Abstract
Canine kobuviruses (CaKoVs) were first identified in diarrhoeic and asymptomatic dogs in 2011 in the USA. Subsequent studies have demonstrated a worldwide distribution of these viruses, but it is not clear if CaKoVs play a role as enteric pathogens of dogs. More recently, CaKoV RNA has been detected in wild carnivores, including red fox, golden jackal, side-striped jackal and spotted hyena. In this study, we addressed the hypothesis that wolves are susceptible to CaKoV infections. A total of 185 wolf stool samples were collected from necropsied animals and from transects in the Liguria, Piemonte and Valle D’Aosta regions of Italy, and CaKoV RNA was identified in two of these specimens. Both samples were obtained from necropsied wolves, with a prevalence rate of 4.9% (2/41). Sequence analysis of the full-length VP1 region showed that these strains displayed the highest nucleotide (nt) sequence identity (86.3-98.5%) to canine strains identified in the UK and Africa, and to kobuviruses that were previously detected in other African wild carnivores. This suggests that genetically related CaKoV strains circulate in domestic and wild carnivores, with interspecies transmission being not uncommon among carnivores of different ecosystems.
Kobuviruses (KoVs) are small non-enveloped RNA viruses belonging to the genus Kobuvirus, family Picornaviridae. The KoV genome is a polyadenylated, single-stranded, positive-sense RNA of 8.2 to 8.4 kb in length, encoding a single polyprotein of approximately 2,432-2,488 amino acids (aa) that is cleaved by the virus-encoded protease to produce a leader protein (L), three structural proteins (VP0, VP3, and VP1), and seven non-structural proteins (2A to 2C and 3A to 3D). Based on sequence comparisons and phylogenetic analysis of the full-length genome, KoVs segregate into at least three species: Aichivirus A (formerly Aichi virus), Aichivirus B (formerly Bovine kobuvirus) and Aichivirus C (porcine kobuvirus) [1]. Human Aichi virus (AiV) A846/88/JP, the prototype strain of the species Aichivirus A, was first recognized in 1989 as the cause of oyster-associated non-bacterial gastroenteritis in humans in Aichi Prefecture, Japan [2]. Since then, several worldwide investigations have revealed that AiV is involved in 0.9-4.1% of sporadic cases of pediatric gastroenteritis [3].
In 2011, KoVs genetically closely related to human AiV were identified in dogs in the USA by metagenomic investigation of the canine fecal virome [4, 5]. Full-genome sequencing, showed that the predicted amino acid sequences of canine KoVs and human AiVs were 79.0-80.0% identical to each other. According to the criteria established by the International Committee on Taxonomy of Viruses (ICTV) (http://www.picornastudygroup.com), the canine KoV (CaKoV) was classified as a distinct genotype (CaKoV type 1) from human AiV (AiV type 1) within the species Aichivirus A [6]. Since their first identification, CaKoVs have been identified in several countries in Europe, the Americas, Africa and Asia [7–13]. Interestingly, similar viruses have been identified recently in the feces of red foxes in Italy, with a prevalence rate of 14.7% [14], and in stool samples from a golden jackal, a side-striped jackal, and a spotted hyena in Africa [12], suggesting that CaKoVs are present in both wild and domestic carnivores. In order to draw a more complete picture of the host-species spectrum of kobuviruses, a surveillance study was initiated by screening fecal samples collected from wolves. Between February 2015 and April 2017, individual rectal swabs were collected from 41 wolves that had been found dead and submitted for necropsy analysis to the National Reference Center for Wild Animal Diseases (CeRMAS) and Torino University, as required by the Wolf Sanitary Surveillance Program established among the regions of Liguria, Piemonte and Valle d’Aosta (Italy). Thirty-three of the carcasses were found in Piemonte, five in Liguria and three in Valle D’Aosta. The most common cause of death (35 free-ranging animals) was trauma, followed by organ rupture and exsanguination. In six wolves, the cause of death remained undetermined due to severe postmortem decomposition. Also, a total of 144 wolf faecal samples were collected from November 2014 to December 2016 along systematic transects in areas of known transit of wolf packs located in the Piemonte region.
To assess the presence of KoVs RNA, the samples were screened using a broadly reactive primer pair, UNIV-kobu-F/UNIV-kobu-R, which amplifies a 217-bp fragment of the 3D region of all kobuviruses, as described previously [15]. In the samples yielding amplicons of the expected sizes, the presence of CaKoVs was confirmed using the specific primers CaKoV/F and CaKoV/R, targeting a 504-bp fragment of the 3D region [8], and primers VP1-F (5’-GCGGGCGAATCCTTCAAC-3’) and VP1-R (5’-GCGACCTTTCGGAGCGCC-3’) to amplify the region encoding the capsid protein VP1 (834 bp) [14].
All of the samples were screened for canine parvovirus (CPV-2) [16], canine adenoviruses (CAdV-1 and CAdV-2) [17], canine enteric coronavirus (CCoV) [18], canine distemper virus (CDV) [19], feline caliciviruses (FCVs) [20] and noroviruses (NoVs) [21] by conventional PCR or RT-PCR. All of the amplicons were purified using a QIAquick Gel Extraction Kit (QIAGEN GmbH, Hilden, Germany) and sequenced directly using BigDye Terminator cycle chemistry and a 3730 DNA Analyzer (Applied Biosystems, Foster City, CA). RT-PCR products for which sequences were not obtained by direct sequencing were cloned into pCR2.1 vector (Invitrogen, Ltd, Paisley, UK). Basic Local Alignment Search Tool (BLAST; http://www.ncbi.nlm.nih.gov) and FASTA (http://www.ebi.ac.uk/Tools/sss/fasta/) with default values were used to find homologous hits. Multiple alignments were performed using the commercially available Geneious software package version 9.1.6 (Geneious software package vers. 9, Biomatters, New Zealand, http://www.geneious.comBiomatters). A phylogenetic tree (neighbor joining and p-distance model) with bootstrap analysis (1,000 replicates) was constructed by using the MEGA software package, version 3.0 [22].
Out of 41 faecal specimens collected from necropsied animals, two (4.9%) were found to contain KoV RNA when using the primer set UNIV-kobu-F/UNIV-kobu-R [15] and the primer sets CaKoV/F-CaKoV/R and VP1-F/R [8, 14]. One additional specimen contained CPV-2 DNA. CaKoV RNA was detected in a 20-month-old female animal (strain CaKoV/6W/ITA from Piemonte) and in a 7-year-old male adult (strain CaKoV/125W/ITA) from Liguria. CPV-2 was identified in a 1-year-old female animal in Liguria (strain CPV/13W/ITA). None of the CaKoV- or CPV-2-infected animals showed macroscopic lesions indicative of enteritis at necropsy. When screening faecal specimens collected from transects, two samples collected in Piemonte were found to be positive for CAdV-2 DNA (2/144; 1.38%). The RNA of CCoV, CDV, FCV and NoV was not detected in rectal swabs collected from necropsied wolves or in transect faecal samples. Sequence analysis of the partial 3D region amplified using the CaKoV-specific primers showed that the two wolf strains shared the closest sequence identity (93.0%) to CaKoVs that had been identified previously in foxes in Italy [14]. Their nucleotide sequences were 89.0-90.0% identical to those of Italian CaKoV strains detected in dogs [8]. Sequence comparison of the full-length VP1 genes showed that strains 6W/ITA and 125W/ITA (GenBank accession numbers MF356367-8) displayed 94.5% nt sequence identity to each other and were most closely related genetically (91.4-93.5% nt sequence identity) to the CaKoV strain UK003 detected in a canine faecal sample in the UK [7] and to fox strains detected in Italy [14]. The nucleotide sequence identity to CaKoVs from golden jackal, side-striped jackal and spotted hyena stool samples [12] ranged from 89.0 to 91.1%, whilst the identity to strains detected in domestic dogs in the USA, Africa, China and Japan ranged from 82.4 to 91% (Table 1). Neighbor-joining phylogenetic analysis was performed with a selection of full-length VP1 sequences available in the databases that are representative of the genus Kobuvirus. Based on inspection of the tree (Fig. 1) and according to the pairwise homology and distance analyses, CaKoV sequences were found to segregate in at least three different genetic clusters. One group included CaKoV strains detected in dogs in China. Another group included three CaKoV strains detected in canine stool samples from Korea [10] and the USA [4, 5]. The wolf CaKoV strains detected in this study segregated within a third genetic group, along with canine strains found in the UK [7] and in Africa [12] and with CaKoV sequences detected in foxes [14], a golden jackal, a side-striped jackal, and a spotted hyena [12]. Overall, these findings indicate that genetically closely related strains circulate in both domestic and wild carnivores, suggesting limited host-species restriction for these viruses among the various carnivores and various ecosystems. Indeed, a high similarity was found between the wolf strains and viruses identified in red fox samples collected between September 2009 and May 2013 in the same Italian regions [14]. Similar results were obtained when comparing CaKoV sequences found in stool samples from dogs and wild carnivores in Africa [12]. This geographically related pattern also suggests that some wild animal species might act as a KoV reservoir for domestic carnivores.
Table 1.
Nucleotide sequence identities in the VP1 gene of the strains Wolf/125W/ITA and Wolf/6W/ITA to other CaKoVs detected in dogs and wild carnivores
| MF356367/Wolf/125W/ITA | MF356368/Wolf/6W/ITA | |
|---|---|---|
| MF356367/Wolf/125W/ITA | 94.5% | |
| MF356368/Wolf/6W/ITA | 94.5% | |
| KC161964/Dog/003/UK | 93.3% | 93.5% |
| KF781164/Fox/14V/ITA | 91.6% | 91.5% |
| KF781165/Fox/18V/ITA | 91.6% | 91.4% |
| KF781166/Fox/26V/ITA | 91.8% | 92.0% |
| KF781163/Fox/8V/ITA | 92.6% | 92.3% |
| KM068051/Spotted hyena/Tanzania | 90.4% | 91.1% |
| KM068050/Golden Jackal/Tanzania | 89.4% | 89.7% |
| KM068049/Side-striped Jackal/Tanzania | 89.0% | 89.4% |
| AB861494/Dog/Y12/Japan | 90.5% | 91.0% |
| KM205027/Dog/DD22/Tanzania | 89.6% | 90.4% |
| JN387133/Dog/AN211D/USA | 88.0% | 88.5% |
| JN088541/Dog/US-PC0082/USA | 88.2% | 88.8% |
| KU513557/Dog/SD2015-3/CH | 83.9% | 84.5% |
| MF062174/Dog/SMCQ-M9/CH | 83.8% | 84.2% |
| KU513558/Dog/FJ2015-4/CH | 84.4% | 84.8% |
| MF062172/Dog/SMCD-25/CH | 82.4% | 83.2% |
| JQ911763/Dog/CH-1/CH | 85.2% | 85.2% |
Fig. 1.
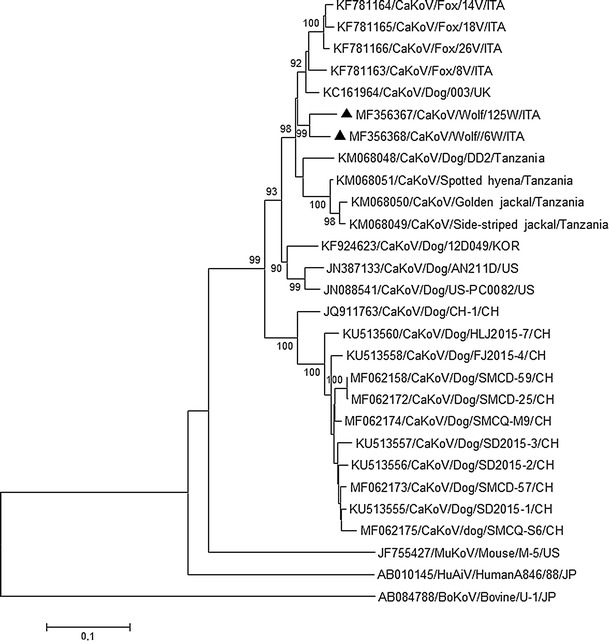
Neighbor-joining phylogenetic tree based on the VP1 gene (~ 800 nt) of the CaKoV strains detected in wolves. The tree was generated using the neighbor-joining method and p-distance correction, with statistical support provided by bootstrapping with 1000 replicates. The scale bar indicates nucleotide substitutions per site. The CaKoV sequences detected in wolves in this study are indicated
Several pieces of historical evidence indicate that wolves and domestic dogs are exposed to common viral infections. In our investigation, this hypothesis was supported by the identification of CPV-2 DNA in a dead adult. Also, two additional stool samples collected from transects were positive for CAdV-2. Circulation of CPV-2 in Italian wolf populations has already been described [23]. Conversely, to our knowledge, this is the first report demonstrating the presence of CAdV-2 in wolf stool samples, whilst CAdV-2 has been already detected in free-ranging red foxes in Italy [24]. Importantly, the clinical relevance of CaKoVs still remains unclear. Epidemiological investigations worldwide have revealed that CaKoVs are common in both symptomatic and asymptomatic dogs [4, 5, 7–13]. The prevalence of CaKoV infections in dogs with clinical signs of gastroenteritis has been estimated to vary from 1.3% [7] to 50.6% [9]. In a previous molecular investigation conducted in Italy [8] on diarrhoeic and healthy young dogs, CaKoVs RNA was identified only in animals with signs of gastroenteritis, either as a single infection or in combination with CCoV and/or CPV, with an overall prevalence rate of 4.37%. Interestingly, CaKoV RNA has also been detected in the liver, lung, brain, and tonsils of a 5-month-old puppy with severe enteritis in coinfection with CDV and CAdV-1 [25], indicating that, in some cases, the virus can spread to and/or replicate in extraintestinal sites.
The presence of CaKoV in healthy dogs has been documented in several studies [5, 12, 13, 26]. In a Japanese study [13], CaKoV was found in 35 of 94 diarrhoeic household dogs (37.2%) and 24 of 50 healthy kennel dogs (48.0%), with no significant differences (P > 0.05) between the two cohorts. In our analysis, CaKoV was detected in two adult wolves that most likely died after traumatic events, without apparent pathological lesions in the enteric tract. Similarly, CaKoVs have been identified in apparently healthy foxes in Italy and in asymptomatic wild carnivores in Tanzania [12, 14]. Accordingly, epidemiological investigations have not been able thus far to gather conclusive information, and it is unclear whether KoVs play a role as enteric pathogens. Asymptomatic viral infections might be due to host-related factors, including genetic resistance and acquired immunity, and to virus-related factors, i.e., phenotype variations among the various viral strains.
In the light of the results obtained in this study, CaKoV should be considered a potential constituent of the wolf enteric virome. As wild animals can serve as reservoirs for a number of viral pathogens of domestic animals and humans, or vice versa, understanding the ecology of viruses requires monitoring of populations of wildlife in an integrated manner.
Acknowledgements
The authors wish to thank Prof. Luca Rossi, Prof. Pier Giuseppe Meneguz, Prof. Ezio Ferroglio (Università degli Studi di Torino, Dipartimento di Scienze Veterinarie), Dr. Francesca Marucco (Centro Conservazione e Gestione Grandi Carnivori, Parco Naturale Alpi Marittime), and Dr. Simona Zoppi (Istituto Zooprofilattico Sperimentale Piemonte, Liguria e Valle d’Aosta) for sample collection.
Compliance with ethical standards
Funding
This work was financed by grants from Progetto di Ricerca Ministeriale Corrente IZS PLV 15/15 “Indagine sulla presenza di virus enterici con potenziale zoonosico in canidi selvatici”. Sandra Bermudez Sanchez received a grant from the Marie Skłodowska-Curie COFUND REP-EAT 713714. Innovative Research and Training Doctoral Programme in the interdisciplinary domain of food and healthy diet.
Conflict of interest
All authors declare that there are no financial or other relationships that might lead to a conflict of interest. All authors have seen and approved the manuscript and have contributed significantly to the work.
Ethics standard
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
References
- 1.Adams MJ, King AM, Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2013) Arch Virol. 2013;158:2023–2030. doi: 10.1007/s00705-013-1688-5. [DOI] [PubMed] [Google Scholar]
- 2.Yamashita T, Sakae K, Tsuzuki H, Suzuki Y, Ishikawa N, Takeda N, Miyamura T, Yamazaki S. Complete nucleotide sequence and genetic organization of Aichi virus, a distinct member of the picornaviridae associated with acute gastroenteritis in humans. J Virol. 1998;72:8408–8412. doi: 10.1128/jvi.72.10.8408-8412.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drexler JF, Baumgarte S, de Souza Luna LK, Eschbach-Bludau M, Lukashev AN, Drosten C. Aichi virus shedding in high concentrations in patients with acute diarrhea. Emerg Infect Dis. 2011;17:1544–1548. doi: 10.3201/eid1708.101556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapoor A, Simmonds P, Dubovi EJ, Qaisar N, Henriquez JA, Medina J, Shields S, Lipkin WI. Characterization of a canine homolog of human Aichivirus. J Virol. 2011;85:11520–11525. doi: 10.1128/JVI.05317-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L, Pesavento PA, Shan T, Leutenegger CM, Wang C, Delwart E. Viruses in diarrhoeic dogs include novel kobuviruses and sapoviruses. J Gen Virol. 2011;92(Pt 11):2534–2541. doi: 10.1099/vir.0.034611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knowles NJ, Hovi T, Hyypiä T, King AMQ, Lindberg AM, Pallansch MA, Palmenberg AC, Simmonds P, Skern T, Stanway G, Yamashita T, Zell R. Picornaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus taxonomy: classification and nomenclature of viruses: ninth report of the international committee on taxonomy of viruses. San Diego: Elsevier; 2012. pp. 855–880. [Google Scholar]
- 7.Carmona-Vicente N, Buesa J, Brown PA, Merga JY, Darby AC, Stavisky J, Sadler L, Gaskell RM, Dawson S, Radford AD. Phylogeny and prevalence of kobuviruses in dogs and cats in the UK. Vet Microbiol. 2013;164:246–252. doi: 10.1016/j.vetmic.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Martino B, Di Felice E, Ceci C, Di Profio F, Marsilio F. Canine kobuviruses in diarrhoeic dogs in Italy. Vet Microbiol. 2013;166:246–249. doi: 10.1016/j.vetmic.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi S, Lim SI, Kim YK, Cho YY, Song JY, An DJ. Phylogenetic analysis of astrovirus and kobuvirus in Korean dogs. J Vet Med Sci. 2014;76:1141–1145. doi: 10.1292/jvms.13-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong N, Zuo Y, Wang Z, Yu H, Zhou EM, Shan T, Tong G. Molecular characterization of new described kobuvirus in dogs with diarrhea in China. Springerplus. 2016;5:2047. doi: 10.1186/s40064-016-3738-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li C, Wei S, Guo D, Wang Z, Geng Y, Wang E, Zhao X, Su M, Wang X, Sun DJ. Prevalence and phylogenetic analysis of canine kobuviruses in diarrhoetic dogs in northeast China. Vet Med Sci. 2016;78:7–11. doi: 10.1292/jvms.15-0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olarte-Castillo XA, Heeger F, Mazzoni CJ, Greenwood AD, Fyumagwa R, Moehlman PD, Hofer H, East ML. Molecular characterization of canine kobuvirus in wild carnivores and the domestic dog in Africa. Virology. 2015;477:89–97. doi: 10.1016/j.virol.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Soma T, Matsubayashi M, Sasai K. Detection of kobuvirus RNA in Japanese domestic dogs. J Vet Med Sci. 2016;78:1731–1735. doi: 10.1292/jvms.16-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Martino B, Di Profio F, Melegari I, Robetto S, Di Felice E, Orusa R, Marsilio F. Molecular evidence of kobuviruses in free-ranging red foxes (Vulpes vulpes) Arch Virol. 2014;159:1803–1806. doi: 10.1007/s00705-014-2109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reuter G, Boldizsár A, Pankovics P. Complete nucleotide and amino acid sequences and genetic organization of porcine kobuvirus, a member of a new species in the genus Kobuvirus, family Picornaviridae. Arch Virol. 2009;154:101–108. doi: 10.1007/s00705-008-0288-2. [DOI] [PubMed] [Google Scholar]
- 16.Buonavoglia C, Martella V, Pratelli A, Tempesta M, Cavalli A, Buonavoglia D, Bozzo G, Elia G, Decaro N, Carmichael L. Evidence for evolution of canine parvovirus type 2 in Italy. J Gen Virol. 2001;82:3021–3025. doi: 10.1099/0022-1317-82-12-3021. [DOI] [PubMed] [Google Scholar]
- 17.Hu RL, Huang G, Qiu W, Zhong ZH, Xia XZ, Yin Z. Detection and differentiation of CAV-1 and CAV-2 by polymerase chain reaction. Vet Res Commun. 2001;25:77–84. doi: 10.1023/A:1006417203856. [DOI] [PubMed] [Google Scholar]
- 18.Pratelli A, Tempesta M, Roperto FP, Sagazio P, Carmichael L, Buonavoglia C. Fatal coronavirus infection in puppies following canine parvovirus 2b infection. J Vet Diagn Investig. 1999;11:550–553. doi: 10.1177/104063879901100615. [DOI] [PubMed] [Google Scholar]
- 19.Kim YH, Cho KW, Youn HY. Detection of canine distemper virus (CDV) through one step RT-PCR combined with nested PCR. J Vet Sci. 2001;2:59–63. [PubMed] [Google Scholar]
- 20.Marsilio F, Di Martino B, Decaro N, Buonavoglia C. A novel nested PCR for the diagnosis of calicivirus infections in the cat. Vet Microbiol. 2005;105:1–7. doi: 10.1016/j.vetmic.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 21.Vennema H, de Bruin E, Koopmans M. Rational optimization of generic primers used for Norwalk-like virus detection by reverse transcriptase polymerase chain reaction. J Clin Virol. 2002;25:233–235. doi: 10.1016/S1386-6532(02)00126-9. [DOI] [PubMed] [Google Scholar]
- 22.Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–1631. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 23.Battilani M, Scagliarini A, Tisato E, Turilli C, Jacoboni I, Casadio R, Prosperi S. Analysis of canine parvovirus sequences from wolves and dogs isolated in Italy. J Gen Virol. 2001;82:1555–1560. doi: 10.1099/0022-1317-82-7-1555. [DOI] [PubMed] [Google Scholar]
- 24.Balboni A, Verin R, Morandi F, Poli A, Prosperi S, Battilani M. Molecular epidemiology of canine adenovirus type 1 and type 2 in free-ranging red foxes (Vulpes vulpes) in Italy. Vet Microbiol. 2013;162:551–557. doi: 10.1016/j.vetmic.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 25.Ribeiro J, Headley SA, Diniz JA, Pereira AH, Lorenzetti E, Alfieri AA, Alfieri AF. Extra-intestinal detection of canine kobuvirus in a puppy from Southern Brazil. Arch Virol. 2017;162:867–872. doi: 10.1007/s00705-016-3164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oem JK, Choi JW, Lee MH, Lee KK, Choi KS. Canine kobuvirus infections in Korean dogs. Arch Virol. 2014;159:2751–2755. doi: 10.1007/s00705-014-2136-x. [DOI] [PMC free article] [PubMed] [Google Scholar]


