Abstract
PA-X, a fusion protein belonging to influenza A viruses (IAVs), integrating the N-terminal 191 amino acids of PA gene and the ribosomal frame-shifting product that lengthens out to 41 or 61 amino acids. Since its discovery in 2012, multiple functions have been attributed to this small protein, including a process, where wide-spread protein synthesis in infected host cells is shut down (called host shutoff), and viral replication, polymerase activity, viral-induced cell apoptosis, PA nuclear localization, and virulence are modulated. However, many of its proposed functions may be specific to strain, subtype, host, or cell line. In this review, we start by describing the well-defined global host-shutoff ability of PA-X and the potential mechanisms underlying it. We move on to the role played by PA-X in modulating innate and acquired immune responses in the host. We then systematically discuss the role played by PA-X in modulating the virulence of influenza viruses of different subtypes and host origins, and finish with a general overview of the research advances made in identifying the host cell partners that interact with PA-X. To uncover possible clues about the differential effects of PA-X in modulating viral virulence, we focus on systemically evaluating polymorphisms in PA-X from various viral subtypes and hosts, including avian and human H5N1, H5N6, H9N2, and H7N9 viruses. Finally, we conclude with a proposition regarding the possible future research directions for this important protein.
Keywords: Influenza virus, PA-X, Host shutoff, Immunomodulatory proteins, Pathogenesis, Virus–host interaction
Introduction
Influenza A virus (IAV) is the most diverse and epidemiologically significant pathogen associated with severe disease manifestations in humans [1]. Wild aquatic birds are the natural reservoirs for IAV, but it can infect a variety of animals, including poultry, aquatic animals (e.g., seals, dolphins, and whales) and terrestrial mammals (e.g., humans, horses, pigs, mink, cats, dogs, and tigers) [2, 3]. Two common mechanisms are used by IAV to cross the host barrier and acquire high virulence in various animals. One mechanism involves the acquisition of adaptive mutations and/or genetic reassortment, a well-known strategy used by several pandemic viruses, including H1N1 (1918 Spanish flu), H2N2 (1957 Asian flu), H3N2 (1968 Hong Kong flu), H1N1 (2009 pandemic), and H7N9 (2013 Chinese epidemic) [4–8]. Another virulence mechanism is where multiple viral accessory proteins are encoded on a single gene segment. The IAV genome comprises eight negative-sense RNA segments that were initially assumed to encode the following ten proteins: polymerase basic proteins 1 (PB1) and 2 (PB2), polymerase acidic protein (PA), nucleoprotein (NP), hemagglutinin (HA), neuraminidase (NA), matrix proteins 1 (M1) and 2 (M2), and non-structural proteins 1 (NS1) and 2 (NS2). However, over the past 16 years, another seven novel proteins have been gradually discovered, including PB1-F2 [9], PB1-N40 [10], PA-X [11], M42 [12], NS3 [13], PA-N155, and PA-N182 [14]. Among these accessory proteins, PB1-F2 and PA-X, have been extensively studied and found to share some similarities in modulating virulence in IAV.
Here, we review current knowledge on the PA-X protein, including the polymorphism characteristics among the different virus subtypes and host species, host-shutoff activity, the role of PA-X in modulating host innate and adaptive responses, the contribution of PA-X to the pathogenesis of IAV, and provide a summary of known PA-X host partners. Finally, we propose several potential research areas that may accelerate understanding about the role played by this novel protein during influenza virus infections, especially for outbreaks of human infections with emerging IAVs, such as H7N9 virus.
Polymorphisms in the PA-X protein from viruses with different subtypes and host origins
The PA-X protein is a fusion protein with an N-terminal 191 amino acid leader sequence originating from PA and a C-terminal region of 61 or 41 codons encoded by an overlapping open-reading frame (ORF) (“X-ORF”), which is accessed by one ribosomal frame shift in the PA gene (Fig. 1). Based on the lineage-specific differences in the distribution of X-ORF lengths, two major X-ORF groups have been classified [11, 15]. About 75% of the isolates have a 61-codon X-ORF, which basically covers all host species and HA/NA subtypes [15]. The remaining 25% carry the truncated form of PA-X, where a 41-codon X-ORF created by nonsense mutations occurs mainly at codon 42 [15]. These truncated PA-X proteins overwhelmingly come from the 2009 human pandemic H1N1 virus, the triple reassortant swine H1N1 virus (a cluster of classic swine H1N1 viruses), equine H7N7, canine H3N8, canine H3N2, and bat influenza virus [15]. Of note, it has been shown that the truncated form of PA-X evolved convergently in viruses from pigs or dogs (H3N2 and H3N8), suggesting that it is associated with the adaptation and emergence of influenza virus in these host species [15].
Fig. 1.

Proposed mechanism of PA-X protein formation and the functional area in PA-X. The PA-X open-reading frame encodes either 61 or 41 amino acids as indicated. In addition, the X-ORF product lies largely within a linker region between the PA N- and C-terminal domains. The frame-shifting motif locates in the sequence of UCC UUU CGUC, after shifting, the sequence changed to UUC GUC. Functional domains are distinguished by shape and color: blue bar (areas that important for host-shutoff activity) and red ellipse (areas that important for endonuclease activity)
Global host-shutoff activity by the PA-X protein
Influenza virus infection results in a rapid decline of global host protein synthesis in infected cells, a process known as host shutoff. This process allows the virus to escape host innate immune recognition and shut down host antigen processing to subvert acquired immunity, enabling it to escape host restriction and promote its multiplication and spread. It has been shown that multiple mechanisms are related to host shutoff in IAV-infected cells. However, recent studies have focused on the following three main mechanisms of global inhibition of host protein expression by IAV infection: (1) blockade of cellular mRNA processing and nuclear export by NS1 [16–19]; (2) degradation of host RNA Pol II by the viral RNA-dependent RNA polymerase RdRp complex (RdRP) [20–22]; and (3) wide-spread host mRNA degradation by the PA-X protein [11, 23–31]. In this review, we focus mainly on the shutoff activity of PA-X protein.
Discovery of host-shutoff ability by the PA-X protein
By co-transfecting reporter plasmids that encode β-galactosidase or the 1918 NP gene together with the wild type or the PA-X-deficient 1918 PA segment, Jagger et al. were the first researchers to discover that PA-X can mediate the suppression of plasmid-driven gene expression [11]. Using global transcriptional profiling, they further demonstrated that down-regulation of PA-X expression leads to an accelerated global host response in virus-infected mouse lungs, notably inflammatory, apoptotic, and T lymphocyte responses [11]. Many later studies have also reported on PA-X-mediated host-shutoff activity in human H1N1 virus [23, 26, 27, 29, 32–34], avian H9N2 virus [35], avian H5N1 virus [24, 36], equine influenza virus (EIV) and canine influenza virus (CIV) of the H3N8 subtype [37], swine H1N2 influenza virus [38], and triple-reassortment (TR) H1N2 swine influenza virus (SIV) [39]. Therefore, the universal expression of PA-X in IAVs and the relatively conservative genetic characteristics of the X-ORF suggest that the shutoff ability of the PA-X protein may also be a common functionality in all influenza virus strains. However, further studies are needed to verify this hypothesis in viruses with different subtypes and in various host cell lines.
Functional domains of the host-shutoff activity in the PA-X protein
The previous studies have shown the involvement of the PA protein in host-shutoff activity; however, the potential function domains could only be mapped after PA-X was discovered [11]. The common mechanism used for host-shutoff activity, as shared by PA and PA-X, has been ascribed to the N-terminal RNA endonuclease domain of these proteins, taking the form of PD(D/E)XK (P107 D108 × 10 E119 K134) in the nuclease family (Fig. 1) [11, 23, 30, 40, 41]. However, over time, other molecular mechanisms also have been shown to be involved in this process. Very early on, Desmet et al. showed that the N-terminal domain of the PA protein is responsible for host shutoff; specifically, helix 4 (amino acids 85, 86, 91, 100, 114, and 186) and the 51–74 amino acid flexible loop (Fig. 1) [23]. These researchers also found that the PA-X C-terminal sequence also plays a role in suppressing expression of the reporter gene [23]. Thereafter, two independent studies demonstrated that the initial 15 amino acids (positions 192–206) in the PA-X C-terminal region are sufficient for the full shutoff activity of PA-X and that six basic amino acids (195R, 198K, 199R, 202K, 203K, and 206K) also play a key role (Fig. 1) [26, 27]. However, differing significantly from these findings, three other studies reported on the contribution made by the last 20 C-terminal residues in PA-X-mediated shutoff activity (Fig. 1). In 2015, Gao et al. reported that amino acids covering 233–252 of the PA-X C-terminus also strongly suppress gene expression and enhance viral replication and virulence in three different strains (human pandemic H1N1 2009, avian H5N1, and avian H9N2) [32]. Simultaneously, Bavagnoli et al. also provided direct evidence that the last 20 amino acids in the PA-X C-terminal region are important for endonuclease activity, and this was assumed to contribute to host shutoff by the virus (Fig. 1) [42]. Moreover, unlike the specific role played by PA in cutting single-stranded (ss)RNA, PA-X is capable of digesting both ssRNA and double-stranded (ds)RNA, with a preference for ssRNA substrates, such as poly r(A) or poly r(U), which suggests that PA-X has a wide-spread degradation ability for various host RNAs [42]. Consistent with the results from both these studies, by comparing the PA-X proteins from EIV and CIV H3N8 viruses, Feng et al. found that position 231 (Ser) and the C-terminal elongated tail both contribute to the stronger host-shutoff ability of EIV PA-X relative to CIV PA-X [37]. Collectively, these studies reveal that both the N- and C-termini of PA-X contribute to its overall shutoff ability. Quite recently, by expressing PA-X in yeast, Oishi et al. further mapped 22 new amino acid mutations (including sites located in the endonuclease active sites, such as P107S, D108N, and E119N) that contribute to a decrease in shutoff activity [43]. However, further studies are needed to elucidate the potential mechanisms related to host-shutoff regulation by these newly identified amino acids.
Potential regulatory mechanism for host shutoff in the PA-X protein
PA-X selectively targets cellular mRNAs while sparing viral mRNAs, thereby ensuring successful viral replication while defeating an effective anti-viral response in the host. The mechanism involved has been partially elucidated by Khaperskyy et al. who found that PA-X selectively degrades host RNA polymerase II (Pol II)-transcribed mRNA by interacting with the host’s 5′->3′ Xrn1 exonuclease activity and that PA-X likely operates in the cell nucleus [29]. Moreover, the shutoff activity is strongly associated with nuclear accumulation of the PA-X protein, with the process majorly mediated by four conserved basic residues (198K, 199R, 202K, and 203K) in X-ORF [29]. In addition, unlike herpes simplex virus 1 (HSV-1) vhs protein [44] and SARS nsp1 protein [45], which all selectively degrade translatable RNA polymerase II (Pol II) transcripts, PA-X also targets Pol II synthesized non-coding RNAs for degradation, highlighting the distinct features of PA-X in mediating host shutoff. Similarly, Hayashi et al. also found that the first nine amino acids are sufficient for nuclear localization and that three basic amino acids (195K, 198K, and 199R) are key residues for host shut off ability and nuclear accumulation in PA-X [27]. Therefore, taking the results from the two studies together, it seems likely that 198K and 199R are the crucial residues controlling the localization of PA-X and both are correlated with host-shutoff function. However, contrasting with the results from the Khaperskyy et al.’s study, Hayashi et al. showed that PA-X can degrade mature mRNAs synthesized in the nucleus and cytoplasm alike and that destruction of mRNA by PA-X in the nucleus was more efficient than in the cytoplasm, thereby revealing the multiple functional sites of action for PA-X-specific mRNA degradation [27].
Concerted effect of PA-X and NS1 in host shutoff and pathogenicity
Another well-characterized host-shutoff mechanism employed by IAV is the NS1-mediated blockade of the cleavage and polyadenylation specificity factor (CPSF) function observed in some strains from humans, which is one of the hallmarks of adaptation to humans by IAV [16–19]. NS1 is involved in the early nuclear phase of the host shutoff, whereas PA-X participates in the late cytoplasmic phase of this process [28]. In the nuclear stages, another mechanism used by IAV is the cap-snatching activity of viral RDRP, which targets host pre-mRNAs to supply capped primers for viral transcription [20–22]. Early on in the viral infection, these three host-shutoff mechanisms act in concert to inhibit the host cell’s ability to initiate an effective anti-viral response. In the late period of the infection, IAV induces host cell apoptosis, thereby completely eliminating the ability of the host cell to synthesize new proteins for its own use. When considering the role of NS1 and PA-X in viral pathogenicity and host shutoff, it is very interesting to see the combined effects of them in modulating viral virulence. Therefore, recently, Nogales et al. investigated the interplay of PA-X and NS1-mediated host shutoff in viral replication and pathogenesis using a temperature-sensitive 2009 pandemic H1N1 virus [31]. These researchers found that viruses that simultaneously encode PA-X and NS1, or are deficient in both proteins, are highly attenuated in vivo, suggesting that there is a strict balance between NS1 and PA-X proteins during host gene expression regulation, viral replication, and pathogenesis. Therefore, it seems likely that optimal control of host protein synthesis by IAV PA-X and/or NS1 protein contributes to efficient virus replication and, consequently, to virulence, a hypothesis strengthened by the findings that wide-spread host mRNA degradation by PA-X only occurs during the late infection stage for IAV [28]. However, additional studies examining the concerted effect of NS1 and PA-X in inhibiting host protein expression in different viral strains need to be undertaken to augment our current understanding of IAV-associated host shutoff and provide new clues about the role played by PA-X in viral replication, pathogenesis, and host adaptation.
Role of PA-X in modulating the host’s innate and acquired immune responses
PA-Xs role in modulating the host’s innate immune response
Jagger et al. were the first researchers to show that down-regulation of PA-X expression markedly elevated the 1918 H1N1-induced immune response in mouse lungs; specifically that IFN-γ, CCR5, CD28, IL-7, and IL-15 signaling pathways, which are associated with lymphocyte activation and/or proliferation or other aspects of cell-mediated immunity, were enhanced [11]. PA-X also markedly suppresses several major histocompatibility complex class I-associated genes and activates many integrins and extracellular matrix components [11]. All of these perturbations in host response pathways may further affect lymphocyte activation and immune cell function, leading to an aberrant immune response and subsequent host immunopathology [46]. Similarly, using microarray analysis, our previous study also found that loss of PA-X expression significantly activates the global host response of the H5N1 influenza virus in chicken lungs, especially the inflammatory and cell death response [24]. Other studies have also shown that decreased PA-X expression clearly elevates the host innate immune response in mouse lungs infected with avian H5N1 virus [36], human 2009 pandemic H1N1 virus [33, 34, 36], and in porcine alveolar macrophages (PAM) cells infected with swine H1N2 virus [39]. In contrast, Gao et al. showed that loss of PA-X expression in H9N2 virus inhibits proinflammatory cytokine and chemokine responses [35]. Elsewhere, by transfecting plasmids expressing only PA-X in cell cultures, Feng et al. found that PA-X markedly elevated a number of genes, notably, innate immune response-related genes, ubiquitin ligases, vesicle transport and budding-related genes, and the genes associated with protein post-translational modifications in the Golgi complex [37]. Therefore, it seems likely that the inhibition effect of PA-X on the innate immune response is strain-specific or subtype-specific.
PA-Xs role in modulating the host’s acquired immune response
To investigate the effect of PA-X on the humoral immune response, Hayashi et al. systematically compared serum antibodies from mice infected with wild-type viruses (A/California/04/09, H1N1, or Cal) and the PA-X-deficient Cal PA-XFS virus. They found that the Cal PA-XFS virus stimulated more neutralizing antibodies and higher levels of anti-HA and anti-NP antibodies than the wild-type virus, suggesting that the shutoff activity of the PA-X protein may also be involved in dampening down the host’s humoral immune response [33].
Other immunomodulatory proteins in influenza viruses
Considering the universal role for PA-X in modulating the host’s innate and adaptive immune responses, undoubtedly, PA-X can be classified as an immunomodulatory protein. As mentioned already, NS1 is another well-characterized immunomodulatory protein [47–50]. NS1 is a relatively small polypeptide with various interesting functions, one of which is its ability to antagonize the innate immune response by inhibiting the type I interferon system at multiple levels. NS1 acts as an antagonist during various stages of the anti-viral response, including (1) pre-transcriptional inhibition of interferon expression by interacting with components of the retinoic acid-induced gene protein I (RIG-I) signaling axis [47, 51–56], (2) co- and post-transcriptional inhibition by limiting host gene expression by blocking the cellular pre-mRNAs 3′ end-processing factor CPSF30 [19, 48, 55, 57–59], and (3) post-translational inhibition of anti-viral genes by antagonizing the PKR Ser/Thr kinase [60–64] and the OAS RNAse L-pathway activator [62].
IAV also encodes several proteins (PB1-F2, PB2, PA, and M2) that have been identified as affecting the host’s innate immune response to a certain degree. PB1-F2 blocks the IFN response by interacting directly with the components (RIG-I/MAVs protein complex) of the interferon pathway [65, 66]. PB1-F2 also promotes the inflammatory response and contributes to viral virulence and the acquisition of secondary bacterial infections [67–71]. As for PB2 and PA, these two proteins affect host cell protein synthesis by facilitating cap snatching from host cell’s mRNAs while indirectly suppressing the host’s anti-viral response [40, 72]. However, PB2 also limits IFN-β expression by interfering with mitochondrial anti-viral signaling [73], or by inhibiting IFN-β promoter activation by regulating RIG-I and interferon beta promoter stimulating factor-1 [74]. Along with its cap-snatching activity [40], PA also plays an important role in inhibiting host type I IFN signaling [75], as well as affecting cytokine production and contributing to virulence in H5N1 viruses [76, 77] and in seasonal H3N2 in mice [78]. As for the M2 ion channel, it has been shown to activate inflammasomes by stimulating the NOD-like receptor family pyrin domain containing 3 protein [79]. Together, these immunomodulatory proteins represent new potential targets for the development of next-generation vaccines, which could exhibit improved duration of protection and protective immunity efficacy; among them, NS1 protein is a well-verified example [80–82].
Role of PA-X in modulating influenza virus virulence in various animal models
Effects of loss of PA-X expression on virulence in influenza virus
After the PA-X protein was discovered in 2012, the role played by it in modulating viral virulence has become gradually clearer. By generating PA-X-deficient viruses, accumulating numbers of studies have defined the important role that PA-X plays in modulating viral replication and virulence in influenza virus. As shown in Table 1, PA-X actively modulates viral pathogenicity in different viral subtypes in various animal models, including mice [11, 24, 32–35, 83], chickens [24], ducks [24], and pigs [38, 39]. In 2012, Jagger et al. were the first group to show that loss of PA-X expression increases host inflammatory and apoptosis responses and enhances virulence in the 1918 pandemic H1N1 virus in mice, but exerts no effect on viral replication in vitro or in vivo [11]. Next, several independent studies revealed that down-regulated PA-X expression enhances viral replication, polymerase activity, apoptosis, inflammatory response, and H5N1 virulence in chickens, ducks, and mice [24, 36, 84]. With the 2009 pandemic H1N1 virus, Gao et al. [36] and Hayashi et al. [33] showed that loss of PA-X expression increases viral virulence in mice, whereas Leea et al. [34] found that down-regulation of PA-X expression attenuates viral replication and virulence in mice. However, although the absence of PA-X in 2009 pandemic H1N1 increased viral pathogenicity in the mice, viral replication became attenuated in both cultured human cells and mice [33].
Table 1.
Effects of loss of PA-X expression on influenza A virus pathogenicity
| Virus subtypes | Replication | RNP activity | Apoptosis | Inflammatory | PA accumulation | Virulence |
|---|---|---|---|---|---|---|
| H1N1(1918 pdm) | Have no effect in MDCK, eggs and mice [11] | –, [11] | Increase in mice [11] | Increase in mice [11] | –, [11] | Increase in mice [11] |
| H5N1(avian) | Increase in various mammalian and avian cells, and in mice, chickens, ducks [24, 35, 84] | Increase in 293T, CEF, DEF cells [24, 35, 84] | Increase in CEF, DEF, MDCK and A549 cells [24, 35, 84] | Increase in chickens, ducks and mice [24, 35, 84] | Increase in MDCK cells [35, 84) and A549 cells (35] | Increase in mice, chickens, ducks [24, 35, 84] |
| H1N1(2009 pdm) | Increase in A549 cells and mice [35]; decrease in Calu-3 cells and mice [33]; decrease in MDCK, A549 and mice [34] | Increase in 293T cells [35]; decrease in 293T cells [34] | Increase in A549 cells [33–35] | Increase in mice [35] | Increase in A549 cells [35] | Increase in mice [33, 35]; decrease in mice [34] |
| H9N2 (avian) | Have no effect in MDCK and A549 cells, decrease in mice [36] | –, [36] | Decrease in A549 cells [36] | Decrease in mice [36] | –, [36] | Decrease in mice [36] |
| H1N1 (swine) | Increase in MDCK, A549 cells and mice [83] | Increase in 293T cells [83] | –, [83] | –, [83] | –, [83] | Increase in mice [83] |
| H1N2 (swine) | Decrease in PK15, PAM cells and pigs [39] | –, [39] | –, [39] | Increase in PAM cells[39] | –, [39] | Decrease in pigs [39] |
–Not carried out
Interestingly, Gao et al. [35] reported that loss of PA-X expression in the H9N2 virus decreased its virulence in mice while exerting no effect on its replication. In swine influenza virus, Gong et al. [83] found that down-regulation of PA-X expression in swine H1N1 virus enhanced the activity of viral polymerase in mammalian cells as well as viral replication and virulence in mice. However, Xu et al. [39] showed that loss of PA-X expression decreased viral replication in PK15, PAM cells, and pigs, and also dampened the virulence of swine H1N2 virus in these animals.
Thus, these studies clearly suggest that the effects of PA-X on viral replication and virulence are strain or host specific. Considering the concerted role for PA-X and NS1 in blockading host gene expression [28, 31], we envisage that the precise impact of PA-X on viral growth and pathogenicity may vary among different viral strains, depending on the specificity and activity of the NS1 protein in each strain.
Effects of the C-terminal 20 amino acids of PA-X on the pathogenicity of influenza virus
The X-ORF from the PA-X protein generally contains either 61 or 41 amino acids. The majority of IAV strains have a full-length 61-codon X-ORF, while around 25% of them (e.g., 2009 human pandemic H1N1 virus, and triple reassortant swine H1N1 viruses) carry a truncated form of PA-X, which contains the 41-codon X-ORF [15]. To investigate the biological significance of the length of PA-X in influenza virus infections, Gao et al. constructed a series of recombinant viruses carrying full or truncated versions of PA-X based on pandemic 2009 H1N1, avian H5N1 and H9N2 viruses and systemically compared their replication and pathogenicity profiles in mice. The results suggested that the last 233–252 amino acids increased viral replication and virulence, strengthened the viral-induced inflammatory response and apoptosis, and elevated the host-shutoff ability by the PA-X protein [32]. Using a molecular evolutionary approach, Xu et al. found that before 1985, all swine influenza viruses (SIVs) possessed full-length PA-X [85]. However, subsequently, the truncated forms of PA-X were continuously detected in SIVs and they gradually replaced the full-length PA-X to become the dominant PA-X phenotype in SIVs. To further explore the potential role of PA-X truncation in the adaptation of influenza virus to pigs, Xu et al. constructed PA-X-lengthened viruses based on the genetic backbone of an H1N2 SIV strain that encodes a truncated PA-X, and assessed their biological characteristics. In contrast to the Gao et al.’s study, Xu et al. found that compared with the whole length PA-X, SIV with truncated PA-X had higher pathogenicity and enhanced viral replication and transmissibility in pigs, and exerted a stronger inhibitory effect on IFN-I mRNA expression. Therefore, it seems likely that truncation of PA-X in SIV contributes to its adaptation in pigs, suggesting that an association between PA-X length and host specificity exists.
Host factors that interact with the PA-X protein
To facilitate viral infection and replication, viral proteins in IAV need to constantly interact with an array of cellular proteins and hijack the host pathways at the helm of cellular responses. Considering the essential role played by PA-X in modulating host responses and viral virulence, it is crucial to identify the host proteins interacting with PA-X to gain better understanding of how PA-X recruits the host cellular machinery for these functions. Using affinity purification and mass spectrometry and a high confidence threshold, Li et al. identified a total of 56 unique proteins physically interacting with PA-X from H5N1 virus in chicken cells [86]. Functional analysis of these proteins revealed the significant enrichment of biological processes, such as those associated with mitochondria and lipid transport, nucleosome assembly, and RNA processing. In addition, among the PA-X-interacting partners that were identified, GNB2L1, YWHAE, RPS20, RPS13, ARF1, and RPLP0 were annotated in the Gene Ontology term ‘poly (A) RNA binding’. APOA1, ATP5B, NCL, NPM1, Thy1, and WDR1 were also reported to contribute to various types of viral infections. For example, APOA1 in hepatitis B virus (HBV) [87] and hepatitis C virus [88], ATP5B in HSV-1 [89]; NCL in influenza virus [90–93], dengue virus [94], parainfluenza virus type 3 [95], respiratory syncytial virus [96], Japanese encephalitis virus [97], Crimean-Congo hemorrhagic fever virus [98] and HIV [99, 100]; NPM1 in HIV [101], Thy1 in HBV [102], and WDR1 in Sendai virus [103]. However, this study only preliminarily described the proteins interacting with the PA-X protein overall in a chicken cell model, and further studies are needed to verify their interactions with PA-X and to investigate the underlying biological significance and mechanisms for these interactions. Moreover, to understand the potential molecular mechanism for PA-X-associated functioning in various hosts, it is also very important to investigate the host’s interactome with PA-X in mammalian cells overall, such as in human or mouse cells.
Polymorphism in PA-X in influenza viruses as a threat to public health
AIVs with various HA subtypes (e.g., H1, H2, H3, H4, H5, H6, H7, H9, H10, and H11) have been circulating in domestic poultry in China [104]. Among them, currently, H5N1, H5N6, H9N2, and H7N9 AIVs are the most prevalent subtypes in many Chinese regions [105, 106]. It is noteworthy that all these subtypes not only cause substantial economic losses to poultry farming but also pose a potential threat to public health [106–112]. To find potential clues about the various effects of PA-X on the modulation of viral virulence, we systematically analyzed the PA-X polymorphisms in four prevalent AIV subtypes. To do this, we thoroughly compared all the available PA-X sequences from the Influenza Research Database (https://www.fludb.org/brc/home.spg?decorator=influenza) and the Global Initiative on Sharing Avian Influenza Data (GISAID) (https://www.gisaid.org/). Differences in the PA-X sequence in various hosts, and in avian and human isolates were separately analyzed. Specifically, the strains from 75 human H5N1, 589 avian H5N1, 19 human H5N6, 651 avian H5N6, 9 human H9N2, 917 avian H9N2, 813 human H7N9, and 437 avian H7N9 were included. Overall, we found that all of the viruses analyzed carry 61 amino acids in the X-ORF of their PA-X genes, and no clear-cut, host-specific genetic characteristics were identified in the avian and human isolates for all subtypes (Tables 2, 3). Next, we further analyzed the specific amino acid variations in the PA-X gene. As shown in Table 2, in the N-terminal of the PA-X gene, a total of 16 amino acid variations were identified in the different subtypes, including 20A, 27D, 37A, 57R, 58G, 61I, 63V, 68P, 70A, 94I, 96N, 100V, 101D, 115N, 129I, and 142K. Interestingly, the RNA endonuclease domain (P107 D108 × 10 E119 K134) was highly conserved among all the viral subtypes (Fig. 1) [40, 41]. Moreover, the helix 4 region (amino acids 85, 86, 91, 100, 114, and 186), which is important for the shutoff activity of the PA-X protein, is also relatively conserved (Fig. 1) [23], except for residue 100 (especially in the H7N9 influenza virus). Of note, the 51–74 amino acid motif in the PA-X protein, which is associated with host-shutoff ability [23], is highly variable and includes 57R, 58G, 61I, 63V, 68P, and 70A.
Table 2.
Statistical analysis of the polymorphism at the N-terminal of PA-X gene
| Sitesa | Number of the analyzed strains and the sequence variation at these residuesb | |||||||
|---|---|---|---|---|---|---|---|---|
| Human H5N1 (75 strains) |
Avian H5N1 (589 strains) |
Human H5N6 (19 strains) |
Avian H5N6 (651 strains) |
Human H9N2 (9 strains) |
Avian H9N2 (917 strains) |
Human H7N9 (813 strains) |
Avian H7N9 (437 strains) |
|
| 20A | 5T (7%) | 51T/1G (9%) | 6T(32%) | 552T (85%) | 1T (11%) | 19T (2%) | 32T (4%) | 5T (1%) |
| 27D | 25N/5S (40%) | 126N/22S/2T/6V/1A (27%) | 6S(32%) | 548S/9N (86%) | 0% | 10N/1K/1S (1%) | 1G (0.1%) | 0% |
| 37A | 0% | 1S/1T (0.3%) | 12S (63%) | 72S/1T (11%) | 6S (67%) | 243A/674S (74%) | 813S (100%) | 437S (100%) |
| 57R | 0% | 12Q (2%) | 0% | 0% | 0% | 29Q/9K/1L (4%) | 45Q (6%) | 2Q (0.4%) |
| 58G | 10S (13%) | 261S/1A (44%) | 0% | 0% | 0% | 19S/3N (2%) | 0% | 0% |
| 61I | 2M/5T (9%) | 64M/17T/4V/1R/1W (15%) | 12T (63%) | 85T/4M (14%) | 6T (67%) | 691T/2K (75%) | 811T (99%) | 1A/434T (100%) |
| 63V | 2I/5A (9%) | 39A/26I/14T/2L (14%) | 12I/6A (95%) | 554A/74I (96%) | 6I (67%) | 669I/3M/2L/1A (74%) | 810I (99%) | 437I (100%) |
| 68P | 2L (3%) | 13Q/1H (2%) | 0% | 1L/1S (0.3%) | 0% | 11Q/3L/1A (2%) | 806P/3L/2T/1S (0.7%) | 0% |
| 70A | 2V (3%) | 19V (3%) | 0% | 47V/1T (7%) | 6V (67%) | 154V/1M/1S/1T (17%) | 95V (12%) | 35V (8%) |
| 94I | 0% | 3V/1T/1M/1L (1%) | 0% | 28V (4%) | 0% | 5L/2V (0.7%) | 4V (0.5%) | 0% |
| 96N | 0% | 4S/1P/1T (1%) | 0% | 0% | 0% | 4H/1T (0.5%) | 43H (5%) | 93H (21%) |
| 100V | 2I (3%) | 8A/1F/7I/ (3%) | 0% | 60A/1I (9%) | 0% | 82A/24I (12%) | 41I (72%) | 181A (41%) |
| 101D | 8E/10N (24%) | 297E/34N/16G/1V (59%) | 13E (68%) | 79E/5G/5N (14%) | 7E (70%) | 870E/13Q/4K/3V (97%) | 4E (100%) | 435E (99%) |
| 115N | 2S (3%) | 30D/8G/5S/3Y (8%) | 0% | 14D/3S (3%) | 0% | 23D/8S/8T/1H/1K/1Y (5%) | 2D/1 (5%) | 340N/93S/2D (22%) |
| 129I | 8T/2V (13%) | 250T/19V/1A/1L/1M (46%) | 0% | 50T (8%) | 0% | 17M/5T/1L (3%) | 4M/1T (0.6%) | 3M/2T (1%) |
| 142K | 5R (7%) | 51R/28N/1Q (14%) | 7R (37%) | 529R/21G (84%) | 0% | 52R/10N/1E (7%) | 5E/2R (0.9%) | 0% |
aIndicates the original residues
bThe number outside the brackets indicates the number of strains that carry certain kind of polymorphism; the number inside the brackets shows the variation rates relative to the original residues
Table 3.
Statistical analysis of the polymorphism at the C-terminal of PA-X gene
| Sitesa | Number of the analyzed strains and the sequence variation at these residuesb | |||||||
|---|---|---|---|---|---|---|---|---|
| Human H5N1 (75 strains) |
Avian H5N1 (589 strains) |
Human H5N6 (19 strains) | Avian H5N6 (651 strains) |
Human H9N2 (9 strains) |
Avian H9N2 (917 strains) |
Human H7N9 (813 strains) |
Avian H7N9 (437 strains) |
|
| 193N | 2S (3%) | 140S/1T/1C (24%) | 13S (68%) | 92S (14%) | 7S (78%) | 793S/8R (87%) | 813S (100%) | 437S (100%) |
| 194P | 2L (3%) | 49L/5Q (9%) | 0% | 5L (0.8%) | 0% | 92L/21Q/6R (13%) | 398L/10Q (50%) | 156L/8Q (38%) |
| 195R | 2K (3%) | 12K (2%) | 12K (63%) | 92K (14%) | 6K (67%) | 670K/1N/1S (73%) | 813K (100%) | 437K (100%) |
| 199R | 2K (3%) | 51K (9%) | 12K (63%) | 81K/1I (14%) | 6K (67%) | 672K/1T (73%) | 811K (99%) | 437K (100%) |
| 204G | 25D/6N/2S (44%) | 235N/4S/1A (79%) | 13D (68%) | 97D/7N (16%) | 9D (100%) | 907D/1N (99%) | 797D/1G/13N/2Y (100%) | 437D (100%) |
| 206K | 0% | 4T/1N/1I/1E (1%) | 0% | 2I/8R (2%) | 0% | 16R/2E/1S/1I (2%) | 3R (0.4%) | 1R (0.2%) |
| 208Q | 8L/2K (13%) | 232L/1K/4P (39%) | 0% | 2K (0.3%) | 0% | 5H/5K/3L/1P (2%) | 1E/3R (0.5%) | 0% |
| 209E | 0% | 4G/1V (0.8%) | 5G (26%) | 12G (2%) | 1G (11%) | 35G/5A/1Q/1V (4%) | 30A/2G/3K/1V (4%) | 2A (0.5%) |
| 210P | 1L (1%) | 12L/2T/1S/17Q (5%) | 0% | 11L/8Q (3%) | 0% | 89Q/35L/1S (14%) | 12Q (1%) | 1Q (0.2%) |
| 213G | 10D/2Y (16%) | 2A/1N/213D (37%) | 0% | 4D/3S (1%) | 0% | 19S/4C/3D (3%) | 0% | 0% |
| 215P | 13L/4Q (23%) | 17Q/1R/1S/78L (16%) | 6L (32%) | 553L (85%) | 1L (11%) | 51L/3Q/1A (0.6%) | 8L (1%) | 1L (0.2%) |
| 218V | 4A (6%) | 88A (15%) | 0% | 4A (0.6%) | 0% | 73A (8%) | 0% | 1A (0.2%) |
| 228I | 8T/2V (13%) | 354T/7N (61%) | 13T (68%) | 98T (15%) | 1P/8T (100%) | 896T/3A/1N (98%) | 813T (100%) | 437T (100%) |
| 248K | 0% | 1I/1Q/7R/1T (2%) | 5R (26%) | 63R (10%) | 0% | 130R (14%) | 530R (65%) | 181R (41%) |
| 251K | 5R (7%) | 76R (13%) | 6R (32%) | 555R (85%) | 0% | 11R/1E/2M (2%) | 0% | 0% |
aIndicates the original residues
bThe number outside the brackets indicates the number of strains that carry certain kind of polymorphism; the number inside the brackets shows the variation rates relative to the original residues
We further analyzed the genetic variation occurring in the C-terminal of the PA-X gene. We found that this region contains much higher variation (24.6%) than the N-terminal (8.38%). Specific variations occurred in positions 193N, 194P, 195R, 199R, 204G, 206K, 208Q, 209E, 210P, 213G, 215P, 218V, 228I, 248K, and 251K (Table 3). Interestingly, six conserved, basic amino acids (195R, 198K, 199R, 202K, 203K, and 206K) (Fig. 1), which were previously identified as contributing to host shutoff by the PA-X protein, also showed variations [26, 27] (e.g., 195R/K/N/S, 199R/K/T, and 206K/R/E/S/I polymorphisms in avian H9N2 viruses, 199R/K/I variations in avian H5N6 viruses, and 206K/T/N/I/E polymorphisms in avian H5N1 viruses). It will be fascinating to determine whether these genetic variations affect the host-shutoff ability and virulence of different viral subtypes.
Polymorphism comparisons of the PA-X proteins from different viral subtypes
To determine whether subtype-specific signatures exist in the PA-X sequence, we compared the variation rates for specific amino acids in PA-X among the different viral subtypes and some specific variations were found to be highly enriched in certain subtypes (Fig. 2). As shown in Fig. 2a, the variation rates at positions 37A, 61I, 101D, 193N, 195R, 199R, and 228I are higher in the H7N9 and H9N2 subtypes than in the H5N1 and H5N6 subtypes. Worth noting is that the variation rates were highest in the H7N9 viruses, reaching almost 100% in the seven sites (Fig. 2a). However, we also noticed that some amino acids (positions 27D, 58G, 129I, 208Q, 213G, and 215P), were more inclined to show variation in H5N1 and H5N6 than in H9N2 and H7N9 viruses, especially in H5N1 viruses (Fig. 2b). Interestingly, another small group of amino acids (20A, 142K, and 251K) showed higher variation rates than the other subtypes, particularly in avian H5N6 viruses (Fig. 2c). Therefore, based on these analyses generally, we found that the pattern of PA-X variation is very similar between H9N2 and H7N9 subtypes, and between H5N1 and H5N6 subtypes. In addition, some variations tended to be enriched in specific viral subtypes. The implementation of further studies investigating the role of PA-X polymorphism in modulating virulence and host-shutoff activity in different virus subtypes and different virus strains will shed further light on the potential mechanisms of pathogenicity in IAV.
Fig. 2.
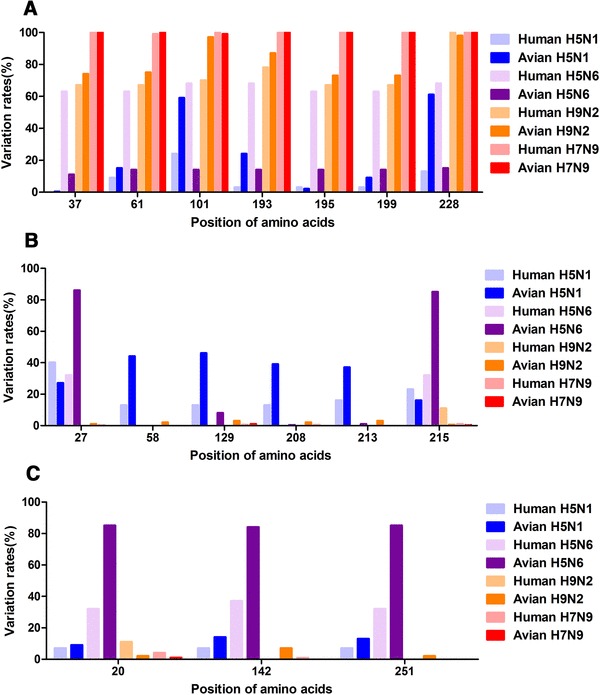
Comparison of the polymorphism of the PA-X protein from viruses of different subtypes. The amino acids (for example, 37A) indicated in the figure are represented as the original residues. Therefore, the calculated mutation rates shown in this figure are compared with the original residues. a Positions of variation rate which are obviously higher in the H7N9 and H9N2 viruses than in H5N1 and H5N6 viruses. b Amino acids those were more inclined to variation in H5N1 and H5N6 viruses than in H9N2 and H7N9 viruses. c Small group amino acids that showed obviously higher variation rates than other subtype viruses, especially in avian H5N6 viruses
Future prospects
To ensure efficient translation of viral mRNAs while constraining host protein expression, IAV employs multiple host-shutoff mechanisms. The PA-X protein is a bona fide host-shutoff endonuclease with host-specific RNA destroying ability. Although accumulating evidence confirms that PA-X-associated host shutoff is related to viral adaptation and pathogenesis, the underlying mechanisms involved remain largely obscure. Further studies to characterize the biogenesis, characteristics, and mechanism of PA-X shutoff in different viral strains and various hosts should help us to unravel the complexity of the influenza virus–host-shutoff mechanisms. Moreover, although a strong connection has been established between PA-X accumulation and host-shutoff activity, further studies are needed to determine whether the C-terminal of PA-X causes nuclear accumulation through its interactions with other nuclear localization signal-containing proteins or whether it contains a functional NLS. Focus should also turn to elucidating the functional differences between the PA-X proteins from different circulating influenza strains, such as H7N9 AIV. Currently, low pathogenic avian influenza (LPAI) and high pathogenic avian influenza (HPAI) H7N9 viruses co-circulate in poultry. Therefore, it will also be very interesting to see whether the PA-X protein elicits similar effects for LPAI and HPAI H7N9 viruses in terms of modulating viral virulence and host responses in avian species and mammals. Finally, the essential role for PA-X in modulating viral replication and host innate immune and adaptive responses makes it highly feasible to explore PA-X as a target for drug development and vaccine design in influenza viruses.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31502076), by the Jiangsu Provincial Natural Science Foundation of China (BK20150444), by the Special Financial Grant from the China Postdoctoral Science Foundation (2016T90515), by the National Key Research and Development Project of China (2016YFD0500202-1 and 2016YFD0501601), by the Earmarked Fund For China Agriculture Research System (CARS-40) by the “Qing Lan Project” of Higher Education Institutions of Jiangsu Province, China, by the “High-end talent support program” of Yangzhou University, China, and by A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Abbreviations
- IAV
Influenza A viruses
- PB2
Polymerase basic protein 2
- PB1
Polymerase basic protein 1
- PA
Polymerase acidic protein
- HA
Hemagglutinin
- NP
Nucleoprotein
- NA
Neuraminidase
- M
Matrix protein
- NS
Non-structural protein
- AIVs
Avian influenza viruses
- GISAID
Global initiative on sharing avian influenza data
- RdRP
RNA-dependent RNA polymerase RdRp complex
- EIV
Equine influenza virus
- CIV
Canine influenza virus
- TR
Triple-reassortment
- SIV
Swine influenza virus
- MHC
Major histocompatibility complex
- RIG-I
Retinoic acid-induced gene protein I
- CPSF30
Cleavage and polyadenylation specificity factor 30
- IPS-1
Interferon beta promoter stimulating factor 1
- NLRP3
NOD-like receptor family pyrin domain containing 3
- AP-MS
Affinity purification and mass spectrometryAP-MS
- HBV
Hepatitis B virus
- HCV
Hepatitis C virus
- HSV-1
Herpes simplex virus 1
- RSV
Respiratory syncytial virus
- JEV
Japanese encephalitis virus
- CCHFV
Crimean-congo hemorrhagic fever virus
- HIV
Human immunodeficiency virus
- HPAIV
Highly pathogenic avian influenza virus
- LPAIV
Low pathogenic avian influenza virus
Author contributions
JH and LX drafted and revised the manuscript; CM contributed to the polymorphism analysis of the PA-X protein. All authors read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interests.
References
- 1.Shindo N, Briand S. Influenza at the beginning of the 21st century. B World Health Organ. 2012;90(4):247–247. doi: 10.2471/Blt.12.104653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cinatl J, Michaelis M, Doerr HW. The threat of avian influenza A (H5N1). Part I: epidemiologic concerns and virulence determinants. Med Microbiol Immun. 2007;196(4):181–190. doi: 10.1007/s00430-007-0042-5. [DOI] [PubMed] [Google Scholar]
- 3.Horimoto T, Kawaoka Y. Influenza: Lessons from past pandemics, warnings from current incidents. Nat Rev Microbiol. 2005;3(8):591–600. doi: 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- 4.Taubenberger JK, Reid AH, Lourens RM, Wang RX, Jin GZ, Fanning TG. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437(7060):889–893. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- 5.Yoon SW, Webby RJ, Webster RG. Evolution and ecology of influenza A viruses. Influenza Pathog Control. 2014;385:359–375. doi: 10.1007/82_2014_396. [DOI] [PubMed] [Google Scholar]
- 6.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu XY, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RAM, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD, Boxrud D, Sambol AR, Abid SH, George KS, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325(5937):197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michaelis M, Doerr HW, Cinatl J. Novel swine-origin influenza A virus in humans: another pandemic knocking at the door. Med Microbiol Immun. 2009;198(3):175–183. doi: 10.1007/s00430-009-0118-5. [DOI] [PubMed] [Google Scholar]
- 8.Wu AP, Su CH, Wang DY, Peng YS, Liu M, Hua S, Li TX, Gao GF, Tang H, Chen JZ, Liu XF, Shu YL, Peng DX, Jiang TJ. Sequential reassortments underlie diverse influenza H7N9 genotypes in China. Cell Host Microbe. 2013;14(4):446–452. doi: 10.1016/j.chom.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Chen WS, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O’Neill R, Schickli J, Palese P, Henklein P, Bennink JR, Yewdell JW. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med. 2001;7(12):1306–1312. doi: 10.1038/Nm1201-1306. [DOI] [PubMed] [Google Scholar]
- 10.Wise HM, Foeglein A, Sun JC, Dalton RM, Patel S, Howard W, Anderson EC, Barclay WS, Digard P. A complicated message: identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J Virol. 2009;83(16):8021–8031. doi: 10.1128/Jvi.00826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science. 2012;337(6091):199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wise HM, Hutchinson EC, Jagger BW, Stuart AD, Kang ZH, Robb N, Schwartzman LM, Kash JC, Fodor E, Firth AE, Gog JR, Taubenberger JK, Digard P. Identification of a novel splice variant form of the influenza A virus M2 ion channel with an antigenically distinct ectodomain. Plos Pathog. 2012 doi: 10.1371/journal.ppat.1002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selman M, Dankar SK, Forbes NE, Jia JJ, Brown EG. Adaptive mutation in influenza A virus non-structural gene is linked to host switching and induces a novel protein by alternative splicing. Emerg Microb Infec. 2012 doi: 10.1038/emi.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muramoto Y, Noda T, Kawakami E, Akkina R, Kawaoka Y. Identification of novel influenza A virus proteins translated from PA mRNA. J Virol. 2013;87(5):2455–2462. doi: 10.1128/Jvi.02656-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi M, Jagger BW, Wise HM, Digard P, Holmes EC, Taubenberger JK. Evolutionary conservation of the PA-X open reading frame in segment 3 of influenza A virus. J Virol. 2012;86(22):12411–12413. doi: 10.1128/Jvi.01677-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeDiego ML, Nogales A, Lambert-Emo K, Martinez-Sobrido L, Topham DJ. NS1 protein mutation I64T affects interferon responses and virulence of circulating H3N2 human influenza A viruses. J Virol. 2016;90(21):9693–9711. doi: 10.1128/JVI.01039-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ayllon J, Domingues P, Rajsbaum R, Miorin L, Schmolke M, Hale BG, Garca-Sastre A. A single amino acid substitution in the novel H7N9 influenza A virus NS1 protein increases CPSF30 binding and virulence. J Virol. 2014;88(20):12146–12151. doi: 10.1128/Jvi.01567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol Cell. 1998;1(7):991–1000. doi: 10.1016/S1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 19.Twu KY, Noah DL, Rao P, Kuo RL, Krug RM. The CPSF30 binding site on the NS1A protein of influenza A virus is a potential antiviral target. J Virol. 2006;80(8):3957–3965. doi: 10.1128/Jvi.80.8.3957-3965.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez A, Perez-Gonzalez A, Nieto A. Influenza virus infection causes specific degradation of the largest subunit of cellular RNA polymerase II. J Virol. 2007;81(10):5315–5324. doi: 10.1128/Jvi.02129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vreede FT, Chan AY, Sharps J, Fodor E. Mechanisms and functional implications of the degradation of host RNA polymerase II in influenza virus infected cells. Virology. 2010;396(1):125–134. doi: 10.1016/j.virol.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Llompart CM, Nieto A, Rodriguez-Frandsen A. Specific residues of PB2 and PA influenza virus polymerase subunits confer the ability for RNA polymerase II degradation and virus pathogenicity in mice. J Virol. 2014;88(6):3455–3463. doi: 10.1128/Jvi.02263-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desmet EA, Bussey KA, Stone R, Takimoto T. Identification of the N-terminal domain of the influenza virus PA responsible for the suppression of host protein synthesis. J Virol. 2013;87(6):3108–3118. doi: 10.1128/Jvi.02826-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu J, Mo YQ, Wang XQ, Gu M, Hu ZL, Zhong L, Wu QW, Hao XL, Hu SL, Liu WB, Liu HM, Liu XW, Liu XF. PA-X decreases the pathogenicity of highly pathogenic H5N1 influenza A virus in avian species by inhibiting virus replication and host response. J Virol. 2015;89(8):4126–4142. doi: 10.1128/Jvi.02132-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi T, Chaimayo C, Takimoto T. Impact of influenza PA-X on host response. Oncotarget. 2015;6(23):19364–19365. doi: 10.18632/oncotarget.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oishi K, Yamayoshi S, Kawaoka Y. Mapping of a region of the PA-X protein of influenza A virus that is important for its shutoff activity. J Virol. 2015;89(16):8661–8665. doi: 10.1128/Jvi.01132-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayashi T, Chaimayo C, McGuinness J, Takimoto T. Critical role of the PA-X C-terminal domain of influenza A virus in its subcellular localization and shutoff activity. J Virol. 2016;90(16):7131–7141. doi: 10.1128/Jvi.00954-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khaperskyy DA, McCormick C. Timing is everything: coordinated control of host shutoff by influenza A virus NS1 and PA-X proteins. J Virol. 2015;89(13):6528–6531. doi: 10.1128/Jvi.00386-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khaperskyy Denys A., Schmaling Summer, Larkins-Ford Jonah, McCormick Craig, Gaglia Marta M. Selective Degradation of Host RNA Polymerase II Transcripts by Influenza A Virus PA-X Host Shutoff Protein. PLOS Pathogens. 2016;12(2):e1005427. doi: 10.1371/journal.ppat.1005427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khaperskyy DA, Emara MM, Johnston BP, Anderson P, Hatchette TF, McCormick C. Influenza A virus host shutoff disables antiviral stress-induced translation arrest. Plos Pathog. 2014 doi: 10.1371/journal.ppat.1004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nogales A, Rodriguez L, DeDiego ML, Topham DJ, Martinez-Sobrido L. Interplay of PA-X and NS1 Proteins in replication and pathogenesis of a temperature-sensitive 2009 pandemic H1N1 influenza A virus. J Virol. 2017 doi: 10.1128/JVI.00720-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao HJ, Sun HL, Hu J, Wang JL, Xiong X, Wang Y, He QM, Lin Y, Kong WL, Seng LG, Pu J, Chang KC, Liu XF, Liu JH, Sun YP. Twenty amino acids at the C-terminus of PA-X are associated with increased influenza A virus replication and pathogenicity. J Gen Virol. 2015;96:2036–2049. doi: 10.1099/vir.0.000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayashi T, MacDonald LA, Takimoto T. Influenza A virus protein PA-X contributes to viral growth and suppression of the host antiviral and immune responses. J Virol. 2015;89(12):6442–6452. doi: 10.1128/Jvi.00319-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leea JW, Yua H, Li YH, Ma JJ, Lang YE, Duff M, Henningson J, Liu QF, Li YH, Nagy A, Bawa B, Li ZJ, Tong GG, Richt JE, Ma WJ. Impacts of different expressions of PA-X protein on 2009 pandemic H1N1 virus replication, pathogenicity and host immune responses. Virology. 2017;504:25–35. doi: 10.1016/j.virol.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao HJ, Xu GL, Sun YP, Qi L, Wang JL, Kong WL, Sun HL, Pu J, Chang KC, Liu JH. PA-X is a virulence factor in avian H9N2 influenza virus. J Gen Virol. 2015;96:2587–2594. doi: 10.1099/jgv.0.000232. [DOI] [PubMed] [Google Scholar]
- 36.Gao HJ, Sun YP, Hu J, Qi L, Wang JL, Xiong X, Wang Y, He QM, Lin Y, Kong WL, Seng LG, Sun HL, Pu J, Chang KC, Liu XF, Liu JH. The contribution of PA-X to the virulence of pandemic 2009 H1N1 and highly pathogenic H5N1 avian influenza viruses. Sci Rep-UK. 2015 doi: 10.1038/Srep08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng KH, Sun M, Iketani S, Holmes EC, Parrish CR. Comparing the functions of equine and canine influenza H3N8 virus PA-X proteins: suppression of reporter gene expression and modulation of global host gene expression. Virology. 2016;496:138–146. doi: 10.1016/j.virol.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu GL, Zhang XX, Sun YP, Liu QF, Sun HL, Xiong X, Jiang M, He QM, Wang Y, Pu J, Guo X, Yang HC, Liu JH. Truncation of C-terminal 20 amino acids in PA-X contributes to adaptation of swine influenza virus in pigs. Sci Rep-Uk. 2016 doi: 10.1038/Srep21845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu GL, Zhang XX, Liu QF, Bing GX, Hu Z, Sun HL, Xiong X, Jiang M, He QM, Wang Y, Pu J, Guo X, Yang HC, Liu JH, Sun YP. PA-X protein contributes to virulence of triple-reassortant H1N2 influenza virus by suppressing early immune responses in swine. Virology. 2017;508:45–53. doi: 10.1016/j.virol.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 40.Dias A, Bouvier D, Crepin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RWH. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 2009;458(7240):914–918. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 41.Yuan PW, Bartlam M, Lou ZY, Chen SD, Zhou J, He XJ, Lv ZY, Ge RW, Li XM, Deng T, Fodor E, Rao ZH, Liu YF. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature. 2009;458(7240):909–912. doi: 10.1038/nature07720. [DOI] [PubMed] [Google Scholar]
- 42.Bavagnoli L, Cucuzza S, Campanini G, Rovida F, Paolucci S, Baldanti F, Maga G. The novel influenza A virus protein PA-X and its naturally deleted variant show different enzymatic properties in comparison to the viral endonuclease PA. Nucleic Acids Res. 2015;43(19):9405–9417. doi: 10.1093/nar/gkv926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oishi K, Yamayoshi S, Kawaoka Y. Identification of novel amino acid residues of influenza virus PA-X that are important for PA-X shutoff activity by using yeast. Virology. 2018;516:71–75. doi: 10.1016/j.virol.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwong AD, Frenkel N. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc Natl Acad Sci USA. 1987;84(7):1926–1930. doi: 10.1073/pnas.84.7.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamitani W, Narayanan K, Huang C, Lokugamage K, Ikegami T, Ito N, Kubo H, Makino S. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc Natl Acad Sci USA. 2006;103(34):12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kash JC, Tumpey TM, Proll SC, Carter V, Perwitasari O, Thomas MJ, Basler CF, Palese P, Taubenberger JK, Garcia-Sastre A, Swayne DE, Katze MG. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature. 2006;443(7111):578–581. doi: 10.1038/nature05181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Talon J, Horvath CM, Polley R, Basler CF, Muster T, Palese P, Garcia-Sastre A. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J Virol. 2000;74(17):7989–7996. doi: 10.1128/Jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noah DL, Twu KY, Krug RM. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3 ’ end processing of cellular pre-mRNAS. Virology. 2003;307(2):386–395. doi: 10.1016/S0042-6822(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 49.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Sousa CRE. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5 ‘-phosphates. Science. 2006;314(5801):997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 50.Opitz B, Rejaibi A, Dauber B, Eckhard J, Vinzing M, Schmeck B, Hippenstiel S, Suttorp N, Wolff T. IFN beta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell Microbiol. 2007;9(4):930–938. doi: 10.1111/j.1462-5822.2006.00841.x. [DOI] [PubMed] [Google Scholar]
- 51.Wang XY, Li M, Zheng HY, Muster T, Palese P, Beg AA, Garcia-Sastre A. Influenza A virus NS1 protein prevents activation of NF-kappa B and induction of alpha/beta interferon. J Virol. 2000;74(24):11566–11573. doi: 10.1128/Jvi.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao S, Peng H, Jiang W, Song L. NS1 protein of avian influenza A virus prevents activation of NF-kappa B through binding to IKK alpha and IKK beta. Int J Infect Dis. 2010;14:E82–E83. doi: 10.1016/j.ijid.2010.02.1672. [DOI] [Google Scholar]
- 53.Ludwig S, Wang XY, Ehrhardt C, Zheng HY, Donelan N, Planz O, Pleschka S, Garcia-Sastre A, Heins G, Wolff T. The influenza A virus NS1 protein inhibits activation of jun N-terminal kinase and AP-1 transcription factors. J Virol. 2002;76(21):11166–11171. doi: 10.1128/Jvi.76.21.11166-11171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hayman A, Comely S, Lackenby A, Murphy S, McCauley J, Goodbourn S, Barclay W. Variation in the ability of human influenza A viruses to induce and inhibit the IFN-beta pathway. Virology. 2006;347(1):52–64. doi: 10.1016/j.virol.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 55.Kochs G, Garcia-Sastre A, Martinez-Sobrido L. Multiple anti-interferon actions of the influenza A virus NS1 protein. J Virol. 2007;81(13):7011–7021. doi: 10.1128/Jvi.02581-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuo RL, Zhao C, Malur M, Krug RM. Influenza A virus strains that circulate in humans differ in the ability of their NS1 proteins to block the activation of IRF3 and interferon-beta transcription. Virology. 2010;408(2):146–158. doi: 10.1016/j.virol.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li YZ, Chen ZY, Wang WR, Baker CC, Krug RM. The 3 ‘-end-processing factor CPSF is required for the splicing of single-intron pre-mRNAs in vivo. Rna. 2001;7(6):920–931. doi: 10.1017/S1355838201010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Das K, Ma LC, Xiao R, Radvansky B, Aramini J, Zhao L, Marklund J, Kuo RL, Twu KY, Arnold E, Krug RM, Montelione GT. Structural basis for suppression of a host antiviral response by influenza A virus. P Natl Acad Sci USA. 2008;105(35):13093–13098. doi: 10.1073/pnas.0805213105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aramini JM, Ma LC, Zhou LG, Schauder CM, Hamilton K, Amer BR, Mack TR, Lee HW, Ciccosanti CT, Zhao L, Xiao R, Krug RM, Montelione GT. Dimer interface of the effector domain of non-structural protein 1 from influenza A virus an interface with multiple functions. J Biol Chem. 2011;286(29):26050–26060. doi: 10.1074/jbc.M111.248765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hatada E, Fukuda R. Binding of influenza A virus NS1 protein to dsRNA in vitro. J Gen Virol. 1992;73(Pt 12):3325–3329. doi: 10.1099/0022-1317-73-12-3325. [DOI] [PubMed] [Google Scholar]
- 61.Lu Y, Wambach M, Katze MG, Krug RM. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology. 1995;214(1):222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 62.Min JY, Krug RM. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2 ‘-5 ’ oligo (A) synthetase/RNase L pathway. P Natl Acad Sci USA. 2006;103(18):7100–7105. doi: 10.1073/pnas.0602184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Min JY, Li SD, Sen GC, Krug RM. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology. 2007;363(1):236–243. doi: 10.1016/j.virol.2007.01.038. [DOI] [PubMed] [Google Scholar]
- 64.Li S, Min JY, Krug RM, Sen GC. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology. 2006;349(1):13–21. doi: 10.1016/j.virol.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 65.Varga ZT, Ramos I, Hai R, Schmolke M, Garcia-Sastre A, Fernandez-Sesma A, Palese P. The Influenza Virus Protein PB1-F2 Inhibits the Induction of Type I Interferon at the Level of the MAVS Adaptor Protein. Plos Pathogens. 2011 doi: 10.1371/journal.ppat.1002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dudek SE, Wixler L, Nordhoff C, Nordmann A, Anhlan D, Wixler V, Ludwig S. The influenza virus PB1-F2 protein has interferon antagonistic activity. Biol Chem. 2011;392(12):1135–1144. doi: 10.1515/Bc-2011-174. [DOI] [PubMed] [Google Scholar]
- 67.McAuley JL, Chipuk JE, Boyd KL, Van De Velde N, Green DR, McCullers JA. PB1-F2 proteins from H5N1 and 20 century pandemic influenza viruses cause immunopathology. PLoS Pathog. 2010;6(7):e1001014. doi: 10.1371/journal.ppat.1001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Le Goffic R, Leymarie O, Chevalier C, Rebours E, Da Costa B, Vidic J, Descamps D, Sallenave JM, Rauch M, Samson M, Delmas B. Transcriptomic analysis of host immune and cell death responses associated with the influenza A virus PB1-F2 protein. Plos Pathog. 2011 doi: 10.1371/journal.ppat.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alymova IV, Green AM, van de Velde N, McAuley JL, Boyd KL, Ghoneim HE, McCullers JA. Immunopathogenic and antibacterial effects of H3N2 influenza A virus PB1-F2 map to amino acid residues 62, 75, 79, and 82. J Virol. 2011;85(23):12324–12333. doi: 10.1128/Jvi.05872-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Le Goffic R, Bouguyon E, Chevalier C, Vidic J, Da Costa B, Leymarie O, Bourdieu C, Decamps L, Dhorne-Pollet S, Delmas B. Influenza A virus protein PB1-F2 exacerbates IFN-beta expression of human respiratory epithelial cells. J Immunol. 2010;185(8):4812–4823. doi: 10.4049/jimmunol.0903952. [DOI] [PubMed] [Google Scholar]
- 71.Krumbholz A, Philipps A, Oehring H, Schwarzer K, Eitner A, Wutzler P, Zell R. Current knowledge on PB1-F2 of influenza A viruses. Med Microbiol Immunol. 2011;200(2):69–75. doi: 10.1007/s00430-010-0176-8. [DOI] [PubMed] [Google Scholar]
- 72.Plotch SJ, Bouloy M, Ulmanen I, Krug RM. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23(3):847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 73.Graef KM, Vreede FT, Lau YF, McCall AW, Carr SM, Subbarao K, Fodor E. The PB2 subunit of the influenza virus RNA polymerase affects virulence by interacting with the mitochondrial antiviral signaling protein and inhibiting expression of beta interferon. J Virol. 2010;84(17):8433–8445. doi: 10.1128/Jvi.00879-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iwai A, Shiozaki T, Kawai T, Akira S, Kawaoka Y, Takada A, Kida H, Miyazaki T. Influenza A virus polymerase inhibits type I interferon induction by binding to interferon beta promoter stimulator 1. J Biol Chem. 2010;285(42):32064–32074. doi: 10.1074/jbc.M110.112458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liedmann S, Hrincius ER, Anhlan D, McCullers JA, Ludwig S, Ehrhardt C. New virulence determinants contribute to the enhanced immune response and reduced virulence of an influenza A virus A/PR8/34 variant. J Infect Dis. 2014;209(4):532–541. doi: 10.1093/infdis/jit463. [DOI] [PubMed] [Google Scholar]
- 76.Sakabe S, Takano R, Nagamura-Inoue T, Yamashita N, Nidom CA, Mai TQL, Iwatsuki-Horimoto K, Kawaoka Y. Differences in cytokine production in human macrophages and in virulence in mice are attributable to the acidic polymerase protein of highly pathogenic influenza A virus subtype H5N1. J Infect Dis. 2013;207(2):262–271. doi: 10.1093/infdis/jis523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hu J, Hu ZL, Song QQ, Gu M, Liu XW, Wang XQ, Hu SL, Chen CY, Liu HM, Liu WB, Chen SJ, Peng DX, Liu XF. The PA-gene-mediated lethal dissemination and excessive innate immune response contribute to the high virulence of H5N1 avian influenza virus in mice. J Virol. 2013;87(5):2660–2672. doi: 10.1128/Jvi.02891-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang CH, Chen CJ, Yen CT, Yu CP, Huang PN, Kuo RL, Lin SJ, Chang CK, Shih SR. Caspase-1 deficient mice are more susceptible to influenza A virus infection with PA variation. J Infect Dis. 2013;208(11):1898–1905. doi: 10.1093/infdis/jit381. [DOI] [PubMed] [Google Scholar]
- 79.Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol. 2010;11(5):404–461. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Talon J, Salvatore M, O’Neill RE, Nakaya Y, Zheng HY, Muster T, Garcia-Sastre A, Palese P. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. P Natl Acad Sci USA. 2000;97(8):4309–4314. doi: 10.1073/pnas.070525997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Burns CC, Shaw J, Campagnoli R, Jorba J, Vincent A, Quay J, Kew O. Modulation of poliovirus replicative fitness in HeLa cells by deoptimization of synonymous codon usage in the capsid region. J Virol. 2006;80(7):3259–3272. doi: 10.1128/Jvi.80.7.3259-3272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nogales A, Baker SF, Ortiz-Riano E, Dewhurst S, Topham DJ, Martinez-Sobrido L. Influenza A virus attenuation by codon deoptimization of the NS gene for vaccine development. J Virol. 2014;88(18):10525–10540. doi: 10.1128/Jvi.01565-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gong XQ, Sun YF, Ruan BY, Liu XM, Wang Q, Yang HM, Wang SY, Zhang P, Wang XH, Shan TL, Tong W, Zhou YJ, Li GX, Zheng H, Tong GZ, Yu H. PA-X protein decreases replication and pathogenicity of swine influenza virus in cultured cells and mouse models. Vet Microbiol. 2017;205:66–70. doi: 10.1016/j.vetmic.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 84.Hu J, Mo YQ, Gao Z, Wang XQ, Gu M, Liang YY, Cheng X, Hu SL, Liu WB, Liu HM, Chen SJ, Liu XW, Peng DX, Liu XF. PA-X-associated early alleviation of the acute lung injury contributes to the attenuation of a highly pathogenic H5N1 avian influenza virus in mice. Med Microbiol Immun. 2016;205(4):381–395. doi: 10.1007/s00430-016-0461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu G, Zhang X, Sun Y, Liu Q, Sun H, Xiong X, Jiang M, He Q, Wang Y, Pu J, Guo X, Yang H, Liu J. Truncation of C-terminal 20 amino acids in PA-X contributes to adaptation of swine influenza virus in pigs. Sci Rep. 2016;6:21845. doi: 10.1038/srep21845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li QH, Yuan XY, Wang Q, Chang GB, Wang F, Liu RR, Zheng MQ, Chen GH, Wen J, Zhao GP. Interactomic landscape of PA-X-chicken protein complexes of H5N1 influenza A virus. J Proteom. 2016;148:20–25. doi: 10.1016/j.jprot.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 87.Zhang J, Fu LL, Tian M, Liu HQ, Li JJ, Li Y, He J, Huang J, Ouyang L, Gao HY, Wang JH. Design and synthesis of a novel candidate compound NTI-007 targeting sodium taurocholate cotransporting polypeptide [NTCP]-APOA1-HBx-Beclin1-mediated autophagic pathway in HBV therapy. Bioorgan Med Chem. 2015;23(5):976–984. doi: 10.1016/j.bmc.2015.01.020. [DOI] [PubMed] [Google Scholar]
- 88.Shi ST, Polyak SJ, Tu H, Taylor DR, Gretch DR, Lai MMC. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology. 2002;292(2):198–210. doi: 10.1006/viro.2001.1225. [DOI] [PubMed] [Google Scholar]
- 89.Zheng SQ, Li YX, Zhang Y, Li X, Tang H. MiR-101 regulates HSV-1 replication by targeting ATP5B. Antivir Res. 2011;89(3):219–226. doi: 10.1016/j.antiviral.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 90.Kumar D, Broor S, Rajala MS. Interaction of host nucleolin with influenza A virus nucleoprotein in the early phase of infection limits the late viral gene expression. PLoS One. 2016;11(10):e0164146. doi: 10.1371/journal.pone.0164146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Murayama R, Harada Y, Shibata T, Kuroda K, Hayakawa S, Shimizu K, Tanaka T. Influenza A virus non-structural protein 1 (NS1) interacts with cellular multifunctional protein nucleolin during infection. Biochem Bioph Res Co. 2007;362(4):880–885. doi: 10.1016/j.bbrc.2007.08.091. [DOI] [PubMed] [Google Scholar]
- 92.Melen K, Tynell J, Fagerlund R, Roussel P, Hernandez-Verdun D, Julkunen I. Influenza A H3N2 subtype virus NS1 protein targets into the nucleus and binds primarily via its C-terminal NLS2/NoLS to nucleolin and fibrillarin. Virol J. 2012 doi: 10.1186/1743-422x-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chan CM, Chu H, Zhang AJ, Leung LH, Sze KH, Kao RYT, Chik KKH, To KKW, Chan JFW, Chen HL, Jin DY, Liu L, Yuen KY. Hemagglutinin of influenza A virus binds specifically to cell surface nucleolin and plays a role in virus internalization. Virology. 2016;494:78–88. doi: 10.1016/j.virol.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 94.Balinsky CA, Schmeisser H, Ganesan S, Singh K, Pierson TC, Zoon KC. Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles. J Virol. 2013;87(24):13094–13106. doi: 10.1128/Jvi.00704-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bose S, Basu M, Banerjee AK. Role of nucleolin in human parainfluenza virus type 3 infection of human lung epithelial cells. J Virol. 2004;78(15):8146–8158. doi: 10.1128/JVI.78.15.8146-8158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tayyari F, Marchant D, Moraes TJ, Duan W, Mastrangelo P, Hegele RG. Identification of nucleolin as a cellular receptor for human respiratory syncytial virus. Nature Med. 2011;17(9):1132–1135. doi: 10.1038/nm.2444. [DOI] [PubMed] [Google Scholar]
- 97.Thongtan T, Wikan N, Wintachai P, Rattanarungsan C, Srisomsap C, Cheepsunthorn P, Smith DR Characterization of putative Japanese encephalitis virus receptor molecules on microglial cells. (Electronic) [DOI] [PubMed]
- 98.Xiao X, Feng Y, Zhu Z, Dimitrov DS. Identification of a putative Crimean-Congo hemorrhagic fever virus entry factor. Biochem Biophys Res Commun. 2011;411(2):253–258. doi: 10.1016/j.bbrc.2011.06.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nisole S, Krust B, Hovanessian AG. Anchorage of HIV on permissive cells leads to coaggregation of viral particles with surface nucleolin at membrane raft microdomains. Exp Cell Res. 2002;276(2):155–173. doi: 10.1006/excr.2002.5522. [DOI] [PubMed] [Google Scholar]
- 100.Nisole S, Said EA, Mische C, Prevost MC, Krust B, Bouvet P, Bianco A, Briand JP, Hovanessian AG. The anti-HIV pentameric pseudopeptide HB-19 binds the C-terminal end of nucleolin and prevents anchorage of virus particles in the plasma membrane of target cells. J Biol Chem. 2002;277(23):20877–20886. doi: 10.1074/jbc.M110024200. [DOI] [PubMed] [Google Scholar]
- 101.Gadad SS, Rajan RE, Senapati P, Chatterjee S, Shandilya J, Dash PK, Ranga U, Kundu TK. HIV-1 infection induces acetylation of NPM1 that facilitates tat localization and enhances viral transactivation. J Mol Biol. 2011;410(5):997–1007. doi: 10.1016/j.jmb.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 102.Lu JW, Chang JG, Yeh KT, Chen RM, Tsai JJP, Hu RM. Overexpression of Thy1/CD90 in human hepatocellular carcinoma is associated with HBV infection and poor prognosis. Acta Histochem. 2011;113(8):833–838. doi: 10.1016/j.acthis.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 103.Irie T, Inoue M, Sakaguchi T. Significance of the YLDL motif in the M protein and Alix/AIP1 for Sendai virus budding in the context of virus infection. Virology. 2010;405(2):334–341. doi: 10.1016/j.virol.2010.06.031. [DOI] [PubMed] [Google Scholar]
- 104.Su S, Bi YH, Wong G, Gray GC, Gao GF, Li SJ. Epidemiology, evolution, and recent outbreaks of avian influenza virus in China. J Virol. 2015;89(17):8671–8676. doi: 10.1128/Jvi.01034-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shi WF, Li W, Li XB, Haywood J, Ma JC, Gao GF, Liu D. Phylogenetics of varied subtypes of avian influenza viruses in China: potential threat to humans. Protein Cell. 2014;5(4):253–257. doi: 10.1007/s13238-014-0036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang XL, Jiang H, Wu P, Uyeki TM, Feng LZ, Lai SJ, Wang LL, Huo X, Xu K, Chen EF, Wang XX, He JF, Kang M, Zhang RL, Zhang J, Wu JB, Hu SX, Zhang HJ, Liu XQ, Fu WJ, Ou JM, Wu SG, Qin Y, Zhang ZJ, Shi YJ, Zhang JJ, Artois J, Fang VJ, Zhu HC, Guan Y, Gilbert M, Horby PW, Leung GM, Gao GF, Cowling BJ, Yu HJ. Epidemiology of avian influenza A H7N9 virus in human beings across five epidemics in mainland China, 2013-17: an epidemiological study of laboratory-confirmed case series. Lancet Infect Dis. 2017;17(8):822–832. doi: 10.1016/S1473-3099(17)30323-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Durand LO, Glew P, Gross D, Kasper M, Trock S, Kim IK, Bresee JS, Donis R, Uyeki TM, Widdowson MA, Azziz-Baumgartner E. Timing of influenza A(H5N1) in poultry and humans and seasonal influenza activity worldwide, 2004–2013. Emerg Infect Dis. 2015;21(2):202–208. doi: 10.3201/eid2102.140877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pan M, Gao RB, Lv Q, Huang SH, Zhou ZH, Yang L, Li XD, Zhao X, Zou XH, Tong WB, Mao SL, Zou SM, Bo H, Zhu XP, Liu L, Yuan H, Zhang MH, Wang DQ, Li ZM, Zhao W, Ma ML, Li YQ, Li TS, Yang HP, Xu JN, Zhou LJ, Zhou XY, Tang W, Song Y, Chen T, Bai T, Zhou JF, Wang DY, Wu GZ, Li DX, Feng ZJ, Gao GF, Wang Y, He SS, Shu YL. Human infection with a novel, highly pathogenic avian influenza A (H5N6) virus: virological and clinical findings. J Infect. 2016;72(1):52–59. doi: 10.1016/j.jinf.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 109.Bi YH, Chen QJ, Wang QL, Chen JJ, Jin T, Wong G, Quan CS, Liu J, Wu J, Yin RF, Zhao LH, Li MX, Ding Z, Zou RR, Xu W, Li H, Wang HJ, Tian KG, Fu GH, Huang Y, Shestopalov A, Li SJ, Xu B, Yu HJ, Luo TR, Lu L, Xu X, Luo Y, Liu YX, Shi WF, Liu D, Gao GF. Genesis, evolution and prevalence of H5N6 avian influenza viruses in China. Cell Host Microbe. 2016;20(6):810–821. doi: 10.1016/j.chom.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 110.Zhang Y, Chen MM, Huang YW, Zhu WF, Yang L, Gao LD, Li XD, Bi FY, Huang CY, Kang N, Zhang HJ, Li Z, Bo H, Wang DY, Shu YL. Human infections with novel reassortant H5N6 avian influenza viruses in China. Emerg Microbes Infec. 2017 doi: 10.1038/emi.2017.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Peiris M, Yuen KY, Leung CW, Chan KH, Ip PLS, Lai RWM, Orr WK, Shortridge KF. Human infection with influenza H9N2. Lancet. 1999;354(9182):916–917. doi: 10.1016/S0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 112.Yuan RY, Liang LJ, Wu J, Kang YF, Song YC, Zou LR, Zhang X, Ni HZ, Ke CW. Human infection with an avian influenza A/H9N2 virus in Guangdong in 2016. J Infection. 2017;74(4):422–425. doi: 10.1016/j.jinf.2017.01.003. [DOI] [PubMed] [Google Scholar]


