/September | 6 Months | Sporadic |
| 121/03 | Ad 5 | Bronchiolitis/Hospitalized | NPS/September | 10 Months | Sporadic |
| 138/03 | Ad 5 | Bronchiolitis/Hospitalized | NPS/October | 4 Months | Sporadic |
| 140/03 | Ad 5 | Bronchiolitis/Hospitalized | NPS/October | 6 Months | Sporadic |
| 201/03 | Ad 6 | Bronchiolitis/Hospitalized | NPS/November | 1 Year | Sporadic |
| 61/05 | Ad D | Bronchiolitis/Hospitalized | NPS/September | 2 Months | Sporadic |
| 7/03 | Ad 48 | URI//Nonhospitalized | NPS/January | 14 Years | Outbreak |
| 17/03 | Ad 3 | URI//Nonhospitalized | NPS/February | 8 Years | Sporadic |
| 20/03 | Ad 3 | URI/Nonhospitalized | NPS/February | 10 Years | Sporadic |
| 136/03 | Ad 5 | URI/Nonhospitalized | NPS/September | 43 Years | Outbreak |
| 139/03 | Ad 5 | URI/Nonhospitalized | NPS/September | 42 Years | Outbreak |
| 141/03 | Ad 5 | URI/Nonhospitalized | NPS/September | 39 Years | Outbreak |
| 143/03 | Ad 5 | URI//Nonhospitalized | NPS/September | 52 Years | Outbreak |
| 149/03 | Ad 5 | URI/Nonhospitalized | NPS/September | 21 Years | Outbreak |
| 43/05 | Ad D | URI/Nonhospitalized | NPS/March | 10 Years | Sporadic |
| 91/05 | Ad D | URI/Nonhospitalized | NPS/May | 18 Years | Sporadic |
| 92/05 | Ad D | URI/Nonhospitalized | NPS/May | 22 Years | Sporadic |
| 1758/05 | Ad D | URI/Hospitalized | NPS/June | 1 Years | Sporadic |
| 264/06 | Ad 17 | URI/Hospitalized | NPS/September | 3 Years | Outbreak |
| 265/06 | Ad 17 | URI/Hospitalized | NPS/September | 9 Years | Outbreak |
| 267/06 | Ad 17 | URI/Hospitalized | NPS/September | 4 Years | Outbreak |
| 268/06 | Ad 17 | URI/Hospitalized | NPS/September | 2 Years | Outbreak |
| 269/06 | Ad 17 | URI/Hospitalized | NPS/September | 12 Years | Outbreak |
| 271/06 | Ad 17 | URI/Hospitalized | NPS September | 20 Years | Outbreak |
| 273/06 | Ad 17 | URI/Hospitalized | NPS/September | 19 Years | Outbreak |
| 274/06 | Ad 17 | URI/Hospitalized | NPS/September | 14 Years | Outbreak |
| 278/06 | Ad 17 | URI/Hospitalized | NPS/September | 11 Years | Outbreak |
| 279/06 | Ad 17 | URI/Hospitalized | NPS/September | 16 Years | Outbreak |
| 77/05 | Ad D | Acute febrile syndrome/Hospitalized | NPS/June | 7 Months | Outbreak |
| 117/05 | Ad D | Acute febrile syndrome/Nonhospitalized | NPS/June | 11 Years | Outbreak |
| 1728/05 | Ad D | Acute febrile syndrome/Hospitalized | NPS/July | 8 Years | Outbreak |
| 1759/05 | Ad D | Acute febrile syndrome/Hospitalized | NPS/July | 5 Months | Outbreak |
| 370/05 | Ad D | Acute febrile syndrome/Nonhospitalized | NPS/July | 3 Years | Outbreak |
| 371/05 | Ad D | Acute febrile syndrome/Nonhospitalized | NPS/July | 2 Years | Outbreak |
| 196/05 | Ad D | Acute febrile syndrome/Nonhospitalized | NPS/July | 3 Years | Outbreak |
| 373/05 | Ad D | Acute febrile syndrome/Nonhospitalized | NPS/July | 1 Year | Outbreak |
| 374/05 | Ad D | Acute febrile syndrome/Nonhospitalized | NPS/July | 18 Years | Outbreak |
| 81/05 | Ad D | Encephalitis/Hospitalized | CSF/April | 64 Years | Sporadic |
| 106/05 | Ad D | AFP/Hospitalized | Feces/June | 1 Year | Sporadic |
| 89/05 | Ad D | ME/Hospitalized | Feces/May | 10 Years | Sporadic |
| 112/05 | Ad D | ME/Hospitalized | Feces/June | 2 Years | Sporadic |
URI upper respiratory infection, AFP acute flaccid paralysis, ME meningoencephalitis, NPS nasopharyngeal swab, CSF cerebrospinal fluid
aClinical sample identification number: number of laboratory and year
The monthly collection of clinical samples is shown in Table 1. Human adenovirus was detected throughout the year; however, 38 samples (77%) were collected between June and September. Two significant outbreaks of HAdV infection were identified, one in July of 2005 and the other in September of 2006.
More than half of the positive samples (53%) were from children younger than 5 years of age, and 50% of them were under 6 months of age. Thirty-two percent were from patients between 5 and 20 years, and 14% were from adults between 21 and 64 years of age. No difference in gender distribution was observed.
The most common diagnosis was upper respiratory infection (44%). Medical records documented the following symptoms at initial presentation: nasal congestion (98%), sore throat (83%) and cough (78%). All patients were febrile. Fourteen (28%) patients had bronchiolitis. Acute febrile syndrome (18%) was defined as the concurrence of fever, cold symptoms, headache, weakness, vomiting and diarrhoea, a common but non-specific symptom. In these cases, the results of diagnostic assays for the detection of other potentially pathogenic enteric microorganisms were negative. An interesting finding was the detection of HAdV in a patient 64 years of age who was admitted with a diagnosis of encephalitis. No human herpesvirus, influenza virus, EV or flavivirus genomes were found in the cerebrospinal fluid of this patient. Human adenovirus DNA was detected in the supernatant of a cell culture infected with viruses obtained from fecal specimens taken from a patient with acute flaccid paralysis (AFP), as well as in two cases of meningoencephalitis. Overall, 73% of HAdV positive patients were hospitalized.
Molecular characterization
Partial hexon nucleotide sequences for 49 different HAdV types were obtained. A BLAST search in the GenBank database for all of the amplicon sequences determined in the present study was done. A 100% agreement with existing GenBank sequences for serotypes 1, 3, 5, and 6 was obtained. An agreement ranging from 99 to 100% was obtained for prototype strains of Human adenovirus D. An alignment of the hexon gene sequences was computed by using sequences from representative serotypes of species B (HAdV 3), C (HAdVs 1, 5, and 6), and D (HAdVs 9, 10, 13,15,17, 19, 23, 25, 28, 30, 32, 33, 37–39, 43, 44, and 47–49). In order to evaluate the phylogenetic relationships between individual prototype viruses and positive cases of HAdV infection by PCR and/or viral culture, phylogenetic trees were constructed using a phylogeny reconstruction algorithm (the neighbour-joining method) and a nucleotide substitution method (Kimura-2p). Phylogenetic and sequence similarity analysis permitted accurate species classification and serotype identification except for species D HAdV, for which serotypes could not be clearly defined.
The resulting phylogenetic tree showed three different clusters, represented by the species B, C and D. The bootstrap values (Fig. 1) were, in general, quite high, and especially at nodes between species. However, these values were less good for some nodes within species D. The assignment of a serotype to a clinical isolate was done by calculating the shortest distance between the isolate and a prototype strain. A total of 31 (63%) HAdVs were serotyped, and typing results yielded some interesting observations.
Correlation of viruses and clinical syndromes
The major species found was HAdV-D, with 29 clinical cases (59%), followed by HAdV-C, with 18 cases (36%), and HAdV-B, with two cases (4%).
In bronchiolitis cases, HAdV-5 (71%) was the major serotype, followed by HAdV-6 (14%) and single cases of HAdV-1 and HAdV-D.
In the group of patients with upper respiratory infection, the major serotype found was HAdV-17 (77%). HAdV-5 (22%) was the second-most common serotype, and five specimens were typed as HAdV-D; one of them recovered in 2003 and four in 2005. Lastly, two specimens recovered in 2003 were HAdV-3. HAdV-D was the species identified in nine (100%) cases detected during an atypical outbreak of acute febrile syndrome in July of 2005. Four isolates from clinical materials obtained from patients with severe disease such as encephalitis, AFP and meningoencephalitis were identified as HAdV-D.
Discussion
HAdVs are categorized by species (HAdV-A, HAdV-B, HAdV-C, HAdV-D, HAdV-E and HAdV-F), and further by serotype (Ad1–Ad51), and cause a wide spectrum of illnesses in both children and adults, including those involving the CNS. A new recently isolated serotype (52) has been proposed to belong to a new species G [3, 13, 21].
More than half of the positive samples (53%) were from children younger than 5 years of age, and 50 % of them were under 6 months of age. The highest incidence of adenoviral infection occurs in children from 6 months to 5 years of age, although outbreaks have been noted when susceptible hosts, such as military recruits, adolescents, visitors to summer camps, and sometimes nursing home residents, congregate together. Explanations for the relative lack of illness in the youngest or in older individuals include the presence of transplacentally acquired maternal antibody in young infants and the development of neutralizing antibody to the most common adenoviral strains in the majority of children older than 5 years old.
The age distribution of HAdV in developing countries appears to be similar to that in developed countries [8]. The results in this study showed that adenovirus infections were more common in children younger than 5 years of age, and 50% of them were under 6 months of age. Most patients were hospitalized (73%); nevertheless, none of these had severe respiratory infections. In Cuba, infants are considered special patients by pediatricians.
This report also describes the seasonal variation of adenovirus infections in Cuba. Respiratory viral agents usually have characteristic seasonal patterns in temperate and tropical climates. In temperate climates, the majority of isolations of respiratory viruses are in the winter. In Central America, there are few studies published about the epidemiological characteristics of adenoviruses. The Cuban island is located in the Caribbean Sea at the entrance of the Gulf of Mexico. The climate of Cuba is semitropical, and two seasons are generally recognized: a rainy season from May to October and dry season from November to April. Cuba is often hit by hurricanes from June to November, resulting in great economic loss and temporarily interrupted sanitary conditions. The average minimum temperature is 21°C (70°F), and the average maximum is 27°C (81°F). The warmest month is July with 30°C (86°F). In this study, adenovirus was detected throughout the year; however, two significant outbreaks of HAdV infection were identified, the first in July of 2005, when Hurricane Dennis hit severely the western part of Cuba, and the second in September of 2006. We believe that the effect of temperature and rainfall appears to be a determinant of the timing of those outbreaks.
Figure 1 displays a phylogenetic tree built from an alignment of the nucleotide sequences of the amplicons obtained from clinical samples and each serotype prototype. As shown in Fig. 1, each serotype is clearly distinct. Serotypes belonging to the same species cluster together.
In Cuba, human adenovirus circulates in the pediatric population throughout the year. Furthermore, members of adenovirus species C of are commonly involved in respiratory diseases in the pediatric population. Two previous studies have been reported on the associations between specific clinical syndromes and HAdV serotypes. The analysis of the occurrence of HAdV-C revealed that HAdV-1 and HAdV-6 were the predominant serotypes in children with acute respiratory diseases, and HAdV-37 was the most prevalent one associated with conjunctivitis [36, 41].
Human adenovirus respiratory infections have usually been associated with species B, C, and E [8]. HAdV-C includes HAdV-1, HAdV-2, HAdV-5, and HAdV-6. These serotypes are commonly associated with febrile respiratory illness in children and are noted to be endemic in certain regions or in epidemics [32].
The significant proportion of infection with HAdV-1, HAdV-3, HAdV-5 and HAdV-6 among patients with respiratory infection is consistent with previous reports [7, 20, 23, 32]. However, we detected HAdV-D in a child with bronchiolitis, and in 15 cases of upper respiratory infection. HAdV-D are rarely isolated in respiratory illness surveys, and strong causal correlations are generally lacking. Nevertheless, this association was previously described by Casas et al. [7]. On the other hand, HAdV-D was identified in a group of patients with acute febrile syndrome and in four isolates from clinical materials obtained from patients with encephalitis, AFP and meningoencephalitis. Sporadic cases or small outbreaks of neurological disease following adenovirus infection are well documented [12, 17, 25, 38]. The high prevalence of infection with HAdV-D in previously healthy patients is not consistent with previous reports. This association was unexpected, and a detailed analysis of this sample set will be published in a separate report.
Human adenovirus is unique among the common respiratory viruses in that it can spread to organs other than the respiratory system, resulting in conjunctivitis, gastroenteritis, acute hemorrhagic cystitis, and meningoencephalitis. The liver, spleen, pancreas, kidney, or heart may also be involved in disseminated infections, both in previously healthy people and in immunocompromised patients (e.g., infants, patients with AIDS, transplant recipients) [33]. HAdV-D has been frequently associated with severe adenovirus infection in these patients. In our study, we reviewed the medical records and did not find that the patients suffered from AIDS or another type of immunodeficiency. However, we thought that is important to take into account that the highest percentage of HAdV-D was identified in children.
The identification of such a large number of HAdV-D infections is extremely interesting and a major and unique finding. We recognize that the limited sequence data analyzed (370 nt in a relatively conserved region of the hexon) for typing did not generate enough results. The sequencing of the hypervariable regions of the hexon gene (HVR7 alone or HVR1-6) and fiber gene of select strains will certainly provide a solid basis for confirming the species/serotype identification [29, 37, 43]. We used the method previously published by Casas et al. [7], to facilitate sequencing and typing of HAdVs using the hexon protein coding region of the HAdV genome. This method was extensively validated using clinical samples and prototype strains, and we also noted unusual associations between specific serotypes and clinical presentation. The primers and PCR conditions have also been extensively validated.
Some years ago, adenovirus infection was considered to have little consequence. However, much has changed since these early epidemiological studies were conducted. In contrast to the modest number of HAdVs recognized 35 years ago, 51 unique serotypes are now recognized, and a new serotype isolated recently (52) has been proposed to belong to a new species G. Different serotypes have been found to have different tissue tropisms that correlate with different clinical manifestations of infection. Partial epidemiological investigations have revealed that, among some specific serotypes, multiple genetic variants exist that often have quite different geographical distributions and associated virulence [19, 21, 28, 30].
Our study expands the range of HAdV serotypes that have been reported elsewhere in association with major clinical syndromes. Further independent testing is needed to verify these associations. However, future studies of HAdV should not exclude these rare serotypes, and they should be kept in mind.
Unfortunately, because adenoviruses can be asymptomatically shed for prolonged periods of time, recovery of HAdV from the upper respiratory tract or stool samples by culture does not confirm it as the cause of a specific disease. Accordingly, recovery of HAdV should always prompt an effort to identify any additional or alternate potential explanations for symptoms present at the time of a positive viral culture [4, 16]. Additional research is needed for the development of better rapid methods to detect and to quantify HAdV in various body fluids (blood, stool, and throat).
Finally, our findings, combined with the existence of several reports concerning the association of HAdV with different syndromes confirm that members of the family Adenoviridae, mainly the serotypes belonging to HAdV-C are common causal agents of respiratory infection. In contrast, serotypes of HAdV-D could be involved in the aetiology of acute respiratory infection and neurological disorders in previously healthy people. However, this observation needs to be confirmed in a larger study.
Our identification of HAdV-D associated with different syndromes may signify the emergence of new genomic variants that have the potential to spread globally. Further studies of such typing with other molecular typing methods are necessary. These reports show that surveillance, serotyping and molecular characterization methods need to be improved in order to identify emerging adenovirus variants.
Acknowledgment
This work was supported in part by the Cuban National Program of Surveillance and Control of Acute Respiratory Infection. The nucleotide sequence was performed in the Instituto de Salud Carlos III, Madrid, Spain, and was funded by the proposal MPY-1177/06 entitled Caracterización molecular de HAdV detectados en muestras clínicas de pacientes con infección respiratoria. The authors wish to thank all of the physicians who provided samples and clinical data. We also acknowledge all technicians and researchers of the Virology Deparment of the Instituto de Medicina Tropical Pedro Kourí, la Habana, Cuba, who collaborated with this investigation. Thanks to Dr Enrique Tabarés, from Universidad Autónoma de Madrid, Spain, for his help.
References
- 1.Avellon A, Perez P, Aguilar JC, Lejarazu R, Echevarria JE. Rapid and sensitive diagnosis of human adenovirus infections by a generic polymerase chain reaction. J Virol Methods. 2001;92:113–120. doi: 10.1016/S0166-0934(00)00269-X. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin A, Kingman H, Darville M, Foot AB, Grier D, Cornish JM, Goulden N, Oakhill A, Pamphilon DH, Steward CG, Marks DI. Outcome and clinical course of 100 patients with adenovirus infection following bone marrow transplantation. Bone Marrow Transpl. 2000;26:1333–1338. doi: 10.1038/sj.bmt.1702716. [DOI] [PubMed] [Google Scholar]
- 3.Benko M, Harrach B, Both GW, Russell CW, Adair BM, Adam E, de Jong JC, Hess M, Johnson M, Kajon A, Kidd A, Lehmkuhl H, Li Q, Mautner V, Pring-Akerblom P, Wadell G. Family Adenoviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus taxonomy. VIIth report of the international committee on taxonomy of viruses. New York: Elsevier; 2005. pp. 213–228. [Google Scholar]
- 4.Brandt CD, Kim HW, Jeffries BC, Pyles G, Christmas EE, Reid JL, Chanock RM, Parrott RH. Infections in 18, 000 infants and children in a controlled study of respiratory tract disease. II. Variation in adenovirus infections by year and season. Am J Epidemiol. 1972;95:218–227. doi: 10.1093/oxfordjournals.aje.a121389. [DOI] [PubMed] [Google Scholar]
- 5.Carter BA, Karpen SJ, Quiros-Tejeira RE, Chang IF, Clark BS, Demmler GJ, Heslop HE, Scott JD, Seu P, Goss JA. Intravenous Cidofovir therapy for disseminated adenovirus in a pediatric liver transplant recipient. Transplantation. 2002;74:1050–1052. doi: 10.1097/00007890-200210150-00027. [DOI] [PubMed] [Google Scholar]
- 6.Casas I, Powell L, Klapper PE, Cleator GM. New method for the extraction of viral RNA andDNA from cerebrospinal fluid for use in the polymerase chain reaction. J Virol Methods. 1995;53:25–36. doi: 10.1016/0166-0934(94)00173-E. [DOI] [PubMed] [Google Scholar]
- 7.Casas I, Avellon A, Mosquera M, Jabado O, Echevarria JE, Campos RH, Rewers M, Pérez-Breña P, Lipkin WI, Palacios G. Molecular identification of adenoviruses in clinical samples by analysing a partial hexon genomic region. J Clin Microbiol. 2005;43:6176–6182. doi: 10.1128/JCM.43.12.6176-6182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng CC, Huang LM, Kao CL, Lee PI, Chen JM, Lu CY, Lee CY, Chang SY, Chang LY. Molecular and clinical characteristics of adenoviral infections in Taiwanese children in 2004–2005. Eur J Pediatr. 2007;167(6):633–640. doi: 10.1007/s00431-007-0562-4. [DOI] [PubMed] [Google Scholar]
- 9.Coiras MT, Pérez-Breña P, García ML, Casas I. Simultaneous detection of influenza A, B and C viruses, respiratory syncytial virus and adenoviruses in clinical samples by multiplex reserve transcription nested-PCR assay. J Med Virol. 2003;69:32–44. doi: 10.1002/jmv.10255. [DOI] [PubMed] [Google Scholar]
- 10.Coiras MT, Aguilar JC, Garcia ML, Casas I, Perez-Brena P. Simultaneous detection of fourteen respiratory viruses in clinical specimens by two multiplex reverse transcription nested-PCR assays. J Med Virol. 2004;72:484–495. doi: 10.1002/jmv.20008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crawford-Miksza L, Schnurr DP. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J Virol. 1996;70(3):1836–1844. doi: 10.1128/jvi.70.3.1836-1844.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Azevedo JP, Nascimento LR, Cortinovis MC, Oliveira SS, da Costa EV, da Silva EE. Characterization of species B adenoviruses isolated from fecal specimens taken from poliomyelitis-suspected cases. J Clin Virol. 2004;31:248–252. doi: 10.1016/j.jcv.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 13.Dubberke ER, Tu B, Rivet DJ, Storch GA, Apisarnthanarak A, Schmidt RE, Weiss S, Polish LB. Acute meningoencephalitis caused by adenovirus serotype 26. J Neurovirol. 2006;12:235–240. doi: 10.1080/13550280600846633. [DOI] [PubMed] [Google Scholar]
- 14.Echavarria M, Forman M, van Tol MJ, Vossen JM, Charache P, Kroes AC. Prediction of severe disseminated adenovirus infection by serum PCR. Lancet. 2001;358:384–385. doi: 10.1016/S0140-6736(01)05580-5. [DOI] [PubMed] [Google Scholar]
- 15.Filho EP, da Costa Faria NR, Fialho AM, de Assis RS, Almeida MM, Rocha M, Galvao M, dos Santos FB, Barreto ML, Leite JP. Adenoviruses associated with acute gastroenteritis in hospitalized and community children up to 5 years old in Rio de Janeiro and Salvador, Brazil. J Med Microbiol. 2007;56:313–319. doi: 10.1099/jmm.0.46685-0. [DOI] [PubMed] [Google Scholar]
- 16.Fox JP, Hall CE, Cooney MK. The seattle virus watch. VII. Observations of adenovirus infections. Am J Epidemiol. 1977;105:362–386. doi: 10.1093/oxfordjournals.aje.a112394. [DOI] [PubMed] [Google Scholar]
- 17.Gabrielson MO, Joseph C, Hsiung GD. Encephalitis associated with adenovirus type 7 occurring in a family outbreak. J Pediatr. 1966;68:142–144. doi: 10.1016/S0022-3476(66)80433-X. [DOI] [PubMed] [Google Scholar]
- 18.Garnett CT, Erdman D, Xu W, Gooding LR. Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J Virol. 2002;76:10608–10616. doi: 10.1128/JVI.76.21.10608-10616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray GC, McCarthy T, Lebeck MG, Schnurr DP, Russell KL, Kajon AE, Landry ML, Leland DS, Storch GA, Ginocchio CC, Robinson CC, Demmler GJ, Saubolle MA, Kehl SC, Selvarangan R, Miller MB, Chappell JD, Zerr DM, Kiska DL, Halstead DC, Capuano AW, Setterquist SF, Chorazy ML, Dawson JD, Erdman DD. Genotype prevalence and risk factors for severe clinical adenovirus infection, United States 2004–2006. Clin Infect Dis. 2007;45:1120–1131. doi: 10.1086/522188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong JY, Lee HJ, Piedra PA, Choi EH, Park KH, Koh YY, Kim WS. Lower respiratory tract infections due to adenovirus in hospitalized Korean children: epidemiology, clinical features, and prognosis. Clin Infect Dis. 2001;32:1423–1429. doi: 10.1086/320146. [DOI] [PubMed] [Google Scholar]
- 21.Jones MS, 2nd, Harrach B, Ganac RD, Gozum MM, Dela Cruz WP, Riedel B, Pan C, Delwart EL, Schnurr DP. New adenovirus species found in a patient presenting with gastroenteritis. J Virol. 2007;81:5978–5984. doi: 10.1128/JVI.02650-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jordan WS. Occurrence of adenovirus infections in civilian populations. Arch Intern Med. 1958;101(1):54–59. doi: 10.1001/archinte.1958.00260130068006. [DOI] [PubMed] [Google Scholar]
- 23.Kajon AE, Wadell G. Molecular epidemiology of adenoviruses associated with acute lower respiratory disease of children in Buenos Aires, Argentina (1984–1988) J Med Virol. 1992;36:292–297. doi: 10.1002/jmv.1890360411. [DOI] [PubMed] [Google Scholar]
- 24.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequence. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 25.Lema CL, Cisterna DM, Freire MC. Neurologic disease due to adenovirus infection. Medicina (B Aires) 2005;65:196–200. [PubMed] [Google Scholar]
- 26.Li L, Shimizu H, Doan LT, Tung PG, Okitsu S, Nishio O, Suzuki E, Seo JK, Kim KS, Muller WE, Ushijima H. Characterizations of adenovirus type 41 isolates from children with acute gastroenteritis in Japan, Vietnam, and Korea. J Clin Microbiol. 2004;42:4032–4039. doi: 10.1128/JCM.42.9.4032-4039.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.López-Huerta MR, Casas I, Acosta-Herrera B, García ML, Coiras MT, Pérez-Breña P. Two RT-PCR based assays to detect human matapneumovirus in nasopharyngeal aspirates. J Virol Methods. 2005;129:1–7. doi: 10.1016/j.jviromet.2005.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Louie JK, Kajon A, Holodniy M, Guardia-LaBar L, Lee B, Petru AM, Hacker JK, Schnurr DP. Severe pneumonia due to adenovirus serotype 14: a new respiratory threat? Clin Infect Dis. 2008;46(12):1935–1936. doi: 10.1086/588558. [DOI] [PubMed] [Google Scholar]
- 29.Lu X, Erdman D. Molecular typing of human adenovirus by PCR and sequencing of a partial region of the hexon gene. Arch Virol. 2006;151:1587–1602. doi: 10.1007/s00705-005-0722-7. [DOI] [PubMed] [Google Scholar]
- 30.Metzgar D, Osuna M, Yingst S, Rakha M, Earhart K, Elyan D, Esmat H, Saad MD, Kajon A, Wu J, Gray GC, Ryan MA, Russell KL. PCR analysis of Egyptian respiratory adenovirus isolates, including identification of species, serotypes, and coinfections. J Clin Microbiol. 2005;43:5743–5752. doi: 10.1128/JCM.43.11.5743-5752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mori T, Aisa Y, Shimizu T, Ikeda Y, Okamoto S, Okada K, Kazuyama Y. Hemorrhagic cystitis caused by adenovirus type 34 after allogeneic bone marrow transplantation. Transplantation. 2005;79:624. doi: 10.1097/01.TP.0000147653.16324.CA. [DOI] [PubMed] [Google Scholar]
- 32.Moura PO, Roberto AF, Hein N, Baldacci E, Vieira SE, Ejzenberg B, Perrini P, Stewien KE, Durigon EL, Mehnert DU, Harsi CM. Molecular epidemiology of human adenovirus isolated from children hospitalized with acute respiratory infection in Sao Paulo, Brazil. J Med Virol. 2007;79:174–181. doi: 10.1002/jmv.20778. [DOI] [PubMed] [Google Scholar]
- 33.Munoz FM, Piedra PA, Demmler GJ. Disseminated adenovirus disease in immunocompromised and immunocompetent children. Clin Infect Dis. 1998;27:1194–1200. doi: 10.1086/514978. [DOI] [PubMed] [Google Scholar]
- 34.Phan TG, Nguyen TA, Yan H, Yagyu F, Kozlov V, Kozlov A, Okitsu S, Muller WE, Ushuijma H. Development of a novel protocol for RT-multiplex PCR to detect diarrheal viruses among infants and children with acute gastroenteritis in Eastern Russia. Clin Lab. 2005;51:429–435. [PubMed] [Google Scholar]
- 35.Potter CW, Shedden WIH. The distribution of adenovirus antibodies in normal children. J Hyg (London) 1963;61:155–160. doi: 10.1017/s0022172400020830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pumariega T, Savon C, Mune M, Cancio R, Gonzalez G, Valdivia A, Gonzalez Z, Goyenechea A. Isolation and identification of adenovirus in hospitalized children, under five years, with acute respiratory disease, in Havana, Cuba. Mem Inst Oswaldo Cruz. 2000;95:859–861. doi: 10.1590/S0074-02762000000600020. [DOI] [PubMed] [Google Scholar]
- 37.Sarantis H, Johnson G, Brown M, Petric M, Tellier R. Comprehensive detection and serotyping of human adenoviruses by PCR and sequencing. J Clin Microbiol. 2004;42:3963–3969. doi: 10.1128/JCM.42.9.3963-3969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Straussberg R, Harel L, Levy Y, Amir J. A syndrome of transient encephalopathy associated with adenovirus infection. Pediatrics. 2001;107:E69. doi: 10.1542/peds.107.5.e69. [DOI] [PubMed] [Google Scholar]
- 39.Takeuchi S, Itoh N, Uchio E, Aoki K, Ohno S. Serotyping of adenoviruses on conjunctival scrapings by PCR and sequence analysis. J Clin Microbiol. 1999;37:1839–1845. doi: 10.1128/jcm.37.6.1839-1845.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tenorio A, Echevarría JE, Casas I, Echevarría JM, Tabarés E. Detection and typing of human herpesviruses by multiplex polymerase chain reaction. J Virol Methods. 1993;44:261–269. doi: 10.1016/0166-0934(93)90061-U. [DOI] [PubMed] [Google Scholar]
- 41.Torres Rojas G, Goyenechea A, Savon C, Valdes O, Oropesa I. The incidence of adenoviruses in viral conjunctivitis. Rev Cubana Med Trop. 1998;50:182–185. [PubMed] [Google Scholar]
- 42.Van der Avoort HG, Wermenbol AG, Zomerdijik TP, Kleijne JA, Van-Asten JA, Jensma P, Osterhaus AD, Kidd AH, De Jong JC. Characterization of fastidious adenovirus type 40 and 41 by DNA restriction analysis and by neutralizing monoclonal antibodies. Virus Res. 1989;12:139–157. doi: 10.1016/0168-1702(89)90060-9. [DOI] [PubMed] [Google Scholar]
- 43.Xu W, McDonough MC, Erdman DD. Species-specific identification of human adenoviruses by a multiplex PCR assay. J Clin Microbiol. 2000;38:4114–4120. doi: 10.1128/jcm.38.11.4114-4120.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


