Abstract
Interferon gamma (IFN-γ) is best known for its ability to regulate host immune responses; however, its direct antiviral activity is less well studied. Transmissible gastroenteritis virus (TGEV) is an economically important swine enteric coronavirus and causes acute diarrhea in piglets. At present, little is known about the function of IFN-γ in the control of TGEV infection. In this study, we demonstrated that IFN-γ inhibited TGEV infection directly in ST cells and intestine epithelial IPEC-J2 cells and that the anti-TGEV activity of IFN-γ was independent of IFN-α/β. Moreover, IFN-γ suppressed TGEV infection in ST cells more efficiently than did IFN-α, and the combination of IFN-γ and IFN-α displayed a synergistic effect against TGEV. Mechanistically, using overexpression and functional knockdown experiments, we demonstrated that porcine interferon regulatory factor 1 (poIRF1) elicited by IFN-γ primarily mediated IFN-γ signaling cascades and the inhibition of TGEV infection by IFN-γ. Importantly, we found that TGEV elevated the expression of poIRF1 and IFN-γ in infected small intestines and peripheral blood mononuclear cells. Thus, IFN-γ plays a crucial role in curtailing enteric coronavirus infection and may serve as an effective prophylactic and/or therapeutic agent against TGEV infection.
Introduction
Type II interferon gamma (IFN-γ), which is primarily produced by NK cells, T cells, and macrophages, is a pleiotropic cytokine that has antiviral activity and immunomodulatory functions [1]. Although IFN-γ was originally discovered due to its ability to “interfere” with virus infection, most studies of IFN-γ have focused on its immunomodulatory effects in both innate and adaptive immunity, such as enhancement of NK- and T-cell-mediated cytotoxicity, B-cell differentiation, surface antigen expression, and macrophage activation [2, 3]. Previous studies have demonstrated that IFN-γ can inhibit infection with Ebola virus (EBOV) [4], Zika virus [5], severe acute respiratory syndrome-associated coronavirus (SARS-CoV) directly [6]. However, little is known about the antiviral activity of IFN-γ in porcine enteric coronavirus infection or the mechanism by which IFN-γ inhibits enteric coronaviruses.
Transmissible gastroenteritis virus (TGEV) is an economically important enteric coronavirus that causes acute viral enteritis [7]. TGEV infection causes vomiting, severe diarrhea, and dehydration, and the disease caused by this virus is the most severe in neonates, resulting in high morbidity and mortality in neonatal piglets [8–10]. Previous reports have shown that TGEV infection in vitro and in vivo induces the production of a significant amount of IFNs including type I IFNs [11–13]. TGEV primarily infects intestinal epithelia in vivo, although infection of dendritic cells and macrophages by TGEV has also been observed [14, 15]. Most of the previous studies focused primarily on the capacity of type I IFNs (IFN-α/β) to control TGEV infection. The ability of type II IFN-γ to inhibit TGEV infection has not been well explored, although many non-lymphoid cells, including epithelia, express IFN-γ receptors (IFNGR) [16]. The heterodimeric IFNGR complex is made up of IFNGR1 and IFNGR2, is highly expressed on myeloid cells and lymphoid cells, and is also widely present in most types of cells, including intestinal epithelia [16, 17]. It is well established that the presence of antigen-specific IFN-γ+ T cells is correlated with protection against rotaviruses, which, like TGEV, primarily infects intestinal epithelia and cause acute diarrhea [18]. IFN-γ is the only cytokine identified in stimulated CD4+ T cell supernatants to inhibit rotavirus infection directly [19]. While studies of rotavirus infection have indicated that IFN-γ plays a crucial role in curtailing enteric viral infection, the importance of IFN-γ in protection against TGEV infection has not been established.
IFN-γ, similar to type I IFNs, stimulates the expression of a number of interferon-stimulated genes (ISGs) directly and exerts antiviral activity [20, 21]. However, many studies have demonstrated that the signaling cascades initiated by IFN-γ and type I IFNs are not identical, although there is a degree of overlap between them [22]. Previous studies have also shown that the mechanism by which IFN-γ exerts its antiviral activity can vary according to the target cell type [23]. Here, we demonstrate that IFN-γ inhibited TGEV infection directly in both ST cells and intestinal epithelial cells and, when combined with IFN-α synergistically suppresses TGEV infection. We also show that porcine IRF1 plays a critical role in IFN-γ-induced signaling and mediates the inhibition of TGEV by IFN-γ. Given the important role of IFN-γ in modulating T cell immunity in addition to its direct antiviral activity, the potential of IFN-γ as an effective therapeutic agent against enteric coronavirus infection may have been underestimated.
Materials and methods
Cell culture, viruses and virus infection
Swine testicular (ST) cells, IPEC-J2 cells, Vero E6 cells and IFNLR knockout Vero E6 cells (knocked out by CRISP-cas9) were grown in Dulbecco’s modified Eagle’s medium (Gibco, USA) supplemented with 8% fetal bovine serum (Gibco) and antibiotics (100 μg of streptomycin and 100 U of penicillin per mL) at 37 °C in a humidified atmosphere of 5% CO2. The H87 strain of TGEV, derived from the virulent strain H16 (GenBank accession no. FJ755618), and the recombinant TGEV infectious clone with green fluorescent protein (GFP) (TGEV-GFP) were propagated and titrated in ST cells. For TGEV infection, ST cells were infected with TGEV at an MOI of 0.01 or mock infected with DMEM. After a 2-h incubation at 37 °C, ST cells were washed three times with PBS to removing the unbound viruses, and the cells were cultured in serum-free DMEM at 37 °C until harvest.
shRNAs, plasmid construction and transfection
Short hairpin RNAs (shRNAs) against porcine IRF1 (shIRF1 s) were synthesized by GenePharma (Shanghai GenePharma, China), and their sequences are listed in Table 1. To construct the pHA-poIRF1 expression vector, the porcine IRF1 gene was amplified by polymerase chain reaction (PCR) and cloned into the pCAGGS-HA vector (Clontech, USA). The pHA-poIRF1 recombinant plasmid was confirmed by sequencing. ST cells were grown in 24-well plates to 70%-80% confluence before transfection with shIRF1 or nontarget control (NC) shRNA using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s protocol.
Table 1.
Sequences of short hairpin (sh) RNAs and primers
| Short hairpin RNA/primer Sequence (5’-3’) |
|---|
| shIRF1#1 GGAAAGAGAGAAAGTCCAAGT |
| shIRF1#2 GGACATTGAACAGGCCCTTAC |
| IRF1-qPCR-F GCAACAGATGAGGACGAG |
| IRF1-qPCR-R GCTTTCAACTTCTGGCTC |
| IRF1-EcoRI-F CCGGAATTCATGCCCATCACTCGGATGC |
| IRF1-NheI –R CTAGCTAGCCTACGGTGCACAAGGAATG |
| CXCL9-qPCR-F ACCTCTTTCTCCCACTC |
| CXCL9-qPCR-R AAGGAACCCTACTAATG |
| CXCL10-qPCR-F TGCCCACATGTTGAGATCAT |
| CXCL10-qPCR-R CGGCCCATCCTTATCAGTAG |
| TGEV-F GCTTGATGAATTGAGTGCTGATG |
| TGEV-R CCTAACCTCGGCTTGTCTGG |
| IFN-γ-qPCR-F GCCAAATTGTCTCCTTC |
| IFN-γ-qPCR-R GTTTCCCAGAGCTACCA |
| IFNAR-qPCR-F GCTGTCGGTATCGGTGCT |
| IFNAR-qPCR-R TGAGTGCTCTGGCTTTGA |
| IFNLR-qPCR-F GCCCGATCTGAACTATGAC |
| IFNLR-qPCR-R GTAGGCTTGGAGAACTTGC |
| IFNGR-qPCR-F CGGGAGCGAGGCTAAGAT |
| IFNGR-qPCR-R TCACCTGGGCGGTAAACA |
RNA isolation, reverse transcription, and qPCR
Total cellular mRNA was extracted using an RNeasy Mini Kit (QIAGEN Sciences, Germany) according to the manufacturer’s protocol. cDNA was synthesized using a PrimeScriptTM II 1st Strand cDNA Synthesis Kit (Takara, China). Quantitative real-time PCR (qPCR) was performed using Power SYBR Green PCR Master Mix reagents (Takara, China). All data were acquired using a LightCycler 480 real-time PCR system (Roche, USA). The sequences of the RT-qPCR primers for amplification of TGEV, CXCL9, CXCL10, poIRF1, IFN-γ, and GAPDH transcripts are listed in Table 1.
Antibodies and IFN-γ
Porcine IFN-γ was purchased from R&D Systems, USA. The antibody against poIRF1 was purchased from Proteintech, USA. An antibody against β-actin was purchased from Sigma-Aldrich, USA. Mouse anti-TGEV nucleocapsid (N) monoclonal antibody was from our laboratory stock.
Preparation of PBMCs
Porcine peripheral blood mononuclear cells (PBMCs) were prepared from 2-week-old specific pathogen-free (SPF) pigs, using lymphocyte separation medium (LSM, 50494X, USA). The PBMCs were resuspended in RPMI-1640 medium (Gibco) supplemented with 5% FBS (Gibco), 2 mM L-glutamine and antibiotics and cultured in the presence or absence of TGEV for 48 h at 37 °C in 96-well plates (100 μL per well at 2 × 107 cells/mL).
Immunofluorescence assay (IFA)
ST cells were fixed with 4% paraformalde for 30 min at 4 °C, permeabilized with 0.2% Triton X-100 for 15 min at room temperature (RT), and blocked with blocking buffer (PBST with 5% milk) for 30 min at 37 °C. The cells were then incubated with anti-HA monoclonal antibody (Sigma-Aldrich) or TGEV N protein antibody at 37 °C for 2 h, followed by labeling with Alexa Fluor 546 goat anti-mouse IgG antibody (Thermo Fisher Scientific) at 37 °C for 1 h. Nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI). The stained cells were visualized using an AMG EVOS F1 fluorescence microscope.
Experimental infection of piglets
Twelve two-day-old SPF piglets were randomly divided into two groups. One group was inoculated orally with a 5-mL suspension containing 1 × 105 of TCID50 TGEV strain H87, and another group was inoculated with DMEM as a mock control. All piglets were euthanized at 48 hpi. Small-intestine samples were collected from the piglets for RT-qPCR. The experiment was approved by the Animal Care and Ethics Committee of Harbin Veterinary Research Institute.
Western blotting
Cells were harvested and treated with NP-40 lysis buffer (Beyotime, China) containing 0.1 mM phenylmethylsulfonyl fluoride (PMSF) (Roche, USA). The protein concentrations were measured by BCA Protein Assay (Beyotime, China). Equal amounts of cells lysates were separated by SDS-PAGE, and the protein in the gels was transferred to nitrocellulose membranes (GE Healthcare, USA). The membranes were blocked with 5% nonfat dry milk in TBST and incubated with poIRF1 or β-actin antibody at 37 °C for 1 h. After washing, the membrane was incubated with HRP-conjugated anti-mouse IgG or HRP-conjugated anti-rabbit IgG (Sigma-Aldrich) in blocking buffer for 1 h at room temperature (RT), and protein bands on the membrane were detected using electrochemiluminescence (ECL) reagent (Thermo Fisher Scientific).
Statistical analysis
All data in the figures are presented as the mean ± standard error of the mean (SEM) from three independent experiments conducted in parallel and were analyzed in GraphPad Prism 7.0 (GraphPad Software, Inc.). Statistical significance was assessed by Student’s t-test. Differences were considered significant if the p-value was < 0.05. p-values are indicated as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; p > 0.05; non-significant (ns).
Results
IFN-γ inhibits TGEV infection in ST cells and small-intestinal epithelial cells
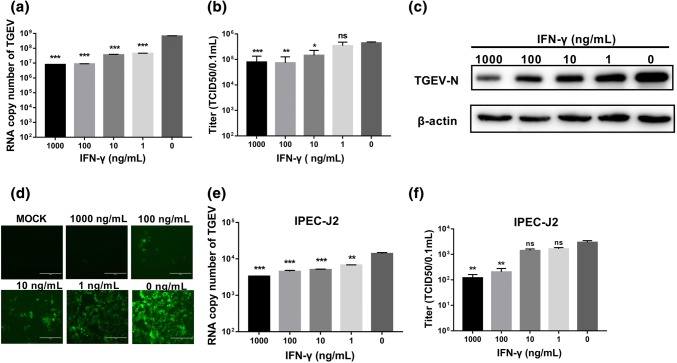
TGEV is generally propagated in ST cells in vitro. To evaluate whether porcine IFN-γ directly inhibits TGEV infection, ST cells were pretreated with different concentrations of IFN-γ and then infected with TGEV. IFN-γ priming significantly reduced both the amount of TGEV genomic RNA and the viral titer when quantified by RT-qPCR and virus titration, respectively (Fig. 1a and b). The suppression of TGEV by IFN-γ was dose dependent, and we found that the amount of TGEV genomic RNA was reduced more than 50-fold at the concentrations of 1000 ng/mL and 100 ng/mL by RT-qPCR. The dose-dependent inhibition of TGEV infection by IFN-γ was confirmed by Western blotting of the TGEV N protein (Fig. 1c). To verify that IFN-γ inhibits TGEV infection directly in ST cells, ST cells were infected with a recombinant TGEV with a green fluorescent protein (TGEV-GFP) marker. IFN-γ almost completely suppressed TGEV-GFP infection at a concentration of 1000 ng/mL, substantially reduced TGEV-GFP infection even at 1 ng/mL, and displayed dose dependence (Fig. 1d). These results demonstrate that IFN-γ inhibits TGEV infection directly in ST cells.
Fig. 1.
IFN-γ inhibits TGEV in a dose-dependent manner. (a, e) ST cells or IEC cells were treated with different concentrations of IFN-γ (1000 ng/mL, 100 ng/mL, 10 ng/mL, 1 ng/mL) for 24 h, followed by infection with TGEV. Cells were harvested at 24 hpi for quantification of the TGEV genome. (c) ST cells were treated with IFN-γ and infected TGEV, and cell lysates were analyzed by Western blotting for detection of the TGEV N protein. (b, f) Cell culture supernatants were collected for virus titration. (d) IFN-γ treatment inhibits infection of ST cells by TGEV-GFP. ST cells were treated with IFN-γ 24 h prior to TGEV-GFP infection. GFP-positive cells at 24 hpi are shown. The data represent the mean ± SEM from three independent experiments. Asterisks indicate statistical significance relative to the no-IFN-γ control. *, p < 0.05; **, p < 0.01; ***, p < 0.001
TGEV primarily infects small-intestinal epithelial cells in vivo. Therefore, we next investigated the antiviral effect of IFN-γ against TGEV infection in intestinal epithelial cells in vitro using the IPEC-J2 cells, which have been reported to be a good in vitro model of porcine enteric infections [24]. Consistent with the results in ST cells, IFN-γ inhibited TGEV infection in IPEC-J2 cells and displayed a dose-dependent effect. At 1000 ng/mL and 100 ng/mL, IFN-γ decreased the TGEV titer more than tenfold when compared with an untreated control (Fig. 1e and f). These data demonstrate that IFN-γ is capable of inhibiting TGEV infection in both ST cells and IPEC-J2 cells.
The inhibition of TGEV infection by IFN-γ does not depend on a type I IFN or type III IFN response
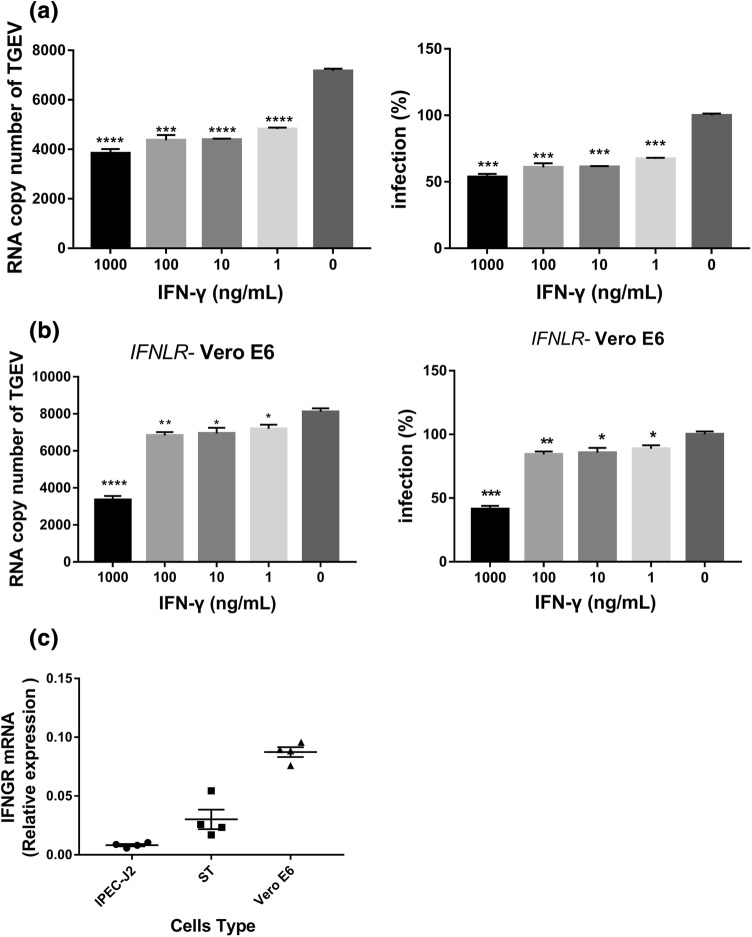
Previous studies have shown that TGEV infection induces significant production of type I IFN-α/β in vitro and in vivo [11, 12, 25], which results in substantial suppression of TGEV infection. To exclude the possibility that the reduction of TGEV infection following IFN-γ stimulation requires the presence of a type I IFN response, we performed IFN-γ anti-TGEV assay in the Vero E6 cell line, which is unable to produce type I IFNs [26]. TGEV infected Vero E6 cells less efficiently than ST cells. Similar to our findings in ST cells and IPEC-J2 cells, IFN-γ pretreatment significantly decreased the amount of TGEV genomic RNA present, even at 1 ng/mL, and displayed a dose-dependent effect compared with the untreated control, indicating that the direct anti-TGEV activity of IFN-γ does not require the response of type I IFNs (Fig. 2a). To exclude the possibility that the TGEV inhibition by IFN-γ is dependent on type III IFN, we performed the inhibition experiment in Vero E6 cells with an IFNLR knockout, making these cells defective for both type I and type III IFNS. We found that IFN-γ still inhibited TGEV infection in the IFNLR knockout Vero E6 cells (Fig. 2b). Taken together, these results indicate that the direct antiviral activity of IFN-γ does not depend on the type I IFN and type III IFN responses.
Fig. 2.
The inhibition of TGEV infection by IFN-γ does not depend on type I IFNs, type III IFN, or the expression of IFNGR in IPEC-J2, ST, and Vero E6 cell lines. (a, b) IFN-γ inhibits TGEV in a dose-dependent manner in Vero E6 cells or IFNLR-knockout Vero E6 cells. Cells were harvested for quantification of the TGEV genome. Asterisks indicate statistical significance relative to the no-IFN-γ control. ***, p < 0.001; ****, p < 0.0001 (c) IPEC-J2, ST, and Vero E6 cell lines were harvested after 24 h for quantification of IFNGR mRNA levels
However, IFN-γ inhibited TGEV less efficiently in Vero E6 cells or IFNLR knockout Vero E6 cells than in ST cells and IPEC-J2 cells. Using 1000 ng of IFN-γ per mL resulted in only about 50% reduction in TGEV (Fig. 2a and b), indicating that the type I IFN and type III IFN response may enhance the antiviral activity of IFN-γ. To exclude the possibility that the observed differences did not result from differences in the expression levels of IFNGR in ST cells, IPEC-J2 cells, and Vero E6 cells, we examined the expression of IFNGR in these three cell lines. We found that all of them expressed IFNGR and that Vero E6 cells had higher levels of IFNGR expression than ST and IPEC-J2 cells (Fig. 2c). However, the inhibition of TGEV infection was less efficient in Vero E6 cells than in ST cells and IPEC-J2 cells, indicating that the differences in inhibition of TGEV infection by IFN-γ were not due to differences in IFNGR expression. Taken together, these results indicate that the direct antiviral activity of IFN-γ does not depend on the type I IFN response.
Effect of poIRF1 on IFN-γ-mediated signaling
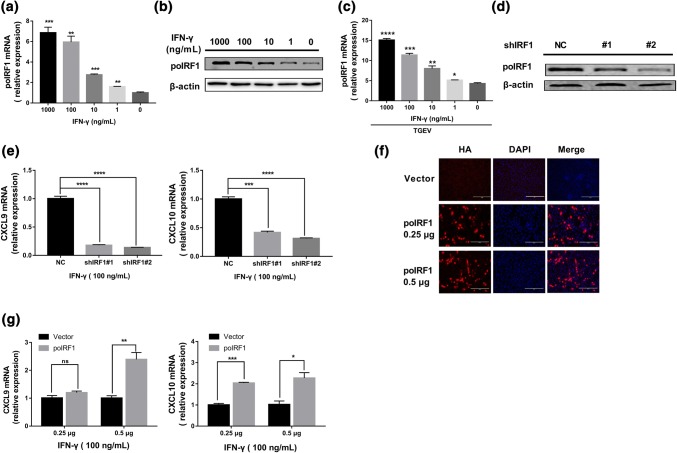
Next, we investigated the mechanism of the anti-TGEV effect of IFN-γ. IFN regulatory factor (IRF) 1 is a crucial regulator of IFN-γ and ISGs [27]. It is constitutively expressed in most cell types, and its transcription is induced by IFN-γ [28]. Moreover, IRF1 is essential for protection of the host against certain classes of viruses [29]. We hypothesized that porcine IRF1 (poIRF1) plays a critical role in IFN-γ-mediated inhibition of TGEV infection in ST cells. Initially, we assessed whether IFN-γ induces poIRF1 expression in ST cells. The levels of poIRF1 mRNA significantly increased following IFN-γ stimulation and displayed a dose-dependent response to IFN-γ (Fig. 3a). The induction of poIRF1 by IFN-γ was verified by Western blotting of poIRF1 (Fig. 3b). Additionally, the expression of poIRF1 also increased after IFN-γ treatment followed by TGEV infection in a manner similar to that observed after IFN-γ treatment alone in ST cells (Fig. 3c). To determine whether poIRF1 is involved in the direct inhibition of TGEV by IFN-γ, we initially evaluated the role of poIRF1 in IFN-γ-induced signaling. The production of CXCL9 and CXCL10 is specifically activated by IFN-γ in different cell types [30]. To determine whether poIRF1 plays an important role in the IFN-γ-mediated induction of CXCL9 and CXCL10, we silenced endogenous poIRF1 expression by using specific shRNAs. ST cells were transfected with poIRF1 shRNA or a non-targeting shRNA (shNC), and the efficiency of poIRF1 knockdown was confirmed by Western blotting (Fig. 3d). shIRF1#1 and #2 led to a 45% and 65% decrease in poIRF1 expression, respectively, when compared with shNC (Fig. 3d). The overexpression of poIRF1 in ST cells was verified by IFA (Fig. 3f). We next examined the mRNA levels of CXCL9 and CXCL10 under conditions of poIRF1 knockdown (Fig. 3e) or overexpression (Fig. 3g) in ST cells following IFN-γ stimulation. IFN-γ substantially upregulated the expression of CXCL9 and CXCL10 in ST cells, and the knockdown of endogenous poIRF1 substantially impaired the IFN-γ-elicited production of CXCL9 and CXCL10 (Fig. 3e), suggesting that poIRF1 plays a crucial role in the induction of CXCL9 and CXCL10 by IFN-γ. Consistent with the results of poIRF1 silencing, the overexpression of poIRF1 significantly increased the expression of CXCL9 and CXCL10 in a dose-dependent manner (Fig. 3g). These data show that poIRF1 plays a vital role in IFN-γ-elicited signaling.
Fig. 3.
IFN-γ elevates expression of poIRF1, and poIRF1 mediates IFN-γ signaling in ST cells. (a, b) Induction of changes in mRNA and protein levels of porcine IRF1 by IFN-γ. ST cells were treated with different concentrations of IFN-γ for 24 h. The level of poIRF1 RNA was measured by RT-qPCR. Cell lysates of ST cells were analyzed by Western blot using an antibody against IRF1. (c) Expression of poIRF1 in TGEV-infected ST cells primed with IFN-γ for 24 h. ST cells pretreated with IFN-γ were infected with TGEV for 24 h, and poIRF1 mRNA was detected by RT-PCR. Significance was determined by Student’s t-test compared to a no-IFN-γ mock control. (d) The knockdown efficiency of poIRF1 by shRNA was determined by Western blotting. (e) Knockdown of poIRF1 suppresses IFN-γ-stimulated genes. ST cells were transfected with NC or poIRF1 shRNA, followed by treatment with 100 ng of IFN-γ per mL, and the expression of CXCL9 and CXCL10 was determined by RT-qPCR. (f) Transient expression of poIRF1 in ST cells. poIRF1 was cloned and expressed using the eukaryotic expression vector pCAGGS-HA. The overexpression efficiency of IRF1 in ST cells was confirmed by IFA. (g) Overexpression of poIRF1 promotes the expression of IFN-γ-stimulated genes. ST cells were transfected with IRF1-HA or empty vector, followed by stimulation with 100 ng of IFN-γ per mL, and the expression of CXCL9 and CXCL10 was determined by RT-qPCR. Data are shown as the mean ± SEM. Significance was determined by Student’s t-test compared to shNC or empty vector. *, p < 0.5; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001
Direct inhibition of TGEV by IFN-γ is mediated through activation of IRF-1 signaling
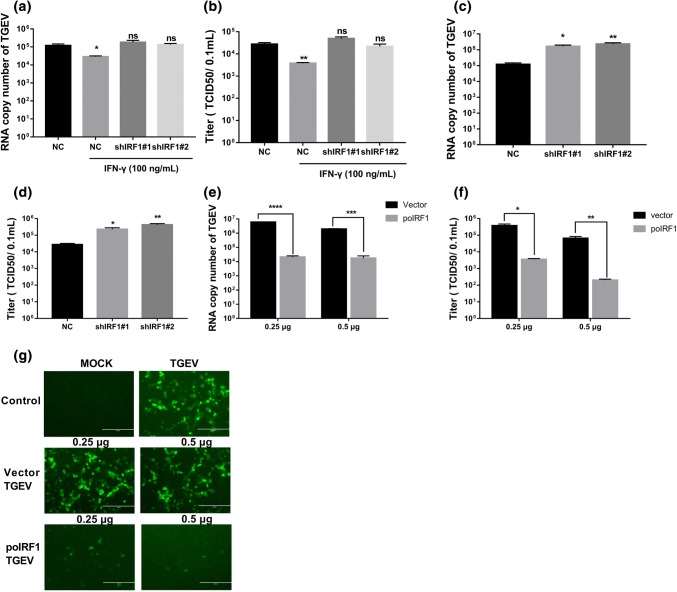
To evaluate whether poIRF1 plays a key role in IFN-γ-mediated TGEV inhibition, we monitored TGEV inhibition of IFN-γ in poIRF1-silenced ST cells. By quantifying TGEV genomic RNA (Fig. 4a) and measuring viral titers (Fig. 4b), we found that IFN-γ significantly inhibited TGEV infection, whereas silencing of endogenous IRF1 by shIRF1 s completely abolished IFN-γ inhibition of TGEV infection, indicating that the direct anti-TGEV activity of IFN-γ is mainly mediated through the poIRF1 signaling pathway. We demonstrated above that IFN-γ treatment substantially increased poIRF1 expression (Fig. 3a and b). To test whether the IFN-γ-mediated TGEV inhibition occurs through the induction of poIRF1, the inhibition of TGEV by poIRF1 itself was evaluated. Knockdown of endogenous poIRF1 by shIRF1#1 and shIRF#2 resulted in more than a tenfold increase in the amount of TGEV genomic RNA (Fig. 4c) and up to a 15-fold elevation of TGEV titers in ST cells (Fig. 4d), indicating that poIRF1 itself is a potent host restrictor of TGEV infection. We next monitored the effect of poIRF1 overexpression on TGEV infection. As expected, poIRF1 overexpression substantially decreased infection by wild-type TGEV (Fig. 4e and f) and reduced TGEV-GFP fluorescence in ST cells, and the reduction of TGEV-GFP fluorescence was related to the expression levels of poIRF1 (Fig. 4g). These data thus indicate that poIRF1 plays a crucial role in protection of the host against TGEV. Collectively, the data show that inhibition by IFN-γ of TGEV infection is mediated primarily through the induction of poIRF1.
Fig. 4.
poIRF1 restricts TGEV infection and plays an essential role in the IFN-γ-mediated inhibition of TGEV infection. (a, b) IRF1 knockdown abolishes IFN-γ-mediated inhibition of TGEV. ST cells were transfected with NC or shIRF1 s and then treated with 100 ng of IFN-γ per mL, followed by infection with TGEV. Cells were harvested at 24 h for quantification of the TGEV genome (a) and determination of the virus titer (b). (c, d) Knockdown of poIRF1 promotes TGEV replication without IFN-γ stimulation. ST cells were transfected with NC or shIRF1 s and subsequently infected with TGEV. After 24 h, cells were harvested for quantification of the TGEV genome (c) and determination of the virus titer (d). (e-g) Transient expression of IRF1 inhibits TGEV infection. ST cells were transfected with poIRF1-HA for 24 h and then infected with wild-type TGEV (e and f) or TGEV-GFP (g). TGEV infection was determined by measuring TGEV genomic RNA (e) and the viral titer (f). GFP-positive cells at 24 h after TGEV-GFP infection are shown (g). Data are shown as the mean ± SEM. Significance was determined by Student’s t-test compared to empty vector. *, p < 0.5; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001
TGEV infection induces the expression of IFN and IRF1 in vivo
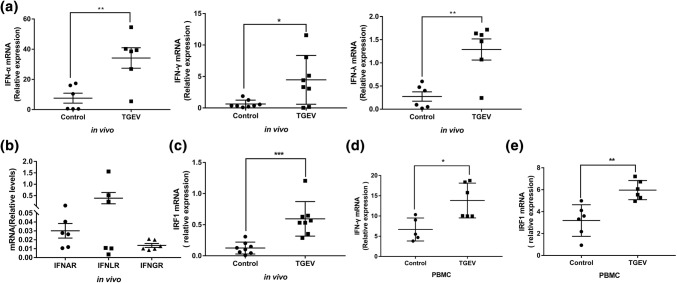
To explore whether IFN-γ potentially contributes to the control of TGEV infection in vivo, we evaluated the expression of IFN in the small intestine after TGEV infection. We found that TGEV infection resulted in a 4.5-fold, 7-fold and 4.7-fold increase in IFN-α,IFN-γ and IFN-λ expression, respectively, in intestine tissue when compared the mock control (Fig. 5a). We also measured the expression of IFN receptors in intestinal tissues and found that all three interferon receptors were expressed. The expression level of IFNLR was higher than that of IFNAR and IFNGR (Fig. 5b). We also examined the expression of IRF1 after TGEV infection in intestinal tissue and found that IRF1 was upregulated after TGEV infection (Fig. 5c). Activated leukocytes are major sources of IFN-γ in vivo [31]. We further investigated whether TGEV can directly stimulate porcine PBMCs to secrete IFN-γ and express IRF1. The results showed that PBMCs exposed to TGEV significantly increased the expression of IFN-γ and IRF1 (Fig. 5d and e). The results suggest that IFN-γ could play an important role in controlling TGEV infection in vivo.
Fig. 5.
TGEV infection induces the expression of IFN-γ and poIRF1 in vivo. (a, c) Piglets were inoculated with 5 mL of a suspension containing 1 × 105 of TCID50 TGEV or DMEM. Total cellular RNA was extracted from the small intestine, and the levels of IFN-α, IFN-γ, IFN-λ and poIRF1 were measured by RT-qPCR. (b) Total cellular RNA was extracted from small intestine cells, and the mRNA levels of IFNAR, IFNLR, and IFNGR were measured by RT-qPCR. (d, e) PBMC were induced to express IFN-γ and poIRF1 by TGEV. Total cellular RNA was collected, and the levels of IFN-γ and poIRF1 were quantified by RT-qPCR. Data are shown as the mean ± SEM. Significance was determined by Student’s t-test compared to the mock control. *, p < 0.5; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001
Enhancement of antiviral effects in ST cells following combined treatment with IFN-γ and IFN-α
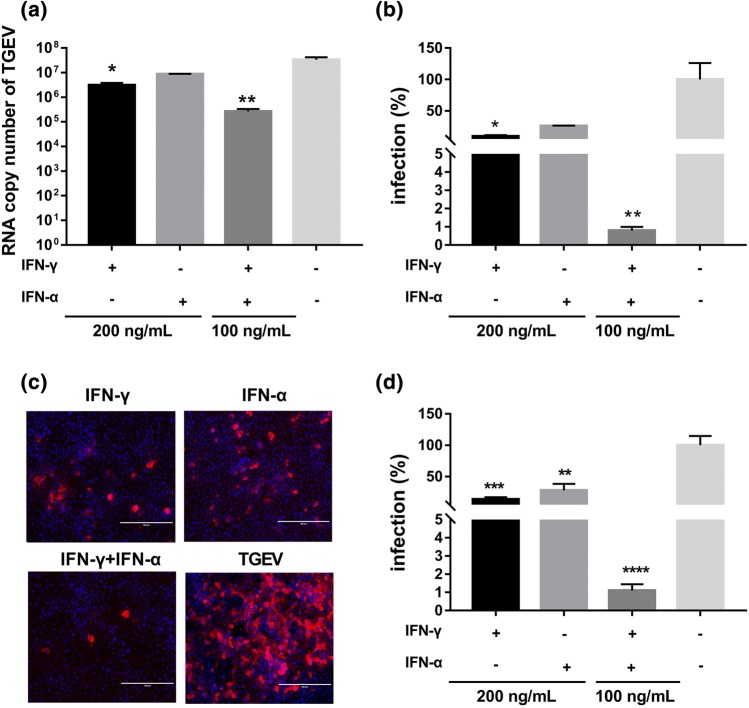
Previous studies and our above results (Fig. 5) have shown that TGEV infection induces both type I IFNs and IFN-γ [12, 13, 25]. The co-existence of IFN-γ and type I IFNs is a common scenario in vivo. To more accurately mimic the in vivo situation, we measured TGEV inhibition after pretreating ST cells with the combination of IFN-α and IFN-γ. IFN-γ or IFN-α alone at a concentration of 200 ng/mL resulted in 10.8-fold or 3.9-fold inhibition of TGEV replication, respectively, when compared to a mock control without IFN treatment. The combination of 100 ng/mL IFN-γ and 100 ng/mL IFN-α inhibited TGEV infection by 126-fold and caused more than 95% inhibition of TGEV (Fig. 6a and b), suggesting that IFN-α and IFN-γ synergistically inhibit TGEV replication. The synergistic antiviral effect of IFN-α and IFN-γ was confirmed by measuring TGEV N protein levels in TGEV-infected cells by IFA (Fig. 6c and d). These results indicate that IFN-γ enhances the anti-TGEV activity of IFN-α and displays a synergistic effect with IFN-α.
Fig. 6.
Synergistic antiviral effect of IFN-γ and IFN-α in ST cells. ST cells were treated with either IFN-γ or IFN-α alone at a concentration of 200 ng/mL, or with a combination of both (100 ng/mL each) and then infected with TGEV at an MOI of 0.01. TGEV infection was detected at 24 hpi by measuring the TGEV genomic RNA (a) or by IFA (c). The percentage of infected cells was calculated after the number of TGEV RNA copies in the IFN-treated groups was normalized to that in the untreated mock control (b). The percentage of fluorescence-positive cells 24 h after infection (c). TGEV-positive cells in six random fields (20X IFA image) were counted, and the percentage of infected cells was calculated after being normalized to the untreated mock control (d). Data are shown as the mean ± SEM. Significance was determined by Student’s t-test compared to the mock control, *, p < 0.5; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001
Discussion
TGEV infection causes a highly contagious swine disease that results in serious economic losses to the pig industry. The development of effective anti-TGEV therapeutics is still very necessary. Here, we demonstrate that IFN-γ treatment directly inhibits the infection of ST cells and IPEC-J2 cells by TGEV. Moreover, in addition to the direct antiviral activity of IFN-γ, IFN-γ enhances the anti-TGEV activity of IFN-α and exhibits a synergistic effect when combined with IFN-α. Since the inhibition of TGEV by IFN-γ occurs largely through the IRF1 pathway, IFN-γ would be predicted to play a vital role in protection of the host against TGEV infection in vivo.
Just as our published results and those of others have indicated that type I IFNs suppress TGEV in vitro [12, 32], IFN-γ was found to inhibit enteric coronavirus TGEV in ST cells and IPEC-J2 cells in vitro. Previous studies have demonstrated that TGEV infection induces a significant amount of type I IFN-α/β in vitro and in vivo [12, 13, 25]. However, we found that IFN-γ inhibited TGEV less efficiently in Vero E6 cells or IFNLR knockout Vero E6 cells than in ST cells and IPEC-J2 cells (Fig. 2), indicating that the type I IFN and type III IFN response may enhance the antiviral activity of IFN-γ. Unlike type I IFNs, TGEV infection did not induce the production of IFN-γ by ST cells or IPEC-J2 cells (data not shown), which is consistent with the fact that IFN-γ is mainly secreted by immune cells. However, we also showed that TGEV infection induces the expression of IFN-γ in infected small intestines and PBMCs (Fig. 5). IFN-γ exerts its direct anti-TGEV activity through a paracrine mechanism by which IFN-γ from immune cells act on TGEV-infected cells in vivo. The co-existence of IFN-γ and IFN-α/β in the local microenvironment of TGEV-infected intestine (Fig. 5a) raises the interesting question of how this affects their anti-TGEV activity. We demonstrated substantial synergy in the antiviral activity of IFN-γ and IFN-α against TGEV infection. The combination of 100 ng/mL IFN-γ and 100 ng/mL IFN-α caused a 126-fold inhibition of TGEV infection, which was more than tenfold higher than that of IFN-γ or IFN-α alone (Fig. 6). This is in agreement with previous studies with herpes simplex virus 1 (HSV-1) and hepatitis C virus (HCV) showing that the combination of type I IFNs and IFN-γ synergistically inhibits viral infection both in vitro and in vivo [6, 33]. In line with the synergetic effect of IFN-γ and IFN-α, we observed reduced anti-TGEV activity of IFN-γ in Vero E6 cells, which do not undergo a type I IFN response, compared with ST or IPEC-J2 cells, which are competent for type I IFN production (Fig. 2). This implies that IFN-γ can play at least a dual role, with a direct anti-TGEV effect and a booster effect for type I IFN antiviral activity in vivo. Given the fact that TGEV infection of ST cells also induces the production of type III IFN, which suppresses TGEV infection (Fig. 5), it will be worthwhile to investigate whether the synergistic effect observed with the combination of type I IFN and type II IFN will also occur with the combination of type II IFN and type III IFN.
We and others have demonstrated that the antiviral activity of IFN-γ does not depend on the presence of type I IFNs (Fig. 2) despite the synergistic effect of IFN-γ and IFN-α [34]. The specific mechanisms of the antiviral activity of IFN-γ are not completely clear. IRF1 is the first identified member of the IRF family of transcription factors and is an important regulator of IFN-γ and ISGs [28]. Consistent with this, we observed that IFN-γ potently elicited expression of poIRF1 in ST cells (Fig. 3a-c), which mediated the induction of the IFN-γ target genes CXCL9 and CXCL10 (Fig. 3e and g), indicating that IRF1 is an important modulator of IFN-γ signaling. In line with this, the knockdown of poIRF1 almost completely abolished the direct antiviral activity of IFN-γ (Fig. 4a and b), indicating that poIRF1 plays a key role in the direct antiviral activity of IFN-γ against TGEV. This is consistent with studies of dengue virus and murine norovirus showing that the antiviral activity of IFN-γ is dependent on IRF1 [27, 34]. IRF1 is a potent host viral restrictor and restricts replication of certain classes of viruses including West Nile virus [35], HCV [36], and vesicular stomatitis virus [37]. In line with a previous study [38], the overexpression of poIRF1 substantially inhibited TGEV infection (up to 287-fold) in ST cells, even in the absence of IFN-γ treatment (Fig. 4e-g), whereas silencing of poIRF1 resulted in a moderate increase of TGEV infection (Fig. 4c and 4d), suggesting that poIRF1 is an important cellular inhibitor of TGEV infection.
In summary, we have demonstrated that IFN-γ inhibits TGEV infection directly in vitro and that the anti-TGEV activity of IFN-γ is dependent on IRF-1 and does not require the presence of type I IFNs. Moreover, IFN-γ was found to enhance the antiviral activity of IFN-α in a synergistic manner. Taken together, these data suggest that IFN-γ could potentially be developed as an effective therapeutic agent against TGEV infection.
Acknowledgements
This work was supported by grants from the National Key R & D Program of China (2017YFD0502200) and the National Natural Science Fund (31772718).
Compliance with ethical standards
Conflict of interest
There is no conflict of interest of any authors in relation to the submission.
Research involving animals
The experiment was approved by the Animal Care and Ethics Committee of Harbin Veterinary Research Institute.
Ethical standards
This research complies with the ethical standards of Archives of Virology.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lingling Shan and Fang Fu contributed equally to this work.
References
- 1.van Boxel-Dezaire AH, Stark GR. Cell type-specific signaling in response to interferon-gamma. Curr Top Microbiol Immunol. 2007;316:119–154. doi: 10.1007/978-3-540-71329-6_7. [DOI] [PubMed] [Google Scholar]
- 2.Barkhouse DA, Garcia SA, Bongiorno EK, et al. Expression of interferon gamma by a recombinant rabies virus strongly attenuates the pathogenicity of the virus via induction of type I interferon. J Virol. 2015;89:312–322. doi: 10.1128/JVI.01572-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schroder K, Hertzog PJ, Ravasi T, et al. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 4.Rhein BA, Powers LS, Rogers K, et al. Interferon-gamma Inhibits Ebola Virus Infection. PLoS Pathog. 2015;11:e1005263. doi: 10.1371/journal.ppat.1005263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaudhary V, Yuen KS, Chan JF, et al. Selective activation of Type II interferon signaling by Zika virus NS5 protein. J Virol. 2017 doi: 10.1128/JVI.00163-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sainz B, Jr, Mossel EC, Peters CJ, et al. Interferon-beta and interferon-gamma synergistically inhibit the replication of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) Virology. 2004;329:11–17. doi: 10.1016/j.virol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin CM, Gao X, Oka T, et al. Antigenic relationships among porcine epidemic diarrhea virus and transmissible gastroenteritis virus strains. J Virol. 2015;89:3332–3342. doi: 10.1128/JVI.03196-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Debouck P, Pensaert M. Experimental infection of pigs with a new porcine enteric coronavirus, CV 777. Am J Vet Res. 1980;41:219–223. [PubMed] [Google Scholar]
- 9.Zhang X, Zhu Y, Zhu X, et al. Identification of a natural recombinant transmissible gastroenteritis virus between Purdue and Miller clusters in China. Emerg Microbes Infect. 2017;6:e74. doi: 10.1038/emi.2017.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brim TA, VanCott JL, Lunney JK, et al. Lymphocyte proliferation responses of pigs inoculated with transmissible gastroenteritis virus or porcine respiratory coronavirus. Am J Vet Res. 1994;55:494–501. [PubMed] [Google Scholar]
- 11.Charley B, Laude H. Induction of alpha interferon by transmissible gastroenteritis coronavirus: role of transmembrane glycoprotein E1. J Virol. 1988;62:8–11. doi: 10.1128/jvi.62.1.8-11.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue M, Fu F, Ma Y, et al. The PERK Arm of the Unfolded Protein Response Negatively Regulates Transmissible Gastroenteritis Virus Replication by Suppressing Protein Translation and Promoting Type I Interferon Production. J Virol. 2018 doi: 10.1128/JVI.00431-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riffault S, Carrat C, van Reeth K, et al. Interferon-alpha-producing cells are localized in gut-associated lymphoid tissues in transmissible gastroenteritis virus (TGEV) infected piglets. Vet Res. 2001;32:71–79. doi: 10.1051/vetres:2001111. [DOI] [PubMed] [Google Scholar]
- 14.Xia L, Yang Y, Wang J, et al. Impact of TGEV infection on the pig small intestine. Virol J. 2018;15:102. doi: 10.1186/s12985-018-1012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzylack-Piriou L, Piersma S, McCullough K, et al. Role of natural interferon-producing cells and T lymphocytes in porcine monocyte-derived dendritic cell maturation. Immunology. 2006;118:78–87. doi: 10.1111/j.1365-2567.2006.02343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bigley NJ. Complexity of Interferon-gamma Interactions with HSV-1. Front Immunol. 2014;5:15. doi: 10.3389/fimmu.2014.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valente G, Ozmen L, Novelli F, et al. Distribution of interferon-gamma receptor in human tissues. Eur J Immunol. 1992;22:2403–2412. doi: 10.1002/eji.1830220933. [DOI] [PubMed] [Google Scholar]
- 18.Wen K, Bui T, Li G, et al. Characterization of immune modulating functions of gammadelta T cell subsets in a gnotobiotic pig model of human rotavirus infection. Comp Immunol Microbiol Infect Dis. 2012;35:289–301. doi: 10.1016/j.cimid.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNeal MM, Stone SC, Basu M, et al. IFN-gamma is the only anti-rotavirus cytokine found after in vitro stimulation of memory CD4+ T cells from mice immunized with a chimeric VP6 protein. Viral Immunol. 2007;20:571–584. doi: 10.1089/vim.2007.0055. [DOI] [PubMed] [Google Scholar]
- 20.Liu SY, Sanchez DJ, Aliyari R, et al. Systematic identification of type I and type II interferon-induced antiviral factors. Proc Natl Acad Sci USA. 2012;109:4239–4244. doi: 10.1073/pnas.1114981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrow AN, Schmeisser H, Tsuno T, et al. A novel role for IFN-stimulated gene factor 3II in IFN-gamma signaling and induction of antiviral activity in human cells. J Immunol. 2011;186:1685–1693. doi: 10.4049/jimmunol.1001359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramana CV, Gil MP, Schreiber RD, et al. Stat1-dependent and -independent pathways in IFN-gamma-dependent signaling. Trends Immunol. 2002;23:96–101. doi: 10.1016/S1471-4906(01)02118-4. [DOI] [PubMed] [Google Scholar]
- 23.Song JH, Wang CX, Song DK, et al. Interferon gamma induces neurite outgrowth by up-regulation of p35 neuron-specific cyclin-dependent kinase 5 activator via activation of ERK1/2 pathway. J Biol Chem. 2005;280:12896–12901. doi: 10.1074/jbc.M412139200. [DOI] [PubMed] [Google Scholar]
- 24.Liu F, Li G, Wen K, et al. Porcine small intestinal epithelial cell line (IPEC-J2) of rotavirus infection as a new model for the study of innate immune responses to rotaviruses and probiotics. Viral Immunol. 2010;23:135–149. doi: 10.1089/vim.2009.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Splichal I, Rehakova Z, Sinkora M, et al. In vivo study of interferon-alpha-secreting cells in pig foetal lymphohaematopoietic organs following in utero TGEV coronavirus injection. Res Immunol. 1997;148:247–256. doi: 10.1016/S0923-2494(97)80866-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desmyter J, Melnick JL, Rawls WE. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero) J Virol. 1968;2:955–961. doi: 10.1128/jvi.2.10.955-961.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlin AF, Plummer EM, Vizcarra EA, et al. An IRF-3-, IRF-5-, and IRF-7-Independent Pathway of Dengue Viral Resistance Utilizes IRF-1 to Stimulate Type I and II Interferon Responses. Cell Rep. 2017;21:1600–1612. doi: 10.1016/j.celrep.2017.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ksienzyk A, Neumann B, Kroger A. IRF-1 is critical for IFNgamma mediated immune surveillance. Oncoimmunology. 2012;1:533–534. doi: 10.4161/onci.19405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura T, Nakayama K, Penninger J, et al. Involvement of the IRF-1 transcription factor in antiviral responses to interferons. Science (New York, NY) 1994;264:1921–1924. doi: 10.1126/science.8009222. [DOI] [PubMed] [Google Scholar]
- 30.Wang F, Cai R, He D, et al. Serum IFN-gamma-inducible chemokines CXCL9 and CXCL10 are elevated in non-immediate drug hypersensitivity reactions. Asian Pac J Allergy Immunol. 2016;34:236–241. doi: 10.12932/AP0679. [DOI] [PubMed] [Google Scholar]
- 31.Farrar MA, Schreiber RD. The molecular cell biology of interferon-gamma and its receptor. Annu Rev Immunol. 1993;11:571–611. doi: 10.1146/annurev.iy.11.040193.003035. [DOI] [PubMed] [Google Scholar]
- 32.Jordan LT, Derbyshire JB. Antiviral action of interferon-alpha against porcine transmissible gastroenteritis virus. Vet Microbiol. 1995;45:59–70. doi: 10.1016/0378-1135(94)00118-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okuse C, Rinaudo JA, Farrar K, et al. Enhancement of antiviral activity against hepatitis C virus in vitro by interferon combination therapy. Antiviral Res. 2005;65:23–34. doi: 10.1016/j.antiviral.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Maloney NS, Thackray LB, Goel G, et al. Essential cell-autonomous role for interferon (IFN) regulatory factor 1 in IFN-gamma-mediated inhibition of norovirus replication in macrophages. J Virol. 2012;86:12655–12664. doi: 10.1128/JVI.01564-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brien JD, Daffis S, Lazear HM, et al. Interferon regulatory factor-1 (IRF-1) shapes both innate and CD8(+) T cell immune responses against West Nile virus infection. PLoS Pathog. 2011;7:e1002230. doi: 10.1371/journal.ppat.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ueki IF, Min-Oo G, Kalinowski A, et al. Respiratory virus-induced EGFR activation suppresses IRF1-dependent interferon lambda and antiviral defense in airway epithelium. J Exp Med. 2013;210:1929–1936. doi: 10.1084/jem.20121401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nair S, Michaelsen-Preusse K, Finsterbusch K, et al. Interferon regulatory factor-1 protects from fatal neurotropic infection with vesicular stomatitis virus by specific inhibition of viral replication in neurons. PLoS Pathog. 2014;10:e1003999. doi: 10.1371/journal.ppat.1003999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Chang H, Yang X, et al. Antiviral activity of porcine interferon regulatory factor 1 against swine viruses in cell culture. Viruses. 2015;7:5908–5918. doi: 10.3390/v7112913. [DOI] [PMC free article] [PubMed] [Google Scholar]