Abstract
Bats play a significant role in maintaining their ecosystems through pollination, dispersal of seeds, and control of insect populations, but they are also known to host many microorganisms and have been described as natural reservoirs for viruses with zoonotic potential. The diversity of viruses in these animals remains largely unknown, however, because studies are limited by species, location, virus target, or sample type. Therefore, the aim of this study was to detect fragments of viral genomes in bat samples. We performed high-throughput sequencing analysis and specific PCR and RT-PCR on pools of anal and oropharyngeal swabs from Artibeus lituratus and Sturnira lilium collected in southern Brazil. As a result, a member of the family Adenoviridae related to human adenovirus C was detected in anal swabs from S. lilium. In addition, we detected a papillomavirus in an anal swab from A. lituratus. Our analyses also allowed the detection of adenoviruses and parvoviruses in oropharyngeal swabs collected from A. lituratus. These results increase our knowledge about viral diversity and illustrate the importance of conducting virus surveillance in bats.
Introduction
Bats (order Chiroptera) are the only mammals with morphological and physiological adaptations that allow them to fly. Bats represent one fifth of the world’s mammal species. They are widely distributed throughout the world and play a significant role in maintaining their ecosystems. Due to their dietary diversity, they are involved in pollination, dispersal of seeds, and control of insect populations. Therefore, they help in the restoration of deforested areas, reproduction of plants and predation of insect pests that harm humans, animals and plants [1].
Bats also have distinctive characteristics that make them reservoirs of various pathogens, especially viruses. They live in large colonies, sometimes with multispecies association, and their long lifespan, possibly associated with specific characteristics of their immune system, favors the maintenance of viral infections. Migration of members of the same species favors dispersal of viruses to different regions. Increased contact with humans, directly or indirectly through saliva, urine or feces, due to habitat sharing increases the risk of virus transmission to humans. Thus, bats play an important role in the maintenance, dispersion, and transmission of viruses that have an impact on public health [2].
A large number of viruses, including important zoonotic viruses such as severe acute respiratory syndrome (SARS)-like coronaviruses [3, 4], filoviruses [5], hantaviruses [6], and henipaviruses [7, 8] have already been found in bats. However, most viruses found in bats have no confirmed zoonotic potential, such as adenoviruses, papillomaviruses and parvoviruses [9]. Adenoviruses, double-stranded DNA (dsDNA) viruses, were detected in bats for the first time in 2006, and since then, adenovirus DNA has been detected in members of different bat species [10]. Currently, seven bat adenoviruses (BtAdVs) have had their genomes nearly fully sequenced: bat mastadenovirus A from Myotis ricketti [11], bat mastadenovirus B from Pipistrellus pipistrellus [12], bat mastadenovirus C from Rhinolophus sinicus [13], bat mastadenovirus D from Miniopterus schreibersi [14], bat mastadenovirus E from M. schreibersi [14], bat mastadenovirus F from Rousettus leschenaultii [14], and bat mastadenovirus G from Corynorhinus rafinesquii [10]. In addition, Ogawa et al. discovered a potential new species, called “bat mastadenovirus H”, in Eidolon helvum [15]. The different BtAdV genome sequences show a high degree of variation.
In Brazil, BtAdV has been detected in Desmodus rotundus, the most common vampire bat in Brazil [16]. Papillomaviruses (PVs), which are also dsDNA viruses, have also been reported in diverse bats species [17–19], but so far, they have not been reported in bats in Brazil. Parvoviruses, a family of single-stranded DNA (ssDNA) viruses that exhibits the highest mutation rate among DNA viruses [9] have been reported worldwide, including in Brazil [20] and have been shown to be related to human-infecting parvoviruses [21–23].
Recently, high-throughput sequencing has been increasingly used for the identification of viruses, because this approach allows the diversity of the whole virus population (virome) present in a biological sample to be examined [24]. For example, in 2013, viruses of the families Adenoviridae, Herpesviridae, Papillomaviridae, Parvoviridae, Picornaviridae, Polyomaviridae, Poxviridae, and Retroviridae were found in African bats using this approach [25]. In 2016, Male et al. described three new cycloviruses, 14 gemycircularviruses and 17 unclassified viruses in Pteropus tonganus feces [26].
In Brazil, adenoviruses, anelloviruses, circoviruses, coronaviruses, polyomaviruses, parvoviruses and rhabdoviruses have already been detected in different bat species [16, 20, 27–30]. However, previous studies were performed using traditional genome amplification methods. The purpose of this study was to analyze and identify fragments of viral genomes present in Artibeus lituratus and Sturnira lilium through high-throughput sequencing and polymerase chain reaction (PCR) or reverse-transcription polymerase chain reaction (RT-PCR).
Materials and methods
Samples
Anal and oropharyngeal swabs from 69 bats of the species A. lituratus and 24 of the species S. lilium were collected from September 2015 to March 2016 near urban centers in south Brazil. A permanent license to collect biological material was granted by Chico Mendes Institute for Biodiversity Conservation (ICMBio) (number: 17869-3; date of issue, 2012), and certification was given by the Ethical Committee on the Use of Animals (CEUA) of the State University of Maringá (number 1697070515; date of issue, 05/05/2015). To capture the specimens, five nylon nets (mist-nest) were used as described by Greenhall and Paradiso [31] and were placed at least 30 meters apart. The capture effort in the forest segment was calculated according to Straube and Bianconi [32], multiplying the area of the nets used (height × length), the number of nets, and the number of hours of total collection (collection period × number of days of collection). The captured animals were hand-held with leather gloves, and biometric data were collected and in addition to the anal and oropharyngeal swabs, taxonomic identification was done according to Vizotto and Taddei [33], Jones and Carter [34], and Gregorin and Taddei [35]. The swabs were stored in vials containing 2 mL of minimum essential medium with 200 U of penicillin (Sigma-Aldrich) and 200 μg of streptomycin (Sigma-Aldrich) per mL and refrigerated at 4 °C until the time of processing. The animals were stored in cotton bags until data collection and collection of the swabs and were subsequently released.
Viral enrichment
Samples were processed in pools of 1 mL for each animal. Twenty mL of sample was vigorously vortexed with glass beads and then centrifuged at 2500×g for 5 min at 4 °C. The supernatant was again centrifuged for 14 min at maximum speed and then filtered through a 0.22-μm syringe filter (MF-Millipore). The viral particles were harvested and pelleted on a 25% sucrose cushion by ultracentrifugation at 190,000×g for 4 h at 4 °C. The pellet was resuspended in TE buffer (10 mM Tris [Invitrogen], pH 7.4; 1 mM EDTA [Invitrogen] pH 8.0) and clarified by emulsifying with 1/1 (v/v) chloroform and centrifugation. In order to remove nucleic acids that were not protected by the capsid, the purified samples were treated with 100 U of DNase I (Roche) and 20 U of RNase (Invitrogen) at 37 °C for 2 h, similar to other studies [36]. Viral genomes were extracted with phenol (Invitrogen) and ethanol precipitated according to Sambrook and Russel [37]. Random amplifications for DNA were performed according to Stang et al. [38] using the primer K-random-s (GAC CAT CTA GCG ACC TCC ACM NN MNM).
High-throughput sequencing
Seventy ng of random amplification products of each sample was used for sequencing. These samples were sent to Institute of Veterinary Research Desidério Finamor (IPVDF), Fepagro Animal Health, Eldorado do Sul, Brazil, and sequencing was performed on an Illumina MiSeq platform using Kit Nextera XT (2 × 150 bp).
Bioinformatics
Reads were trimmed using PRINSEQ (prinseq.sourceforge.net), and the quality of the sequences was analyzed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Trimmed reads were assembled de novo using St. Petersburg genome assembler (SPAdes) 3.10.1 (http://cab.spbu.ru/software/spades/). The resulting contigs (> 100 bp) were subjected to a blastx search against the viral protein database, using the software Blas2GO (https://www.blast2go.com/). Viral contigs were then checked manually using a blastx search against the National Center for Biotechnology Information (NCBI, www.ncbi.nlm.nih.gov) non-redundant database (nr). Amino acid sequence alignment was performed using the software MEGA 7 (http://www.megasoftware.net/), and phylogenetic trees were constructed with the software MrBayes 3.2.6 (http://mrbayes.sourceforge.net/), using evolutionary and substitution models predicted by ProtTest 3.4.2 (https://github.com/ddarriba/prottest3). Bacteriophage contigs were not analyzed further.
Polymerase chain reaction (PCR)
Nucleic acids from pooled samples of oral swabs from 24 S. lilium and 58 A. lituratus bats were extracted using an RTP DNA/RNA Virus Mini Kit (Stratec). The nucleic acids were then subjected to polymerase chain reaction (PCR) or reverse transcription polymerase chain reaction (RT-PCR), depending on the virus being investigated. The amplification reactions targeted genomic fragments of members from the families Rhabdoviridae, Herpesviridae, Adenoviridae and Paramyxoviridae as described by Carnieli et al. [39], Razafindratsimandresy et al. [40], Li et al. [11], and Tong et al. [41], respectively.
Results
All samples tested by PCR and RT-PCR targeting members of the families Rhabdoviridae, Herpesviridae, Adenoviridae and Paramyxoviridae gave negative results. However, high-throughput sequencing resulted in 1,345,784 total reads and 12,073 total contigs after trimming and assembly. Eleven percent (1,303) yielded matches against viral protein databases in Blast2go analysis, where 99.6% were bacteriophages, 0.15% were adenoviruses, 0.15% were parvoviruses and 0.1% were papillomaviruses (Table 1). Adenovirus contigs were identified in an A. lituratus oropharyngeal swab (ALOS) and S. lilium anal swabs (SLAS), while papillomavirus and parvoviruses were identified in A. lituratus anal swabs (ALAS) and ALOS, respectively. Bacteriophages were detected in all samples, and only viral components were found in S. lilium oropharyngeal swabs (SLOS). For further analyses, only adenoviruses, papillomaviruses and parvoviruses contigs were used.
Table 1.
Sequencing results by sample
| Sample | Reads | Contigs | Viral contigs | Bacteriophage contigs | Parvovirus contigs | Adenovirus contigs | Papillomavirus contigs |
|---|---|---|---|---|---|---|---|
| ALAS | 152,788 | 2,839 | 406 | 405 | - | - | 1 |
| ALOS | 277,710 | 3,042 | 387 | 384 | 2 | 1 | - |
| SLAS | 539,804 | 1,998 | 163 | 162 | - | 1 | - |
| SLOS | 375,482 | 4,205 | 347 | 347 | - | - | - |
| TOTAL | 1,345,784 | 12,084 | 1,303 | 1,298 | 2 | 2 | 1 |
ALAS, A. lituratus anal swab; ALOS, A. lituratus oropharyngeal swab; SLAS, S. lilium anal swab; SLOS, S. lilium oropharyngeal swab
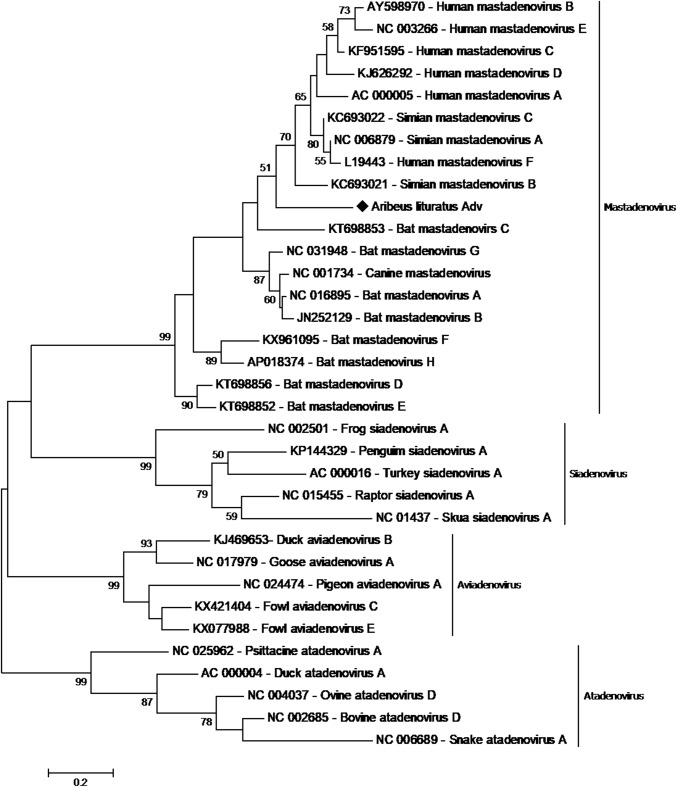
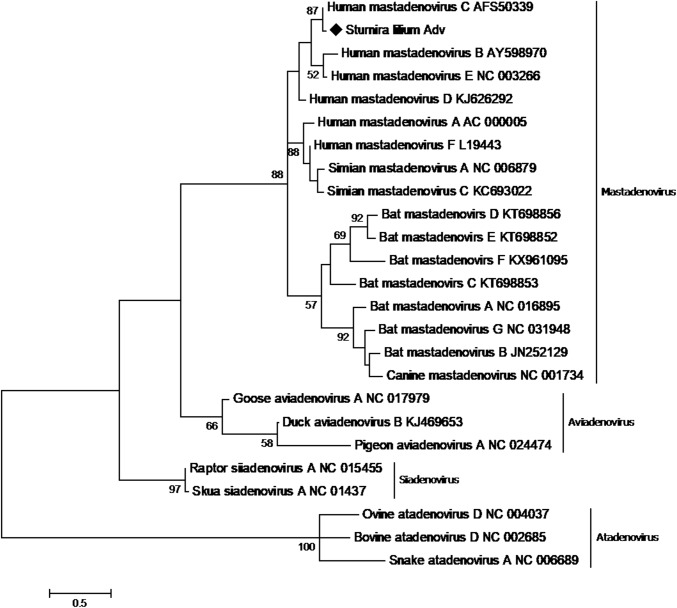
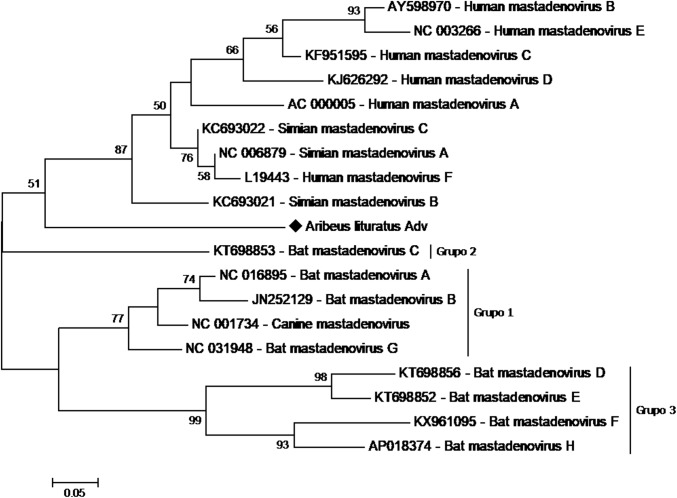
To confirm the sequences identities, contigs were manually submitted to NCBI blastx (Table 2). The Artibeus lituratus adenovirus (AlAdV) genomic fragment is 312 nucleotides (nt) long and displays 71.2% sequence identity to the DNA terminal protein of Equine adenovirus (Genbank accession number YP_009272543). The Sturnira lilium adenovirus (SlAdV) fragment is 177 nt long and displays 98% sequence identity to the human adenovirus C L4 encapsidation protein (GenBank accession number AFS50339). Phylogenetic analysis of both fragments indicated that these viruses belong to the genus Mastadenovirus (Fig. 1 and Fig. 2).
Table 2.
Contigs (> 150 bp) with significant blastx hits to known eukaryotic viruses obtained from the Brazilian bats S. lilium and A. lituratus
| Sample | Name | Length (nt) | Protein | Best hit | GenBank ID | Identity | e-value |
|---|---|---|---|---|---|---|---|
| ALSA | Artibeus lituratus papillomavirus | 549 | L1 protein | Rhinolophus ferrumequinum papillomavirus | AHJ81407 | 68.9% | 3e−89 |
| ALSO | Artibeus lituratus parvovirus NS1 | 1170 | Nonstructural protein 1 | Desmodus rotundus parvovirus | YP_009328889 | 70.9% | 3e−173 |
| Artibeus lituratus parvovirus capsid | 858 | Capsid protein | Desmodus rotundus parvovirus | YP_009328890 | 80.0% | 3e−157 | |
| Artibeus lituratus adenovirus | 312 | DNA terminal protein | Equine adenovirus | YP_009272543 | 71.2% | 9e−50 | |
| SLSA | Sturnira lilium adenovirus | 177 | L4 encapsidation protein | Human adenovius C | AFS50339 | 98% | 3e−34 |
ALAS, A. lituratus anal swab; ALOS, A. lituratus oropharyngeal swab; SLAS, S. lilium anal swab
Fig. 1.
Phylogenetic tree of Artibeus lituratus adenovirus. The tree was constructed based on a partial sequence of the DNA terminal protein of members of the family Adenoviridae. The A. lituratus adenovirus detected in this study is represented by ♦. The tree was constructed using the maximum-likelihood method with a JTT substitution matrix as predicted by ProtTest and 1000 bootstrap replicates
Fig. 2.
Phylogenetic tree of Sturnia lilium adenovirus. The tree was constructed based on a partial sequence of the L4 encapsidation protein of members of the family Adenoviridae. The S. lilium adenovirus detected in this study is represented by ♦. The tree was constructed using the maximum-likelihood method with a JTT substitution matrix as predicted by ProtTest and 1000 bootstrap replicates
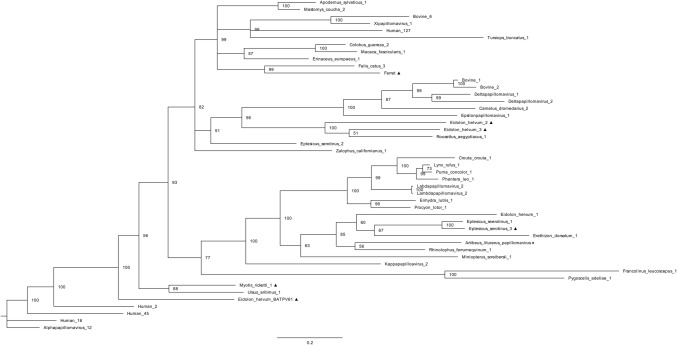
The Artibeus lituratus papillomavirus (AlPV) fragment was 549 nt long and displayed 68.9% identity to the L1 protein of Rhinolophus ferrumequinum papillomavirus (RferPV1) (GenBank accession number AHJ81407), which belongs to the genus Treisdeltapapillomavirus. In an attempt to determine the genus to which AlPV belongs, we constructed a phylogenetic tree including members of the family Papillomaviridae (Fig. 3). AlPV and RferPV1 clustered together, suggesting that they are phylogenetically related; however, the branch distance between them may indicate that they do not belong to the same genus. It was thus concluded that AlPV belongs to an undetermined genus in the family Papillomaviridae.
Fig. 3.
Phylogenetic tree of Artibeus lituratus papillomavirus. The tree was constructed based on a partial sequence of the L1 protein of members of the family Papillomaviridae. The A. lituratus papillomavirus detected in this study is represented by ♦. Samples that remain unclassified are represented by ▲. The phylogenetic tree was constructed using Bayesian inference with an LG substitution matrix and gamma distribution with a portion of invariable sites as predicted by ProtTest. The number of generations was 1,000,000, and two chains were used
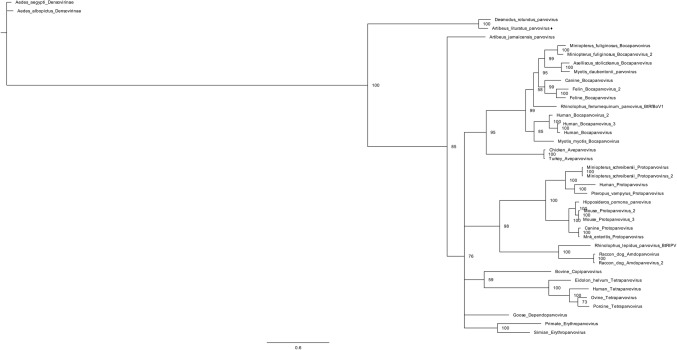
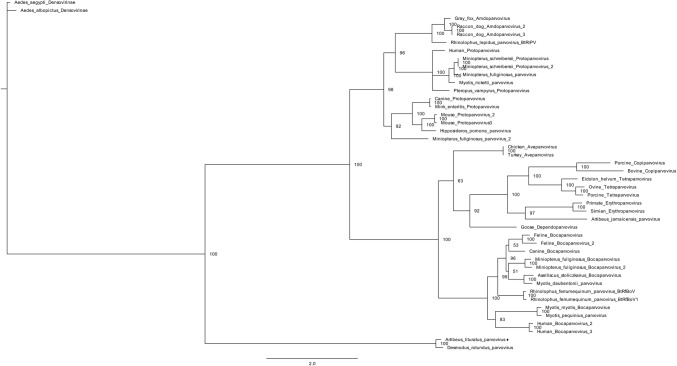
Both Artibeus lituratus parvovirus (AlPrV) fragments aligned with Desmodus rotundus parvovirus (DrPV-1). The first fragment, which is 1170 nt long, displays 70.9% sequence identity to nonstructural protein 1 (NS1) (GenBank accession number YP_009328889). The second fragment, which is 858 nt long, displays 80.0% sequence identity to the capsid protein (GenBank accession number YP_009328890). Although both AlPrV and DrPrV-1 grouped in the same cluster, the phylogenetic distance between these sequences may indicate that they belong to different genera (Fig. 4 and Fig 5).
Fig. 4.
Phylogenetic tree of Artibeus lituratus parvovirus. The tree was constructed based on a partial sequence of the NS1 protein of members of the family Parvoviridae. The A. lituratus parvovirus detected in this study is represented by ♦. Aedes aegypti parvovirus and Aedes albopictus parvoviruses, subfamily Densovirinae, were used as outgroups. The phylogenetic tree was constructed using Bayesian inference with an RtREV substitution matrix and gamma distribution as predicted by ProtTest. The number of generations was 1,000,000, and two chains were used
Fig. 5.
Phylogenetic tree of Artibeus lituratus parvovirus. The tree was constructed based on a partial sequence of the capsid protein of members of the subfamily Parvovirinae. The A. lituratus parvovirus detected in this study is represented by ♦. Aedes aegypti parvovirus and Aedes albopictus parvovirus, subfamily Densovirinae, were used as outgroups. The phylogenetic tree was constructed using Bayesian inference with an LG substitution matrix, gamma distribution, and empirical amino acid frequency as predicted by ProtTest. The number of generations was 1,000,000, and two chains were used
Discussion
Bats are recognized as potential hosts of many zoonotic viruses. To assess the viral diversity within two bat species in southern Brazil, anal and oropharyngeal swabs were collected from 69 A. lituratus and 24 S. lilium bats in Maringa, Paraná state, Brazil, and subjected to Illumina MiSeq sequencing. From the generated contigs, only 11% matched viral proteins. Of these, about 1% belonged to eukaryotic viruses from distinct families. The diversity of viral contigs was lower than previously described in other bat species [25, 26, 42]. As it is unlikely that viral diversity differs greatly among different species of bats, we believe that environmental features, such as different feeding habits, geographical location, and contact with other species of bats [26, 42–44], might have contributed to these different results. In addition, the nucleic acid extraction and enrichment techniques may have also influenced the results.
Adenoviruses genome fragments were detected in both species in this study: from A. lituratus oropharyngeal swabs and from S. lilium anal swabs. Although not totally unexpected, this is the first study to describe the presence of adenovirus DNA in samples from these two species. Although adenoviruses have already been identified in Brazilian bats [15], the previous study described a different DNA fragment from the one found in our study, making any comparison between them impossible. The Artibeus lituratus adenovirus (AlAdV) fragment displayed 71.2% sequence identity to the DNA terminal protein of equine adenovirus and was compared with 32 sequences of different adenovirus species, including the seven BatAdV species recognized by the ICTV and the species newly proposed by Ogawa et al. [15]. As expected, this sample clustered with mastadenovirus-genus members, which have specificity for mammals. A Mastadenovirus specific analysis demonstrated that the BatAdV samples formed three distinct clusters (Fig. 6), similar to those obtained by Ogawa et al. when analyzing complete genome sequences [15]. Interestingly, AlAdV clustered in a distinct group with primate adenoviruses. Just as the close relationship between some adenoviruses of canines and bats indicates the possibility of interspecific transmission [12], the fact that AlAdV and primate AdVs occupy a monophyletic cluster indicates a potential zoonotic risk.
Fig. 6.
Phylogenetic tree of members of the genus Mastadenovirus. The tree was constructed based on a partial sequence of the DNA terminal protein of members of the family Adenoviridae. The A. lituratus adenovirus detected in this study is represented by ♦. The tree was constructed using the maximum-likelihood method with a JTT substitution matrix as predicted by ProtTest and 1000 bootstrap replicates
Sturnira lilium adenovirus (SlAdV) showed 98% sequence identity to the human adenovirus C (HAdV-C) L4 encapsidation protein. Studies have already demonstrated the presence of HAdV-C in water samples [45, 46], and therefore this virus might have been a contaminant in the sample. However, all pools were processed together, and the biosecurity measures adopted make this hypothesis unlikely. Another possible explanation for the presence of HAdV in bat specimens is accidental ingestion through food or water, since AdV infections may be caused by water consumption or inhalation of contaminated aerosols [47]. In addition, the presence of AdV in feces might be related to its frequent presence in environment [48]. Although adenoviruses infections are usually species specific, recent findings have suggested that cross-species transmission can occur. In 2011, Chen et al, described an adenovirus in a titi monkey (TMAdV) which, due to its aggressiveness in this species, resulted from a spillover event [49]. Finally, SlAdV displayed high sequence similarity to human adenovirus C, indicating that direct or indirect contact between bats and humans might allow interspecies transmission.
Papillomaviruses (PVs) were detected only in A. lituratus anal swabs. The sequence fragment that was obtained was 183 amino acids (aa) long and displayed 68.9% identity to the Rhinolophus ferrumequinum papillomavirus (RferPV1) L1 protein, which was first described in Spain by Garcia-Pérez et al. [17]. This is the first detection of a papillomavirus in A. lituratus and the first PV described in Brazilian bats. Phylogenetic analysis showed that Artibeus lituratus papillomavirus (AlPV) and RferPV1 (which belongs to the genus Treisdeltapapillomavirus) clustered independently. Nevertheless, due to the fragment sizes, it was not possible to assign AlPV to a genus. In bats, PVs have already been detected in oropharyngeal and anal swabs in distinct species [17, 43, 50, 51]. Although most mammalian PV infections are asymptomatic, some reports have associated the presence of these viruses with different types of human cancer. However, papillomavirus-infection-associated cancers have also been described in bats [52]. Although transmission between species rarely occurs, the high genomic diversity of these viruses, associated with ecological characteristics of bats, can intensify their spread and increase the chances of interspecies transmission.
Two DNA fragments derived from parvoviruses (PrVs) were detected in A. lituratus oropharyngeal swabs. The first one aligns with nonstructural protein 1 (NS1), and the second one corresponds to the capsid protein; both are related to Desmodus rotundus parvovirus (DrPV-1), which was detected previously in Brazil [19]. The Artibeus lituratus parvovirus (AlPrV) NS1 fragment is 390 aa long and displays 70.9% sequence identity to the DrPV-1 NS1 protein, while the AlPrV capsid fragment is 286 aa long and displays 80.0% identity to the DrPV-1 capsid protein. Although Artibeus jamaicensis bat parvovirus 1 (Aj-BtPV-1) has been identified previously in a sample collected from A. lituratus, phylogenetic analysis of both fragments showed that AlPrV and DrPV-1 are in the same cluster [53]. The cluster distances between these two viruses and between them and other members suggest that AlPrV belongs to a new genus in the subfamily Parvovirinae. Although it was not the first PrV detected in A. lituratus, AlPrV differs significantly from Aj-BtPV-1.
Different metagenomic studies have already identified adenoviruses, papillomaviruses, and parvoviruses in a number of bat species [25, 42, 43, 51, 54]. However, differences in sample collection, sample types, viral nucleic acid isolation methodologies, nucleic acid amplification and/or high-throughput sequencing platforms may account for the diverse results found in different studies, preventing a direct comparison between studies. This work highlights the importance of investigating viruses in bats and indicates that high-throughput sequencing can be more suitable than PCR or RT-PCR to explore the viral diversity in bats. The successful detection of adenovirus by high-throughput sequencing but not by PCR may have been due to concentration of the virus in sample pools in the random-enrichment process, whereas without pooling of samples, the viral load might have been below the PCR detection limit.
The results provide information about the diversity of viruses carried by bats; however, the number of species analyzed and the geographical regions targeted were limited. Such studies should be continued to gain further knowledge about the viral diversity in bats and to identify infectious agents that might cause zoonotic infections, thereby facilitating decision-making concerning the strategies that should be adopted to combat emerging viruses.
Acknowledgements
We thank Dra. Fabiana Quoos Mayer from Instituto de Pesquisas Veterinárias Desidério Finamor. All authors have seen and approved the manuscript and have contributed significantly to the work.
Funding
This study was supported by a grant from FAPESP, Brazil, in 2016 (Project Code No. 2015/25367-0) and the Secretary of State for Health of São Paulo. Part of this work was supported by CNPq and CAPES. A. C. F. and P. M. R. are CNPq fellows.
Compliance with ethical standards
Conflict of interest
All authors declare that there are no financial or other relationships that might lead to a conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Allocati N, Petrucci AG, Di Giovanni P, Masulli M, Di Ilio C, De Laurenzi V. Bat–man disease transmission: zoonotic pathogens from wildlife reservoirs to human populations. Cell Death Discov. 2016 doi: 10.1038/cddiscovery.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serra-Cobo J, López-Roig M. Bats and emerging infections: an ecological and virological puzzle. Adv Exp Med Biol. 2016 doi: 10.1007/5584_2016_131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, Wong SS, Leung SY, Chan KH, Yuen KY. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S, Wang LF. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 5.Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Délicat A, Paweska JT, Gonzalez JP, Swanepoel R. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:557–575. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 6.Guo WP, Lin XD, Wang W, Tian JH, Cong ML, Zhang HL, Wang MR, Zhou RH, Wang JB, Li MH, Xu J, Holmes EC, Zhang YZ. Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathog. 2013;9:e1003159. doi: 10.1371/journal.ppat.1003159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halpin K, Hyatt AD, Fogarty R, Middleton D, Bingham J, Epstein JH, Rahman SA, Hughes T, Smith C, Field HE, Daszak P, Henipavirus Ecology Research Group Pteropid bats are confirmed as the reservoir hosts of henipaviruses: a comprehensive experimental study of virus transmission. Am J Trop Med Hyg. 2011;85:946–951. doi: 10.4269/ajtmh.2011.10-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith I, Wang LF. Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr Opin Virol. 2013;3:84–91. doi: 10.1016/j.coviro.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geoghegan JL, Duchêne S, Holmes EC. Comparative analysis estimates the relative frequencies of co-divergence and cross-species transmission within viral families. PLoS Pathog. 2017;13(2):e1006215. doi: 10.1371/journal.ppat.1006215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hackenbrack N, Rogers MB, Ashley RE, Keel MK, Kubiski SV, Bryan JA, Ghedin E, Holmes EC, Hafenstein SL, Allison AB. Evolution and cryo-electron microscopy capsid structure of a North American bat adenovirus and its relationship to other mastadenoviruses. J Virol. 2017;91:e01504–e01516. doi: 10.1128/JVI.01504-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Ge X, Zhang H, Zhou P, Zhu Y, Zhang Y, Yuan J, Wang LF, Shi Z. Host range, prevalence, and genetic diversity of adenoviruses in bats. J Virol. 2010;84:3889–3897. doi: 10.1128/JVI.02497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohl C, Vidovszky MZ, Mühldorfer K, Dabrowski PW, Radonic A, Nitsche A, Wibbelt G, Kurth A, Harrach B. Genome analysis of bat adenovirus 2: indications of interspecies transmission. J Virol. 2012;86:1888–1892. doi: 10.1128/JVI.05974-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan B, Yang XL, Ge XY, Peng C, Zhang YZ, Zhang LB, Shi ZL. Novel bat adenoviruses with an extremely large E3 gene. J Gen Virol. 2016;97:1625–1635. doi: 10.1099/jgv.0.000470. [DOI] [PubMed] [Google Scholar]
- 14.Tan B, Yang XL, Ge XY, Peng C, Liu HZ, Zhang YZ, Zhang LB, Shi ZL. Novel bat adenoviruses with low G+C content shed new light on the evolution of adenoviruses. J Gen Virol. 2017;98:739–748. doi: 10.1099/jgv.0.000739. [DOI] [PubMed] [Google Scholar]
- 15.Ogawa H, Kajihara M, Nao N, Shigeno A, Fujikura D, Hang’Ombe BM, Mweene AS, Mutemwa A, Squarre D, Yamada M, Higashi H, Sawa H, Takada A. Characterization of a novel bat adenovirus isolated from straw-colored fruit bat (Eidolon helvum) Viruses. 2017;9(12):1–16. doi: 10.3390/v9120371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lima FES, Cibulski SP, Elesbao F, JrP Carnieli, Batista HBCR, Roehe PM, Franco AC. First detection of adenovirus in the vampire bat (Desmodus rotundus) in Brazil. Virus Genes. 2013 doi: 10.1007/s11262-013-0947-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.García-Pérez R, Ibáñez C, Godínez JM, Aréchiga N, Garin I, Pérez-Suárez G, de Paz O, Juste J, Echevarría JE, Bravo IG. Novel papillomaviruses in free-ranging iberian bats: no virus–host co-evolution, no Strict host specificity, and hints for recombination. Genome Biol Evol. 2014;6(1):94–104. doi: 10.1093/gbe/evt211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu D, Zhu C, Wang Y, Ai L, Yang L, Ye F, Ding C, Chen J, He B, Zhu J, Qian H, Xu W, Feng Y, Tan W, Wang C. Virome analysis for identification of novel mammalian viruses in bats from Southeast China. Sci Rep. 2017;7(1):10917. doi: 10.1038/s41598-017-11384-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salmier A, Tirera S, de Thoisy B, Franc A, Darcissac E, Donato D, Bouchier C, Lacoste V, Lavergne A. Virome analysis of two sympatric bat species (Desmodus rotundus and Molossus molossus) in French Guiana. PLoS One. 2017;12(11):e0186943. doi: 10.1371/journal.pone.0186943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Souza WM, Romeiro MF, Fumagalli MJ, Modha S, de Araujo J, Queiroz LH, Durigon EL, Figueiredo LTM, Murcia PR, Gifford RJ. Chapparvoviruses occur in at least three vertebrate classesand have a broad biogeographic distribution. J Gen Virol. 2017;98:225–229. doi: 10.1099/jgv.0.000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kemenesi G, Dallos B, Görföl T, Estók P, Boldogh S, Kurucz K, Oldal M, Marton S, Bányai K, Jakab F. Genetic diversity and recombination within bufaviruses: detection of a novel strain in Hungarian bats. Infect Genet Evol. 2015 doi: 10.1016/j.meegid.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau SKP, Ahmed SS, Yeung HC, Li KS, Fan RY, Cheng TY, Cai JP, Wang M, Zheng BJ, Wong SS, Woo PC, Yuen KY. Identification and interspecies transmission of a novel bocaparvovirus among different bat species in China. J Gen Virol. 2016;97:3345–3358. doi: 10.1099/jgv.0.000645. [DOI] [PubMed] [Google Scholar]
- 23.Sasaki M, Gonzalez G, Wada Y, Setiyono A, Handharyani E, Rahmadani I, Taha S, Adiani S, Latief M, Kholilullah ZA, Subangkit M, Kobayashi S, Nakamura I, Kimura T, Orba Y, Ito K, Sawa H. Divergent bufavirus harboured in megabats represents a new lineage of parvoviruses. Sci Rep. 2016;6:24257. doi: 10.1038/srep24257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moratelli R, Calisher CH. Bats and zoonotic viruses: can we confidently link bats with emerging deadly viruses? Mem Inst Oswaldo Cruz. 2015;10(1):1–22. doi: 10.1590/0074-02760150048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker KS, Leggett RM, Bexfield NH, Alston M, Daly G, Todd S, Tachedjian M, Holmes CE, Crameri S, Wang LF, Heeney JL, Suu-Ire R, Kellam P, Cunningham AA, Wood JL, Caccamo M, Murcia PR. Metagenomic study of the viruses of African straw-coloured fruit bats: detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virology. 2013;441(2):95–106. doi: 10.1016/j.virol.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Male MF, Kraberger S, Stainton D, Kami V, Varsani A. Cycloviruses, gemycircularviruses and other novel replication-associated protein encoding circular viruses in Pacific flying fox (Pteropus tonganus) faeces. Infect Genet Evol. 2016;39:279–292. doi: 10.1016/j.meegid.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 27.Cibulski SP, Teixeira TF, Lima FES, do Santos HF, Franco AC, Roehe PM. A novel Anelloviridae species detected in Tadarida brasiliensis bats: first sequence of a chiropteran Anellovirus. Genome Announc. 2014;2(5):e01028-14. doi: 10.1128/genomeA.01028-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lima FES, Cibulski SP, Witt AA, Franco AC, Roehe PM. Genomic characterization of two novel polyomaviruses in Brazilian insectivorous bats. Arch Virol. 2015 doi: 10.1007/s00705-015-2447-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asano KM, Hora AS, Scheffer KC, Fahl WO, Iamamoto K, Mori E, Brandão PE. Alphacoronavirus in urban Molossidae and Phyllostomidae bats, Brazil. Virol J. 2016;24(13):110. doi: 10.1186/s12985-016-0569-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Góes LGB, Campos ACA, de Carvalho C, Ambar G, Queiroz LH, Cruz-Neto AP, Munir M, Durigon EL. Genetic diversity of bats coronaviruses in the Atlantic Forest hotspot biome, Brazil. Infect Genet Evol. 2016;44:510–513. doi: 10.1016/j.meegid.2016.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenhall GG, Paradiso JL (1968) BaLs and hal handing. Washington, U.S. Dep. In: Fish and wildlife service. Bureau 01’ Sport Fisheries and Wildlife, p 45
- 32.Straube FC, Bianconi GV. Sobre a grandeza e a unidade utilizada para estimar esforço de captura com utilização de redes-de-neblina. Chiroptera Neotropical Brasília. 2002;8(1–2):150–152. [Google Scholar]
- 33.Vizotto LD, Taddei VA. Chave para determinação de quirópteros brasileiros. Gráfica Francal: São José do Rio Preto; 1973. p. 72p. [Google Scholar]
- 34.Jones JK, Carter DC (1976) Annotated checklist, with keys to subfamilies and genera. In: Baber RJ, Jones Jr., JK, Carter DC (eds) Biology of bats of the new world family Phyllostomidae, part I. Lubbock: Museum Texas Tech. University, pp 7–38 (Special Publications)
- 35.Gregorin R, Taddei VA. Chave artificial para identificação de molossídeos brasileiros (Mammalia, Chiroptera) Mastozoologia Neotropical. 2002;1(9):13–32. [Google Scholar]
- 36.Kluge M, Campos FS, Tavares M, de Amorim DB, Valdez FP, Giongo A, Roehe PM, Franco AC. Metagenomic survey of viral diversity obtained from feces of subantarctic and south american fur seals. PLoS One. 2016;11(3):e0151921. doi: 10.1371/journal.pone.0151921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 38.Stang A, Korn K, Wildner O, Uberla K. Characterization of virus isolates by particle-associated nucleic acid PCR. J Clin Microbiol. 2005;43:716–720. doi: 10.1128/JCM.43.2.716-720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.JrP Carnieli, Brandão PE, Carrieri ML, Castilho JG, Macedo CI, Machado LM, Rangel N, de Carvalho RC, de Carvalho VA, Montebello L, Wada M, Kotait I. Molecular epidemiology of rabies virus strains isolated from wild canids in Northeastern Brazil. Virus Res. 2006;120(1–2):113–120. doi: 10.1016/j.virusres.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 40.Razafindratsimandresy R, Jeanmaire EM, Counor D, Vasconcelos PF, Sall AA, Reynes JM. Partial molecular characterization of alphaherpesviruses isolated from tropical bats. J Gen Virol. 2009;90(Pt 1):44–47. doi: 10.1099/vir.0.006825-0. [DOI] [PubMed] [Google Scholar]
- 41.Tong S, Chern SW, Li Y, Pallansch MA, Anderson LJ. Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses. J Clin Microbiol. 2008;46(8):2652–2658. doi: 10.1128/JCM.00192-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He B, Li Z, Yang F, Zheng J, Feng Y, Guo H, Li Y, Wang Y, Su N, Zhang F, Fan Q, Tu C. Virome profiling of bats from Myanmar by metagenomic analysis of tissue samples reveals more novel Mammalian viruses. PLoS One. 2013;8(4):e61950. doi: 10.1371/journal.pone.0061950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, Moore NE, Murray ZL, McInnes K, White DJ, Tompkins DM, Hall RJ. Discovery of novel virus sequences in an isolated and threatened bat species, the New Zealand lesser short-tailed bat (Mystacina tuberculata) J Gen Virol. 2015;96:2442–2452. doi: 10.1099/vir.0.000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yinda CK, Conceição-Neto N, Zeller M, Heylen E, Maes P, Ghogomu SM, Ranst MV, Matthijnssens J. Novel highly divergent sapoviruses detected by metagenomics analysis in straw-colored fruit bats in Cameroon. Emerg Microbes Infect. 2017 doi: 10.1038/emi.2017.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bibby K, Peccia J. Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ Sci Technol. 2013;47(4):1945–1951. doi: 10.1021/es305181x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staggemeier R, Heck TM, Demoliner M, Ritzel RG, Röhnelt NM, Girardi V, Venker CA, Spilki FR. Enteric viruses and adenovirus diversity in waters from 2016 Olympic venues. Sci Total Environ. 2017;586:304–312. doi: 10.1016/j.scitotenv.2017.01.223. [DOI] [PubMed] [Google Scholar]
- 47.Jiang SC. Human adenoviruses in water: occurrence and health implications: a critical review. Environ Sci Technol. 2006;40(23):7132–7140. doi: 10.1021/es060892o. [DOI] [PubMed] [Google Scholar]
- 48.Barardi CRM, Viancelli A, Rigotto C, Correa AA, Moresco V, Souza DSM, ElMahdy MEI, Fongaro G, Pilotto MR, Nascimento MA. Monitoring viruses in environmental samples. Intern J Environ Sci Eng. 2012;3:62–67. [Google Scholar]
- 49.Chen EC, Yagi S, Kelly KR, Mendoza SP, Maninger N, Rosenthal A, Spinner A, Bales KL, Schnurr DP, Lerche NW, Chiu CY. Cross-species transmission of a novel adenovirus associated with a fulminant pneumonia outbreak in a new world monkey colony. PLoS Pathog. 2011;7(7):e1002155. doi: 10.1371/journal.ppat.1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tse H, Tsang AKL, Tsoi HW, Leung ASP, Ho CC, Lau SKP, Woo PCY, Yuen KY. Identification of a novel bat papillomavirus by metagenomics. PLoS One. 2012;7(8):e43986. doi: 10.1371/journal.pone.0043986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu Z, Ren X, Yang L, Hu Y, Yang J, He G, Zhang J, Dong J, Sun L, Du J, Liu L, Xue Y, Wang J, Yang F, Zhang S, Jin Q. Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. J Virol. 2012;86(20):10999–11012. doi: 10.1128/JVI.01394-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rector A, Mostmans S, Van Doorslaer K, McKnight CA, Maes RK, Wise AG, Kiupel M, Van Ranst M. Genetic characterization of the first chiropteran papillomavirus, isolated from a basosquamous carcinoma in an Egyptian fruit bat: the Rousettus aegyptiacus papillomavirus type 1. Vet Microbiol. 2006;117(2–4):267–275. doi: 10.1016/j.vetmic.2006.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Canuti M, Eis-Huebinger AM, Deijs M, de Vries M, Drexler JF, Oppong SK, Müller MA, Klose SM, Wellinghausen N, Cottontail VM, Kalko EK, Drosten C, van der Hoek L. Two novel parvoviruses in frugivorous New and Old World bats. PLoS One. 2011;6(12):e29140. doi: 10.1371/journal.pone.0029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ge X, Li Y, Yang X, Zhang H, Zhou P, Zhang Y, Shia Z. Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in china. J Virol. 2012 doi: 10.1128/jvi.06671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]