Abstract
Since 2010, continual outbreaks of highly virulent variants of porcine epidemic diarrhea virus (PEDV) belonging to genotype GII have led to serious economic losses for the Chinese swine industry. To better understand the biological characteristics and pathogenicity of the current prevalent Chinese PEDV field strains, in this study, a highly virulent Chinese genotype GIIa PEDV strain, CH/HBXT/2018, was isolated and serially propagated using Vero cells. Sequencing and phylogenetic analysis showed that strain CH/HBXT/2018 contained novel insertion and deletion mutations in the S gene region relative to the classical strain and belonged to the genotype GIIa, similar to other recently isolated PEDV strains from China and the United States. Pig infection studies indicated that the CH/HBXT/2018 strain was highly virulent in suckling piglets, and the median pig diarrhea dose (PDD50) was 8.63 log10PDD50/3 mL at 7 days postinfection (DPI). The results of the present study are important for future PEDV challenge studies and the development of new PEDV vaccines based on prevalent field strains for the prevention and control of PED in China.
Introduction
Porcine epidemic diarrhea (PED) is caused by porcine epidemic diarrhea virus (PEDV) and is a highly contagious disease characterized by watery diarrhea, dehydration, vomiting, enteritis and weight loss. PEDV infects pigs of all ages, and the mortality rate in neonatal piglets is as high as 80-100% [1, 2]. PED was first reported in the United Kingdom in 1971, and the causative agent CV777 strain was isolated in 1978 [3]. Through the end of the 1990s, PEDV outbreaks occurred in many European countries [4, 5]. In October 2010, a widespread outbreak of PED caused by highly virulent PEDV variants that differed from the classical European strain CV777 occurred in China and resulted in high mortality and huge economic losses. Since then, PED outbreaks have increased significantly and spread rapidly nationwide [6, 7]. PEDV variants emerged in the United States starting in May 2013 and then spread rapidly throughout the nation. Many American, Asian and European countries, including Canada, Mexico, Colombia, Japan, South Korea, the Philippines, Thailand, Germany, the Netherlands and Switzerland have also reported outbreaks of PED [6, 8–13].
PEDV is an enveloped, single-stranded, positive-sense RNA virus that belongs to the order Nidovirales, family Coronaviridae and genus Alphacoronavirus. The PEDV genome is approximately 28 kb in length and is composed of a 5´ untranslated region (UTR), seven open reading frames (ORFs), and a 3´UTR [14]. The seven ORFs encode three non-structural proteins, namely ORF1a, ORF1b and ORF3, and four structural proteins, designated receptor-binding spike glycoprotein (S), envelope protein (E), membrane glycoprotein (M) and nucleocapsid protein (N) [15, 16]. ORF1a and ORF1b play vital roles in viral replication and transcription and may be related to cell adaptability and virulence [17]. The spike protein is a type I membrane glycoprotein consisting of S1 and S2 domains, which play important roles in receptor binding, cell membrane fusion, induction of host immune responses and virus neutralization by antibodies [18]. The E protein is important for viral budding, and loss of the E protein reduces viral virulence. The M protein can induce neutralizing antibodies and is the most abundant constituent of viral particles [19]. PEDV N binds to RNA, and capped N gene transcripts can improve replication rates during virus rescue [20].
Recently, many studies have demonstrated that commercially available PEDV vaccines derived from classical strains of PEDV cannot provide effective protection against highly virulent PEDV variant infections in China, resulting in high morbidity and mortality in neonatal piglets [21, 22]. Furthermore, almost all of the currently prevalent PEDV variants in China belong to genotype GII, particularly genotype GIIa [23]. Therefore, the development of new vaccines based on the currently prevalent PEDV variants is urgent. To achieve this objective, a current, prevalent, and highly virulent Chinese genotype GIIa PEDV strain, CH/HBXT/2018 (GenBank accession number: MH816969), was isolated, and the biological characteristics and pathogenicity of this virus strain were investigated in this study. Furthermore, the infectious titer (PDD50) of CH/HBXT/2018 was determined. These results provide very helpful information for future pig challenge studies of PEDV.
Materials and methods
Cells and clinical samples
Vero cells (ATCC, CCL-81) were grown in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, USA) containing 10% heat-inactivated fetal bovine serum (FBS; Invitrogen, Australia) and 1% antibiotics and antimycotics (10,000 units of penicillin, 10,000 µg of streptomycin, and 25 µg of Fungizone® per mL) (Gibco™, USA) and cultured at 37 °C with 5% CO2. Fecal samples were collected from diarrheal piglets that displayed acute watery diarrhea, dehydration, and extreme emaciation in Hebei Province, China, in March 2018. These samples were confirmed to be PEDV positive by reverse transcription PCR (RT-PCR) and real-time PCR (developed in our lab). The intestinal contents were diluted tenfold in virus growth medium, which consisted of DMEM supplemented with antibiotics (100 units of penicillin and 100 mg of streptomycin [Gibco™] per mL), 0.3% TPB (Sigma), and 20 μg of trypsin (Gibco™, USA) per mL, and then vortexed and centrifuged at 3,500 rpm for 30 min at 4 °C. The supernatant was filtered through a 0.22-μm-pore-size filter (Merck Millipore, Germany) and stored at -70 °C for use as virus stock.
Virus isolation and propagation
Vero cells were seeded in T-25 culture flasks and placed in a 37°C incubator with 5% CO2 until a 100% confluent Vero cell monolayer formed. The culture medium was then removed, and the cells were washed three times with sterile phosphate-buffered saline (PBS; pH 7.2, Gibco™, USA). Subsequently, 1 mL of prepared virus stock was inoculated into flasks with 20 μg of trypsin (Gibco™, USA) per mL. After incubation for 1 h at 37 °C with 5% CO2, virus growth medium was added and cell culture was continued at 37 °C and 5% CO2, with daily observation for a cytopathic effect (CPE). When more than 90% of cells showed CPE, the supernatant and cells were harvested, frozen and thawed three times and stored at -80 °C for the next virus passage.
Tissue culture infectious dose (TCID50) assay
A 50% (TCID50) assay was used to determine the virus titer. Vero cells were seeded into 96-well plates until the wells were covered with monolayers, and tenfold serial dilutions of virus were then inoculated onto the cell monolayers with 20 μg of trypsin per mL and incubated for 3-5 days at 37°C with 5% CO2. Virus-induced CPE was observed and expressed as TCID50/mL according to the Reed and Muench method.
Viral replication kinetics
Virus with a multiplicity of infection (MOI) of 0.01 was inoculated onto Vero cell monolayers in T-25 culture flasks with 20 μg of trypsin per mL. Viral cultures were harvested at 0, 12, 24, 36, 48, 60 and 72 hours postinfection (hpi). The number of viral genome copies was determined by real-time PCR (developed in our lab).
Electron microscopy
PEDV-infected Vero cells were harvested when more than 90% of the culture showed CPE. After being frozen and thawed three times, the supernatant and cells were collected and centrifuged at 10,000 rpm for 1 h and then filtered with a 0.22-μm-pore-size filter (Merck Millipore, Germany) to remove the cell debris. Subsequently, the supernatant was mixed with polyethylene glycol 8000 (PEG-8000; Solarbio, China) at a 10% final concentration overnight. After centrifugation at 12,000 rpm and 4 °C for 2 h, viral particles were resuspended in Tris-buffered saline solution (TBS) and then negatively stained with 2% phosphotungstic acid and examined using a transmission electron microscope (JEOL, JEM-1200EX, Japan). Virions on infected Vero cell surfaces were imaged according to methods described in a previous study [24].
Immunofluorescence analysis
Vero cells in 6-well plates were inoculated with CH/HBXT/2018 at passage 10 (P10) at an MOI of 0.01 using virus growth medium as a negative control. At 6, 12, 24, and 36 h after inoculation, the medium was removed and cells were washed three times with PBS. The cells were fixed with 4% paraformaldehyde for 30 min at 4°C, and then 1 mL of penetrant (0.25% Triton X-100; Solarbio, China), was added for 10 min at room temperature. After washing three times with PBS, the cells were blocked with 5% bovine serum albumin (BSA; Solarbio, China) for 1 h. The plates were reacted with a 1:2,000 dilution of mouse anti-PEDV-N protein MAbs #12 (prepared and stored in our laboratory) followed by a dilution of Alexa Fluor® 488-conjugated goat anti-mouse IgG (Abcam, UK) at 37°C for 1 h. Finally, cells were stained with 4’,6-diamidino-2-phenylindole (DAPI; Vectorlabs, USA) for 5 min and washed with PBS. The samples were examined under a fluorescence microscope (Olympus, Japan).
Sequence analysis and phylogenetic tree construction
Viral RNA was extracted from intestinal contents using an RNeasy Mini Kit (QIAGEN, USA). Reverse transcription and amplification of the PEDV S gene were carried out using AMV reverse transcriptase (Promega, USA) and PrimeSTAR® GXL DNA polymerase (TaKaRa, Dalian, China), respectively, with the primers 5´-GCAACACTATGCATGCCAAT-3´ and 5´-TGTTGCACACTTATTGGCAGG-3´. PCR products were isolated by agarose gel electrophoresis and then purified using a DNA Gel Extraction Kit. The target fragment was cloned into the vector included in the TOPO™ XL-2 Complete PCR Cloning Kit (Invitrogen, USA) and the identity of the plasmids was confirmed by restriction enzyme digestion and sequencing. The sequence was assembled and analyzed using DNAStar 7.0 and BioEdit software, respectively. Forty-eight PEDV reference strains that contained five different PEDV genotypes (Ia, Ib, IIa, IIb and S-INDEL) were obtained from the GenBank database for phylogenetic analysis. A phylogenetic tree was constructed with MEGA6.0 software using the neighbor-joining method with bootstrap values calculated for each node from 1,000 replicates.
Pig infection experiments
A total of 25 4-day-old conventional suckling piglets were used to determine the virulence and infectious titer (the median pig diarrhea dose, PDD50) of the CH/HBXT/2018 strain according to a method used in our previous study [25]. In detail, all piglets were randomly divided into four experimental groups (G1-G4) and one mock-infected group (G5) (Table 1). Each piglet was housed in an individual steel cage, and each group of pigs was housed in a different room. Before inoculation, we collected fecal samples from all piglets and tested the umbilical cord blood of sows, using a commercial enzyme-linked immunosorbent assay (ELISA) kit (Biovet, Canada) to ensure that none of the piglets were infected with PEDV. The piglets in groups 1 to 4 were each inoculated orally with 3 mL of tenfold serially diluted (from 10−6 to 10−9) P4 CH/HBXT/2018 (4.33 log10 TCID50/mL), and the mock-infected group was inoculated with 3 mL of PBS. After inoculation, clinical symptoms were observed and scored daily for fecal consistency (scores: 0 = normal, 1 = pasty, 2 = semiliquid, and 3 = liquid). At the same time, rectal swabs were collected daily to test fecal viral RNA shedding by real-time PCR. Intestinal samples were collected for hematoxylin and eosin (HE) staining and immunohistochemistry (IHC) examination. The PDD50 was determined as the reciprocal of the virus dilution at which 50% of pigs developed watery diarrhea at a given time point, using the Reed and Muench method.
Table 1.
Summary of pig groups, inoculum and diarrhea outcomes after infection with PEDV in suckling pigs
| Pig group | Pig numbers | Inoculuma | Calculated inoculum infectious titers (log10 TCID50/mL)c | Tested inoculum RNA titers (CT value)d | Diarrhea (percent)e | Fecal virus RNA shedding (CT value) |
|---|---|---|---|---|---|---|
| G1 | 5 | 10-6 diluted P4b | − 2.33 | 31.66 | 5/5 (100) | 17.49–26.65 |
| G2 | 5 | 10-7 diluted P4 | − 3.33 | 31.92 | 5/5 (100) | 19.02–25.00 |
| G3 | 5 | 10-8 diluted P4 | − 4.33 | 31.36 | 5/5 (100) | 22.27–28.27 |
| G4 | 5 | 10-9 diluted P4 | − 5.33 | 31.55 | 1/5 (20) | 21.67–35.89 |
| Mock | 5 | PBS | – | – | 0/5 (0) | –f |
aAll piglets were orally inoculated with 3 mL of inoculum
bG6-G9 were infected with P4 cell culture-adapted CH/HBXT/2018
cTiters were calculated based on the titer of the original P4 virus (104.33 TCID50/mL) and dilution times
dCT value: the mean cycle threshold value. A critical point was set at 35; CT values greater than 35 were considered negative
eFecal scores of 3 as determined at 7 DPI for pigs in G6-G9 and the control group
fNegative or below the detection limit of real-time PCR
Histopathology and IHC
At necropsy, small-intestine tissue specimens were examined and collected from each piglet. After 48 h of fixation in 4% paraformaldehyde solution at room temperature, tissue specimens were processed and embedded in paraffin. The paraffin-embedded tissues were cut into thick sections on a microtome (Leica, Germany), deparaffinized with xylene, and washed in decreasing concentrations of ethanol. Then, the intestinal tissue specimens were routinely stained with HE (Baso, China) for histopathology or subjected to IHC using PEDV-N-specific monoclonal antibodies (MAbs) (prepared in our laboratory).
Results
Biological characterization of the CH/HBXT/2018 strain
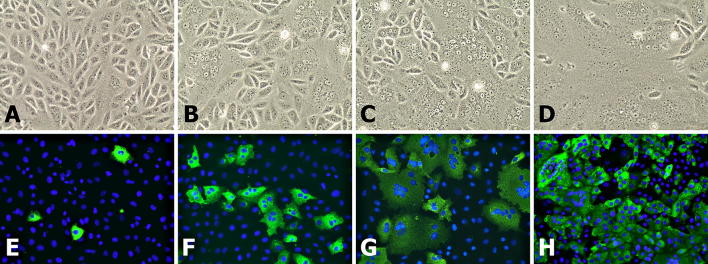
Typical PEDV CPE, including cell rounding, syncytial, vacuoles and, eventually, detachment was observed in cells infected with the P3 CH/HBXT/2018 strain. Typical CPE caused by P5 is shown in comparison to uninoculated cells in Fig. 1. CPE was clearly observed at 6 hpi (Fig. 1B), and more than 95% CPE was observed at 36 hpi (Fig. 1D). Immunofluorescence assay results indicated that the CH/HBXT/2018 strain could be detected with PEDV-N-specific MAbs. A small amount of fluorescent signal was detected at 6 hpi for P10 virus at an MOI of 0.01 (Fig. 1E), and the signals gradually increased from 12 to 36 h (Fig. 1F, G, and H).
Fig. 1.
Isolation and detection of the PEDV CH/HBXT/2018 strain in Vero cells. The upper and lower panels show light and immunofluorescence images, respectively, of Vero cells infected with the PEDV CH/HBXT/2018 strain. (A) Vero cells were seeded in T-25 culture flasks and infected with P5 virus at an MOI of 0.01. CPE was observed at 6 hpi under a microscope (400×). (B-D) CPE was observed at 12, 24 and 36 hpi, respectively. (E) Immunofluorescence assay results for the PEDV CH/HBXT/2018 strain at P10 in infected Vero cells at 6 hpi (400×). PEDV antigens and nuclei were detected with mouse anti-PEDV N protein MAbs and DAPI, respectively; (F-H) green fluorescent signals were observed at 12, 24 and 36 hpi, respectively
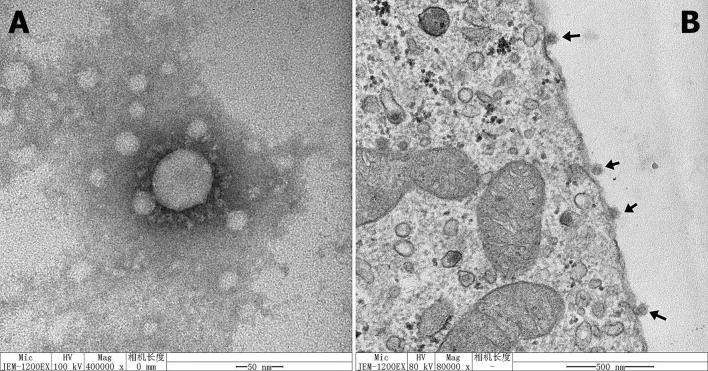
The viral replication kinetics of the CH/HBXT/2018 strain in Vero cells were examined, and the results showed that the number of viral genome copies continued to increase after inoculation (Fig. 2). The viral titers of the serially passaged PEDV CH/HBXT/2018 strain at P1 and P10 were 4.33 log10 TCID50/mL and 5.33 log10 TCID50/mL, respectively. Virions of the CH/HBXT/2018 strain were imaged by electron microscopy (EM), and the results showed that they were typical coronavirus particles approximately 80-120 nm in diameter with a crown shape (Fig. 3A). These particles were also observed on surface sections of PEDV-infected Vero cells (Fig. 3B).
Fig. 2.
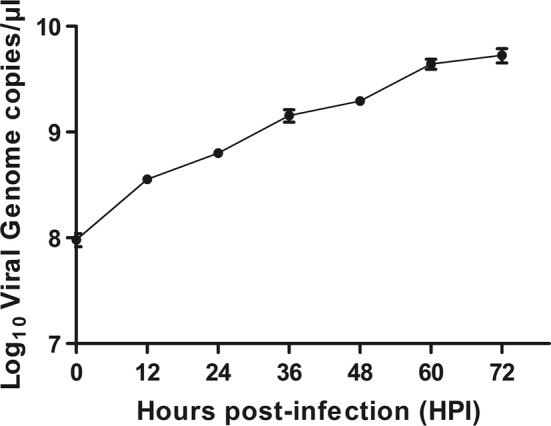
Viral replication kinetics of the CH/HBXT/2018 strain in Vero cells. Vero cells in T25 flasks were infected with each PEDV isolate at an MOI of 0.01. After incubation at 37 °C for various time periods, cells and supernatants were harvested, and the number of viral genome copies was determined by real-time PCR
Fig. 3.
Virions of the CH/HBXT/2018 strain in cell culture medium from infected Vero cells or on the surface of infected Vero cells were observed by EM. (A) Images of PEDV virions in cell culture medium from Vero cells infected with the PEDV CH/HBXT/2018 strain, as indicated by the arrow. Scale bar = 50 nm. (B) Images of a PEDV-infected Vero cell. PEDV particles (arrowheads) on the surface of an infected Vero cell, as shown by the arrow. Scale bar = 500 nm
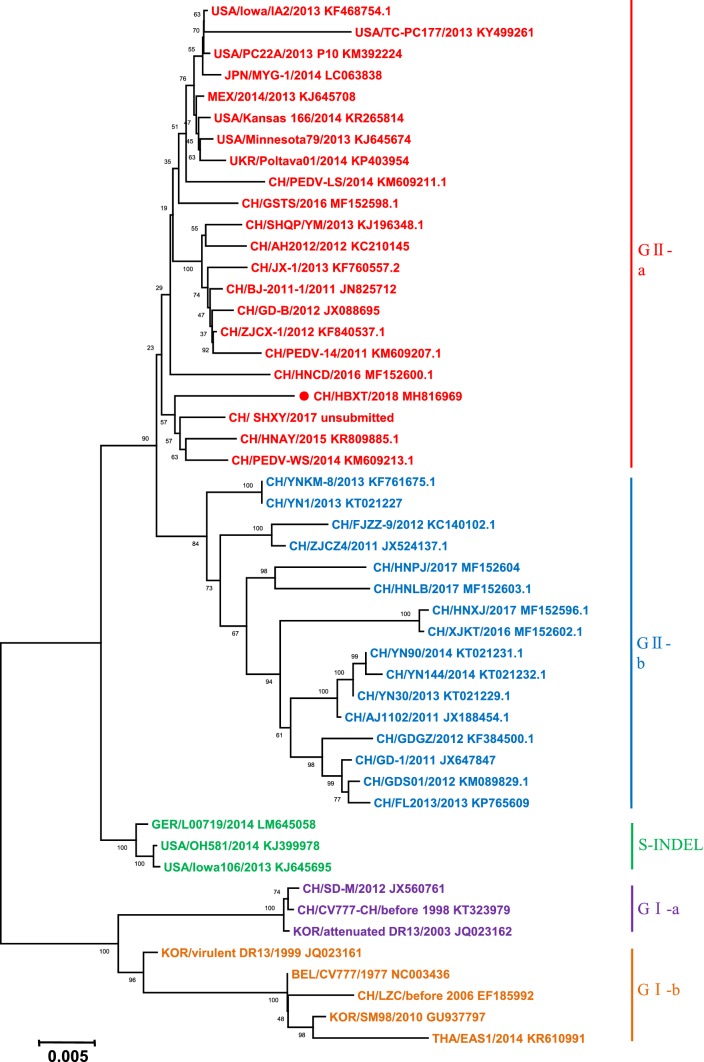
Sequence analysis of the S protein showed that compared with classical or attenuated PEDVs, such as the CV777 strain, the CH/HBXT/2018 strain contained novel insertions and deletions. A 4-aa insertion (QGVN) and a 1-aa insertion (N) were found at positions 58-59 and 139-140, respectively, and two deletions were detected at positions 163-164 (a 2-aa deletion of DI) and 1199 (a 1-aa deletion of N). Phylogenetic analysis based on the S gene indicated that the CH/HBXT/2018 strain belonged to genogroup GIIa (Fig. 4), along with other recently isolated PEDV strains from China and the United States.
Fig. 4.
Phylogenetic analysis based on the S gene of the CH/HBXT/2018 strain. The tree was constructed by the neighbor-joining method, and bootstrap values from 1,000 resamplings are shown for each node
Determination of the infectious titer and virulence of the CH/HBXT/2018 strain
To determine the infectious titer and virulence of the CH/HBXT/2018 strain, the PDD50 was determined using 4-day-old conventional suckling pigs. Before inoculation, all piglets were lively, showed no clinical symptoms, and had no PEDV genetic material detected in their fecal samples by PEDV-specific real-time PCR. During the experiment, none of piglets in the negative control group developed diarrhea, and all of their rectal swab samples were negative for PEDV RNA by real-time PCR. After infection, clinical symptoms, including watery diarrhea, vomiting, weight loss and depression, were observed in G6 to G9, while clinical signs and fecal shedding of PEDV RNA were negative in the control group. By 7 days post-inoculation (dpi), viral RNA fecal shedding in samples from rectal swabs or intestinal contents became positive in G1-G4 piglets (10−6-10−9 diluted virus), with cycle threshold (CT) values ranging from 17.49 to 32.98 (Table 1). In detail, 100% (5/5) of pigs in G1, G2 and G3 and 20% (1/5) of pigs in G4 had diarrhea, and no pigs (0/5) in the mock-infected group had diarrhea (Table 1). The cutoff time point was set as 7 dpi for determination of the PDD50, which was 8.63 log10 PDD50/3 mL. This value was very different from the TCID50 titer (4.33 log10 TCID50/mL).
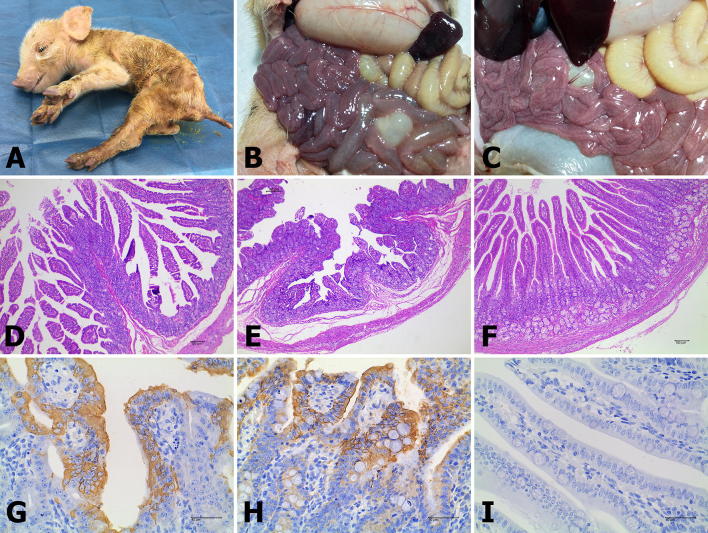
All of the diarrheal piglets were anorexic and depressed and displayed weight loss (Fig. 5A). At necropsy, typical macroscopic PED-like lesions were found in the small intestines; the intestinal tract was distended, transparent and filled with yellow fluid, and mesenteric congestion was present (Fig. 5B and C). The results of histopathological examination showed viral enteritis in all diarrheal piglets, including shortening, fusion and sloughing of the small intestinal villi (Fig. 5D and E). The piglets in the control group exhibited normal intestinal histopathology (Fig. 5F). Furthermore, immunohistochemical examination revealed that PEDV antigens were predominant in the cytoplasm of the epithelial cells of atrophied villi in some segments of the small intestines (Fig. 5G and H). No PEDV antigens were present in the small intestines of any piglets in the mock-infected control group (Fig. 5I).
Fig. 5.
Clinical symptoms, necropsy examinations, histopathology and IHC of the small intestines of piglets inoculated with P4 of the CH/HBXT/2018 strain (104.33 TCID50/mL). (A) The infected pigs were anorexic and depressed and had severe diarrhea. (B-C) Necropsy examinations of CH/HBXT/2018-infected piglets. Severe hyperemia, swelling and transparency were present in the intestinal tissues, especially in the small intestines. (D-F) HE-stained tissue sections of jejunum from CH/HBXT/2018-infected and mock control piglets (100 × magnification). (D) Severe villous abruption was observed in the infected piglets. (E) Severe villous atrophy and fusion, degeneration and necrosis of mucosal epithelial cells were observed in the infected piglets. (F) The normal villous epithelium of the jejunum from mock control piglets. (G-I) Detection of PEDV antigen by IHC analysis of jejunal tissue sections from CH/HBXT/2018-infected and mock-infected control piglets (400× magnification). (G-H) PEDV antigen signals appear brown and were detected in jejunal epithelial cells from CH/HBXT/2018-infected piglets. (I) No PEDV antigen was detected in jejunum from mock-infected control piglets
Discussion
PED was first reported in England in 1971 and then spread to China in 1986 [5]. From 1986 to 2010, PED caused by the classical European strain CV777 was mostly regional and occurred sporadically in China but did not cause significant economic losses to the swine industry [22]. In 2010, a large-scale epidemic caused by a PEDV strain with sequences that were distinct from those of CV777 caused outbreaks in China resulting in high morbidity and mortality and serious economic losses [22]. In the past, many commercial vaccines, such as killed/inactivated or attenuated PEDV vaccines based on the classical CV777 strain, were widely used on Chinese pig farms and played an important role in the prevention and control of PED [2]. However, since 2010, new highly virulent PEDV variants characterized by partial insertion and deletion mutations in the S gene relative to classical strains became prevalent in China. These new highly virulent PEDV variants belong to the GII genogroup and cause 50-100% mortality. Commercial vaccines based on the classical strain do not provide effective protection against these variants [26]. Therefore, there is an urgent need to develop new vaccines based on current, prevalent, highly virulent PEDV variants. Improved understanding of the biological characteristics and pathogenicity of currently prevalent Chinese PEDV variants is very important for the prevention and control of PED in China. For this purpose, we isolated and passaged a current, prevalent, highly virulent Chinese genotype GIIa PEDV strain, CH/HBXT/2018 (GenBank accession number MH816969), in Vero cells, and we investigated its biological characteristics and pathogenicity.
Phylogenetic analysis based on the S gene showed that the CH/HBXT/2018 strain belongs to genotype GIIa, along with other highly virulent strains isolated in recent years, including the US PC22A strain and Chinese AH2012/12 strain (Fig. 4). Compared to the classical strains CV777 and DR13, the highly virulent PEDV variants of the GII genogroup show insertion, deletion and substitution mutations in the S gene, suggesting that the increased virulence of PEDV variants is mainly due to mutations in the S gene [15, 25, 26]. Analysis of the S protein sequence of CH/HBXT/2018 showed that it contains typical insertions and deletions that have also been found in other recently isolated highly virulent PEDV variants from China and other countries. The consistency of the S gene sequence between the CH/HBXT/2018 strain and currently, prevalent, highly virulent PEDV variants suggests that the CH/HBXT/2018 strain may provide better protection than classical strains against currently, prevalent, highly virulent PEDV variants. However, this hypothesis requires further evaluation.
In previous animal infection studies of highly virulent PEDV variants, such as the PC22A [25], BJ1102C [18] and AH2012/12 strains [27], all strains caused watery diarrhea, dehydration, variable vomiting, and high mortality and morbidity in newborn pigs. In this study, typical PEDV clinical symptoms and intestinal pathological damage were observed in pigs infected with strain CH/HBXT/2018, and the PDD50 of the CH/HBXT/2018 strain was determined to be 8.63 PDD50/3 mL at 7 dpi. Compared to the PC22A PEDV strain (7.83 log10 PDD50/3 mL, 24 hpi) [25], the infectious titer of the CH/HBXT/2018 strain was higher (8.63 log10 PDD50/3 mL, 7 dpi). These results indicated that the CH/HBXT/2018 strain is highly virulent.
Conclusions
In this study, a highly virulent Chinese genotype GIIa PEDV strain, CH/HBXT/2018, was successfully isolated and serially propagated using Vero cells. The biological characteristics and pathogenicity of the virus were further elucidated. The information provided by this study is useful for future pig challenge studies and the development of new effective vaccines against PEDV in China.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant no. 31602095), the National Key Research and Development Program (2016YFD0501505), the China Agriculture Research System (CARS-35) and the Central Public Interest Scientific Institution Basal Research Fund (Y2016CG23).
Author contributions
XL, YW and YZ conceived and designed the experiments. XL wrote the manuscript and analyzed the data. XL performed the sample collection. XL, LZ, QZ, PZ, YF, ZD, DZ, WL, and JF performed the experiments. All authors read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval
All piglets used in the present study were humanely bred during the experiment and euthanized at the end of the experiment. Animal care and use protocols were reviewed and approved by the Institutional Animal Use and Care Committee of Lanzhou Veterinary Research Institute.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Liping Zhang, Xinsheng Liu and Qiaoling Zhang contributed equally to this work.
Contributor Information
Liping Zhang, Email: zhanglp2319@163.com.
Xinsheng Liu, Phone: 09318342665, Email: liuxinsheng@caas.cn.
Qiaoling Zhang, Email: 876835192@qq.com.
Peng Zhou, Email: zhoupeng02@caas.cn.
Yuzhen Fang, Email: fangyuzhen@caas.cn.
Zhaoliang Dong, Email: dzlgsdx@163.com.
Donghong Zhao, Email: zhaodonghong123@163.com.
Weiyan Li, Email: liweiyan160129@163.com.
Jiaxin Feng, Email: fengjiaxin0524@163.com.
Yongguang Zhang, Phone: 09318342537, Email: zhangyongguang@caas.cn.
Yonglu Wang, Phone: 09318343796, Email: wangyonglumd@hotmail.com.
References
- 1.Guo J, Fang L, Ye X, Chen J, Xu S, Zhu X, Miao Y, Wang D, Xiao S. Evolutionary and genotypic analyses of global porcine epidemic diarrhea virus strains. Transbound Emerg Dis. 2018 doi: 10.1111/tbed.12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun D, Wang X, Wei S, Chen J, Feng L. Epidemiology and vaccine of porcine epidemic diarrhea virus in China: a mini-review. J Vet Med Sci. 2016;78(3):355–363. doi: 10.1292/jvms.15-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pensaert MB, de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch Virol. 1978;58(3):243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choudhury B, Dastjerdi A, Doyle N, Frossard JP, Steinbach F. From the field to the lab—an European view on the global spread of PEDV. Virus Res. 2016;226:40–49. doi: 10.1016/j.virusres.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin CM, Saif LJ, Marthaler D, Wang Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016;226:20–39. doi: 10.1016/j.virusres.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paraguison-Alili R, Domingo CY. Phylogenetic tracking of current porcine epidemic diarrhea virus (PEDV) strains in the Philippines. Arch Virol. 2016;161(9):2601–2604. doi: 10.1007/s00705-016-2938-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan Y, Tian X, Li W, Zhou Q, Wang D, Bi Y, Chen F, Song Y. Isolation and characterization of a variant porcine epidemic diarrhea virus in China. Virol J. 2012;9:195. doi: 10.1186/1743-422X-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park S, Kim S, Song D, Park B. Novel porcine epidemic diarrhea virus variant with large genomic deletion, South Korea. Emerg Infect Dis. 2014;20(12):2089–2092. doi: 10.3201/eid2012.131642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamamoto R, Soma J, Nakanishi M, Yamaguchi R, Niinuma S. Isolation and experimental inoculation of an S INDEL strain of porcine epidemic diarrhea virus in Japan. Res Vet Sci. 2015;103:103–106. doi: 10.1016/j.rvsc.2015.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vui DT, Thanh TL, Tung N, Srijangwad A, Tripipat T, Chuanasa T, Nilubol D. Complete genome characterization of porcine epidemic diarrhea virus in Vietnam. Arch Virol. 2015;160(8):1931–1938. doi: 10.1007/s00705-015-2463-6. [DOI] [PubMed] [Google Scholar]
- 11.Puranaveja S, Poolperm P, Lertwatcharasarakul P, Kesdaengsakonwut S, Boonsoongnern A, Urairong K, Kitikoon P, Choojai P, Kedkovid R, Teankum K, Thanawongnuwech R. Chinese-like strain of porcine epidemic diarrhea virus, Thailand. Emerg Infect Dis. 2009;15(7):1112–1115. doi: 10.3201/eid1507.081256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crawford K, Lager KM, Kulshreshtha V, Miller LC, Faaberg KS. Status of vaccines for porcine epidemic diarrhea virus in the United States and Canada. Virus Res. 2016;226:108–116. doi: 10.1016/j.virusres.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Lara-Romero R, Gomez-Nunez L, Cerriteno-Sanchez JL, Marquez-Valdelamar L, Mendoza-Elvira S, Ramirez-Mendoza H, Rivera-Benitez JF. Molecular characterization of the spike gene of the porcine epidemic diarrhea virus in Mexico, 2013–2016. Virus Genes. 2018;54(2):215–224. doi: 10.1007/s11262-017-1528-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarvis MC, Lam HC, Zhang Y, Wang L, Hesse RA, Hause BM, Vlasova A, Wang Q, Zhang J, Nelson MI, Murtaugh MP, Marthaler D. Genomic and evolutionary inferences between American and global strains of porcine epidemic diarrhea virus. Prev Vet Med. 2016;123:175–184. doi: 10.1016/j.prevetmed.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrence PK, Bumgardner E, Bey RF, Stine D, Bumgarner RE. Genome sequences of porcine epidemic diarrhea virus: in vivo and in vitro phenotypes. Genome Announc. 2014 doi: 10.1128/genomea.00503-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng FM, Huo JY, Zhao J, Chang HT, Wang XM, Chen L, Wang CQ. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea virus field strains in central China during 2010–2012 outbreaks. Bing du xue bao Chin J Virol. 2013;29(2):197–205. [PubMed] [Google Scholar]
- 17.Kaewborisuth C, He Q, Jongkaewwattana A. The accessory protein ORF3 contributes to porcine epidemic diarrhea virus replication by direct binding to the spike protein. Viruses. 2018 doi: 10.3390/v10080399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang D, Ge X, Chen D, Li J, Cai Y, Deng J, Zhou L, Guo X, Han J, Yang H. The S gene is necessary but not sufficient for the virulence of porcine epidemic diarrhea virus novel variant strain BJ2011C. J Virol. 2018 doi: 10.1128/jvi.00603-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi D, Lv M, Chen J, Shi H, Zhang S, Zhang X, Feng L. Molecular characterizations of subcellular localization signals in the nucleocapsid protein of porcine epidemic diarrhea virus. Viruses. 2014;6(3):1253–1273. doi: 10.3390/v6031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim SH, Cho BH, Lee KY, Jang YS. N-terminal domain of the spike protein of porcine epidemic diarrhea virus as a new candidate molecule for a mucosal vaccine. Immune Netw. 2018;18(3):e21. doi: 10.4110/in.2018.18.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia A, Feng X, Liu Q, Zhou R, Wang G. Complete genome sequence of CHYJ130330, a highly virulent strain of porcine epidemic diarrhea virus in South China. Genome Announc. 2014;2(2):e00165-14. doi: 10.1128/genomeA.00165-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Li H, Liu Y, Pan Y, Deng F, Song Y, Tang X, He Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg Infect Dis. 2012;18(8):1350–1353. doi: 10.3201/eid1803.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Q, Liu X, Fang Y, Zhou P, Wang Y, Zhang Y. Detection and phylogenetic analyses of spike genes in porcine epidemic diarrhea virus strains circulating in China in 2016-2017. Virol J. 2017;14(1):194. doi: 10.1186/s12985-017-0860-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oka T, Saif LJ, Marthaler D, Esseili MA, Meulia T, Lin CM, Vlasova AN, Jung K, Zhang Y, Wang Q. Cell culture isolation and sequence analysis of genetically diverse US porcine epidemic diarrhea virus strains including a novel strain with a large deletion in the spike gene. Vet Microbiol. 2014;173(3–4):258–269. doi: 10.1016/j.vetmic.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Lin CM, Annamalai T, Gao X, Lu Z, Esseili MA, Jung K, El-Tholoth M, Saif LJ, Wang Q. Determination of the infectious titer and virulence of an original US porcine epidemic diarrhea virus PC22A strain. Vet Res. 2015;46:109. doi: 10.1186/s13567-015-0249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang D, Fang L, Xiao S. Porcine epidemic diarrhea in China. Virus Res. 2016;226:7–13. doi: 10.1016/j.virusres.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan B, Yu Z, Pang F, Xu X, Zhang B, Guo R, He K, Li B. Characterization of a pathogenic full-length cDNA clone of a virulent porcine epidemic diarrhea virus strain AH2012/12 in China. Virology. 2017;500:50–61. doi: 10.1016/j.virol.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]