Abstract
Little is known about the neutralizing epitopes in turkey coronavirus (TCoV). The spike (S) protein gene of TCoV was divided into 10 fragments to identify the antigenic region containing neutralizing epitopes. The expression and antigenicity of S fragments was confirmed by immunofluorescence antibody (IFA) assay using an anti-histidine monoclonal antibody or anti-TCoV serum. Polyclonal antibodies raised against expressed S1 (amino acid position 1 to 573 from start codon of S protein), 4F/4R (482-678), 6F/6R (830-1071), or Mod4F/Epi4R (476-520) S fragment recognized native S1 protein and TCoV in the intestines of TCoV-infected turkey embryos. Anti-TCoV serum reacted with recombinant 4F/4R, 6F/6R, and Mod4F/Epi4R in a western blot. The results of a virus neutralization assay indicated that the carboxyl terminal region of the S1 protein (Mod4F/Epi4R) or the combined carboxyl terminal S1 and amino terminal S2 protein (4F/4R) possesses the neutralizing epitopes, while the S2 fragment (6F/6R) contains antigenic epitopes but not neutralizing epitopes.
Keywords: Porcine Epidemic Diarrhea Virus, Infectious Bronchitis Virus, Virus Neutralization, Neutralize Epitope, Antigenic Domain
Introduction
Turkey coronaviral enteritis is characterized by diarrhea, anorexia, depression, ruffled feathers, decreased weight gain and increased mortality [1]. Increased mortality and uneven flock growth have caused significant economic loss in the turkey industry in the United States [2], Canada [3, 4], Europe [5], and Brazil [6]. Infection with TCoV can induce humoral and cellular immune responses in infected turkeys [7, 8]. TCoV-specific antibodies can be detected as early as 7 days postinfection and reach to a peak at 21 days postinfection. A virus-specific lymphocyte proliferation response in spleen cells from TCoV-infected turkeys is also stimulated significantly [8]. Poults exposed to TCoV were shown to develop a protective immune response against challenge with virulent TCoV 20 days after the first infection, and they showed no clinical signs or shedding of TCoV in feces [7]. Although TCoV was identified as the causative agent in the 1970s, there are still no effective vaccines or treatment to prevent or control TCoV infection.
Turkey coronavirus is an enveloped virus with a linear positive-sense, single-stranded RNA genome and belongs to the species Avian coronavirus in the genus Gammacoronavirus, subfamily Coronavirinae, family Coronaviridae and order Nidovirales. In 2009, the International Committee on Taxonomy of Viruses (ICTV) replaced the old classification of coronavirus groups 1, 2, and 3 with the new designation of three genera in the subfamily Coronavirinae [9]. The genus Alphacoronavirus is composed of eight species: Human coronavirus 229E, Human coronavirus NL63, Porcine epidemic diarrhea virus (PEDV), Miniopterus bat coronavirus 1 (bat-CoV 1), Miniopterus bat coronavirus HKU8, Rhinolophus bat coronavirus HKU2, Scotophilus bat coronavirus 512, and Alphacoronavirus 1, which includes transmissible gastroenteritis virus (TGEV), feline coronavirus (FCoV), and canine coronavirus (CCoV). The genus Betacoronavirus consists of seven species: Betacoronavirus 1, Human coronavirus HKU1, Murine coronavirus, Severe acute respiratory syndrome-related coronavirus (SARS-CoV), Pipistrellus bat-CoV HKU5, Rousettus bat coronavirus HKU9, and Tylonycteris bat coronavirus HKU4. In addition to the species Avian coronavirus, which includes infectious bronchitis virus (IBV) and TCoV, the genus Gammacoronavirus contains the species Beluga whale coronavirus SW1 [9, 10].
The major structural proteins of TCoV include the nucleocapsid (N) protein, S protein, small envelope (E) protein and membrane (M) protein [11]. The S protein is the largest structural protein of coronaviruses and a major inducer of neutralizing antibodies [12, 13]. Antibodies to the S protein expressed in mammalian cells, insect cells, Escherichia coli, or with other viral vectors can neutralize coronavirus in vitro and provide protection against lethal virus challenge [13–17]. The spike structure of coronaviruses is a homotrimer of the S protein, which is composed of an N-terminal subunit (S1) and a C-terminal subunit (S2) [18, 19]. S1 forms the globular head and contains the receptor-binding domain (RBD) for the determination of host and tissue tropism [20–23]. S2 forms the stalk of the spike homotrimer, which anchors it to the coronavirus envelope through a transmembrane domain containing heptad repeat and fusion peptides [24, 25]. Due to variations in the S proteins of different coronaviruses and the methods used to find neutralizing epitopes on the S protein, no conclusion has been made about the locations of neutralizing epitopes except that S1 carries more neutralizing epitopes [26–32] than S2 [33–36] in members of many species.
Many approaches have been used to locate the neutralizing epitopes recognized by neutralizing antibodies on the S proteins of coronavirus. Most of them have involved the characterization of neutralizing monoclonal antibodies (MAbs) by using competition enzyme-linked immunosorbent assays (ELISAs) with a panel of synthesized peptides [29], by screening phage display libraries [27, 37], or by characterizing recombinant fragments [28, 30, 33]. In some studies, neutralizing epitopes were found by analyzing neutralizing-MAb-resistant virus variants [31, 32, 38, 39]. Some neutralizing epitopes appear to be composed of a consecutive sequence [29], and some epitopes can only be brought into close proximity by the folding of the protein [28, 37]. A single amino acid change within an antigenic domain of the S1 protein of bovine coronavirus (BCoV) allows the altered BCoV to escape neutralization [32], and single amino acid substitutions in the S1 and S2 subunits of SARS-CoV have resulted in a neutralization-escape phenotype [39]. In some studies, the whole sequence of the target protein was scanned to locate antigenic domains [27], and in other studies, a specific fragment of the target protein for epitope mapping was chosen based on previous references [29], the reactivity of MAb library [40] or the antigenicity of amino acid sequences calculated by prediction software [28]. Immunodominant fragments containing epitopes have been determined based on their reactivity with mono- or polyclonal antibodies.
Identification of neutralizing epitopes on the surface of the immunodominant S protein is pivotal not only for the understanding of humoral immune responses induced by TCoV but also for the development of diagnostic reagents and effective vaccines for the prevention and control of turkey coronaviral enteritis. Epitopes can be used as diagnostic markers for detection of specific antibodies produced by virus infection [41]. Furthermore, epitopes can be applied to many different forms of vaccines, including peptide vaccine conjugated with carrier protein and adjuvant [29] or recombination with different vaccine vectors, such as attenuated salmonella [42], adenovirus [43–45] or poxvirus [46, 47]. Because they do not contain the whole virus, epitope-based vaccines do not carry the risk of virulence reversion that can occur with a live attenuated vaccine.
Our knowledge of antigenicity and neutralization of structural proteins of TCoV is limited due to difficulties in growing TCoV in a cell culture system [48, 49]. In this paper, the antigenic domains containing neutralizing epitopes in the S protein of TCoV are identified and characterized.
Materials and methods
Turkey eggs and poults
Turkey eggs and 1-day-old turkey poults (British United Turkey of America, BUTA) of both sexes were obtained from Perdue Farm (Washington, IN, USA). Turkey eggs were incubated in an incubator (Jamesway, Indian Trail, NC). Turkey poults were housed in isolated floor pens. Feed and water were provided ad libitum. The protocol for care and use of turkey eggs and turkey poults in the present study was approved and supervised by the Purdue University Animal Care and Use Committee.
Virus stock
TCoV 540 was isolated from the intestines of 28-day-old turkey poults from outbreaks of acute enteritis in Indiana. Affected intestines were homogenized with a 5-fold volume of phosphate-buffered saline (PBS), clarified by centrifugation at 3000 rpm for 10 min at 4°C, and 22-day-old embryonated turkey eggs were inoculated with 200 μl of the filtrate via the amniotic route. The embryo intestines harvested after 3 days of incubation were prepared as a 20% suspension in PBS and homogenized. After centrifugation at 3,000 rpm for 10 min at 4°C, the supernatant was layered on the top of 30% and 60% sucrose and clarified by ultracentrifugation in an SW28 rotor at 24,000 rpm for 3 hours at 4°C in an Optima XL-100K ultracentrifuge (Beckman Coulter, Fullerton, CA, USA). The interface between the 30% sucrose and 60% sucrose layers was collected, placed on the top of a continuous 40-60% sucrose gradient, and clarified by ultracentrifugation at 24,000 rpm for 20 hours at 4°C. A band of buoyant density 1.16-1.24 g/ml containing the virus was collected and saved as a virus stock at −80°C and in liquid nitrogen.
RNA and cDNA
Viral RNA was extracted from the purified virus using RNApure™ reagent (GenHunter, Nashville, TN, USA) and chloroform and precipitated with cold isopropyl alcohol and ethanol. cDNA was obtained from the extracted RNA by reverse transcription, which included 10 min of incubation with the first reaction mixture of RNA, random primers (100 ng/μl), and 10 mM dNTPs at 70°C, 1 min on ice, 5 min of incubation at 25°C after adding the second reaction mixture of 5x first-strand buffer, 0.1 M of DDT, 1U of SuperScript™ III reverse transcriptase, and 40U of RNaseOUT™ (Invitrogen, Carlsbad, CA, USA) in the first reaction mixture, 1 hour of incubation at 50°C, and 15 min of inactivation at 70°C.
Prediction of antigenic sites on the TCoV S protein
Potential antigenic regions on the S protein of TCoV540 were predicted using the EMBOSS:Antigenic program [50, 51], which uses theoretical calculations to estimate the physicochemical properties of amino acid residues. The scores are related to the probability that the amino acid sequence is an antigenic determinant on the basis of empirical data and the distribution of amino acids.
Construction of TCoV S fragments
Ten sets of sense and antisense primers were synthesized (Sigma-Aldrich, St. Louis, MO, USA) to amplify fragments covering the whole S protein gene of TCoV540 (Fig. 1). PCR products amplified using different sets of primers from the cDNA of TCoV540 were purified using a DNA Clean & Concentrator-5 kit (Zymo Research, Orange, CA, USA) and cloned into the vector pTriEx3 (Novagen, Madison, WI, USA) to yield the constructs pTriEx3-S/1R, pTriEx3-12F/3R, pTriEx3-Mod4F/Epi4R, pTriEx3-4F/4R, pTriEx3-5F/5R, pTriEx3-6F/6R and pTriEx3-S1, or into vector pcDNA3.1 (Invitrogen, Carlsbad, CA, USA) to yield the construct pcDNA3.1-Mod4F/Epi4R. To clone the PCR product into the vector pTriEx3 or pcDNA3.1, the PCR products and the vector pTriEx3 or pcDNA3.1 were digested with the restriction enzymes NcoI and KpnI. The digested products were purified and ligated to each other using T4 ligase (Invitrogen, Carlsbad, CA, USA). Competent Top 10 cells were transformed with the cloned plasmid by heat shock at 42°C for 30 sec. After adding super optimal broth with catabolite repression (S.O.C.) medium, the culture was incubated at 37°C for 1 hour and then spread on an LB plate with 100 μg/ml ampicillin. Colonies were screened by PCR, and the colony containing the plasmid was amplified and purified using a QIAprep Spin Miniprep Kit (QIAGEN, Valencia, CA, USA).
Fig. 1.
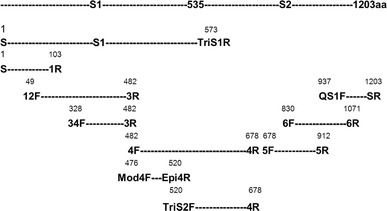
Positions of primers used to amplify fragments covering the whole spike (S) protein of turkey coronavirus (TCoV) isolate 540. The S protein of TCoV has 1203 amino acids and contains an N-terminal subunit (S1) and a C-terminal subunit (S2). The predicted cleavage site is located at amino acid 535. The amino acid (aa) position presented in the figure is from the start codon on the S protein gene of TCoV isolate 540 (GenBank accession number EU022525). The numbers above the names of the forward and reverse primers represent the position of the first or the last amino acid in the expressed S fragments
Characterization of constructed plasmids in vitro
COS-7 cells grown in RPMI-1640 medium with 10% fetal bovine serum were transfected with the constructed plasmids using FuGENE® HD transfection reagent (Roche, Indianapolis, IN, USA) at a ratio of 1:3 of DNA (μg) to reagent (μl). The transfected cells were harvested after incubation at 37°C for 72 hours, fixed in acetone, and washed with PBS. To confirm the expression of cloned S fragments in transfected COS-7 cells, the cells were incubated with anti-His monoclonal antibodies (MAbs) (EMD, San Diego, CA, USA) diluted 1:40, followed by fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (H+L) (KPL, Gaithersburg, MD, USA) diluted 1:100. To determine the antigenicity of the expressed S fragment, the cells were incubated with turkey anti-TCoV serum diluted 1:40, followed by FITC-conjugated goat anti-mouse IgG (H+L) (KPL, Gaithersburg, MD, USA) diluted 1:100.
Expression of recombinant S proteins in E. coli
Recombinant 4F/4R, 6F/6R, and Mod4F/Epi4R S fragments were expressed in transformed E. coli induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 4 hours at 250 rpm in a shaker at 37°C. The cells were harvested, and the proteins were extracted using BugBuster reagent with benzonase and lysozyme (EMD, San Diego, CA, USA). Recombinant proteins that had aggregated in inclusion bodies were dissolved in 6 M urea and purified on Ni2+-charged His-Bind columns (EMD, San Diego, CA, USA). To confirm their size and purity, the purified 4F/4R and 6F/6R S fragments were dissolved in reducing SDS-PAGE sample buffer with β-mercaptoethanol, subjected to 12.5% glycine SDS-PAGE, and transferred onto a nitrocellulose membrane (Millipore, Billerica, MA, USA). Protein Mod4F/Epi4R, with an expected size of about 10 kD for the dimer, was treated with reducing sample buffer and loaded into a 16%/6 M urea tricine SDS-PAGE gel and transferred to a nitrocellulose membrane. The blots were blocked with 1% bovine serum albumin (BSA)-PBS, stained with turkey anti-TCoV serum diluted at a 1:400 dilution at room temperature for 1 hour, and inoculated with 1:400 of horseradish peroxidase (HRP)-conjugated goat anti-turkey IgG (H+L) antibodies at room temperature for 1 hour. The substrate, diaminobenzidine (DAB), was added until the protein bands were visible, and the reaction was stopped by washing the membrane with distilled water.
Generation of polyclonal antibodies to TCoV S fragments
Three experiments were conducted to collect antiserum against individual S fragments recognizing TCoV540. Sera were collected for the determination of the antibody level by ELISA and the virus neutralization (VN) titer by VN assay. In experiment 1, antiserum against 4F/4R (482-687) or S1 (1-573) fragment were collected. For the production of antiserum against the 4F/4R S fragment, two one-week old specific-pathogen-free (SPF) chickens were inoculated intramuscularly (IM) with 1 mg of pTriEx3-4F/4R DNA weekly for 9 weeks and boosted 4 times with the purified 4F/4R S fragment with Freud’s adjuvant one week after the final inoculation with pTriEx3-4F/4R DNA. Three one-week-old SPF chickens were inoculated IM 9 times with the purified 4F/4R S fragment with Freud’s adjuvant at various intervals for a total of 20 weeks. To produce an antiserum against the S1 fragment, one-week-old SPF chickens were injected IM 10 times with 1 or 2 mg of pTriEx3-S1 DNA. In experiment 2, antiserum against 4F/4R, 6F/6R (830-1071) or Mod4F/Epi4R (476-520) S fragment was collected. Two 5-week-old turkeys were inoculated IM 3 times bi-weekly with 500 μg or 1 mg of pTriEx3-6F/6R DNA or pTriEx3-4F/4R with the transfection reagent Lipofectin (Invitrogen, Carlsbad, CA, USA) and boosted 2 times with purified 4F/4R or the 6F/6R S fragment with Freud’s adjuvant to produce antiserum against 6F/6R or 4F/4R S fragment. For production of antiserum against the Mod4F/Epi4R S fragment, one 5-week-old turkey was inoculated IM 4 times with pcDNA3.1-Mod4F/Epi4R DNA. In experiment 3, more antiserum against 4F/4R, 6F/6R, or Mod4F/Epi4R S protein was collected. Two-week-old turkeys were inoculated IM 4 times biweekly with pTriEx3-4F/4R, pTriEx3-6F/6R, or pcDNA3.1-Mod4F/Epi4R DNA with Lipofectin and boosted IM once with purified 4F/4R, 6F/6R, or Mod4F/Epi4R S fragment two weeks after the final inoculation with plasmid DNA.
Enzyme-linked immunosorbent assay (ELISA) for antibodies to TCoV S fragments
An ELISA based on recombinant 4F/4R (482-678) S fragment was used to determine the specific antibody titer to the Mod4F/Epi4R (476-520) S fragment, 4F/4R S fragment, or S1 fragment (1-573), and an ELISA using recombinant 6F/6R (830-1071) S fragment for coating was used to measure the level of antibody to the 6F/6R S fragment. Each well of a 96-well NUNC MaxiSorp™ high-protein-binding-capacity ELISA plate (eBioscience, San Diego, CA, USA) was coated with 2 μg of purified 4F/4R or 6F/6R S fragment diluted with PBS buffer. The positive control (PC) serum was turkey anti-TCoV serum and the negative control (NC) serum was a pool of samples collected from normal turkeys raised in an isolation room. All serum samples were diluted 1:200 in dilution buffer (150 mM phosphate buffer, 0.85% NaCl, 1% BSA, and 0.02% Tween-20) and 100 μl of each diluted serum sample was added to duplicate wells and incubated at 37°C for 1 hour. After washing five times with PBS buffer with 0.05% Tween-20 (PBS-T), 100 μl of HRP-conjugated goat anti-turkey or anti-chicken IgG (L+H) (KPL, Gaithersburg, MD, USA) diluted 1:40,000 in dilution buffer was added to each well, and the plate was incubated at 37°C for 1 hour. The plates were then washed five times with PBS-T, 100 μl/well of tetramethyl benzidine (TMB, Sigma-Aldrich, St. Louis, MO, USA) was added. After 30 min of incubation at room temperature, 100 μl/well of 2 N HCl was added to terminate the color development. The antibody titer was expressed as the absorbance value measured at 450 nm (OD450nm) using a spectrophotometer (Vmax™ kinetic microplate reader, Molecular Devices Corporation, Menlo Park, CA, USA).
Titration by immunofluorescence antibody (IFA) assay on TCoV-infected intestines
Acetone-fixed frozen sections of TCoV-infected intestines were incubated with serially diluted serum against the S1, 4F/4R, Mod4F/Epi4R, or 6F/6R S fragment, followed by a 1:100 dilution of FITC-conjugated goat anti-chicken or turkey IgG (L+H) antibodies. The highest dilution of antibody giving a positive IFA result is the IFA titer of the antiserum to TCoV.
Virus neutralization (VN) assay for antibodies to TCoV S fragments
The serum to be tested was inactivated at 55°C for 30 min and diluted twofold or fourfold serially. The diluted serum was incubated with 20 EID50 of TCoV at 37°C for 1 hour and sequentially used to inoculate 22-day-old turkey embryos. After three days, the embryo intestines were collected and used for IFA with anti-TCoV serum to determine the infectivity of TCoV. Embryonated eggs inoculated with serially diluted anti-TCoV serum were used as a positive control, embryonated eggs without viral inoculation were used as a negative control, and embryonated eggs were inoculated with virus only were used as an inoculation control. Three to five eggs were used for each dilution of each antibody. The accumulated infected ratio and percentage infected were calculated based on the results of TCoV detection by IFA in turkey embryo intestines. The VN titer is the dilution of antibody that can neutralize TCoV and inhibit infection with TCoV in 50% of embryonated eggs.
Results
Antigenicity of TCoV S protein fragments
Amino acid sequence alignment showed that the TCoV S protein has low homology to those of other coronaviruses, even to IBV, which belongs to the same groups (group 3) of coronaviruses. Using the EMBOSS:Antigenic program, the top 10 predicted antigenic sites were identified in both the S1 and S2 regions (Table 1). Therefore, 10 fragments covering the whole S protein gene were designed and cloned into plasmid vectors. The expression of each cloned S protein gene fragment with a His-tag in plasmid-transfected COS-7 cells was confirmed by positive immunofluorescent signals with anti-His monoclonal antibody. The protein expressed in COS-7 cells from transfected plasmid DNA coding for the S1 fragment (1-573),4F/4R (carboxyl terminus of S1 and amino terminus of S2 protein, 482-678), Mod4F/Epi4R (carboxyl terminus of S1 protein, 476-519), and 6F/6R (S2 protein, 830-1071) showed positive reactivity with anti-TCoV serum in the IFA, indicating that the fragments S1, 4F/4R, Mod4F/Epi4R, and 6F/6R contain antigenic epitopes (Fig. 2). Recombinant 4F/4R (26kD), 6F/6R (31 kD), and Mod4F/Epi4R (10 kD for the dimer) S fragments fused with C-terminal His-tag expressed in E. coli and purified by His-Bind columns were detected using anti-TCoV serum (Fig. 3).
Table 1.
Antigenic regions predicted by EMBOSS: Antigenic regions in the spike (S) protein of turkey coronavirus (TCoV) isolate 540. The scores are based on the possibility that the amino acid sequence is an antigenic determinant, calculated by a semi-empirical method using physicochemical properties of amino acid residues. The amino acid sequence was deduced from the S sequence of TCoV 540 (GenBank accession number EU022525)
| Predicted antigenic regions in the spike protein of TCoV540 | |||
|---|---|---|---|
| Rank | Score | Region | Length |
| 1 | 1.302 | 376–391 | 16 |
| 2 | 1.262 | 4–25 | 22 |
| 3 | 1.258 | 1133–1177 | 45 |
| 4 | 1.204 | 957–985 | 29 |
| 5 | 1.185 | 476–511 | 36 |
| 6 | 1.181 | 570–602 | 33 |
| 7 | 1.177 | 604–645 | 42 |
| 8 | 1.175 | 361–368 | 8 |
| 9 | 1.172 | 735–765 | 31 |
| 10 | 1.171 | 32–45 | 14 |
Fig. 2.
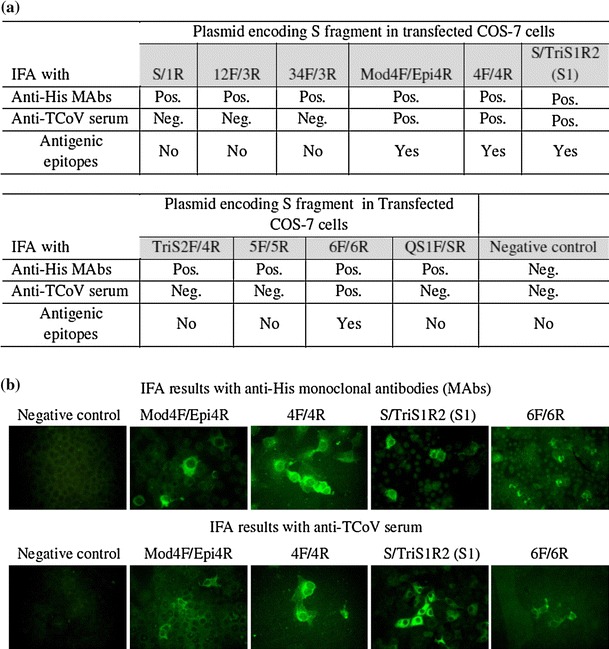
Antigenicity of spike (S) protein fragments of turkey coronavirus (TCoV). IFA results with untransfected COS-7 cells as a negative control or cells transfected with various plasmids encoding fragments covering the whole S protein of TCoV are shown. The positive IFA result obtained with mouse anti-His monoclonal antibodies (Anti-His MAbs) indicates expression of recombinant protein fused with a His-tag from the transfected plasmid in COS-7 cells. The positive IFA result obtained with anti-TCoV serum indicates that the expressed S fragment from the transfected plasmid contained antigenic epitopes. (a) Summary of IFA results for anti-His MAbs or anti-TCoV serum reacting with COS-7 cells transfected with various plasmids encoding TCoV S fragments. (b) Photomicrography of untransfected COS-7 cells as negative acontrol or cells transfected with various plasmids encoding TCoV S fragments in an IFA with anti-His MAbs or anti-TCoV serum. Magnification, 400x
Fig. 3.
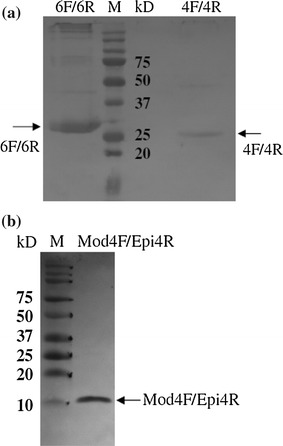
Recombinant 4F/4R (amino acid position 482-678), 6F/6R (830-1071), and Mod4F/Epi4R (476-520) spike (S) proteins of turkey coronavirus (TCoV) expressed in E. coli and purified on His columns can be recognized by anti-TCoV serum in a western blot assay. (a) Recombinant 6F/6R and 4F/4R S proteins transferred from a 12.5% glycine SDS-PAGE gel to a nitrocellulose membrane were stained with 1:400-diluted anti-TCoV serum, followed by 1:400-diluted horseradish peroxidase (HRP)-conjugated goat anti-turkey IgG (H+L) secondary antibodies. The arrows point to 6F/6R S protein at 31 kD and 4F/4R S protein at 26 kD. (b) Recombinant Mod4F/Epi4R S protein transferred from a 16%/6 M urea Tricine SDS-PAGE gel to a nitrocellulose membrane was stained with 1:400-diluted anti-TCoV serum, followed by 1:400 HRP-conjugated goat anti-turkey IgG (H+L) secondary antibodies. The arrow points to a dimer of Mod4F/Epi4R S protein at 10 kD
Antibody generation by priming with TCoV S-fragment-expressing DNA and/or boosting with the corresponding recombinant protein
Turkeys and chickens were immunized with TCoV S fragments that reacted to anti-TCoV serum, and the level of antibodies to TCoV S fragment in their sera was tested weekly by ELISA. The purified 4F/4R S fragment induced the highest level of antibody when compared to that induced by immunization with pTriEx3-S1 DNA or pTriEx3-4F/4R alone in SPF chickens. After boosting with purified 4F/4R S fragment in the chicken inoculated previously with pTriEx3-4F/4R DNA, the antibody level increased dramatically to a level similar to that elicited by the purified 4F/4R S fragment. The ELISA OD450nm value of antibody reached and remained at the highest point two weeks after two doses of purified 4F/4F S fragment in experiment 1. The highest OD450nm value of TCoV 4F/4R or 6F/6R S fragment-coated ELISA was obtained from turkeys primed with pTriEx3-4F/4R DNA, pTriEx3-6F/6R, or pcDNA3.1-Mod4F/Epi4R and boosted once with purified 4F/4R, 6F/6R, or Mod4F/Epi4R S fragment in experiments 1 and 2. Immunization six times with pcDNA3.1-Mod4F/Epi4R DNA induced antibodies at only a slightly higher level than the negative control.
Characterization of polyclonal antibodies to TCoV S protein fragments
The polyclonal antibodies produced by priming with plasmid DNA encoding the TCoV S fragment and/or boosting with the corresponding purified S fragment were characterized by western blotting and IFA. The denatured form of 4F/4R, Mod4F/Epi4R, or 6F/6R S fragments purified from E. coli reacted with the antiserum induced by priming with DNA pTriEx3-4F/4R and boosting with the 4F/4R S fragment, pcDNA3.1-Mod4F/Epi4R with the Mod4F/Epi4R S fragment, or pTriEx3-6F/6R with the 6F/6R S fragment in a western blot. The native form of the S1 protein expressed in COS-7 cells transfected with DNA plasmid pTriEx3-S1 could be recognized by the antiserum raised by priming with DNA pTriEx3-S1 or pTriEx3-4F/4R, with or without the boost using the 4F/4R S fragment. Neither the transfected nor the untransfected cells treated with a normal serum showed a positive signal (Table 2a). Positive fluorescent signals were found in the epithelial cells of TCoV 540-infected turkey intestines prepared in frozen sections after incubation with the antiserum with the highest ELISA OD450nm value. The IFA titer is the highest dilution of the antiserum that can recognize TCoV in the sections of frozen intestines from TCoV-infected turkey embryos. In experiment 1, the IFA titer of antiserum raised by priming with pTriEx3-4F/4R DNA and boosting with 4F/4R S protein, pTriEx3-4F/4R DNA alone, or pTriEx3-S1 DNA alone was 640, 80, and 160, respectively. In experiment 2, the antiserum produced by priming with pTriEx3-4F/4R DNA and boosting with the 4F/4R S fragment, pcDNA3.1-Mod4F/Epi4R DNA alone, and priming with pTriEx3-6F/6R DNA and boosting with the 6F/6R S fragment had the same IFA titer, 640. In experiment 3, the antiserum produced by priming with pTriEx3-4F/4R DNA and boosting with the 4F/4R S fragment had a higher IFA titer (400) than the IFA titer (200) of antiserum produced by priming with pTriEx3-6F/6R DNA or pcDNA3.1-Mod4F/Epi4R and boosting with 6F/6R or Mod4F/Epi4R S fragment (Table 2b). This indicates that antibody to 4F/4R, 6F/6R, or Mod4F/Epi4R S protein can recognize TCoV.
Table 2.
Characterization of polyclonal antibodies induced by the TCoV S fragments S1, 4F/4R, Mod4F/Epi4R, and 6F/6R
| (a) | ||||||
|---|---|---|---|---|---|---|
| Serum used in IFA | COS-7 | COS-7 with pTriEx3-S1 | ||||
| Normal serum | Negative | Negative | ||||
| Anti-TCoV serum | Negative | 1:40 – Positive | ||||
| Anti-DNA pTriEx3-4F/4R | Negative | 1:20 – Positive | ||||
| Anti-DNA pTriEx3-4F/4R & 4F/4R S protein | Negative | 1:40 – Positive | ||||
| Anti-DNA pTriEx3-S1 | Negative | 1:40 – Positive | ||||
| (b) | ||||||
|---|---|---|---|---|---|---|
| Serum | Dilution | |||||
| Trial 1 | 1280 | 2560 | 5120 | 10240 | ||
| Anti-TCoV serum | + | + | + | − | ||
| 40 | 80 | 160 | 320 | 640 | 1280 | |
| Anti-DNA pTriEx3-4F/4R & 4F/4R protein | + | + | + | + | + | − |
| Anti-DNA pTriEx3-4F/4R | + | + | − | − | − | − |
| Anti-DNA pTriEx3-S1 | + | + | + | − | − | − |
| Trial 2 | 1280 | 2560 | 5120 | 10240 | ||
| Anti-TCoV serum | + | + | + | − | ||
| 40 | 80 | 160 | 320 | 640 | 1280 | |
| Anti-DNA pTriEx3-4F/4R & 4F/4R protein | + | + | + | + | + | − |
| Anti-DNA pTriEx3-6F/6R & 6F/6R protein | + | + | + | + | + | − |
| Anti-DNA pcDNA3.1-Mod4F/Epi4R | + | + | + | + | + | − |
| Trial 3 | 50 | 100 | 200 | 400 | 800 | 1600 |
| Anti-TCoV serum | + | + | + | + | + | − |
| Anti-DNA pTriEx3-4F/4R & 4F/4R protein | + | + | + | + | − | − |
| Anti-DNA pTriEx3-6F/6R & 6F/6R protein | + | + | + | − | − | − |
| Anti-DNA pcDNA3.1-Mod4F/Epi4R & Mod4F/Epi4R protein | + | + | + | − | − | − |
Neutralization activity of polyclonal antibodies to TCoV S protein fragments
The neutralization activity of polyclonal antibodies was determined by VN assay to see if the antibodies could inhibit the infection of TCoV in turkey embryonic intestines. The antiserum against 4F/4R or the Mod4F/Epi4R S fragment showed neutralization activity, while the antiserum against the 6F/6R S fragment and normal turkey serum could not inhibit the infection of TCoV in turkey embryonic intestines. To normalize for variations between the different batches of embryonated turkey eggs used in the VN assay, anti-TCoV serum used as the assay control serum was tested every time, and the ratio of VN titer in log4 between the anti-TCoV serum and the antiserum against the S fragments was calculated and compared between assays. The highest ratio of VN titer in log4 against TCoV was 0.93 between the anti-TCoV serum and the antiserum against the 4F/4R S fragment, and 0.81 between the anti-TCoV serum and the antiserum against the Mod4F/Epi4R S fragment collected in experiment 2 (Table 3). This indicates that there are neutralizing epitopes within deduced amino acid position 476 to 519 from the start codon of the TCoV S protein.
Table 3.
Neutralization activities of polyclonal antibodies raised against S1, 4F/4R, Mod4F/Epi4R, and 6F/6R S fragments
| Serum | VN titera | Log4 (VN titer) | Ratio of log4 VN titerb |
|---|---|---|---|
| Trial 1 | |||
| Anti-TCoV serum | 16 | 2 | 1 |
| Anti-DNA pTriEx3-4F/4R & 4F/4R protein | 2 | 0.5 | 0.25 |
| Anti-4F/4R protein | 3.09 | 0.81 | 0.41 |
| Anti-DNA pTriEx3-S1 | 1.7 | 0.38 | 0.19 |
| Trial 2 | |||
| Anti-TCoV serum | 9.44 | 1.62 | 1 |
| Anti-DNA pTriEx3-4F/4R & 4F/4R protein | 8 | 1.5 | 0.93 |
| Anti-DNA pcDNA3.1-Mod4F/Epi4R | 6.2 | 1.32 | 0.81 |
| Anti-DNA pTriEx3-6F/6R & 6F/6R protein | <1 | 0 | 0 |
| Normal serum | <1 | 0 | 0 |
| Trial 3 | |||
| Anti-TCoV serum | 32 | 2.5 | 1 |
| Anti-DNA pTriEx3-4F/4R & 4F/4R protein | 8 | 1.5 | 0.6 |
| Anti-DNA pcDNA3.1-Mod4F/Epi4R | 7.14 | 1.42 | 0.57 |
| Anti-pcDNA3.1-Mod4F/Epi4R & Mod4F/Epi4R protein | 11.43 | 1.76 | 0.7 |
| Anti-DNA pTriEx3-6F/6R & 6F/6R protein | <1 | 0 | 0 |
| Normal serum | <1 | 0 | 0 |
aTheVN titer is the highest dilution of antiserum that can neutralize TCoV and inhibit the infection of TCoV in 50% of 22-day-old embryonated turkey eggs
bThe ratio of log4 VN titer is between the log4 value of the VN titer in the tested serum and the log4 value of the VN titer in the assay control serum, anti-TCoV serum, used in each assay
Discussion
By screening S protein fragments of TCoV expressed in eukaryotic cells using polyclonal antibodies against TCoV S fragments, a region at the carboxyl terminus of the S1 protein (Mod4F/Epi4R) was found to contain neutralizing epitopes. The antiserum raised in turkeys using Mod4F/Epi4R S fragment recognized the denatured S protein and the whole virus in turkey intestines infected with TCoV. To the best of our knowledge, this is the first report to address the S protein domain containing the neutralizing epitopes in TCoV.
B-cell antigenic epitopes on the S protein of IBV have been identified, and these epitopes induce virus-neutralizing antibodies and confer protection against virulence viruses [29, 31]. Since the genetic identity between S protein of TCoV and S protein of IBV is low [52], the information about antigenic epitopes on the S protein of IBV may not applicable for TCoV. From the findings of the present study, the antigenic domain containing the neutralizing epitopes was located at the carboxyl terminus of the S1 protein, similar to what was found with IBV [29]. This implies that the S protein of TCoV must have a structure similar to that of the S protein of IBV, although the amino acid sequences in the antigenic region containing the neutralizing epitopes only shared 30% genetic identity between TCoV and IBV. However, the amino acid identity of the same region is more than 90% among TCoV isolates from Indiana, Minnesota, North Carolina, and Texas [52]. In a previous antigenicity study of 18 different TCoV isolates from various geographic areas in the U.S., TCoV isolates reacted to polyclonal antibodies to different TCoV isolates and the IBV Massachusetts strain as well as monoclonal antibodies to the M protein of IBV, but not to the S protein of IBV [2]. However, virus cross-neutralization assays with TCoV/IN-517/94, TCoV/VA-74/03, and TCoV/TX-1038/98 showed very little cross-neutralization activity of antiserum to heterogeneous TCoV [53], while there was only one amino acid differences between TCoV/IN-517/94 and TCoV/TX-1038/98, and three amino acid differences between TCoV/IN-517/94 and TCoV/VA-74/03 or between TCoV/TX-1038/98 and TCoV/VA-74/03 in the antigenic domain identified in the present study (Fig. 4). Since three different amino acids in TCoV isolates had more cross-neutralization activity than one different amino acid, all three amino acids may be critical residues for neutralization.
Fig. 4.

Results of deduced amino acid sequence alignment in the antigenic domain containing neutralizing epitopes (Mod4F/Epi4R S fragment) from TCoV isolates. TCoV 540, TCoV/TX-1038/98, TCoV/IN-517/94, and TCoV/VA-74/03 showed four different amino acid differences among four TCoV isolates and three differences among TCoV isolates excluding TCoV 540. The sequences of TCoV 540, TCoV/TX-1038/98, TCoV/IN-517/94, and TCoV/VA-74/03 are from GenBank accession numbers EU022525, GQ427176, GQ427175, and GQ4271731, respectively
The antigenic domains identified in the present study, Mod4F/Epi4R (476-520), 4F/4R (482-678), and 6F/6R (830-1071) were included in the fourth to seventh regions predicted by the EMBOSS:Antigenic program. There were two predicted antigenic regions located in the 4F/4R S fragment. These fragments have hydrophobic residues on the surface of protein. Nevertheless, the highly antigenic 6F/6R S fragment induced no neutralizing antibodies.
The conformation and glycosylation of the S protein have been shown to be important for the induction of neutralization antibodies [31, 54]. A eukaryotic expression system can provide posttranslational modification to the expressed protein, like glycosylation, and the expressed protein may have a more native conformation than the same protein expressed in a bacterial or insect cell expression system [15]. Therefore, DNA plasmids encoding TCoV S fragments encompassing the entire S protein of TCoV were expressed in COS-7 cells in the present study to ensure the correct conformation and modification of expressed fragments. While it is convenient to screen the antigenicity of S fragments with anti-TCoV serum, the yield of the expressed protein is relatively low, and the cost of production is relatively high for a mammalian cell system compared to bacteria or insect cells. Although the immunogenicity of the receptor-binding domain of SARS-CoV expressed in the E. coli system was lower than that observed using a mammalian or insect cell system, a high level of neutralizing antibodies was still induced to provide protection against infection by SARS-CoV, especially for the conformation-independent neutralization epitopes present in the receptor domain [15]. By expression in the E. coli system, a sufficient amount of S fragment was produced and used to generate antiserum for ELISA, IFA, and VA assays to characterize each antigenic fragment. Because the Mod4F/Epi4R and 4F/4R S fragments expressed in E. coli showed strong immunogenicity and neutralizing activity, the neutralization epitopes in these antigenic domains are considered conformation and glycosylation independent.
Because polyclonal antibodies generated by priming with DNA encoding TCoV 4F/4R or Mod4F/Epi4R S fragment and boosting with the corresponding S fragment expressed in E. coli were able to neutralize TCoV infection in embryonated turkey eggs, such a prime-boost approach may offer a potential vaccination strategy against TCoV infection. It has been shown that subunit vaccine containing a coronaviral S fragment with the neutralizing epitopes induced antibody production, providing protection against IBV in chickens or SARS-CoV in mice [28, 29]. A DNA vaccine encoding a coronaviral S fragment containing neutralizing epitopes has been shown to enhance the induction of a protective immune response [55, 56]. Mice primed intramuscularly with S1 DNA and boosted with recombinant S1 protein produced in E. coli produced a higher titer of neutralizing antibody against SARS-CoV infection than a DNA vaccine or S1 protein alone [57]. Therefore, a DNA prime-protein boost regimen may elicit a stronger immune response by combining the advantages of a DNA vaccine and a protective subunit vaccine. While DNA vaccines have the ability to mimic the effects of a live attenuated vaccine and induce humoral and cellular immune responses, highly immunogenic subunit vaccines can overcome the low immunogenicity observed with DNA vaccination [57–59].
In the present study, we have identified one antigenic domain containing neutralization epitopes by using both eukaryotic and prokaryotic expression systems. Further localization of critical residues in neutralization epitopes can be achieved by screening peptide libraries with neutralizing MAbs generated from the identified domain or point mutations on further identified peptides recognized by neutralizing MAbs. Although neutralizing antibodies can be raised using the Mod4F/Epi4R fragment, it remains to be confirmed that this fragment can induce protective immunity and serve as a potential subunit vaccine against TCoV infection. Additional studies are needed to fully understand the neutralization mechanism of anti-TCoV antibodies and the immunoprotective nature of the S protein.
Acknowledgments
The authors thank Perdue Farm (Thorntown, IN, USA) for providing turkey eggs and turkey poults for study.
References
- 1.Guy JS. Turkey Coronavirus Enteritis. In: Saif YM, Barnes HJ, Glisson JR, Fadly AM, McDougald LR, Swayne DE, editors. Disease of poultry. 11. Ames: Iowa State University Press; 2003. pp. 300–307. [Google Scholar]
- 2.Lin TL, Loa CC, Wu CC, Bryan T, Hooper T, Schrader D. Antigenic relationship of turkey coronavirus isolates from different geographic locations in the United States. Avian Dis. 2002;46:466–472. doi: 10.1637/0005-2086(2002)046[0466:AROTCI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 3.Dea S, Marsolais G, Beaubien J, Ruppanner R. Coronaviruses associated with outbreaks of transmissible enteritis of turkeys in Quebec: hemagglutination properties and cell cultivation. Avian Dis. 1986;30:319–326. doi: 10.2307/1590535. [DOI] [PubMed] [Google Scholar]
- 4.Gomaa MH, Yoo D, Ojkic D, Barta JR. Seroprevalence of turkey coronavirus in North American turkeys determined by a newly developed enzyme-linked immunosorbent assay based on recombinant antigen. Clin Vaccine Immunol. 2008;15:1839–1844. doi: 10.1128/CVI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavanagh D, Mawditt K, Sharma M, Drury SE, Ainsworth HL, Britton P, Gough RE. Detection of a coronavirus from turkey poults in Europe genetically related to infectious bronchitis virus of chickens. Avian Pathol. 2001;30:355–368. doi: 10.1080/03079450120066368. [DOI] [PubMed] [Google Scholar]
- 6.Teixeira MC, Luvizotto MC, Ferrari HF, Mendes AR, da Silva SE, Cardoso TC. Detection of turkey coronavirus in commercial turkey poults in Brazil. Avian Pathol. 2007;36:29–33. doi: 10.1080/03079450601102939. [DOI] [PubMed] [Google Scholar]
- 7.Gomaa MH, Yoo D, Ojkic D, Barta JR. Virus shedding and serum antibody responses during experimental turkey coronavirus infections in young turkey poults. Avian Pathol. 2009;38:181–186. doi: 10.1080/03079450902751863. [DOI] [PubMed] [Google Scholar]
- 8.Loa CC, Lin TL, Wu CC, Bryan T, Thacker HL, Hooper T, Schrader D. Humoral and cellular immune responses in turkey poults infected with turkey coronavirus. Poult Sci. 2001;80:1416–1424. doi: 10.1093/ps/80.10.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.http://www.ictvonline.org/virusTaxonomy.asp?version=2009. Accessed 29 March 2011
- 10.Woo PC, Lau SK, Huang Y, Yuen KY. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med. 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 11.Lin TL, Loa CC, Tsai SC, Wu CC, Bryan TA, Thacker HL, Hooper T, Schrader D. Characterization of turkey coronavirus from turkey poults with acute enteritis. Vet Microbiol. 2002;84:179–186. doi: 10.1016/S0378-1135(01)00447-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ignjatovic J, Galli L. The S1 glycoprotein but not the N or M proteins of avian infectious bronchitis virus induces protection in vaccine chickens. Arch Virol. 1994;138:117–134. doi: 10.1007/BF01310043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taguchi F, Kubo H. Localization of neutralizing epitopes and receptor-binding site in murine coronavirus spike protein. Adv Exp Med Biol. 1995;380:359–365. doi: 10.1007/978-1-4615-1899-0_58. [DOI] [PubMed] [Google Scholar]
- 14.Daniel C, Talbot PJ. Protection of mice from lethal coronavirus MHV-A59 infection by monoclonal affinity-purified spike glycoprotein. Adv Exp Med Bio. 1990;276:205–210. doi: 10.1007/978-1-4684-5823-7_28. [DOI] [PubMed] [Google Scholar]
- 15.Du L, Zhao G, Chan CC, Sun S, Chen M, Liu Z, Guo H, He Y, Zhou Y, Zheng BJ, Jiang S. Recombinant receptor-binding domain of SARS-CoV spike protein expressed in mammalian, insect and E. coli elicits potent neutralizing antibody and protective immunity. Virology. 2009;393:144–150. doi: 10.1016/j.virol.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishi K, Hasegawa H, Nagata N, Ami Y, Fukushi S, Taguchi F, Tsunetsugu-Yokota Y. Neutralizing antibody against severe acute respiratory syndrome (SARS)-coronavirus spike is highly effective for the protection of mice in the murine SARS model. Microbiol Immunol. 2009;53:75–82. doi: 10.1111/j.1348-0421.2008.00097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson MA, Pooley C, Ignjatovic J, Tyack SG. A recombinant fowl adenovirus expression the S1 gene of infectious bronchitis virus protects against challenge with infectious bronchitis virus. Vaccine. 2003;21:2730–2736. doi: 10.1016/S0264-410X(03)00227-5. [DOI] [PubMed] [Google Scholar]
- 18.Delmas B, Laude H. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J Virol. 1990;64:5367–5375. doi: 10.1128/jvi.64.11.5367-5375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spiga O, Bernini A, Ciutti A, Chiellini S, Menciassi N, Finetti F, Causarono V, Anselmi F, Prischi F, Niccolai N. Molecular modeling of S1 and S2 subunits of SARS coronavirus spike glycoprotein. BBRC. 2003;310:78–83. doi: 10.1016/j.bbrc.2003.08.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godet M, Grosclaude J, Delmas B, Laude H. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J Virol. 1994;68:8008–8016. doi: 10.1128/jvi.68.12.8008-8016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tresnan DB, Holmes KV. Feline aminopeptidase N is a receptor for all group I coronaviruses. Adv Exp Med Biol. 1998;440:69–75. doi: 10.1007/978-1-4615-5331-1_9. [DOI] [PubMed] [Google Scholar]
- 22.Ohtsuka N, Taguchi F. Mouse susceptibility to mouse hepatitis virus infection is linked to viral receptor genotype. J Virol. 1997;71:8860–8863. doi: 10.1128/jvi.71.11.8860-8863.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao J, Wang W, Yuan Z, Jia R, Zhao Z, Xu X, Lv P, Zhang Y, Jiang C, Gao XM. A study on antigenicity and receptor-binding ability of fragment 450–650 of the spike protein of SARS coronavirus. Virology. 2007;359:362–370. doi: 10.1016/j.virol.2006.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madu IG, Roth SL, Belouzard S, Whittaker GR. Characterization of highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J Virol. 2009;83:7411–7421. doi: 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He Y, Zhou Y, Wu H, Luo B, Chen J, Li W, Jiang S. Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: implication for developing SARS diagnostics and vaccines. J Immunol. 2004;173:4050–4057. doi: 10.4049/jimmunol.173.6.4050. [DOI] [PubMed] [Google Scholar]
- 27.Hu H, Li L, Kao RY, et al. Screening and identification of linear B-cell epitopes and entry-blocking peptide of severe acute respiratory syndrome (SARS)-associated coronavirus using synthesized overlapping peptide library. J Comb Chem. 2005;7:648–656. doi: 10.1021/cc0500607. [DOI] [PubMed] [Google Scholar]
- 28.Zhou T, Wang H. An exposed domain in the severe acute respiratory syndrome coronavirus spike protein induces neutralizing antibodies. J Virol. 2004;78:7217–7226. doi: 10.1128/JVI.78.13.7217-7226.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ignjatovic J, Sapats S. Identification of previously unknown antigenic epitopes on S and N proteins of avian infectious bronchitis virus. Arch Virol. 2005;150:1813–1831. doi: 10.1007/s00705-005-0541-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubo H, Yamada YK, Taguchi F. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J Virol. 1994;68:5403–5410. doi: 10.1128/jvi.68.9.5403-5410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore KM, Jackwood MW, Hilt DA. Identification of amino acids involved in a serotype and neutralization specific epitope within the s1 subunit of avian infectious bronchitis virus. Arch Virol. 1997;142:2249–2256. doi: 10.1007/s007050050239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoo D, Deregt D. A single amino acid change within antigenic domain II of the spike protein of bovine coronavirus confers resistance to virus neutralization. Clin Diagn Lab Immunol. 2001;8:297–302. doi: 10.1128/CDLI.8.2.297-302.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keng CT, Zhang A, Shen S, Lip KM, Fielding BC, Tan TH, Chou CF, Loh CB, Wang S, Fu J, Yang X, Lim SG, Hong W, Tan YJ. Amino acids 1055 to 1192 in the S2 region of severe acute respiratory syndrome coronavirus S protein induce neutralizing antibodies: implications for the development of vaccines and antiviral agents. J Virol. 2005;79:3289–3296. doi: 10.1128/JVI.79.6.3289-3296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Wang G, Li J, Nie Y, Shi X, Lian G, Wang W, Yin X, Zhao Y, Qu X, Ding M, Deng H. Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies. J Virol. 2004;78:6938–6945. doi: 10.1128/JVI.78.13.6938-6945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun D, Feng L, Shi H, Chen J, Cui X, Chen H, Liu S, Tong Y, Wang Y, Tong G. Identification of two novel B cell epitopes on porcine epidemic diarrhea virus spike protein. Vet Microbiol. 2008;131:73–81. doi: 10.1016/j.vetmic.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu MW, Scott JK, Fournier A, Talbot PJ. Characterization of murine coronavirus neutralization epitopes with phage-displayed peptides. Virology. 2000;271:182–196. doi: 10.1006/viro.2000.0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhong L, Haynes L, Struble EB, Tamin A, Virata-Theimer ML, Zhang P. Antibody-mediated synergy and interference in the neutralization of SARS-CoV at an epitope cluster on the spike protein. Biochem Biophys Res Commun. 2009;390:1056–1060. doi: 10.1016/j.bbrc.2009.10.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kida K, Hohdatsu T, Fuji K, Koyama H. Selection of antigenic variants of the S glycoprotein of feline infectious peritonitis virus and analysis of antigenic sites involved in neutralization. J Vet Med Sci. 1999;61:935–938. doi: 10.1292/jvms.61.935. [DOI] [PubMed] [Google Scholar]
- 39.Mituki YY, Ohnishi K, Takgi H, Oshima M, Yamamoto T, Mizukoshi F, Terahara K, Kobayashi K, Yamamoto N, Yamaoka S, Tsunetsugu-Yokota Y. A single amino acid substitution in the S1 and S2spike protein domains determines the neutralization escape phenotype of SARS-CoV. Microbes Infect. 2008;10:908–915. doi: 10.1016/j.micinf.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niesters HG, Bleumink-Pluym NM, Osterhaus AD, Horzinek MC, van der Zeijst BA. Epitopes on the peplomer protein of infectious bronchitis virus strain M41 as defined by monoclonal antibodies. Virology. 1987;161:511–519. doi: 10.1016/0042-6822(87)90145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu L, Manopo I, Leung BP, Chng HH, Ling AE, Chee LL, Ooi EE, Chan S-W, Kwang J. Immunological characterization of the spike protein of the severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2004;42:1570–1576. doi: 10.1128/JCM.42.4.1570-1576.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen H, Schifferli DM. Construction, characterization, and immunogenicity of an attenuated Salmonella enterica serovar typhimurium pgtE vaccine expressing fimbriae with integrated viral epitopes from the spiC promoter. Infect Immun. 2003;71:4664–4673. doi: 10.1128/IAI.71.8.4664-4673.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Callebaut P, Enjuanes L, Pensaert M. An adenovirus recombinant expressing fimbriae with integrated viral epitopes from the spic promoter. Infect Immun. 1996;71:309–313. [Google Scholar]
- 44.Wesseling JG, Godeke GJ, Schijns VE, Prevec L, Graham FL, Horzinek MC, Rottier PJ. Mouse hepatitis virus spike and nucleocapsid proteins expressed by adenovirus vectors protect mice against a lethal infection. J Gen Virol. 1993;74:2061–2069. doi: 10.1099/0022-1317-74-10-2061. [DOI] [PubMed] [Google Scholar]
- 45.Zakhartchouk AN, Viswanathan S, Mahony JB, Gauldie J, Babiuk LA. Severe acute respiratory syndrome coronavirus nucleocapsid protein expressed by an adenovirus vector is phosphorylated and immunogenic in mice. J Gen Virol. 2005;86:211–215. doi: 10.1099/vir.0.80530-0. [DOI] [PubMed] [Google Scholar]
- 46.Hebben M, Duguesne V, Cronier J, Rossi B, Aubert A. Modified vaccinia virus Ankara as a vaccine against feline coronavirus: immunogenicity and efficacy. J Feline Med Surg. 2004;6:111–118. doi: 10.1016/j.jfms.2003.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Schnitzlein WM, Tripathy DN, Girshick T, Khan MI. Construction and immunogenicity studies of recombinant fowl poxvirus containing the S1 gene of Massachusetts 41 strain of infectious bronchitis virus. Avian Dis. 2002;46:831–838. doi: 10.1637/0005-2086(2002)046[0831:CAISOR]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 48.Breslin JJ, Smith LG, Guy JS. Baculovirus expression of turkey coronavirus nucleocapsid protein. Avian Dis. 2001;45:136–143. doi: 10.2307/1593020. [DOI] [PubMed] [Google Scholar]
- 49.Guy JS, Barnes HJ, Smith LG, Breslin J. Antigenic characterization of a turkey coronavirus identified in poult enteritis- and mortality syndrome-affected turkeys. Avian Dis. 1997;41:583–590. doi: 10.2307/1592148. [DOI] [PubMed] [Google Scholar]
- 50.Kolaskar AS, Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276:172–174. doi: 10.1016/0014-5793(90)80535-Q. [DOI] [PubMed] [Google Scholar]
- 51.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 52.Loa CC, Wu CC, Lin TL. Comparison of 3′-end encoding regions of turkey coronavirus isolates from Indiana, North Carolina, and Minnesota with chicken infectious bronchitis coronavirus strains. Intervirology. 2006;49:230–238. doi: 10.1159/000091470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jackwood MW, Boynton TO, Hilt DA, McKinley ET, Kissinger JC, Paterson AH, Robertson J, Lemke C, McCall AW, Williams SM, Jackwood JW, Byrd LA. Emergence of a group 3 coronavirus through recombination. Virology. 2009;398:98–108. doi: 10.1016/j.virol.2009.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lenstra JA, Kusters JG, Koch G, van der Zeijst BAM. Antigenicity of the peplomer protein of infectious bronchitis virus. Mol Immunol. 1989;25:7–15. doi: 10.1016/0161-5890(89)90014-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan B, Wang H, Shang L, Yang T. Coadministration of chicken GM-CSF with a DNA vaccine expressing infectious bronchitis virus (IBV) S1 glycoprotein enhances the specific immune response and protects against IBV infection. Arch Virol. 2009;154:1117–1124. doi: 10.1007/s00705-009-0424-7. [DOI] [PubMed] [Google Scholar]
- 56.Yang ZY, Kong WP, Huang Y, Roberts A, Murphy BR, Subbarao K, Nabel GJ. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woo PC, Lau SK, Tsoi HW, Chen ZW, Wong BH, Zhang L, Chan JKH, Wong L, He W, Ma C, Chan K, Ho DD, Yuen K. SARS coronavirus spike polypeptide DNA vaccine priming with recombinant spike polypeptide from Escherichia coli as booster induces high titer of neutralizing antibody against SARS coronavirus. Vaccine. 2005;23:4959–4968. doi: 10.1016/j.vaccine.2005.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laddy DJ, Weiner DB. From plasmids to protection: a review of DNA vaccines against infectious diseases. Int Rev Immunol. 2006;25:99–123. doi: 10.1080/08830180600785827. [DOI] [PubMed] [Google Scholar]
- 59.Coban C, Koyama S, Takeshita F, Akira S, Ishii KJ. Molecular and cellular mechanisms of DNA vaccines. Hum Vaccines. 2008;4:453–456. doi: 10.4161/hv.4.6.6200. [DOI] [PubMed] [Google Scholar]


