Abstract
Neonatal calf diarrhea (NCD) is a major cause of morbidity, mortality and economic losses in the beef and dairy industries. This study was conducted to investigate the existence of enteric viruses in two Egyptian farms with a history of recurrent diarrhea. Fecal samples were collected from 25 diarrheic calves. RNA was extracted and tested by reverse transcription polymerase chain reaction (RT-PCR) for the presence of rotavirus, norovirus, astrovirus, torovirus, coronavirus and bovine viral diarrhea virus. Overall, 76 % (19/25) of samples tested positive for one or more viruses. Rota-, noro- and astroviruses were detected in 48 %, 24 % and 32 % of tested samples, respectively. About 37 % (7/19) of positive samples had two different viruses. One-month-old calves were the group most vulnerable to infections. Based on phylogenetic analysis, bovine rotaviruses were of genotypes G6 and G10, bovine noroviruses were in GIII.2, and bovine astroviruses were in the BAstV lineage 1. Astrovirus sequences showed a high level nucleotide sequence similarity with the Brazilian BAstV sequences available in GenBank. We believe this is the first report of bovine norovirus and bovine astrovirus circulating among calves in Egypt. Further epidemiological studies are recommended to investigate their presence on a wider scale, to predict their association with NCD, and to design appropriate diagnostic and control methods.
Electronic supplementary material
The online version of this article (doi:10.1007/s00705-016-3088-0) contains supplementary material, which is available to authorized users.
Keywords: Reverse Transcription Polymerase Chain Reaction, Bovine Viral Diarrhea Virus, Enteric Virus, Reverse Transcription Polymerase Chain Reaction Reaction, Diarrheic Calf
Introduction
Neonatal calf diarrhea (NCD) is one of the most common causes of morbidity and mortality in cattle. In the dairy industry, approximately 50 % of deaths in one-month-old calves have been attributed to diarrhea caused by bacterial, viral or parasitic pathogens [1]. Bovine rotavirus and bovine coronavirus (BRV and BCV) are the leading causes of viral diarrhea [2], although bovine viral diarrhea virus (BVDV), bovine norovirus (BNoV), bovine astrovirus (BAstV) and bovine torovirus (BToV) have also been implicated. Co-infection with two or more viruses is also common and often aggravates the diarrheal symptoms [3]. Both BCR and BToV are members of the family Coronaviridae. They are frequently associated with acute diarrheal and respiratory infections in calves of different ages in many countries [4]. BVDV is a member of the genus Pestivirus, family Flaviviridae. BVDV is classified molecularly into two species (Bovine viral diarrhea virus 1, and Bovine viral diarrhea virus 2). It was first identified in Egypt in 1972, and BVDV-1b was circulating in the Ismailia province of Egypt during 2014 [5].
Members of the genus Rotavirus, family Reoviridae, are medium-sized, non-enveloped viruses. Their dsRNA genome consists of 11 RNA segments that encode six structural (VP1-4, VP6 and VP7) and six non-structural proteins (NSP1-6) [6]. Based on their common group antigen (VP6), rotaviruses are classified into eight antigenically distinct groups (from A to H). Group A rotaviruses are usually incriminated in human and animal cases of gastroenteritis [7]. Since capsid proteins VP4 and VP7 autonomously trigger neutralizing antibodies, group A rotaviruses are further classified into P (for protease sensitive) and G (for glycoprotein) types, respectively. At least 37 P-types and 27 G-types have been reported (http://rotac.regatools.be/classificationinfo.html), of which G6, G8, G10, P1, P5 and P11 are usually associated with most cases of bovine diarrhea [8].
Norwalk-like viruses (noroviruses) are known to infect humans and animals and cause epidemic and sporadic gastroenteritis. As members of the family Caliciviridae, they are small, non-enveloped, positive-sense, ssRNA viruses. Their viral genome is 7.3 to 8.3 kb in size, excluding the 3′ poly(A) tail and consists of three open reading frames (ORFs). ORF1 encodes the non-structural polyprotein, which is cleaved by 3C-like protease into six proteins, including the RNA-dependent RNA polymerase (RdRp). ORF2 and ORF3 encode the structural capsid proteins VP1 and VP2, respectively [9]. Based on phylogenetic analysis, noroviruses are classified into at least six genogroups (GI to GVI). A tentative genogroup VII has also been proposed. The bovine noroviruses are clustered in genogroup GIII, which is further divided into two distinct genotypes (GIII.1 and GIII.2) [10]. Initially GIII.1 (prototype Bo/Jena/80/DE) was identified in Germany in 1980 while GIII.2 (prototype Bo/Newbury-2/76/UK) was discovered in England in 1978 [11, 12]. The lack of a culture system for in vitro propagation of noroviruses, recombination events, and the presence of the virus in fecal samples of both diarrheic and healthy animals are major obstacles to determine their impact on the cattle industry, especially in developing countries.
Astroviruses are small, star-shaped, non-enveloped viruses with a positive-sense ssRNA. The family Astroviridae consists of two genera, Mamastrovirus and Avastrovirus, whose members infect mammals and birds, respectively [13]. The genome of astroviruses is 6.4–7.9 kb long and consists of three ORFs in addition to a 5′ UTR, a 3′ UTR and a polyadenylated tail. ORF1a encodes the non-structural polyprotein, ORF1b encodes the RdRp, and ORF2 encodes the structural capsid protein. ORF2 is the most divergent part of the genome, while ORF1b is the least divergent [14]. BAstV was first reported in England in 1978 from calves suffering from acute enteritis. Initially, it was considered avirulent [15], but subsequently, it was proven to be a pathogen, especially in calves that are co-infected with BRV or BToV. The astroviruses have also sometimes been detected in association with bovine encephalitis [16]. Based on serological assays, bovine astroviruses are classified into two serotypes, BAstV-1 and BAstV-2 [17].
Materials and methods
Samples and RNA extraction
A total of 25 fecal samples were randomly collected from two private cattle farms in Sharkia and Cairo provinces in early 2015. The calves had diarrhea, fever and variable degrees of dehydration and weakness. Diseased calves did not respond to antibiotic therapy. No mortalities were found at the time of sample collection. The average age of animals ranged from three weeks to ten months. The calves on one farm were vaccinated against BRV and BCV. The viral RNA was extracted from a 20 % fecal suspension using a Blood/Liquid Sample Total RNA Rapid Extraction Kit (Bioteke Corporation, China) according to the manufacturer’s instructions.
Reverse transcription polymerase chain reaction (RT-PCR)
The RNA extracts were screened by RT-PCR for the detection of BRV, BCV, BVDV, BToV, BNoV and BAstV. Reactions were performed in a 25 µl volume using a One Step RT-PCR Kit (QIAGEN, Valencia, CA, USA). Reverse transcription was done at 50 °C for 30 min, followed by PCR activation at 95 °C for 15 min and 35 cycles of 94 °C for 1 min for denaturation, the appropriate temperature and time for annealing (Table 1) and 72 °C for 1 min for extension. The final extension was done at 72 °C for 10 min. For the G and P typing of BRV-positive samples, RT-PCR reactions were performed using specific primers for VP7 and VP4, respectively. The primers used in this study are listed in Table 1. The PCR products were confirmed by analysis in an ethidium-bromide-stained agarose gel followed by visualization under a UV transilluminator.
Table 1.
Primers used and their annealing temperatures
| Virus | Primer sequence (5′-3′) | Amplicon size (bp) | Target gene | Annealing temperature/time | References |
|---|---|---|---|---|---|
| BRV | F: GGCTTTWAAACGAAGTCTTC | 928 | Group (A)–VP6 | 52 °C/45 s | [29] |
| R: GGYGTCATATTYGGTGG | [30] | ||||
| BRV | F: ATGTATGGTATTGAATATACCAC | 881 | VP7 | 52 °C/1 min | [31] |
| R: AACTTGCCACCATTTTTTCC | |||||
| BRV | F: TAT GCT CCA GTN AAT TGG | 633 | VP4 | 50 °C/1 min | [32] |
| R: ATT GCA TTT CTT TCC ATA ATG | |||||
| BAstV | F: GAYTGGACBCGHTWTGATGG | 432 | RdRp | 48 °C/1 min | [33] |
| R: KYTTRACCCACATNCCAA | |||||
| BNoV | CBECu-F: AGTTAYTTTTCCTTYTAYGGBGA | 532 | RdRP | 51 °C/45 s | [34] |
| CBECu-R: AGTGTCTCTGTCAGTCATCTTCAT | |||||
| BCoV | F: CCGATCAGTCCGACCAATC | 460 | N gene | 55 °C/45 s | [35] |
| R: TAGTCGGAATAGCCTCATCGC | |||||
| BToV | F: TTCTTACTACACTTTTTGGA | 602 | Matrix gene | 45 °C/45 s | [36] |
| R: ACTCAAACTTAACACTAG AC | |||||
| BVDV | F: ACAAACATGGTTGGTGCAACTGGT | 820 | Polyprotein | 54 °C/ 45 s | [37] |
| R: CAGACATATTTGCCTAGGTTCCA |
BRV bovine rotavirus, BAstV bovine astrovirus, BNoV bovine norovirus, BCoV bovine coronavirus, BToV bovine torovirus, BVDV bovine viral diarrhea virus, F forward primer, R reverse primer
Sanger sequencing
The PCR products were purified using a QIAquick PCR purification kit (QIAGEN, Valencia, CA, USA). The purified DNA was sequenced at the University of Minnesota Genomics Center (UMGC) using the same forward and reverse primers as were used for RT-PCR. Forward and reverse sequences were aligned together to generate a consensus sequence using Sequencher software version 5.1 (http://genecodes.com/). Also the newly obtained sequences were compared with existing sequences in the GenBank database using online BLAST search tool (http://www.ncbi.nlm.nih.gov/).
Sequencing and phylogenetic analysis
The newly obtained nucleotide sequences were deposited in the GenBank database under accession numbers KX268316 to KX268321 for BRV, KX268306 to KX268309 for BNoV, and KX268310 to KX268315 for BAstV. Using MEGA 6.0 software, a comparative alignment was done using the Clustal W method. The evolutionary distances were calculated using the pairwise (p) distance method (percent of nucleotide [nt] and amino acid [aa] sequence identity and divergence). The phylogenetic trees were constructed using the neighbor-joining method in MEGA 6.0 software [18], and the tree topology was evaluated by 1,000 bootstrap replicates.
Results
Detection of enteric viruses in fecal samples
The RT-PCR results of the tested RNA showed that 48 % (95 % CI: 28.42 % to 67.58 %) were BRV positive, 24 % (95 % CI: 7.26 % to 40.74 %) were BNoV positive, and 32 % (95 % CI: 13.71 % to 50.29 %) were BAstV positive. About 37 % (7/19) of positive samples had two different viruses (Table 2). Products of 928 bp, 881 bp, 532 bp, and 432 bp were visualized on a 1.2 % agarose gel for BRV-VP6, BRV-VP7, BNoV-RdRp and BAstV-RdRp, respectively (Supplementary Fig. 1a–d). When data were correlated to the age of the tested calves, one-month-old calves showed the highest rate of infections (Fig. 1). Results were negative for BCV, BVDV and BToV.
Table 2.
Detection rates of different viruses
| Virus | No. (%) of positive samples | L. CI (%) | U. CI (%) |
|---|---|---|---|
| BRV group (A) | 9 (36 %) | 17.18 | 54.82 |
| BNoV | 1 (4 %) | 0 | 13.54 |
| BAstV | 2 (8 %) | 0 | 18.63 |
| BRV and BNoV | 1 (4 %) | 0 | 13.54 |
| BRV and BAstV | 2 (8 %) | 0 | 18.63 |
| BNoV and BAstV | 4 (16 %) | 1.63 | 30.37 |
L.CI lower confidence interval, U.CI upper confidence interval
Fig. 1.
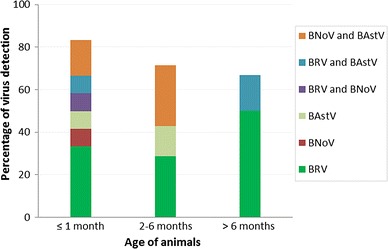
Virus detection in various age groups
Sequencing and phylogenetic analysis
The phylogenetic analysis of the BRV-VP7 coding sequences revealed that they clustered with the G6 and G10 genotypes of group A rotaviruses (http://rotac.regatools.be/) [19] (Fig. 2). At both the nt and aa level, the G10 sequences were 100 % identical and the G6 sequences showed 99.5–100 % identity to each other. A comparison of the G10 sequences with published sequences in the GenBank database showed overall identities of 93.7–95.9 % and 97.1–99.2 % in nt and aa, respectively. The same comparison using G6 sequences showed identities of 93.1–94.7 % at the nt level and 95.5–96.8 % at the aa level. On P typing, only one isolate was found to be P11, while all remaining samples were non-typable.
Fig. 2.
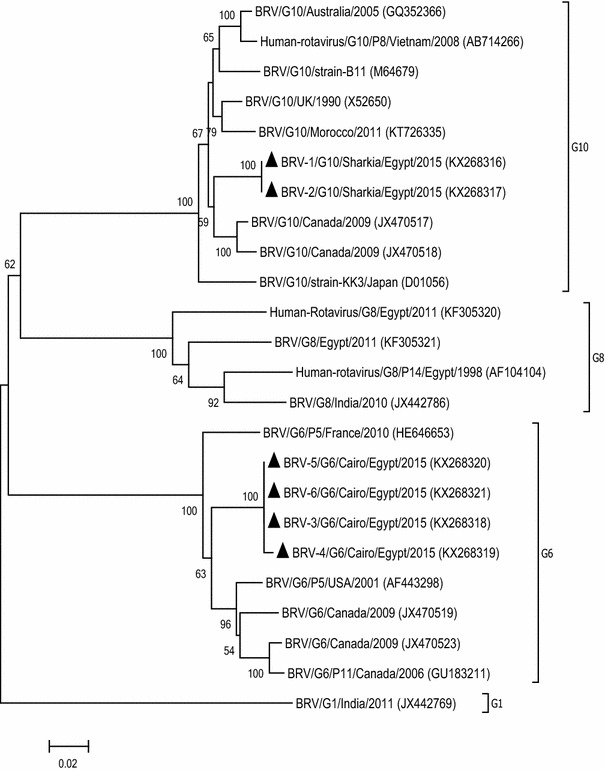
Neighbor-joining phylogenetic tree of BRV based on partial nucleotide sequences (881 bp) of the VP7 gene. Bootstrap values (>50 %) are shown above the branches. The isolates from this study are indicated by a solid triangle
The norovirus sequences of this study showed identities of 87.5–99.8 % and 96.7–100 % at nt and aa levels, respectively. A phylogenetic tree illustrated that all samples were related to genotype GIII.2 of BNoV (Bovine/Newbury2/UK strain-like) (Fig. 3). Three sequences (BNoV-1, BNoV-2, and BNoV-3) grouped together with 99.3–99.8 % nt and 100 % aa identity with each other and maximum identity with previously published sequences from the USA (Nov-6 and Nov-45), the UK (Penrith5500) and Tunisia (MonastirB139). One of the study sequences (BNoV-4) grouped differently from the remaining sequences, with 87.5 % and 96.7 % nt and aa identity, respectively, and showed maximum identity to isolates from Italy (BEC195), the UK (Aberystwyth24) and Belgium (Florennes-B242).
Fig. 3.
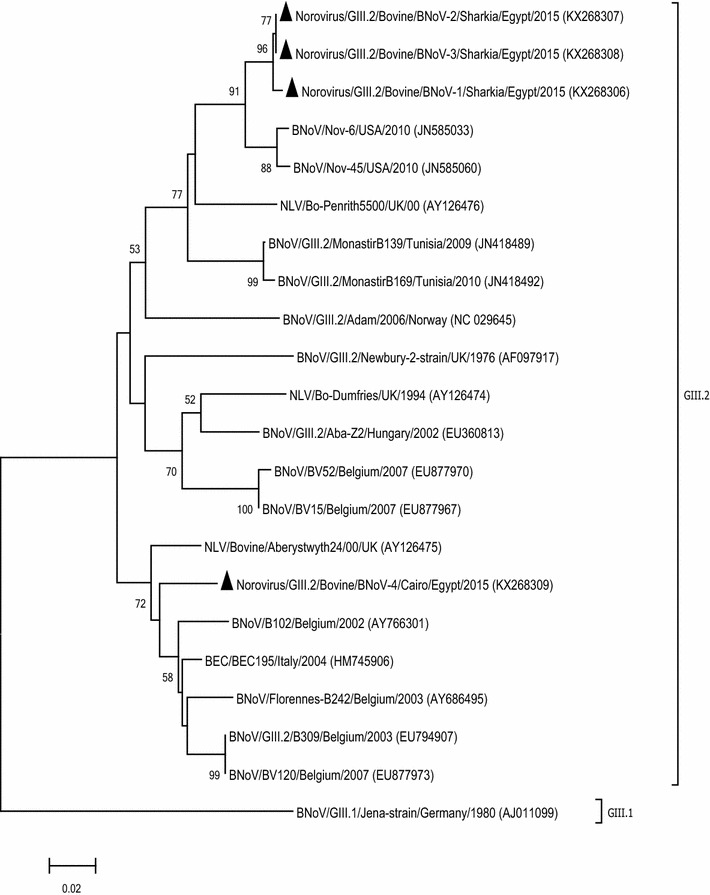
Neighbor-joining phylogenetic tree of BNoV isolates based on partial nucleotide sequences of the RdRp gene (532 bp). BNoV GIII.I (Jena strain) was selected as an out-group. Bootstrap values (>50 %) are shown above the branches. The isolates from this study are indicated by a solid triangle
The BAstV study sequences were compared with each other as well as with previously published BAstV sequences available in GenBank (Fig. 4) and clustered with BAstV lineage 1. Except BAstV-3, all study sequences grouped together with 98.2–100 % nt and 98.5–100 % aa identity to each other. These sequences showed maximum sequence similarity to BoAstV-199-BRA (Brazil) and Ishikawa9728 (Japan). The BAstV-3 showed 87.3 % nt and 95.3 % aa sequence identity to the rest of the study sequences and maximum identity to Kagoshima2-24 and Hokkaido12-27 (Japan).
Fig. 4.
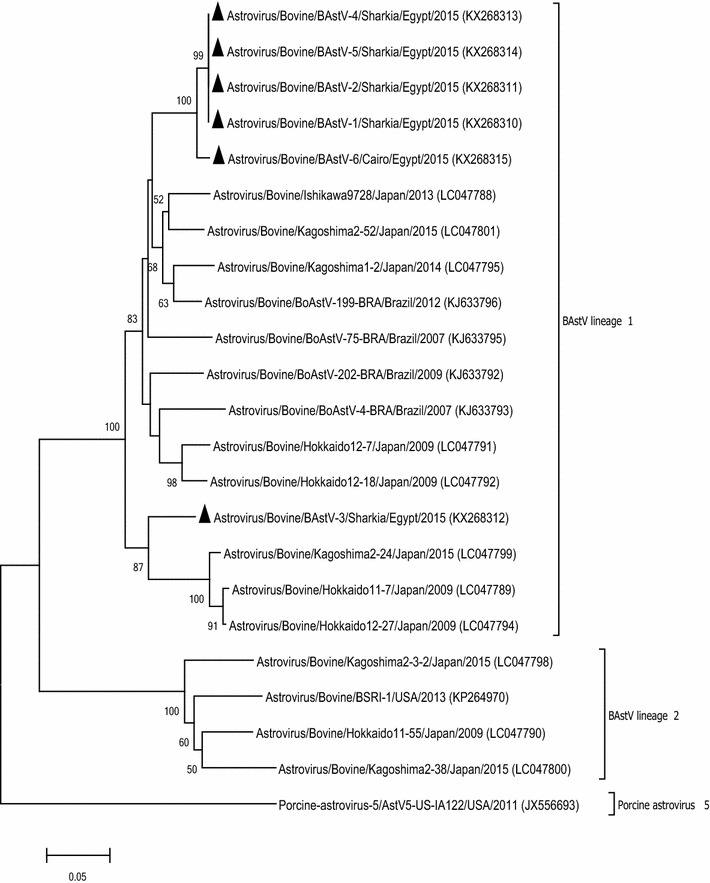
Neighbor-joining phylogenetic tree based on partial RdRp gene sequences (432 bp) of BAstV. An isolate of porcine astrovirus was selected as an out-group. Bootstrap values (>50 %) are shown above the branches. The isolates from this study are indicated by a solid triangle
Discussion
Calves within their first month are highly susceptible to viral diarrhea probably due to suckling milk, which neutralizes the acidic pH of their digestive tract and in turn allows several pathogens to survive [2, 38]. Several improvements in vaccination, medication and management have been implemented to reduce the incidence of NCD. However, NCD is still persistent because it is complex and multifactorial and can be triggered by several different infectious and non-infectious causes [2]. NCD has a devastating economic impact on cattle-raising businesses worldwide [2, 20, 21]. Direct losses are due to dehydration, reduced growth rate, and high morbidity and mortality. Indirect losses are due to the imposition of trade restrictions and increased costs of management and veterinary care as well as animal suffering. In Egypt, the diarrhea control programs are completely dependent on mass vaccination of animals using commercially available inactivated vaccines. Data on the detection, prevalence and typing of infectious diarrheal causes are not available, and this study was undertaken in an attempt to fill that gap.
Rotaviruses are the main causes of diarrhea in many animal and human species. The G6, G8 and G10 types of group A rotaviruses are usually found in cases of NCD. We detected rotaviruses in 48 % of the tested samples (66.6 % for G6 and 33.3 % for G10). This could be a preliminary indication of the types of bovine rotaviruses currently circulating in Egypt. Nearly the same rates of G6 and G10 were recorded by Caruzo et al. [22], while the overall rotavirus detection rate was 9.9 %, probably due to a larger sample size. Only one of the BRV isolates in our study was P typable (BRV-1/Sharkia/2015) and was found to be P11, which is the most dominant P type. Also, the G10-P11 genotype is a common combination worldwide [22]. In Egypt, BRV was first diagnosed in the early 1980s. The BRV-G8 type was detected by Merwad et al. [23]. Some studies have found evidence of viral reassortment and the possibility of interspecies transmissions, including to humans [24]. Our results might help in future studies on rotaviruses in Egypt.
BNoV was molecularly detected (24 %) either alone or as a co-infection with other enteric viruses (BRV and BAstV). All BNoV sequences were found to be phylogenetically related to genotype GIII.2, which is known to be distributed globally. Ferragut et al. [25] discussed an interesting finding about the Argentinean strain B4881 (tentatively classified as GIII.4), which grouped with GIII.2 based on partial ORF1 sequence analysis but showed a significant divergence from previously described genotypes based on ORF2/3 sequence analysis. Molecular investigations in several countries have revealed a number of potential recombinant strains, which emphasizes the importance of studying complete genome sequences; the use of only a single primer set (CBECu-F and R) may limit the opportunity to detect novel divergent/recombinant strains.
The connection between bovine astroviruses and enteric diseases in not obvious; some researchers maintain that BAstV is not associated directly with severe diarrhea under natural conditions [12], while others believe that it can happen [26]. We reported BAstV in feces of calves with clinical diarrhea (32 %), which clustered with BAstV lineage 1 [27]. Lineage 1 involves one and nine strains of genogroup G2 and G3, respectively [28], in addition to five Korean strains within genogroup G1 [26]. The Egyptian BAstV strains were closely related to isolates from Brazil and Japan. This further supports the assumption of Nagai et al. [27] that lineage 1 might be the most prevalent genogroup of BAstV in the world. Efforts should be made to develop specific criteria for the classification of BAstVs as lineages or genogroups. Our BAstV sequences showed 87.1–100 % nt sequence identity, in contrast to those in a Brazilian BAstV study in which the identity was 48.9–80.6 % [28]. This may be due to the small sample size from two Egyptian provinces, while the Brazilian study was based on 272 samples from seven different states.
Conclusions
To the best of our knowledge, this is the first report on the existence of bovine noroviruses and bovine astroviruses in Egypt. The association of these viruses with the pathogenesis of severe diarrhea is still inconclusive. The genetic diversity and evolution of these enteric viruses should be monitored regularly in domestic and wild animal populations as well as in humans. Our results will contribute to better understanding of these challenging gastrointestinal viruses and may help to spur studies designed to prevent and control calf diarrhea in Egypt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1: Agarose gel electrophoresis of RT-PCR products (A) Detection of BRV-VP6 positive samples (product size 928 bp). (B) Detection of BRV-VP7 positive samples (product size 881 bp). (C) Detection of BNoV positive samples (product size 532 bp). (D) Detection of BAstV positive samples (product size 432 bp) (DOC 494 kb)
Acknowledgments
The authors thank the Ministry of Higher Education and Scientific Research (MHESR) in Egypt for providing a visiting grant to FFM. We also thank Dr. Hany Abdalla (Department of Theriogenology, Faculty of Veterinary Medicine, Zagazig University, Egypt) for help in collection of samples and historical data.
Compliance with ethical standards
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors confirm that this article content has no conflict of interest.
References
- 1.Lorenz I, Fagan J, More SJ. Calf health from birth to weaning. II. Management of diarrhoea in pre-weaned calves. Ir Vet J. 2011;64:1. doi: 10.1186/2046-0481-64-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho YI, Yoon KJ. An overview of calf diarrhea-infectious etiology, diagnosis, and intervention. J Vet Sci. 2014;15:1–17. doi: 10.4142/jvs.2014.15.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharp CP, Gregory WF, Mason C, Barend M, Beard PM. High prevalence and diversity of bovine astroviruses in the faeces of healthy and diarrhoeic calves in South West Scotland. Vet Microbiol. 2015;178:70–76. doi: 10.1016/j.vetmic.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lojkić I, Krešić N, Šimić I, Bedeković T. Detection and molecular characterisation of bovine corona and toroviruses from Croatian cattle. BMC Vet Res. 2015;11:1. doi: 10.1186/s12917-014-0312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soltan MA, Wilkes RP, Elsheery MN, Elhaig MM, Riley MC, Kennedy MA. Circulation of bovine viral diarrhea virus–1 (BVDV-1) in dairy cattle and buffalo farms in Ismailia Province, Egypt. J Infect Dev Ctries. 2015;9:1331–1337. doi: 10.3855/jidc.7259. [DOI] [PubMed] [Google Scholar]
- 6.Greenberg HB, Estes MK. Rotaviruses: from pathogenesis to vaccination. Gastroenterology. 2009;136:1939–1951. doi: 10.1053/j.gastro.2009.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Bányai K, Brister JR, Buesa J, Esona MD, Estes MK, Gentsch JR, Iturriza-Gómara M. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG) Arch Virol. 2011;156:1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Midgley SE, Bányai K, Buesa J, Halaihel N, Hjulsager CK, Jakab F, Kaplon J, Larsen LE, Monini M, Poljšak-Prijatelj M, Pothier P. Diversity and zoonotic potential of rotaviruses in swine and cattle across Europe. Vet Microbiol. 2012;156:238–245. doi: 10.1016/j.vetmic.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 9.Green KY. Caliciviridae: the noroviruses. Fields Virol. 2007;5:949–979. [Google Scholar]
- 10.Vinjé J. Advances in laboratory methods for detection and typing of norovirus. J Clin Microbiol. 2015;53:373–381. doi: 10.1128/JCM.01535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Günther H, Otto P. Diarrhea in young calves. 7. “Zackenvirus” (Jena agent 117/80)–a new diarrhea pathogen in calves. Arch Exp Veterinarmed. 1987;41:934–938. [PubMed] [Google Scholar]
- 12.Woode GN, Bridger JC. Isolation of small viruses resembling astroviruses and caliciviruses from acute enteritis of calves. J Med Microbiol. 1978;11:441–452. doi: 10.1099/00222615-11-4-441. [DOI] [PubMed] [Google Scholar]
- 13.De Benedictis P, Schultz-Cherry S, Burnham A, Cattoli G. Astrovirus infections in humans and animals–molecular biology, genetic diversity, and interspecies transmissions. Infect Genet Evol. 2011;11:1529–1544. doi: 10.1016/j.meegid.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strain E, Kelley LA, Schultz-Cherry S, Muse SV, Koci MD. Genomic analysis of closely related astroviruses. J Virol. 2008;82:5099–5103. doi: 10.1128/JVI.01993-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bridger JC, Hall GA, Brown JF. Characterization of a calici-like virus (Newbury agent) found in association with astrovirus in bovine diarrhea. Infect Immun. 1984;43:133–138. doi: 10.1128/iai.43.1.133-138.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlottau K, Schulze C, Bilk S, Hanke D, Höper D, Beer M, Hoffmann B. Detection of a novel bovine astrovirus in a cow with encephalitis. Transbound Emerg Dis. 2016;63:253–259. doi: 10.1111/tbed.12493. [DOI] [PubMed] [Google Scholar]
- 17.Woode GN, Gourley NE, Pohlenz JF, Liebler EM, Mathews SL, Hutchinson MP. Serotypes of bovine astrovirus. J Clin Microbiol. 1985;22:668–670. doi: 10.1128/jcm.22.4.668-670.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maes P, Matthijnssens J, Rahman M, Van Ranst M. RotaC: a web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol. 2009;9:1. doi: 10.1186/1471-2180-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hur TY, Jung YH, Choe CY, Cho YI, Kang SJ, Lee HJ, Ki KS, Baek KS, Suh GH. The dairy calf mortality: the causes of calf death during ten years at a large dairy farm in Korea. Korean J Vet Res. 2013;53:103–108. doi: 10.14405/kjvr.2013.53.2.103. [DOI] [Google Scholar]
- 21.Østerås O, Gjestvang MS, Vatn S, Sølverød L. Perinatal death in production animals in the Nordic countries–incidence and costs. Acta Vet Scand. 2007;49:1. doi: 10.1186/1751-0147-49-S1-S14. [DOI] [Google Scholar]
- 22.Caruzo TAR, Brito WMEDD, Munford V, Rácz ML. Molecular characterization of G and P-types bovine rotavirus strains from Goiás, Brazil: high frequency of mixed P-type infections. Mem Inst Oswaldo Cruz. 2010;105:1040–1043. doi: 10.1590/S0074-02762010000800014. [DOI] [PubMed] [Google Scholar]
- 23.Merwad A, Abdallah F, Saber T. Close relationship of group A rotaviruses between bovine and human based onVP7 gene sequence in Egypt. Pak Vet J. 2014;34:391–393. [Google Scholar]
- 24.El Sherif M, Esona MD, Wang Y, Gentsch JR, Jiang B, Glass RI, Baker SA, Klena JD. Detection of the first G6P [14] human rotavirus strain from a child with diarrhea in Egypt. Infect Genet Evol. 2011;11:1436–1442. doi: 10.1016/j.meegid.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 25.Ferragut F, Vega CG, Mauroy A, Conceição-Neto N, Zeller M, Heylen E, Uriarte EL, Bilbao G, Bok M, Matthijnssens J, Thiry E. Molecular detection of bovine noroviruses in Argentinean dairy calves: circulation of a tentative new genotype. Infect Genet Evol. 2016;40:144–150. doi: 10.1016/j.meegid.2016.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oem JK, An DJ. Phylogenetic analysis of bovine astrovirus in Korean cattle. Virus Genes. 2014;48:372–375. doi: 10.1007/s11262-013-1013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagai M, Omatsu T, Aoki H, Otomaru K, Uto T, Koizumi M, Minami-Fukuda F, Takai H, Murakami T, Masuda T, Yamasato H. Full genome analysis of bovine astrovirus from fecal samples of cattle in Japan: identification of possible interspecies transmission of bovine astrovirus. Arch Virol. 2015;160:2491–2501. doi: 10.1007/s00705-015-2543-7. [DOI] [PubMed] [Google Scholar]
- 28.Candido M, Alencar AL, Almeida-Queiroz SR, Buzinaro Mda G, Munin FS, de Godoy SH, Livonesi MC, Fernandes AM, de Sousa RL. Molecular detection and phylogenetic analysis of bovine astrovirus in Brazil. Arch Virol. 2015;160:1519–1525. doi: 10.1007/s00705-015-2400-8. [DOI] [PubMed] [Google Scholar]
- 29.Matthijnssens J, Rahman M, Van Ranst M. Two out of the 11 genes of an unusual human G6P [6] rotavirus isolate are of bovine origin. J Gen Virol. 2008;89:2630–2635. doi: 10.1099/vir.0.2008/003780-0. [DOI] [PubMed] [Google Scholar]
- 30.Fukuda M, Kuga K, Miyazaki A, Suzuki T, Tasei K, Aita T, Mase M, Sugiyama M, Tsunemitsu H. Development and application of one-step multiplex reverse transcription PCR for simultaneous detection of five diarrheal viruses in adult cattle. Arch Virol. 2012;157:1063–1069. doi: 10.1007/s00705-012-1271-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gómara MI, Cubitt D, Desselberger U, Gray J. Amino acid substitution within the VP7 protein of G2 rotavirus strains associated with failure to serotype. J Clin Microbiol. 2001;39:3796–3798. doi: 10.1128/JCM.39.10.3796-3798.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, Das BK, Bhan MK. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992;30:1365–1373. doi: 10.1128/jcm.30.6.1365-1373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tse H, Chan WM, Tsoi HW, Fan RY, Lau CC, Lau SK, Woo PC, Yuen KY. Rediscovery and genomic characterization of bovine astroviruses. J Gen Virol. 2011;92:1888–1898. doi: 10.1099/vir.0.030817-0. [DOI] [PubMed] [Google Scholar]
- 34.Smiley JR, Hoet AE, Tråvén M, Tsunemitsu H, Saif LJ. Reverse transcription-PCR assays for detection of bovine enteric caliciviruses (BEC) and analysis of the genetic relationships among BEC and human caliciviruses. J Clin Microbiol. 2003;41:3089–3099. doi: 10.1128/JCM.41.7.3089-3099.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsunemitsu H, Smith DR, Saif LJ. Experimental inoculation of adult dairy cows with bovine coronavirus and detection of coronavirus in feces by RT-PCR. Arch Virol. 1999;144:167–175. doi: 10.1007/s007050050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park SJ, Jeong C, Yoon SS, Choy HE, Saif LJ, Park SH, Kim YJ, Jeong JH, Park SI, Kim HH, Lee BJ. Detection and characterization of bovine coronaviruses in fecal specimens of adult cattle with diarrhea during the warmer seasons. J Clin Microbiol. 2006;44:3178–3188. doi: 10.1128/JCM.02667-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Givens MD, Heath AM, Carson RL, Brock KV, Edens MSD, Wenzel JGW, Stringfellow DA. Analytical sensitivity of assays used for detection of bovine viral diarrhea virus in semen samples from the Southeastern United States. Vet Microbiol. 2003;96:145–155. doi: 10.1016/S0378-1135(03)00213-X. [DOI] [PubMed] [Google Scholar]
- 38.Dhama K, Chauhan RS, Mahendran M, Malik SVS. Rotavirus diarrhea in bovines and other domestic animals. Vet Res Commun. 2009;33:1–23. doi: 10.1007/s11259-008-9070-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Agarose gel electrophoresis of RT-PCR products (A) Detection of BRV-VP6 positive samples (product size 928 bp). (B) Detection of BRV-VP7 positive samples (product size 881 bp). (C) Detection of BNoV positive samples (product size 532 bp). (D) Detection of BAstV positive samples (product size 432 bp) (DOC 494 kb)


