Abstract
Eighteen canine distemper virus (CDV) isolates were obtained from clinical samples in Henan province, China, between 2012 and 2016. These viruses could not be recognized by 1A4, a monoclonal antibody specific for the H protein of CDV vaccine strains. The complete haemagglutinin (H) genes of all 18 isolates were sequenced, and phylogenetic analysis showed that they segregated into two clusters within the Asia-1 genotype. Moreover, the H genes of four viruses were found to lack a potential N-glycosylation site at position 309, which is the most conserved site within the Asia-1 genotype of CDV, and a novel potential N-glycosylation site (amino acids 517–519) was found in strain HL013, which has not been reported previously. These results will help in achieving a better understanding of the evolution of CDV in China.
Electronic supplementary material
The online version of this article (10.1007/s00705-019-04298-7) contains supplementary material, which is available to authorized users.
Canine distemper virus (CDV) belongs to the genus Morbillivirus in the family Paramyxoviridae and causes an acute-to-subacute highly contagious disease in domestic dogs that presents a variety of clinical signs [1]. Domesticated dogs are the main reservoir of CDV, which is a host to several pathogens, such as canine parvovirus (CPV), canine adenovirus (CAV), and canine parainfluenza virus (CPIV) [2]. The diseases associated with these pathogens have been controlled for many years using multiple live attenuated vaccines [1]. However, in many cases, vaccinated dogs, minks, foxes and raccoon dogs have recently been reported to show classical clinical signs of CDV infection [3, 4]. It remains unknown whether it is due to vaccination failure or the ability of new virulent CDV strains to escape vaccine-induced immunity. In addition, there have been reports of antigenic differences between vaccine strains and field strains [5, 6]. The haemagglutinin (H) protein, which is a structural protein of the virion, is a major target of the immune system and plays a key role as a virus ligand in mediating attachment to cellular receptors and entry into host cells. The H gene shows great genetic variability and is often used for investigating the relatedness of CDV isolates and for molecular epidemiological studies [3–7].
In a previous study, 10 field strains of CDV with amino acid changes at residues 542 and 549, which generated a novel N-glycosylation site in the H protein, were reported in vaccinated minks, foxes and raccoon dogs in China [4]. To expand our knowledge of CDV prevalence and to gain a better understanding of the molecular epidemiology of CDV in domestic dogs, an epidemiological study was conducted between 2012 and 2016 in Henan province, China.
Eighteen samples were collected from dying domestic pet dogs with suspected CDV infection. Collected samples included whole blood and nasal discharges from live dogs and brain, lung, liver, spleen and intestinal tissues were collected from euthanized dogs after necropsy. Tissues were suspended in cold phosphate-buffered saline (PBS) with antibiotics and were ground into homogenates. Homogenized samples were centrifuged at 3000 × g for 10 min at 4 °C. The supernatants were collected and centrifuged for an additional 10 min at 3000 × g. Peripheral blood mononuclear cells (PBMCs) from sick dogs were isolated from whole peripheral blood using dog lymphocyte separation medium (TBD, Tianjin, China) according to the manufacturer’s protocols. Canine distemper virus (CDV), canine coronavirus (CCV) and canine parainfluenza virus (CPIV) infection were identified by laboratory diagnosis using reverse transcription polymerase chain reaction (RT-PCR) and CDV/CCV/CPIV colloidal gold test strips (Bionote, Inc., South Korea). Canine adenovirus (CAV) and canine parvovirus (CPV) infections were diagnosed by PCR and CAV or CPV colloidal gold test strips (Bionote, Inc., South Korea). Pairs of primers for CDV, CCV, CPIV, CAV and CPV were designed based on reported sequences in the GenBank database and are listed in Supplementary Table 1 [8]. Eighteen clinical samples were confirmed to be positive for CDV by RT-PCR and sequencing. Of these, eight cases were single CDV infections, and 10 were CDV co-infections with other canine viruses, such as CAV-2, CPIV, CPV, or CCV (Supplementary Table 2).
A canine ??signaling lymphocyte activation molecule?? (SLAM) gene lacking a signal sequence was inserted into a pCAGGS (Neo) construct containing the immunoglobulin Igκ leader sequence (GAGACAGACACACTCCTGCTATGGGTACTGCTGCTCTGGGTTCCAGGTTCCACTGGTGAC) and the influenza virus hemagglutinin (HA) epitope (TATCCATATGATGTTCCAGATTATGCT) to generate pCAGDog-SLAM [9, 10], which was then introduced into Vero cells by transfection using 800 μg of G418 per ml. The resulting stable cloned cell lines from passages 8 and 40 were tested by immunofluorescence staining with anti-HA monoclonal antibody (Supplementary Fig. 1).
Vero/DogSLAM cells were inoculated with lung tissues (16 samples) or PBMCs (2 samples) from the CDV-positive samples. PBMCs were stimulated overnight in the presence of 6 μg of ??concanavalin A?? (ConA) [7]. The stimulated PBMCs were then co-cultivated with Vero/Dog SLAM cells until a cytopathic effect (CPE) was observed. Lung tissue homogenate supernatants were filtered through 0.45-μm filters (Millipore, Bedford, MA, USA) and then inoculated onto Vero/Dog SLAM cells. At 30 h post-inoculation, typical CPE formation of (large syncytia) was observed (Supplementary Fig. 2B).
In previous reports, the isolation rate in Vero/Dog SLAM cells inoculated with homogenates of spleen samples from CDV-infected dogs was 71% (5/7), and that in B95a cells was 43% [10]. By contrast, the isolation rate of CDV in Vero/DogSLAM cells inoculated with lung homogenate supernatants in this study was 100% (16/16). Therefore, in addition to expression of the SLAM receptor for CDV and the defect in IFN-β production in the parental Vero cells [10], selection of appropriate clinical specimens (such as lung samples) may contribute to a higher rate of isolation of CDV. Six of the lung samples (HL006,HL009,HNly150403,HL010,HL011,HL012) were found to be coinfected with CPIV or/and CAV-2 (Supplementary Table 2). Vero cells were inoculated with these samples, and the other two viruses were isolated using at least 10 blind passages. Only one strain of CPIV (HeN0718) was successfully isolated after eight blind passages before obvious CPE appeared [11]. Thus, for all of the lung samples tested on Vero/Dog SLAM cells, the formation of large syncytia was caused by CDV rather than the other viruses.
After the third passage, the presence of CDV was tested by indirect immunofluorescence assay (IFA). CDV-infected Vero/Dog SLAM cells were fixed with acetone for 30 min at room temperature, followed by incubation in a 1:1000 dilution of monoclonal mouse anti-CDV-F antibody (1G5) at 37 °C [12]. After washing with PBS, cells were incubated with a 1:500 dilution of fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG (Sigma) (Biomedical Technologies Inc., Madrid, Spain) for 1 h at 37 °C. After washing with PBS, fluorescence was observed using a fluorescence microscope (Olympus). Strong and specific positive staining was observed by IFA using an anti-CDV-F protein monoclonal antibody (Supplementary Fig. 2D).
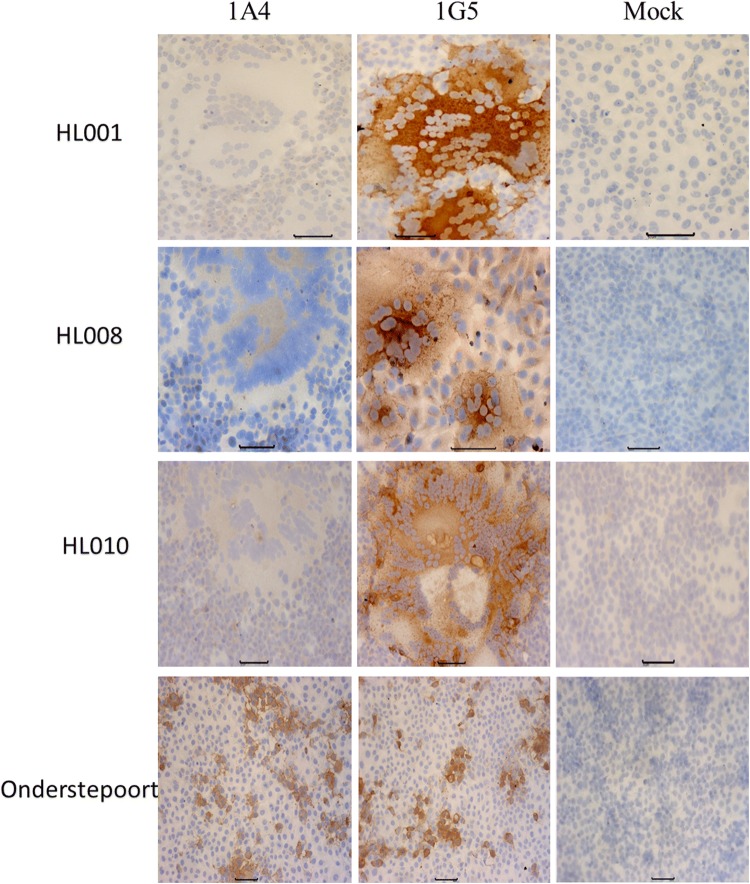
Nine of the 18 dogs in this study had received one or more vaccinations with the Onderstepoort or Snyder Hill strain CDV vaccines (Supplementary Table 2). The isolation of new virulent CDV strains from these vaccinated dogs indicates that the commercial vaccines did not confer full protection. Recently, a research group in Japan reported that eight CDV field strains could not be completely neutralized by anti-CDV (vaccine strain, Onderstepoort) canine plasma [3]. To investigate if the current Chinese CDV field strains could escape vaccine-induced immunity, two well-defined CDV monoclonal antibodies (mAb) were used. The mAb 1A4 recognizes the H proteins of vaccine strains such as Onderstepoort and Snyder Hill, but not those of field CDV strains, and 1G5 recognizes the F proteins of both vaccine and field strains of CDV [12]. As shown in Fig.1, none of the field strains of CDV isolated in this study were recognized by mAb 1A4, as demonstrated by immunoperoxidase monolayer assay (IPMA). By contrast, the strains HL001, HL008, HL010 and Onderstepoort were detected by 1G5 mAb, as indicated by brown staining in the IPMA (Fig. 1). The other 15 Chinese field strains isolated in this study also reacted with 1G5 (not shown). These results are consistent with those of a previous study [3]. ??Mori et al.?? showed that eight field isolates of CDV from dogs in Japan were also not recognized and neutralized by an anti-CDV-H antibody (d-7) that showed neutralizing activity against a vaccine strain and laboratory-adapted strains [3]. These results indicated that the new circulating CDV strains have lost or do not expose some epitopes that are present or exposed in the vaccine strains and thus escape binding by MAb 1A4 or d-7.
Fig. 1.
Reactivity of MAbs (1A4 and 1G5) with three representative Chinese field strains isolated in this study and the CDV vaccine strain Onderstepoort. None of the other 15 Chinese field strains isolated in this study reacted with 1A4, but they reacted with 1G5 (not shown)
The complete H gene of each CDV isolate from this study was amplified by PCR using 2× TransStart FastPfu PCR SuperMix (TransGen Biotech, Beijing, China), and the entire open reading frame (ORF) was amplified using a single pair of primers (CDV-HF/HR). After purification, the amplified segments were cloned into pEASY-Blunt vector (TransGen Biotech, Beijing, China) and sequenced (GENEWIZ, Beijing, China). The nucleotide sequences were analyzed using DNAStar and were deposited in the GenBank database (Supplementary Table2). The maximum-likelihood method was used to construct phylogenetic tree from aligned amino acid sequences in MEGA 6.0 software. The sequences of the 18 CDV isolates were 97.28% to 100% identical. Comparison with previously reported strains [4, 6, 13, 14] showed that the H gene sequences of these field strains were 90.43-93.76%, 85.2-86.7% and 88.8-91.4% identical to those of Asia-2 genotype (AB040767/HM-3Dog/2002/JP, EU252148/SeoulDog/ 2007/KOR), Asia-3 genotype (EU7439 23/HLJ1Dog/ 2004/China, EU743935 /HLJ1Fox/2005/ China) and vaccine (Snyder Hill, Onderstepoort, Lederle, Convac) strains, respectively. All 18 strains segregated into the Asia-1 genotype and grouped into two clusters along with other Chinese field strains of CDV (Fig. 2). One cluster was composed of 10 field strains (strains HL002, HL004-009, HL012-014) with Chinese isolates from domestic dogs from different areas, and the other cluster was composed of the remaining eight strains (HL001, HL010, HL011, HL015, HL016, HNly150326, HNly150403, and HNly150520B) with Chinese strains from domestic dogs, minks, foxes and raccoon dogs (Fig. 2). The two clusters showed 95.74%-98.40% identity at the amino acid level. In previous reports, 42 Chinese field strains of CDV isolated from breeding foxes, minks and raccoon dogs in different areas of China during 2004-2008 and 2012-2013 grouped together in one cluster within the Asia-1 genotype [4, 14]. Our results and those in the previous report demonstrate the constant evolution of CDV field strains in China.
Fig. 2.
Phylogenetic relationship of the CDV isolates from the study to other reported viruses based on the full-length H gene. The phylogenetic tree was constructed using the maximum-likelihood method in MEGA6
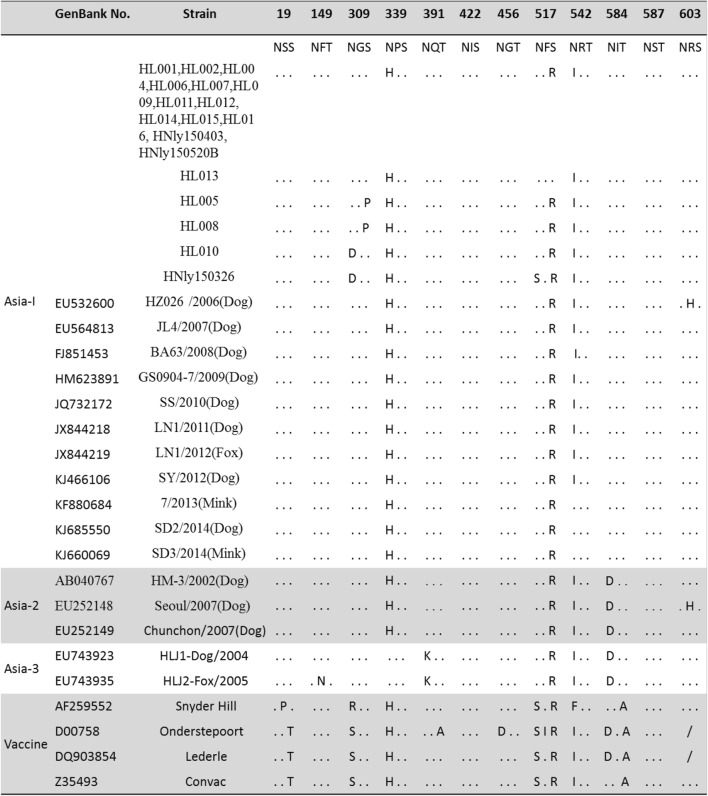
Eight to 10 potential N-glycosylation sites (19-21, 149-151, 309-311, 339-341, 391-393, 422-424, 456-458, 542-544, 584-586, 587-589, 603-605) are conserved in the H protein of Asia-1 strains [4, 6, 13, 14]. However, four of these sites are present in Onderstepoort vaccine strain, and seven are present in Snyder Hill and Convac vaccine strains [13, 14]. The H protein sequences of the 18 CDV field strains described in this study were aligned with those of other Asia-1 strains (Table 1). The predicted H protein of 14 strains in this study contained nine potential N-glycosylation sites at amino acid positions 19-21, 149-151, 309-311, 391-393, 422-424, 456-458, 584-586, 587-589 and 603-605 (Table 1) as described previously for Chinese field isolates within the Asia-1 group [4, 6, 13, 14], and a novel potential N-glycosylation site (amino acids 517-519) was found in strain HL013, which has not been reported previously (Table 1). The number of N-glycosylation sites in the H protein has been associated with the virulence of CDV [15]. The presence of a potential N-glycosylation site (amino acids 517-519) in strain HL003 might affect the virulence. Furthermore, it has been suggested that extensive masking of antigenic epitopes by sugar moieties could prevent neutralizing antibodies from binding the H protein [16]. By contrast, a potential N-glycosylation site (amino acids 309-311) that is unique to field strains [13] was lacking in the H protein of four CDV field strains (HL005, HL008, HL010 and HNly150326) (Table 1). The N-glycosylation site at position 309-311 was absent from the vaccine strains and the America-1 field strains [17], but its absence has not been reported previously in Asia-1 genotype isolates from dogs. In our study, the four strains that lacked this N-glycosylation site were isolated from vaccinated puppies, three of which died of CDV infection, suggesting that these strains were highly pathogenic. This finding is different from previous reports that the generation of a novel N-glycosylation site in the H protein of CDV strains was associated with CDV vaccine failures [4]. The effects of different N-glycosylation sites in the H protein of CDV need further exploration.
Table 1.
Potential N-linked glycosylation sites and their ??amino acid positions in the H protein?? of Chinese field strains and vaccine strains of canine distemper virus (CDV). Dots (.) indicate identical amino acids
It has been suggested that changes at amino acid positions 519, 530 and 549 of the H gene are associated with host specificity [18–20]. The host range of CDV strains and the outcome of infection depend on virus traits other than those affecting virus binding to SLAM receptors. The strain type 519R/549Y of the CDV H gene is typical of dog strains worldwide, and highest performance has been observed in cells expressing dog SLAM receptors [20]. With the exception of HL013, the isolates from this study encoded 519 R/549 Y. CDV strains with histidine at position 549 have been shown to be highly virulent for raccoons [17]. The HL013 strain of CDV contains a specific substitution of tyrosine to histidine at 549. Prior to 2015 in China, only the Liaoning and Shandong CDV strains isolated from minks and foxes possessed the Y549H mutation [4]. The H sequence of HL013 is 95.71-97.40% identical to those of these non-canine strains.
In summary, 18 CDVs were successfully isolated in this study. These viruses clustered into two different subgenotypes within the Asia-1 genotype, and these new isolates had different antigenic properties from the vaccine strains used in China. The conserved N-glycosylation sites are also undergoing changes in these new virus isolates, indicating that CDV is constantly evolving in domestic dogs in China.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the Ten-Thousand Talents Program (Dr. Xiangdong Li) and the Luoyang Heluo Talent Plan (Dr. Kegong Tian).
Compliance with ethical standards
Ethical approval
All animal trials were approved by the Animal Care and Ethics Committee of the China National Research Center for Veterinary Medicine.
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Yuxiu Liu and Caihong Liu contributed equally to this study.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiangdong Li, Phone: + (86) 10-59198895, Email: xiaonanzhong@163.com.
Kegong Tian, Phone: + (86) 10-59198895, Email: tiankg@263.net.
References
- 1.Appel MJ. Virus infections of carnivores. Virus Infect Verteb I. 1987;1987:p516. [Google Scholar]
- 2.Damián M, Morales E, Salas G, Trigo FJ. Immunohistochemical detection of antigens of distemper, adenovirus and parainfluenza viruses in domestic dogs with pneumonia. J Comp Pathol. 2005;133:289–293. doi: 10.1016/j.jcpa.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Mori T, Shin YS, Okita M, Hirayama N, Miyashita N, Gemma T, Kai C, Mikami T. The biological characterization of field isolates of canine distemper virus from Japan. J Gen Virol. 1994;75(Pt 9):2403. doi: 10.1099/0022-1317-75-9-2403. [DOI] [PubMed] [Google Scholar]
- 4.Zhao J, Zhang H, Bai X, Martella V, Hu B, Sun Y, Zhu C, Zhang L, Liu H, Xu S. Emergence of canine distemper virus strains with two amino acid substitutions in the haemagglutinin protein, detected from vaccinated carnivores in North-Eastern China in 2012–2013. Vet J. 2014;200:191–194. doi: 10.1016/j.tvjl.2014.01.028. [DOI] [PubMed] [Google Scholar]
- 5.Hashimoto M, Une Y, Mochizuki M. Hemagglutinin genotype profiles of canine distemper virus from domestic dogs in Japan. Arch Virol. 2001;146:149–155. doi: 10.1007/s007050170198. [DOI] [PubMed] [Google Scholar]
- 6.Wei S, Zhou S, Zhao W, Cui SJ. A multiplex reverse transcription-nested polymerase chain reaction for detection and differentiation of wild-type and vaccine strains of canine distemper virus. Virol J. 2010;7:86. doi: 10.1186/1743-422X-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harder TC, Kenter M, Vos H, Siebelink K, Huisman W, Van AG, Orvell C, Barrett T, Appel MJ, Osterhaus AD. Canine distemper virus from diseased large felids: biological properties and phylogenetic relationships. J Gen Virol. 1996;77:397–400. doi: 10.1099/0022-1317-77-3-397. [DOI] [PubMed] [Google Scholar]
- 8.Hu RL, Huang G, Qiu W, Zhong ZH, Xia XZ, Yin Z. Detection and differentiation of CAV-1 and CAV-2 by polymerase chain reaction. Vet Res Commun. 2001;25:77–84. doi: 10.1023/A:1006417203856. [DOI] [PubMed] [Google Scholar]
- 9.Nakano H, Kameo Y, Andoh K, Ohno Y, Mochizuki M, Maeda K. Establishment of canine and feline cells expressing canine signaling lymphocyte activation molecule for canine distemper virus study. Vet Microbiol. 2009;133:179–183. doi: 10.1016/j.vetmic.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seki F, Ono N, Yamaguchi R, Yanagi Y. Efficient isolation of wild strains of canine distemper virus in vero cells expressing canine SLAM (CD150) and Their adaptability to marmoset B95a cells. J Virol. 2003;77:9943. doi: 10.1128/JVI.77.18.9943-9950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu C, Li X, Zhang J, Yang L, Fan L, Deng J, Tan F, Ming S, Liu Y, Tian K. Isolation and genomic characterization of a canine parainfluenza virus type 5 strain in China. Arch Virol. 2017;162:1–8. doi: 10.1007/s00705-016-3065-7. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Hao L, Li X, Wang L, Zhang J, Deng J, Tian K. Development and characterization of canine distemper virus monoclonal antibodies. Monoclon Antib Immunodiagn Immunother. 2017;36:119. doi: 10.1089/mab.2017.0012. [DOI] [PubMed] [Google Scholar]
- 13.Iwatsuki K, Miyashita N, Yoshida E, Gemma T, Shin YS, Mori T, Hirayama N, Kai C, Mikami T. Molecular and phylogenetic analyses of the haemagglutinin (H) proteins of field isolates of canine distemper virus from naturally infected dogs. J Gen Virol. 1997;78:373–380. doi: 10.1099/0022-1317-78-2-373. [DOI] [PubMed] [Google Scholar]
- 14.Zhao JJ, Yan XJ, Chai XL, Martella V, Luo GL, Zhang HL, Han G, Liu YX, Xue B, Lei Z. Phylogenetic analysis of the haemagglutinin gene of canine distemper virus strains detected from breeding foxes, raccoon dogs and minks in China. Vet Microbiol. 2010;140:34–42. doi: 10.1016/j.vetmic.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 15.Sawatsky B, Von MV. Canine distemper viruses expressing a hemagglutinin without N-glycans lose virulence but retain immunosuppression. J Virol. 2010;84:2753–2761. doi: 10.1128/JVI.01813-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashiguchi T, Kajikawa M, Maita N, Takeda M, Kuroki K, Sasaki K, Kohda D, Yanagi Y, Maenaka K. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. PNAS. 2007;104:19535–19540. doi: 10.1073/pnas.0707830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lednicky JA, Dubach J, Kinsel MJ, Meehan TP, Bocchetta M, Hungerford LL, Sarich NA, Witecki KE, Braid MD, Pedrak C. Genetically distant American Canine distemper virus lineages have recently caused epizootics with somewhat different characteristics in raccoons living around a large suburban zoo in the USA. Virol J. 2004;1:2. doi: 10.1186/1743-422X-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ke GM, Ho CH, Chiang MJ, Sannoduanda B, Chung CS, Lin MY, Shi YY, Yang MH, Tyan YC, Liao PC. Phylodynamic analysis of the canine distemper virus hemagglutinin gene. BMC Vet Res. 2015;11:164. doi: 10.1186/s12917-015-0491-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mccarthy AJ, Shaw MA, Goodman SJ. Pathogen evolution and disease emergence in carnivores. Proc R Soc Lond B Biol. 2007;274:3165. doi: 10.1098/rspb.2007.0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikolin VM, Olarte-Castillo XA, Osterrieder N, Hofer H, Dubovi E, Mazzoni CJ, Brunner E, Goller KV, Fyumagwa RD, Moehlman PD. Canine distemper virus in the Serengeti ecosystem: molecular adaptation to different carnivore species. Mol Ecol. 2016;26:7. doi: 10.1111/mec.13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.